Abstract
Rhythmic frq transcription is essential for the function of the Neurospora circadian clock. Here we show that there is a circadian histone occupancy rhythm at the frq promoter that is regulated by FREQUENCY (FRQ). Using a combination of forward genetics and genome sequencing, we identify Clock ATPase (CATP) as an essential clock component. Our results demonstrate that CATP associates with the frq locus and other WCC target genes and promotes histone removal at these loci to allow circadian gene transcription. These results indicate that the rhythmic control of histone occupancy at clock genes is critical for circadian clock function.
Keywords: circadian clock, nucleosome, chromatin structure, Neurospora
INTRODUCTION
Eukaryotic circadian oscillators are auto-regulatory circadian feedback loops that are based on regulation of transcription and translation [1, 2]. The mechanism of the circadian oscillator of the filamentous fungus Neurospora crassa is remarkably similar to those of higher eukaryotes [3–5]. In the Neurospora clock, the heterodimeric white collar complex (WCC) formed by two PAS-domain transcription factors WHITE COLLAR-1 (WC-1) and WC-2 binds to the promoter of the frequency (frq) gene and activates its transcription [6–13]. FRQ and its protein partner FRQ-interacting RNA helicase (FRH) form a complex called FFC that inhibits the expression of frq both transcriptionally and post-transcriptionally [8, 9, 14, 15, 16, 17, 18]. The inhibition of frq transcription by FFC is mediated by FRQ-dependent WC phosphorylation, which represses WC DNA binding activity and promotes its cytoplasmic localization [8, 9, 18, 19, 20, 21, 22]. The rhythmic activation and repression of frq transcription allow the endogenous rhythmic expression of frq.
DNA bound by core histones forms nucleosomes, which are the fundamental units of eukaryotic chromatin. Post-translational modifications of histones are the basis of epigenetic regulation that can have important roles in controlling gene transcription. Rhythmic histone acetylation, and histone methylation of clock gene loci and of many clock-controlled genes have been shown [23, 24]. In addition to histone modifications, nucleosome occupancy also impacts gene expression [25, 26]. In Neurospora, two ATP-dependent chromatin-remodelling factors, CLOCKSWITCH (CSW-1) and chromodomain helicase DNA-binding-1 (CHD1), have been shown to be involved in the clock function by regulating frq transcription [10, 27]. These proteins presumably function by regulating the chromatin status of the frq locus, but how they act is not clear. In addition, a histone H3K4 methyltransferase is required for normal circadian rhythms [28]. Still, how nucleosome occupancy is controlled by circadian clocks and whether it has a role in the circadian control of gene expression are not clear.
In this study, we show that there is a circadian nucleosome occupancy rhythm at the frq promoter that corresponds to the activation and repression of frq transcription. By combining forward genetic- and genome-sequencing approaches, we identified Clock ATPase (CATP), a highly conserved eukaryotic protein, as an essential circadian clock component in Neurospora. We showed that CATP controls frq transcription and WCC binding to the frq promoter by regulating the nucleosome occupancy and chromatin status of the frq locus and other WCC target genes.
RESULTS AND DISCUSSION
Rhythmic nucleosome occupancy at the frq locus
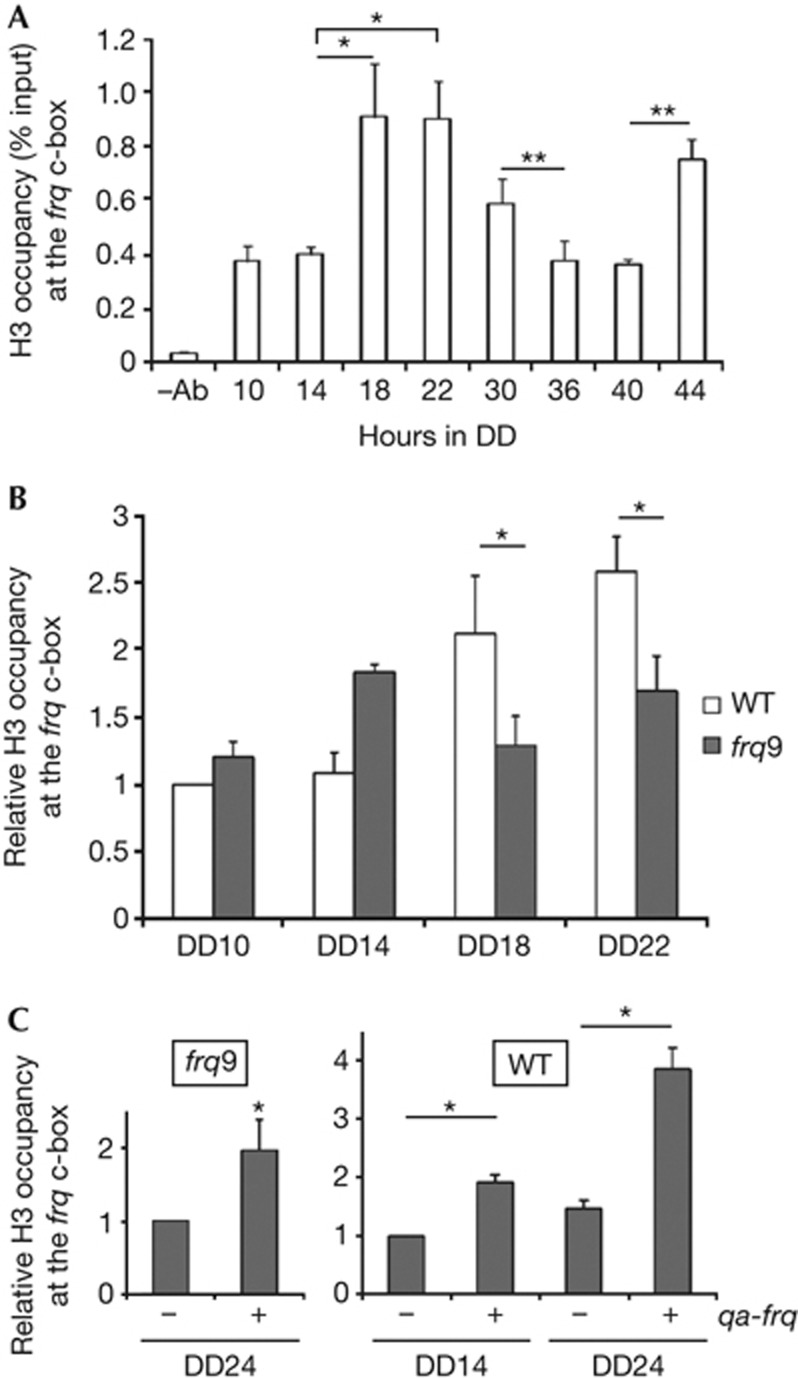
To investigate the involvement of chromatin structure in the regulation of frq transcription, we examined the nucleosome occupancy at the c-box, a WCC-binding site in the frq promoter, using a chromatin immunoprecipitation (ChIP) assay with a histone H3 antibody. As shown in Fig 1A, the histone H3 occupancy was rhythmic in constant darkness (DD): low around DD14 and high around DD22. This result indicates that nucleosome occupancy rhythm is anti-phase to the known rhythm of WCC binding at the frq c-box [8, 10]. Nucleosome occupancy is low when the WCC binding is high and the frq transcription is activated, and is high when the WCC binding is low and the frq transcription is repressed.
Figure 1.
Nucleosome occupancy at the frq c-box shows a circadian rhythm. (A) Histone H3 ChIP assay with the WT strain in DD. (B) H3 ChIP assay in the WT and frq9 strains. (C) H3 ChIP assays of the frq9 and WT strains with or without QA-induced FRQ. Mean with standard errors (n=3). *P<0.05, **P<0.01 (paired student’s t-test). ChIP, chromatin immunoprecipitation; DD, constant darkness; QA, quinic acid; WT, wild type.
To further confirm this result, we performed the histone H3 ChIP assays with the frq9 strain, in which a premature stop codon results in a truncated FRQ protein and the loss of circadian rhythm [29]. The H3 occupancy rhythm was lost in the frq9 strain; no histone occupancy increase was observed at DD18 or 22 (Fig 1B). Because FRQ protein levels peak at these time points when frq transcription is repressed, this result suggests that FRQ promotes nucleosome formation at the frq locus. To test this hypothesis, we induced FRQ expression in frq9 and wild-type strains that harbour a quinic acid-inducible frq construct. As expected, ectopic expression of FRQ resulted in a significant increase in H3 occupancy at the frq c-box in both strains (Fig 1C). These results indicate that FRQ is required for high nucleosome occupancy on its own promoter in the repressive phase. Nucleosome occupancy is important in regulating transcription: gene expression is inhibited when nucleosome occupancy is high [25, 26]. Therefore, the rhythmic nucleosome occupancy at the frq locus likely has a critical role in the circadian negative feedback process.
Identification of a new Neurospora clock mutant
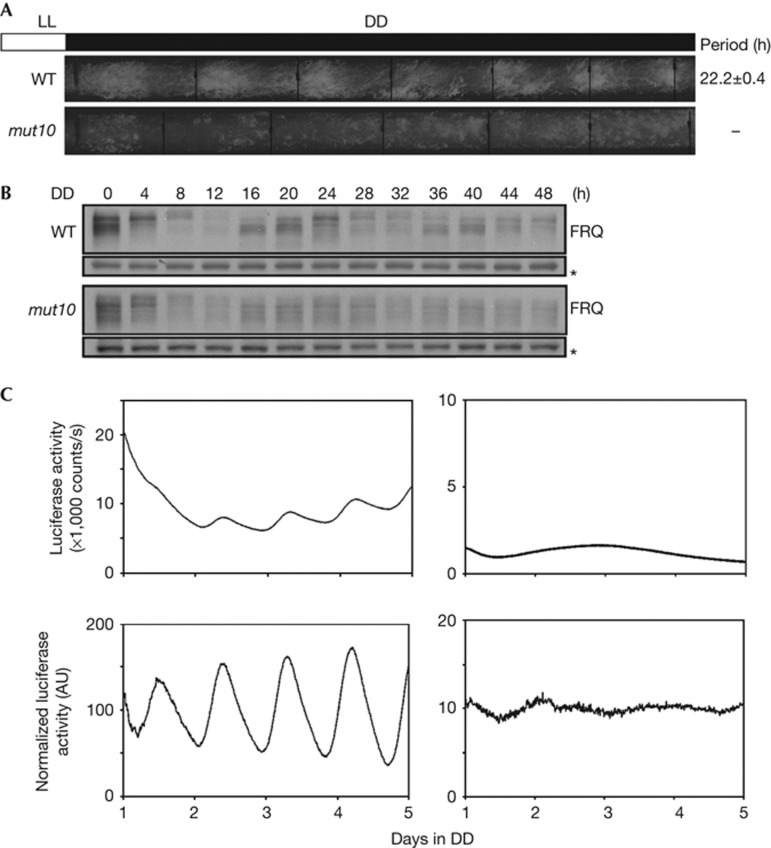
In our analyses of different progenies from a routine cross between Neurospora strains, we identified a clock mutant strain, mut10 that arose from a spontaneous mutation. As shown by race tube assay, mut10 has lost the normal circadian conidiation rhythm: this strain showed a low-amplitude long-period rhythm for the first 2 days and then became arrhythmic (Fig 2A). In addition, the circadian conidiation rhythm was also abolished in the bd mut10 double mutant (supplementary Fig S1 online). To determine the molecular phenotype of the mutant, we examined the FRQ expression profile at different time points in DD. As shown previously [30], FRQ oscillated in both its level and its phosphorylation profile in a wild-type strain. In contrast, the mut10 strain only exhibited a very low-amplitude oscillation after the light-to-dark transition, and there was little change in FRQ phosphorylation after DD12 (Fig 2B; supplementary Fig S2A online). Furthermore, we introduced a bioluminescence reporter construct (frq-luc) [31], in which luciferase expression is controlled by the frq promoter, into the mutant. As shown in Fig 2C, a robust circadian rhythm of luciferase activity was observed in the wild-type strain, but no rhythmic expression of luciferase was observed in the mut10 strain, indicating that the normal clock function is abolished in the mut10 strain.
Figure 2.
Loss of normal circadian rhythms in the mut10 strain. (A) Race tube analysis showing the conidiation rhythms in DD. Period of the WT strain is shown as average±s.e.m. (n=6). (B) Western blot analyses showing FRQ expression in the mut10 strain in DD. Asterisks indicate nonspecific bands. (C) Luciferase reporter assay showing the frq promoter activity in the WT (left) and mut10 (right) strains in DD. Raw data (above) were normalized to subtract the baseline calculated by LumiCycle Analysis software. WT, wild type.
CATP is an essential clock component
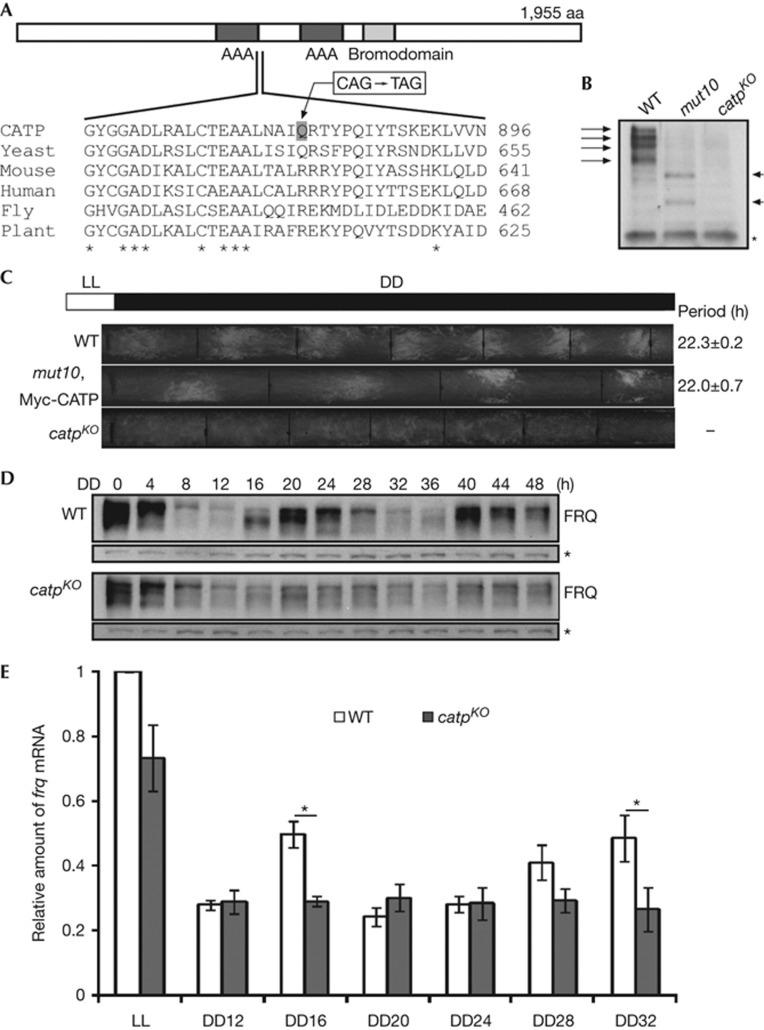
To identify the mutation responsible for the clock phenotype in the mut10 strain, we crossed it with a Mauriceville strain and performed race tube assays to examine the resulting progenies. We then performed cleaved amplified polymorphic sequence analyses utilizing the previously identified cleaved amplified polymorphic sequence markers in the genome [32]. The mutation was mapped to an ∼1-Mbp region in the right arm of linkage group III. To identify the mutation, whole-genome sequencing of the mut10 strain was performed. Comparison of the genome sequence of a wild-type strain with that of mut10 within the genetically mapped region revealed a single nucleotide change within the open reading frame of NCU06484 (Fig 3A). This mutation results in a nonsense mutation that changes Glu880 to a premature stop codon. The predicted protein product of NCU06484 is highly conserved in eukaryotic genomes from fungi to human and contains two ATPase domains and a non-canonical bromodomain (Fig 3A). Because of its role in the clock and its ATPase domains, this gene was named catp, for Clock ATPase.
Figure 3.
CATP is a critical component of the Neurospora circadian clock. (A) Domain structure of CATP and amino-acid sequence alignment of CATP homologues. (B) Western blot analysis showing CATP expression (indicated by arrows) in different strains. (C) Race tube analysis showing the circadian conidiation phenotype of different strains. Periods are shown as average±s.e.m. (n=6). (D) Western blot analysis showing FRQ expression in DD. (E) qRT–PCR results showing that rhythmic expression of frq is abolished in the catpKO strain. Mean with standard errors (n=3). *P<0.05 (paired student’s t-test). CATP, Clock ATPase; DD, constant darkness; KO, knockout; qRT–PCR, quantitative reverse transcriptase polymerase chain reaction; WT, wild type.
The premature stop codon in the mut10 mutant occurs immediately after the first ATPase domain. Thus mut10 should express a truncated CATP protein without the second ATPase domain and the bromodomain. We generated an antibody against amino terminus of CATP and examined CATP expression in wild-type, mut10, and a catpKO strain was obtained from the Neurospora crassa knockout library [33]. As shown in Fig 3B, the CATP antibody recognized four high-molecular-weight bands in the wild-type strain but not in the catpKO strain, indicating that they are CATP protein products. In the mut10 mutant, the wild-type CATP bands were absent and two main bands of lower molecular weight were observed, indicating that the premature stop codon resulted in truncated CATP products.
As shown in Fig 3C, the race tube phenotype of the catpKO strain is very similar to that of the mut10 strain with the circadian conidiation rhythm abolished after 2 days in DD. Furthermore, the circadian conidiation and slower growth phenotype of mut10 was rescued by introduction of a construct in which wild-type capt was expressed under the control of the quinic acid-inducible qa-2 promoter (Fig 3C). We tried to obtain the catpKO ras-1bd double mutant and were not successful, suggesting that the catpKO strain is sterile probably because of meiotic silencing. Together, these results indicate that CATP is a critical component in the Neurospora circadian clock.
CATP positively regulates frq expression
We compared FRQ expression in DD in the wild-type strain to that in the catpKO strain (Fig 3D; supplementary Fig S2B online). Unlike the robust oscillation of FRQ levels in the wild-type strain, the catpKO strain had a very low-amplitude FRQ fluctuation after the light-to-dark transition with no obvious changes in FRQ phosphorylation profiles. In addition, the overall FRQ levels were lower in the mutant. We next examined frq mRNA levels in DD by quantitative reverse transcriptase polymerase chain reaction analysis (Fig 3E). The catpKO strain exhibited lower frq mRNA levels in DD than in the wild-type strain, as the FRQ protein. These lower levels of frq mRNA in the catpKO strain are consistent with the results of expression of luciferase from the frq promoter in the mut10 strain (Fig 2C). These results indicate that CATP positively regulates frq transcription.
We also checked the light-induced gene expression in the catpKO strain and found that the light-induction of frq, al-1 and vvd expression was near normal (supplementary Fig S3 online), suggesting that the role of CATP is specific for circadian control of gene expression.
CATP is essential for the nucleosome occupancy rhythm
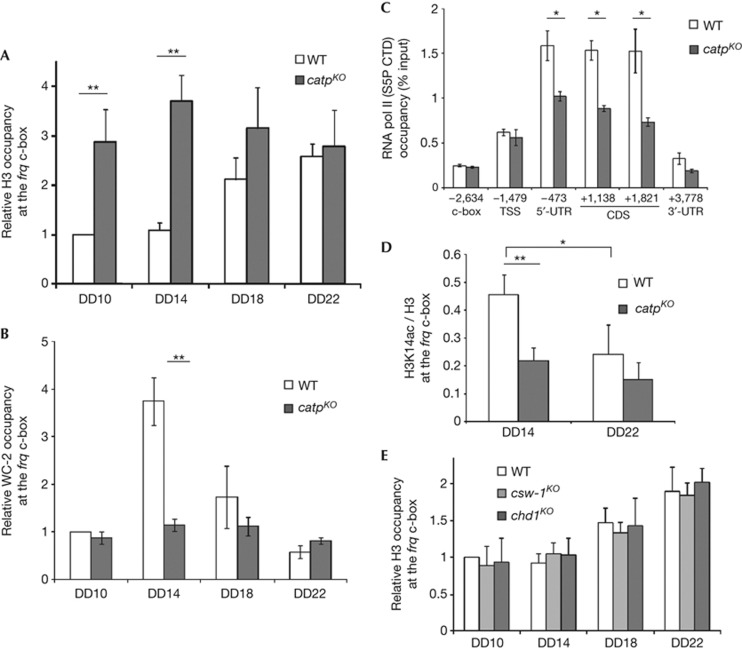
Yta7, the yeast homologue of CATP, is thought to function at the boundary of silent and active chromatin states and interacts with histones as well as histone chaperones [34, 35]. To understand the role of CATP, we examined nucleosome occupancy of the frq c-box in the catpKO strain. In contrast to the rhythmic H3 occupancy in the wild-type strain, the H3 occupancy level in the catpKO strain was arrhythmic and constantly high in DD (Fig 4A). The biggest difference in nucleosome occupancy between the two strains was observed at DD14, a time point when frq transcription peaks in the wild-type strain.
Figure 4.
CATP regulates chromatin structure at the frq locus. (A) H3 ChIP assay showing histone occupancy in DD. (B) WC-2 ChIP assays with the WT and the catpKO strains. (C) ChIP assays of the indicated strains using an S5P CTD antibody at DD14. Primer sets corresponding to different frq regions with indicated distances from the start codon (+1) were used to analyse pol II occupancies. (D) ChIP assays showing H3K14 acetylation at the frq c-box. IP/input values of H3K14ac ChIP assays were divided by those of H3 ChIP assays to calculate the ratio of acetylated H3K14. (E) H3 ChIP assays with indicated strains. Mean with standard errors (n=3). *P<0.05, **P<0.01 (paired student’s t-test). CATP, Clock ATPase; ChIP, chromatin immunoprecipitation; DD, constant darkness; IP, immunoprecipitation; KO, knockout; TSS, transcription start site; UTR, untranslated region; WT, wild type.
A ChIP assay using a WC-2 antibody showed that WCC-binding rhythm at the frq c-box was abolished and remained low throughout DD in the catpKO strain (Fig 4B). In addition, ChIP assays using an antibody specific for the phosphorylated Ser5 of the carboxy-terminal domain of the largest subunit of RNA polymerase II (pol II) showed that levels of the phosphorylated pol II were significantly reduced in the frq open reading frame in the catpKO strain compared with those in the wild-type strain (Fig 4C). Because pol II Ser5 phosphorylation is a marker for the completion of transcriptional initiation [36], this result indicates that CATP promotes transcription initiation of frq. It was previously shown that histone H3 acetylation, a marker for active chromatin, fluctuates at the frq locus [10]. As shown in Fig 4D, the rhythmic histone H3 Lys14 acetylation was abolished and the level of H3 Lys14 acetylation was reduced in the catpKO strain. Together, these results indicate that CATP regulates the rhythmic frq transcription by controlling nucleosome occupancy and chromatin structure to allow rhythmic activation and repression of frq transcription.
To examine whether CATP functions in the same pathway as CSW-1 and CHD1 to regulate frq transcription, we examined the histone occupancy at the frq promoter in the csw-1 and chd1 knockout strains. As shown in Fig 4E, the H3 occupancy levels were similar in the wild-type strain and these two mutant strains, indicating that the role of CATP in regulating histone occupancy at the frq promoter is independent of these two chromatin-remodelling enzymes.
CATP associates with chromatin at the frq locus
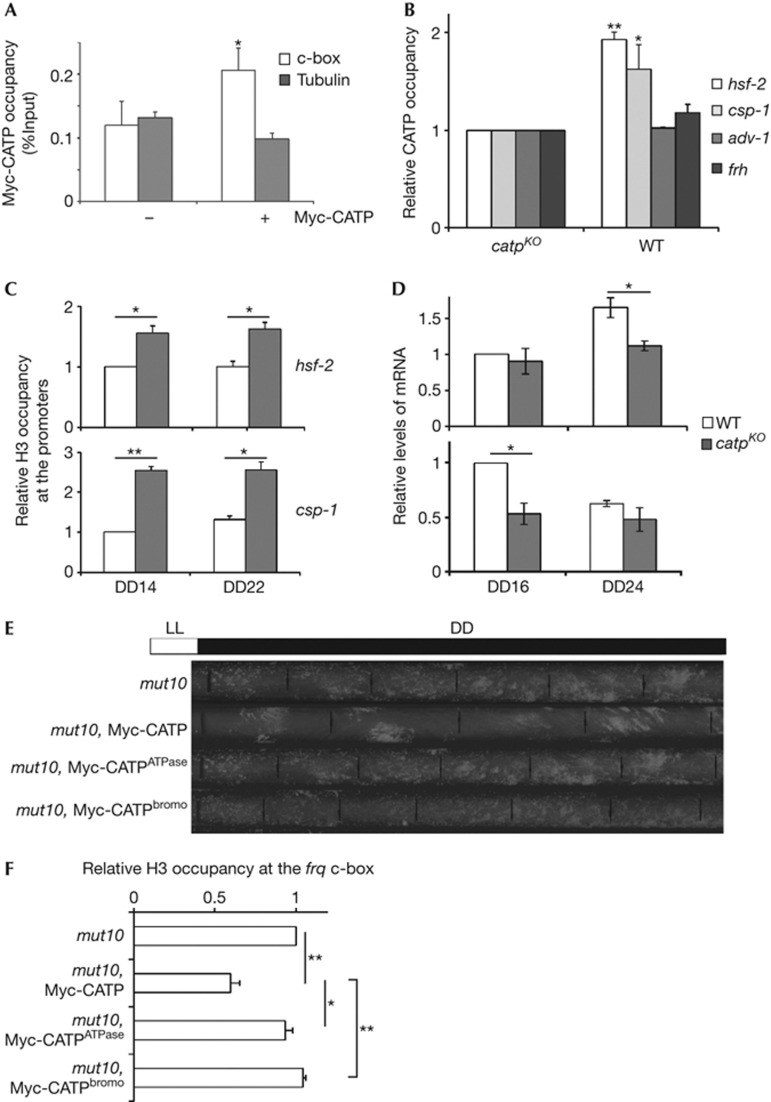
ChIP assays using both the Myc-tagged CATP and the antibody against endogenous CATP showed that it binds to the frq promoter and open reading frame regions (Fig 5A; supplementary Fig S4A online). These results indicate that CATP associates with chromatin at the frq locus, probably by interacting with the histones [34, 37]. In addition, these results indicate that the role of CATP in regulating frq transcription is specific and is not because of a nonspecific effect of CATP on general transcription.
Figure 5.
CATP associates with chromatin and requires its conserved domains for its function. (A) c-Myc ChIP assays with the Myc-tagged CATP strain at DD18. (B) ChIP assays using anti-CATP antibody to analyse CATP binding to the indicated loci. (C) H3 ChIP assays for hsf-2 and csp-1 loci in a WT strain at the indicated time points. (D) qRT–PCR analyses of hsf-2 and csp-1. (E) Race tube assays showing the circadian conidiation rhythms of CATP mutants. (F) H3 ChIP assays of the indicated strains in DD14. Mean with standard errors (n=3). *P<0.05, **P<0.01 (paired student’s t-test). CATP, Clock ATPase; ChIP, chromatin immunoprecipitation; DD, constant darkness; KO, knockout; qRT–PCR, quantitative reverse transcriptase PCR; WT, wild type.
We then examined whether CATP also acts on other WCC target genes. hsf-2, csp-1 and adv-1 are directly regulated by WCC [38]. In contrast, frh expression is not influenced by WCC [15]. Myc-tagged CATP showed nonspecific binding to the tubulin gene (Fig 5A), so it was used as the internal negative control for the ChIP assays (supplementary Figs S4A and S5B online). As shown in Fig 5B, there was significant CATP binding in the promoter regions of hsf-2 and csp-1 but not at the adv-1 and frh loci. Consistent with this result, histone H3 ChIP showed that the nucleosome occupancy at the hsf-2 and csp-1 loci, but not at adv-1 and frh loci, were significantly higher in the catpKO strain than in the wild-type strain (Fig 5C; supplementary Fig S4B online). Furthermore, quantitative reverse transcriptase polymerase chain reaction analyses showed that the mRNA levels of hsf-2 and csp-1 but not if adv-1 and frh, were downregulated in the catpKO strain (Fig 5D; supplementary Fig S4C online). Together, these results indicate that, in addition to frq, CATP affects the chromatin structures to regulate transcription of some WC target genes.
Conserved domains of CATP required for its function
The ATPase domains and the non-canonical bromodomain are conserved in all CATP eukaryotic homologues. We generated Myc-tagged CATP constructs that contain point mutations in the ATPase domain or bromodomain (CATPATPase and CATPbromo). Although the wild-type CATP construct could rescue the circadian conidiation phenotype of the mut10 strain, the mutant CATP constructs could not (Fig 5E). H3 ChIP assays showed that the nucleosome occupancy at the frq c-box was significantly reduced in the wild-type CATP strain but remained at high levels in the CATPATPase and CATPbromo strains (Fig 5F). These results demonstrate that the ATPase domain and the bromodomain of CATP are essential for its function in the clock.
Concluding remarks
In this study, we identified CATP as the critical clock component that regulates the nucleosome occupancy at the frq locus. Our results indicate that the nucleosome occupancy rhythm is important for clock function. They indicate that CATP promotes the removal of nucleosomes at the frq locus to enhance WCC binding. In contrast, FRQ promotes an increase of nucleosome occupancy at the c-box. Although the expression of CATP is not circadian (supplementary Fig S5 online), the counter balance of these two effects, therefore, results in rhythmic nucleosome occupancy at the frq promoter, which allows rhythmic binding of the WCC and rhythmic activation and repression of frq transcription. The regulation of nucleosome occupancy at the frq locus is, thus, part of the circadian negative feedback process that permits rhythmic transcription.
CATP specifically associates with the frq locus, suggesting that CATP regulates nucleosome occupancy directly on chromatin. In addition, CATP also associates with the WCC target genes, hsf-2 and csp-1 [38], and modulates the histone occupancy at these loci. Thus, CATP also participates in the circadian output pathway, but it remains to be demonstrated whether CATP acts directly or indirectly on nucleosomes.
Two chromatin remodelers, CSW-1 and CHD1, were previously shown to affect clock function by regulating the chromatin structure at the frq locus [10, 27]. CHD1 appears to affect the frq chromatin structure near the 3′ UTR region, and DNA methylation at the frq locus is altered in the chd1 mutant. On the other hand, micrococcal nuclease I assays showed that there is a rhythm of nuclease sensitivity near the c-box region of the frq promoter and CSW-1 appeared to promote the nuclease resistance in this region [10]. However, as the nuclease accessibility rhythms were present in both the wild-type and the csw-1KOstrains, an additional chromatin-remodelling factor is likely involved in regulating nucleosome occupancy at the frq promoter [10]. Interestingly, using ChIP assays, we found that the H3 occupancy at the frq c-box region is not altered in either csw-1 or chd1 mutants, suggesting that CATP acts differently from CSW-1 and CHD1. The difference between our results and the earlier findings might be because of the use of different assays. Nonetheless, the combination of these results indicates that CATP, CSW-1 and CHD1 act together to maintain the normal chromatin structure at the frq locus. In addition, we showed that histone H3K14 acetylation is also reduced in the catp mutant (Fig 4D), suggesting that changes in nucleosome occupancy and histone modifications act together to regulate the chromatin state of the frq locus.
The mammalian homologue of CATP, ANCCA, is required for the chromatin association of transcriptional activators [39, 40]. Mutations in the yeast homologue of CATP, yta7, further result in increased nucleosome occupancy at some genetic loci [41]. These results indicate that the role of CATP homologues in regulating nucleosome occupancy to permit transcription factor binding is conserved. How CATP and its homologues regulate nucleosome occupancy is not clear. It is possible that its non-canonical bromodomain interacts with histones and the ATPase domains generate energy to promote removal or degradation of nucleosomes. Consistent with this model, both Yta7 and ANCCA associate with histones [35, 40]. The conservation of circadian clock mechanisms from Neurospora to animals suggests that the circadian control of histone occupancy by CATP homologues might also be a critical process of circadian process in other organisms.
METHODS
Luciferase reporter assay. pfrq-luc-I (a generous gift from Dr Jay Dunlap) was transformed into wild-type and mut10 strains at the his-3 locus. The luciferase assay was performed and analysed as previously described [31, 42].
ChIP assays. ChIP assays were performed as previously described [22]. Bound DNA was eluted using Chelex 100 resin (Bio-Rad) and subjected to quantitative reverse transcriptase polymerase chain reaction. Occupancies were normalized by the ratio of ChIP to input (histone H3 ChIP assays) or the relative binding to β-tubulin gene (internal negative control for WC-2 and CATP ChIP assays).
Construction of point mutations of CATP. To generate mutant constructs of CATP, site-directed mutagenesis was carried out as previously described [43]. The mutated residues 700GTGKT704 in CATPATPase were changed to ATAEA; in CATPbromo,1226DPNF1229 was mutated to AAAA. The constructs were transformed into a mut10, his-3 strain at the his-3 locus.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Hai-Yan Yuan and Qianhong Ye for technical assistance and Dr Bing Li for valuable comments on histone ChIP assays. This work was supported by grants from the National Institutes of Health (GM068496 & GM062591) to Y.L. and from the Welch Foundation (I-1560) to Y.L.
Author contributions: J.C. and Y.L. designed the experiments and wrote the paper; J.C. and M.Z. performed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Liu Y (2007) The Neurospora crassa circadian clock. Adv Genet 58: 25–66 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bell-Pedersen D (2006) Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell 5: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC (2006) Proteins in the Neurospora circadian clockworks. J Biol Chem 281: 28489–28493 [DOI] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Liu Y (2001) Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc Natl Acad Sci USA 98: 7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ (1997) Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science 276: 763–769 [DOI] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee H, Yang Y, Liu Y (2006) CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20: 2552–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu Y (2005) Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev 19: 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC (2007) Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell 25: 587–600 [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Loros JJ, Dunlap JC (2003) Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci USA 100: 5914–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y (2002) PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol 22: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC (2001) Genetic and molecular analysis of circadian rhythms in NEUROSPORA. Annu Rev Physiol 63: 757–794 [DOI] [PubMed] [Google Scholar]
- Aronson B, Johnson K, Loros JJ, Dunlap JC (1994) Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science 263: 1578–1584 [DOI] [PubMed] [Google Scholar]
- Cheng P, He Q, He Q, Wang L, Liu Y (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 19: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Chen S, Li S, Cha J, Long C, Li L, He Q, Liu Y (2007) Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev 21: 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Cheng P, Yuan H, Liu Y (2009) The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138: 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T, Haase A, Kaldi K, Scholz J, Fuchs M, Brunner M (2005) Transcriptional feedback of neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122: 235–246 [DOI] [PubMed] [Google Scholar]
- Cha J, Chang SS, Huang G, Cheng P, Liu Y (2008) Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. Embo J 27: 3246–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, Dunlap JC (2008) Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev 22: 3196–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T, Diernfellner A, Schafer A, Dintsis O, Neiss A, Brunner M (2008) Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes Dev 22: 3397–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Shu H, Cheng P, Chen S, Wang L, Liu Y (2005) Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J Biol Chem 280: 17526–17532 [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182 [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Jansen A, Verstrepen KJ (2011) Nucleosome positioning in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 75: 301–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Lewis ZA, Selker EU, Loros JJ, Dunlap JC (2011) CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet 7: e1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raduwan H, Isola AL, Belden WJ (2013) Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 is needed for normal clock gene expression. J Biol Chem 288: 8380–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Johnson KA, Dunlap JC (1994) The circadian clock locus frequency: a single ORF defines period length and temperature compensation. Proc Natl Acad Sci USA 91: 7683–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N, Liu Y, Loros JJ, Dunlap JC (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476 [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC (2008) Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambreghts R et al. (2009) A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181: 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT (2005) Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol 169: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A et al. (2009) A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol Cell Biol 29: 4604–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P (2005) A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Gradolatto A, Rogers RS, Lavender H, Taverna SD, Allis CD, Aitchison JD, Tackett AJ (2008) Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM et al. (2010) Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot Cell 9: 1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW (2007) ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci USA 104: 18067–18072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko AS, Kalashnikova EV, Gemo AT, Zou JX, Chen HW (2010) Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol Cell Biol 30: 5260–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Ellahi A, Rine J (2011) Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc Natl Acad Sci USA 108: E1302–E1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Yuan H, Liu Y (2011) Regulation of the activity and cellular localization of the circadian clock protein FRQ. J Biol Chem 286: 11469–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.