Abstract
The human endogenous retrovirus family HERV-K(HML-2) Rec protein is an RNA transport factor that enhances nuclear export of intron-containing retroviral transcripts. Using the yeast two-hybrid approach, we have newly identified human Staufen-1 as a Rec-interacting protein. The interaction was confirmed by coimmunoprecipitation experiments, and the relevant site in Staufen-1 has been mapped to double-stranded RNA binding domain 4 (RBD4). Staufen-1 is in several aspects functionally related to retroviral RNA transport proteins. It binds mRNAs and targets its ribonuclear cargo to polysomes for efficient translation. We observed an accumulation of Staufen-1 in the nucleus of Rec-expressing cells and colocalization in the nucleoli as well as in the cytoplasm. Overexpression of Staufen-1 resulted in a 5-fold enhancement in nuclear export and/or translation of unspliced HERV-K(HML-2) viral RNAs in the presence of Rec and its Rec-responsive element (RcRE) binding site together with a clear increase in virus production. Staufen-1 was previously shown to interact with the Gag protein of HIV-1, promoting Gag oligomerization and RNA encapsidation. We demonstrate here that Staufen-1 also binds to the Gag protein of HERV-K(HML-2). Under stress conditions, Rec colocalizes with Staufen-1 in stress granules in cells that express viral RNA but not in mRNA-decay-related processing bodies. Our results suggest a new role for Staufen-1 as a cellular Rec and HERV-K(HML-2) Gag cofactor.
INTRODUCTION
Over millions of years of coevolution, retroviruses have developed strategies to overcome the tight connection between splicing and nuclear mRNA export to allow translation of intron-containing viral mRNAs and incorporation of full-length primary transcripts into particles in the cytoplasm. To achieve this, simple retroviruses rely on cis-acting sequence regions, known as constitutive export elements (CTEs), that depend entirely on factors encoded by the host cell genome. Complex retroviruses, on the other hand, encode accessory proteins that bind to intron-containing viral transcripts to facilitate their nucleocytoplasmic transport (1). These proteins include the Rev protein of HIV, the Rex protein of human T cell leukemia virus (HTLV), and the Rec protein of the human endogenous retrovirus HERV-K. These proteins bind selectively to intricate stem-loop structures in the terminal 3′ region of retroviral transcripts, the so-called responsive elements (REs), and use the host protein Crm-1 to support nuclear exit through the pore complex. In addition to their regulatory roles in nuclear mRNA export, there is evidence that Rev and the other related retroviral proteins facilitate translation of the transported mRNAs by enhancing the association with polysomes and accelerating the encapsidation of viral transcripts (2–4).
The functions of the HERV-K Rec protein in this regard are poorly understood. HERV-K is a group of endogenized betaretroviruses whose early germ line integrations occurred as much as 40 million years ago (5, 6). The youngest elements belong to the HERV-K(HML-2) family, with several integrations having occurred during the last 3 million years. Although all of the known proviruses have accumulated inactivating mutations or deletions during their germ line residence, open reading frames for functional proteins are preserved in many of the younger elements (7–9), including that coding for the HERV-K(HML-2) Rec protein (10). In healthy differentiated tissues, the expression of endogenous retroviruses is largely blocked by epigenetic factors such as promoter methylation. Germ cell tumors, melanomas, mammary carcinomas, and other malignancies frequently express these proteins, resulting in immune responses and virion assembly (11–17). The expression levels of HERV-K(HML-2) proteins and particles are also strongly elevated in HIV patients (18). To date, however, none of the viral particles produced have been shown to be replication competent (5, 9, 19). However, infectivity has been demonstrated using consensus sequences of the best-preserved HERV-K(HML-2) elements and using interelement recombinants (9, 19). Although regarded as unlikely, it is conceivable that replication-competent HERV-K(HML-2) elements are still present at a low frequency in the human population or that they sporadically reemerge by recombination (9, 20, 21).
The HERV-K(HML-2) Rec protein is encoded by two exons and consists of 105 amino acids. The 14-kDa protein accumulates primarily in the nucleoli but can also be found in the cytoplasm (10). It contains an arginine-rich nuclear location signal (NLS), which interacts with Importin-β, mediating entry into the nucleus. The NLS is also responsible for the recognition of the highly structured Rec-responsive element (RcRE) within the viral RNAs (vRNAs). It was recently shown that Rec forms tetramers, and presumably, three tetramers bind to purine-rich stretches within the RcRE (22). Rec contains a leucine-rich nuclear export signal (NES) for nuclear export of Rec/vRNA complexes. In addition to its function as an RNA transport protein, we have recently identified a transactivation activity of the viral protein in cells expressing the androgen receptor (10). HERV-K(HML-2) does not encode a Tat- or Tax-related accessory protein.
Many nuclear and cytoplasmic Rev and Rex cofactors have been identified, allowing several of their functions to be characterized (23, 24). In contrast, only a few Rec-interacting proteins have been identified so far. These include the promyelocytic leukemia zinc finger protein (PLZF), which plays a role in germ line stem cell pluripotency, and the related testicular zinc finger protein (TZFP) (25, 26). Interactions with these proteins and with human small glutamine-rich tetratricopeptide repeat-containing protein (hSGT) are all implicated in the proposed oncogenic activity of Rec that is absent in Rev and Rex (10, 27).
In the work presented here, we identified a new Rec-interacting protein, Staufen-1, which is known to be involved in nucleocytoplasmic RNA transport and which participates in “decisions” regarding the fate and utilization of the cargo RNA for efficient transport, transcription, particle encapsidation, or transport into stress granules.
MATERIALS AND METHODS
Plasmid construction.
The pcDNA3.1/wtRec-V5 mammalian expression plasmid was constructed by amplifying spliced rec RNA from cells transfected with the molecular bacterial artificial chromosome (BAC) clone of HERV-K113 (17) with the primer pair 5′-ACGTCGAAGCTTGCCATGAACCCATCGGAGATGCAAAG-3′ and 5′-TGCAGTCGGCGGCCGCTGGCCCGTTCTCGATGGTCGCTGTC-3′, with subsequent cloning into pcDNA3.1-V5 (Invitrogen) via its HindIII/NotI sites. Enhancement of expression was achieved by codon optimization of the rec gene. The codon-optimized rec sequence (coRec) was synthesized by fusion PCR using overlapping primers. CoRec-V5 and oricoRec-V5 (codon-optimized and V5-tagged reconstituted original rec sequence) were described previously (10). For yeast two-hybrid (YTH) screens, the wild-type rec sequence (wtRec) and oricoRec were cloned in frame into the LEX-A gene in pEG202 (OriGene) by amplifying the rec sequence with primers containing EcoRI/NotI sites. For immunofluorescence experiments, oricoRec was cloned into the pEGFP vector in frame with the enhanced green fluorescent protein gene (egfp) (Clontech) via HindIII/ApaI restriction sites. OricoRec-V5 expression in bacteria was carried out with a pET16b (Novagen) construct expressing the protein with an N-terminal 6×His tag. The NdeI/XhoI restriction sites were used for cloning. A FLAG-oricoRec construct was used for converse coimmunoprecipitation experiments. OricoRec was cloned behind the N-terminal FLAG tag using the HindIII/XhoI restriction sites. To avoid promoter-specific titration effects of translation factors, a pBud/oricoRec-V5 construct carrying an EF-1α promoter was used for the luciferase reporter assays. OricoRec-V5 was cloned into pBud (Invitrogen) behind a Kozak consensus sequence by using the KpnI/XhoI restriction sites.
To clone staufen-1 into expression vectors, cellular RNA from HEK 293T cells was isolated with an RNA extraction kit (Qiagen) and amplified with the OneStep RT kit (Qiagen) using the staufen-1 outer primers 5′-CTTCGTCCCTTCTTCCTCTCC-3′ and 5′-CTCGGCCCACTGGAGGTATC-3′. A subsequent nested PCR was performed by using the inner primers 5′-GATTAGATTGCAAGCTTATGAAACTTGGAAAAAAACCAATGT-3′ and 5′-GTGCGAGCTCCTCGAGTCAGCACCTCCCACACACAG-3′. staufen-1 was cloned into pCMVTag2B (Stratagene) via the HindIII/XhoI restriction sites. The Staufen-1–Cherry expression vector was constructed by cloning staufen-1 into the pmCherry N1 expression plasmid (Clontech) using the XhoI/HindIII restriction sites. The double-stranded RNA binding domain 4 (RBD4) deletion mutant was made by looping out amino acids 205 to 275 of staufen-1 within the pCMVTag2B vector by using a site-directed mutagenesis kit (Stratagene).
For pulldown experiments and production of recombinant proteins, staufen-1 was cloned into pGex-5x-1 (Amersham Bioscience) via the BamHI/NotI restriction sites. Alternatively, stop codons were introduced to truncate the expression construct. The following expression constructs were generated (mutagenesis primer sequences are in parentheses): pGex/Stau1, pGex/StauRBD2,3 (5′-GCAGTTGAACGAGTATAGCCTAGAATCAAAAAG-3′), pGex/StauΔ256-400 (5′-GGAACGGGCACCAACTAGTAGGTGGCCAAGCGCAAT-3′), pGex/StauRBD2,3,4 (5′-GGTTTCAAAGTCCCGTAGGCGCAGCCCACCAAA-3′), and pGexStauΔ306-400 (5′-GAAAAGTAACCTTTTTTTAACCTGGCTCTGGGGATGA-3′).
The efficiency of Rec-mediated RNA transport was measured by using firefly luciferase-based shuttle constructs (Shuttle and ShuttleRcRE) that were cloned into pcDNA4 (Invitrogen). This construct consists of the HERV-K113 5′ untranslated region (UTR) (amplified by using primers 5′-AGCTAGCGATGGCTAGCGGTTCCCCGGTTCCCCTTATTTC-3′ and 5′-CTCGTATCATGCGGTACCTATCACCCTAGCTTCTTCCGAG-3′), the coding sequence for firefly luciferase (5′-CAGGTGATAGGGTACCATGGAAGACGCCAAAAACAT-3′ and 5′-CAGGTGATAGGGATCCTTACACGGCGATCTTTCCGC-3′), HERV-K113 gag (5′-AGTGTGGGAAATTGTCGATAATAG-3′ and 5′-CAGGTGATAGGAATTCCTACTGCTGCACTGCCGCTTG-3′), and HERV-K113 env (5′-CCACAAGCGGCAGTGCACAGCAGTAGGAATTCG-3′ and 5′-GCATGATACGAGGGGCCCCTACACAGACACAGTAACAATC-3′). The ShuttleRcRE construct contains, besides the env open reading frame (ORF), the complete C-terminal coding sequence, including the RcRE. This was amplified and cloned by using the primers 5′-CCACAAGCGGCAGTGCACAGCAGTAGGAATTCG-3′ and 5′-GATGACTAGATGCGGCCGCTACACACCTGTGGGTGTTTCTCGT-3′. Intrinsic expression of Rec was prevented in both constructs by the introduction of stop codons in the rec ORF at nucleotide (nt) 16, nt 88, nt 109, and nt 244. The mutagenesis primers used were 5′-CATGAACCCATCGGAGTAGCAAAGAAAAGCACCTC-3′, 5′-TGACTCACAAGATGAACAAATAGGTGACGTCAGAAGAACAGTAGAAGTTGCCATCCACCAAGAAG-3′, and 5′-CTGCTTGCAGCCTTGTAGATTGTATCAATGGT-3′. Plasmids pBSK_oriHERV-K113 and pcDNA_CMVoricoHERV-K113 were used to express HERV-K113 virus-like particles with reconstituted proteins (5, 28, 29). pEYFP-TIA-1 and pmRFP-Dcp1a were generously provided by N. Kedersha and P. Anderson (Brigham and Women's Hospital, Boston, MA).
To downregulate staufen-1 expression by short hairpin RNA (shRNA), the following target sequences were cloned into the pLVTHM vector (Addgene): 5′-GGAGGTGAATGGAAGAGAATC-3′, 5′-GCCTGCAGTTGAACGAGTAAA-3′, 5′-GCCACAGACAAGCCCAGAATA-3′, 5′-GCTGCGCTGAACATCTTAAAG-3′, and 5′-GCAGGGAAGACAACAGAAACA-3′.
Yeast two-hybrid screen.
Two-hybrid experiments were performed by using the DupLex-A system (OriGene). To identify proteins interacting with Rec, Saccharomyces cerevisiae strain EGY48 was cotransformed with the pEG202/wtRec bait construct, the lacZ reporter plasmid pSH18-34 (OriGene), and a commercially available random-primed spleen cDNA library cloned into the pYESTrp2 vector (Invitrogen).
The transformants were plated onto yeast nitrogen base lacking Leu, Trp, Ura, and His and containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and were grown at 30°C for 3 to 4 days. All media were purchased from Clontech. Plasmid DNA from potentially positive blue colonies was isolated and transformed into Escherichia coli strain KC8 (Clontech) to recover the library plasmid. To confirm the screening results, recovered plasmids were reintroduced into competent EGY48 yeast cells containing pSH18-34 plus pEG202wtRec, pEG202oricoRec, or negative-control bait vectors. At least three colonies from each transformation were plated onto glucose- or galactose-containing selective media, as recommended by the manufacturer. Those library plasmid inserts that were shown to again interact (blue yeast colonies) were sequenced.
Recombinant proteins.
Recombinant 10×His-tagged Rec, glutathione S-transferase (GST)-tagged Staufen-1, and the different truncation mutants were produced in E. coli strain BL21(DE3) Codon Plus (Novagen). After transformation of pGex, pGex/Staufen-1, pGex/StauRBD2,3, pGex/StauΔ256-400, pGex/StauRBD2,3,4, pGexStauΔ306-400, or pET16b/oricoRec, exponentially growing cultures were induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG; Roth) for 3 h at 30°C and 37°C or overnight at room temperature. Bacterial cell pellets containing recombinant GST fusion proteins were resuspended in 50 ml phosphate-buffered saline (PBS) containing 1% Triton X-100, 1 mg lysozyme, 5 to 10 units of benzonase, and a protease inhibitor mixture (Roche Applied Science). The extracts were sonicated, incubated on ice for 30 min, and cleared by centrifugation. Glutathione-Sepharose 4B (Amersham Biosciences) was added to bind the soluble GST fusion protein. After extensive washing with PBS, the suspension was transferred to a column, and recombinant protein was eluted with GST elution buffer (10 mM reduced glutathione in 50 mM Tris-HCl [pH 8.0]), followed by dialysis in PBS. Bacterial pellets containing 10×His-tagged proteins were resuspended in 30 ml lysis buffer containing 50 mM Tris-HCl, 5% glycerol, 50 mM NaCl, 1 mg lysozyme, 5 to 10 units of benzonase, and a protease inhibitor mixture (Roche). The extracts were sonicated for 3 min on ice and cleared by centrifugation. Recombinant proteins were extracted from the lysate by nickel nitrilotriacetic acid (Ni-NTA)-agarose (Qiagen), followed by washing with lysis buffer containing 5 mM imidazole and 0.5% Triton X-100 (pH 7.2). The fusion protein was eluted in four fractions with increasing imidazole concentrations from 50 mM up to 1 M and was dialyzed against PBS.
In vitro binding assay (pulldown).
Three micrograms of GST fusion proteins and 3 μg of 10×His-tagged proteins were diluted in 500 μl GST binding buffer (1% Triton X-100, 100 mM NaCl, 10 mM MgCl2, 0.1% bovine serum albumin, and protease inhibitor mix [Roche Applied Science]). Glutathione-Sepharose 4B was added, and after 2 h of incubation at 4°C on a rotator, the beads were extensively washed with binding buffer containing 0.02% SDS and no bovine serum albumin. Finally, the beads were resuspended in sample buffer, and bound proteins were detected by Western blotting using an anti-His-horseradish peroxidase (HRP) antibody (Sigma), an anti-GST antibody (Sigma), and enhanced chemiluminescence (ECL) detection reagent (Pierce). Proteins carrying an additional V5 tag were detected by using an anti-V5-HRP antibody (Invitrogen).
Immunization.
To obtain a Staufen-1-specific antiserum, the coding mRNA was inserted into the pET16b vector (Novagen) and expressed in E. coli BL21(DE3) Codon Plus cells. Recombinant proteins were affinity purified on a Ni-NTA column. Proteins were eluted in 8 M urea and dialyzed against PBS. Wistar rats received four immunizations of 100 μg recombinant Staufen-1 protein over a period of 12 weeks, and sera were collected throughout. Animal experiments were performed according to institutional and state guidelines (§10a of German Tierschutzgesetz). The Staufen-1-specific sera generated were used at dilutions of 1:5,000 in immunoblots and 1:4,000 in immunofluorescence assays.
Cell culture.
HEK 293T and Cf2Th cells were grown in Dulbecco's modified Eagle's medium (D-MEM; Gibco) containing 10% fetal bovine serum supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine (TPP; Biochrome). The cells were maintained at 37°C in a 5% CO2 atmosphere. In stress experiments, cells were treated with 0.5 mM sodium arsenite for 1 h at 37°C prior to further analysis.
Coimmunoprecipitation and immunoblotting.
HEK 293T cells were transfected in 100-mm plates using either Polyfect transfection reagent (Qiagen) according to the manufacturer's instructions or calcium phosphate. For immunoprecipitation, transfected cells were treated with 500 μl lysis buffer (1% Triton X-100, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, and protease inhibitor mix), and lysates were cleared from cell debris by centrifugation. Antibodies covalently linked to Sepharose (Sigma) were used to precipitate proteins carrying V5 and FLAG tags. The mixture was incubated on a rotator at 4°C for 4 h and subsequently washed 10 times with lysis buffer supplemented with 0.02% SDS to reduce nonspecific binding.
The precipitates were separated on SDS-polyacrylamide gels, transferred onto a polyvinylidene difluoride (PVDF) membrane (Roth), and detected by standard Western blotting using ECL (Pierce and USB).
Cell fractionation for protein analysis.
To analyze intracellular protein distribution, transfected HEK 293T cells from three 10-cm cell culture dishes were gently detached, pooled in PBS, and pelleted at 1,000 rpm at 4°C. The cells were carefully resuspended in 300 μl hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], and protease inhibitor [Roche]) on ice. Twenty microliters of 10% NP-40 (Sigma-Aldrich) was added to dissolve the outer cell membrane, and the samples were mixed gently by inversion before centrifugation at 800 rpm for 5 min at 4°C. The clear supernatants containing the soluble cytosolic proteins were carefully removed and aliquoted for further analysis. The remaining cell pellet containing both nuclear and insoluble proteins was washed with 1 ml hypotonic buffer. After centrifugation at 800 rpm for 5 min, the nuclei were cracked by resuspending the cell pellets in 300 μl nuclear lysis buffer I (hypotonic buffer containing additionally 0.1% SDS, 400 mM ammonium sulfate, and 100 mM sodium sulfate) at room temperature. The viscous samples were treated with benzonase (Novagen) to fragment DNA and RNA until the samples were liquid. After centrifugation, the soluble nuclear fraction was removed for further analysis. The remaining pellet containing insoluble nuclear and membrane proteins was dissolved in 300 μl nuclear lysis buffer II (8 M urea, 100 mM NaH2PO4, 10 mM Tris [pH 4.5]). The purity of the fractions was confirmed by Western blotting using monoclonal mouse antitubulin and rabbit anti-histone H3 antibodies (both from Epitomics). Staufen-1 and HERV-K113 capsids were quantified both by Western blotting and by dot blotting of serial 1:2 dilutions (for Staufen-1) of each sample by using the Odyssey imaging system (Li-Cor). P values were estimated from three independent experiments using analysis of variance (ANOVA) with subsequent Bonferroni tests or a t test.
Immunofluorescence microscopy.
Cells (1 × 105) were grown in chamber slides or on glass plates in six-well plates (Nalge Nunc and Biochrom) and transfected by using Effectene or Polyfect reagent (Qiagen). At 24 h or 48 h posttransfection, cells were fixed with 2% paraformaldehyde (Roth) in PBS for 30 min, rinsed briefly in PBS, permeabilized with 0.5% Triton X-100 in PBS for 15 min, and washed 3 times with PBS. After incubation in Marvel (1% skim milk in PBS) for 20 min, cells were incubated with primary antibodies diluted in Marvel for 60 min at 37°C. The slides were washed 3 times with PBS, and the fluorophore-conjugated secondary antibodies were added for 30 min. Primary antibodies used were mouse anti-V5 (Serotec), rabbit anti-FLAG (Sigma), rat anti-Staufen-1, and the direct fluorophore-conjugated mouse anti-V5-Cy3 antibody (Sigma). Anti-mouse IgG-Cy3, anti-mouse IgG-Alexa 647, anti-rabbit IgG-Alexa 647, anti-rat IgG-Alexa 543, and anti-rabbit IgG-fluorescein isothiocyanate (FITC) antibodies were used as secondary antibodies as indicated and were purchased from Sigma-Aldrich, Invitrogen, and Jackson Immuno Research. The nucleus/chromatin was stained by 4′,6-diamidino-2-phenylindole (DAPI). After extensive washing with PBS, the cells were mounted in Mowiol, and images were obtained by using a Zeiss confocal laser scanning microscope (LSM780).
Luciferase and RT assay.
A dual-luciferase reporter assay from Promega was used to measure luciferase activity. Transfected HEK 293T cells were grown in 96-well plates and lysed at 48 h posttransfection with 40 μl passive lysis buffer (Promega). Cell debris was removed by centrifugation, 10 μl of the supernatant was transferred onto a white optical 96-well plate (Nunc), and activity was measured in a Bertholdt luminometer: 75 μl of luciferase reagent was added, and after a delay of 2 s, activity was measured for 10 s.
To quantify particles by reverse transcriptase (RT) activity, HEK 293T cells were seeded into 6-well plates and transfected with pcDNA_CMVoricoHERV-K113 and the indicated vectors. The amount of plasmid DNA was maintained by the addition of empty vectors. At 48 h posttransfection, supernatants were filtered (0.45 μm), and RT activity was measured by using the HS-Mg RT activity kit (Cavidi, Uppsala, Sweden).
Ultracentrifugation.
Viral particles were concentrated by ultracentrifugation of cell supernatants through a 20% sucrose cushion for 3 h at 4°C and 175,000 × g. The viral pellet was resuspended in 50 μl of 0.05 M HEPES, pH 7.2, before Western blot analysis.
RESULTS
Staufen-1 interacts with HERV-K(HML-2) Rec in YTH analyses.
Our first aim was to identify novel Rec-interacting proteins to gain insight into its role during HERV-K(HML-2) replication. We therefore performed YTH analyses with the HERV-K113 sequence reported previously by Turner and coworkers (17), here termed wtRec, and a random-primed human spleen library. After cultivation on different dropout media with and without X-Gal, screening of the spleen library yielded 20 potentially positive clones. The prey-bait interaction in 15 of these clones was confirmed by a retransformation assay. Restriction analysis revealed four different restriction patterns, and subsequent sequence analysis showed that 4 of these 15 clones carried a sequence comprising the C-terminal 251 amino acids of the human Staufen-1 protein (Fig. 1A). During the screening assays, we became aware that the HERV-K113 wtRec sequence contains two presumed postinsertional mutations: T89A and D96E (10). We therefore reversed these mutations by site directed mutagenesis in a codon-optimized version to reconstitute the original Rec (oricoRec) protein coded for by the virus at the time of integration. The amino acid sequence of the reconstituted HERV-K113 Rec matches the Rec consensus sequence of HERV-K(HML-2) (10). A YTH analysis with oricoRec as bait and Staufen-1 as prey in EGY48 yeast cells demonstrated that the interaction is not associated with the two postinsertional mutations in wtRec (data not shown). We used the oricoRec sequence for efficient expression of the viral protein in all subsequent experiments.
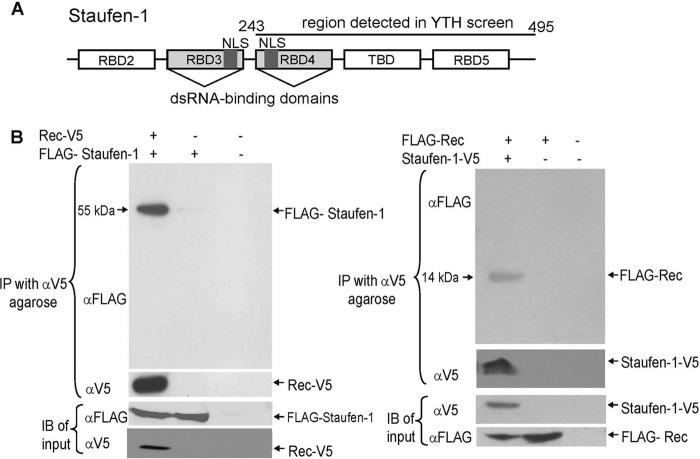
Fig 1.
Staufen-1 interacts with HERV-K(HML-2) Rec. (A) Schematic overview of the domain structure of Staufen-1. The region present in the Rec-interacting YTH clones (amino acids 243 to 495) is indicated. dsRNA, double-stranded RNA. (B) Coimmunoprecipitation experiments. (Left) V5-tagged HERV-K(HML-2) Rec and FLAG-tagged Staufen-1 or appropriate empty control vectors (−) were cotransfected into HEK 293T cells as indicated. By V5-mediated immunoprecipitation (IP), FLAG–Staufen-1 was detectable only in samples expressing Rec-V5. Expression of both proteins was confirmed by immunoblotting (IB). (Right) Converse experiment with Staufen-1-V5 coprecipitating FLAG-tagged Rec.
The Rec/Staufen-1 interaction is independent of yeast-specific factors.
To confirm the specificity of the interaction between Rec and Staufen-1, we performed coimmunoprecipitation experiments in which FLAG-tagged Staufen-1 and Rec-V5 or control vectors were cotransfected into HEK 293T cells. Cells were lysed, and the soluble cytosolic supernatants were incubated with anti-V5-Sepharose. Staufen-1 was detectable by immunoblotting (using an anti-FLAG antibody) only in samples with both proteins (Rec-V5 and FLAG–Staufen-1). No Staufen-1 was observable in controls transfected with the empty pcDNA3.1-V5 vector in place of pcDNA3.1/oricoRec-V5 (Fig. 1B, left, second lane) or with two empty plasmids (third lane). The specificity of this interaction was corroborated by converse experiments using Staufen-1–V5 and FLAG-Rec (Fig. 1B, right). These results indicate the existence of a protein complex consisting of Rec and Staufen-1 in a mammalian system that does not require the presence of any yeast-specific factors.
The Rec-interacting domain of Staufen-1 is located within the RBD4 region.
Human Staufen-1 is a protein consisting of five functional subdomains: RBD2, RBD3, RBD4, the C-terminal tubulin binding domain (TBD), and RBD5 (Fig. 1A and 2A). The YTH screen demonstrated that the Rec-interacting region is located in the C-terminal part of the protein. To determine the precise interacting region, we constructed a subset of C-terminally truncated Staufen-1 mutants that carry an N-terminal GST tag (Fig. 2A). These truncated proteins as well as the His-tagged Rec-V5 protein were expressed in E. coli BL21(DE3) and used for in vitro pulldown experiments with Ni-NTA–Sepharose. RNAs and DNAs were digested by benzonase.
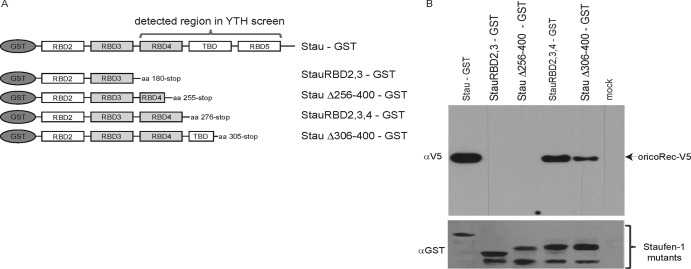
Fig 2.
The Rec-interacting region of Staufen-1 is located within RBD4. (A) Schematic overview of GST-tagged Staufen-1 mutants used for pulldown experiments with Rec-V5. All proteins were produced in E. coli. aa, amino acids. (B) Pulldown experiments using anti-GST agarose directed against GST-tagged Staufen-1 deletion mutants. Binding of full-length Rec-V5 was determined by using an anti-V5 antibody. (Top) Detection of coprecipitated Rec-V5 protein. (Bottom) Immunoblot of GST–Staufen-1 mutants used for pulldown.
These pulldowns revealed that the C-terminal part (amino acids 255 to 276) of the RBD4 region of Staufen-1 is essential for Rec binding (Fig. 2B). Both mutants lacking this amino acid stretch from positions 255 to 276 (StauRBD2,3 and StauΔ256-400) were unable to bind Rec (Fig. 2B, second and third lanes). Interestingly, the StauΔ256-400 mutant still harbors both functional RNA binding sites within the RBD3 and RBD4 regions as well as the nuclear location signals (indicated in Fig. 1A). RBD3 was previously shown to bind to the trans-activating response element (TAR) region on HIV-1 transcripts, enhancing translation and mediating the incorporation of 2 to 10 Staufen-1 molecules per virion (30–33). As the recombinant Rec and Staufen-1 proteins had been treated with benzonase prior to Ni-NTA or GST purification and mixing of the purified proteins in the pulldown assay, RNA-mediated coprecipitation can be virtually excluded. Moreover, in some assays, we added benzonase to the binding reaction mixture without affecting pulldown efficiency (data not shown). Furthermore, RNA binding regions of Staufen-1 (e.g., RBD3) were still present in the nonbinding Staufen-1 deletion mutants.
Rec expression traps Staufen-1 in the nucleus.
We next examined a putative subcellular colocalization of Rec-V5 and FLAG-tagged Staufen-1 in cotransfected HEK 293T cells by laser scanning confocal microscopy. In agreement with previous reports (10, 34–37), we observed HERV-K(HML-2) Rec in both the nucleoli and the cytoplasm, whereas FLAG–Staufen-1 was located exclusively in the cytoplasm, despite having a functional NLS, in cells that were not cotransfected with the Rec-V5 vector (Fig. 3A, top). These observations were verified and extended by cell fractionation experiments separating cytosolic and nuclear fractions from insoluble proteins. The majority of Rec was present in the insoluble fraction, indicating that Rec is predominantly in a tight association with nucleolar components or subsists in an aggregated form. Staufen-1 was found predominantly in the cytosolic fraction (Fig. 3A, bottom).
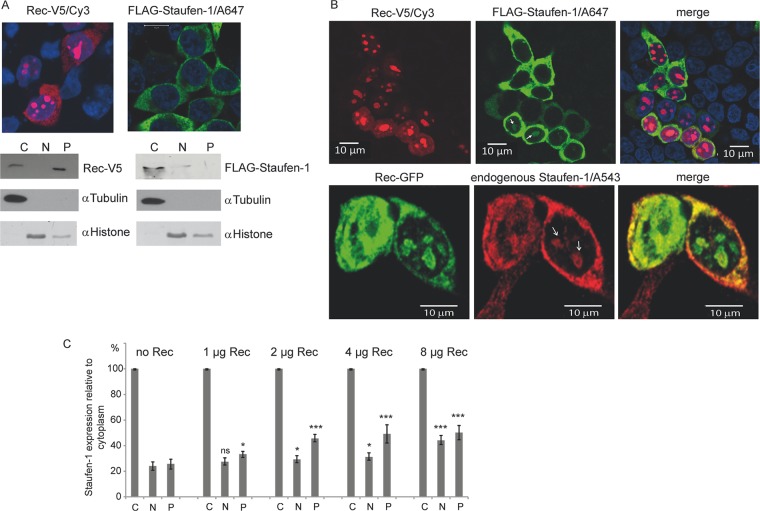
Fig 3.
Subcellular localization of Rec and Staufen-1. HEK 293T cells were transfected to express Rec-V5, FLAG–Staufen-1, or both proteins. (A) Immunofluorescence (top) and cell fractionation studies (bottom) of cells expressing each protein alone. The proteins were separated into cytoplasmic (C), nuclear (N), and insoluble pellet (P) fractions. Fractionation efficiency was confirmed with antibodies against tubulin as a cytoplasmic marker and histone as a nuclear protein. (B) Immunofluorescence of cells transfected with Rec-V5 and FLAG–Staufen-1 or Rec-GFP only. Colocalization of Rec with ectopically expressed (top) or endogenous (bottom) Staufen-1 in the cytosol and in bulky nucleoli of some cells is evident (indicated by white arrows). A647, Alexa 647. (C) Analysis of the Rec-dependent localization of endogenous Staufen-1 by cell fractionation into the cytoplasmic fraction, soluble nuclear fraction, and insoluble pellet from dot blot experiments using the Odyssey imaging system for quantification. The signal of the cytoplasmic fraction of each sample was set at 100% to estimate the change in endogenous Staufen-1 distribution in the other fractions. P values were calculated by comparing the signals from either the nuclear fractions or the protein pellets with those from cells without Rec. P values are indicated as ∗ (P < 0.05), ∗∗ (P < 0.01), ∗∗∗ (P < 0.001), and ns (not significant).
However, when coexpressed with Rec, the distribution of ectopically expressed FLAG–Staufen-1 and endogenous Staufen-1 was altered. Indeed, colocalization of both proteins in the cytoplasm was observed, but immunofluorescence staining also showed Staufen-1 in the nucleus colocalizing with Rec (Fig. 3B). The generally observed cytoplasmic localization of Staufen-1 was probably due to a rapid nuclear export and strong retention of the protein in the cytoplasm mediated by RBD2 and RBD3 of Staufen-1 (33). As shown here, expression of Rec partially overcomes this phenomenon, and a presumed interaction between Rec and Staufen-1 in the nucleoli seems to hold the protein in place or at least delay its export from the nucleus into the cytoplasm.
We followed this observation by performing additional cell fractionation experiments comparing the degree of endogenous Staufen-1 localization in each fraction with the amount of transfected Rec. Cytoplasmic Staufen-1 was set at 100%, and the relative amount of Staufen-1 in the nucleus and in the insoluble fraction was quantified by dot blotting. Staufen-1 was found to be located mainly in the cytoplasm but was also detected to a much lesser extent in the nuclear and insoluble protein fractions, as shown in Fig. 3C. However, higher Rec expression levels resulted in an increase in the amount of Staufen-1 detected relative to that in the cytoplasmic pool, in both the nuclear and the insoluble fractions (Fig. 3C). In particular, the latter fraction harbors nuclear Rec protein and other nucleolar proteins (38).
Expression of Rec-dependent viral RNAs is enhanced by Staufen-1.
When comparing the functional domains and activities of Rec and Staufen-1 within the cell, several similarities stand out. Both molecules contain a nuclear location sequence and an export signal. Both proteins also recognize and selectively shuttle RNAs containing distinctive secondary loop structures. To examine whether Staufen-1 modulates Rec-dependent viral RNA transport, two shuttling constructs were designed for transfection experiments using various combinations of Rec and Staufen-1 plasmids. These constructs consist of the HERV-K113 5′ UTR with the major splice donor site, a firefly luciferase gene, and the HERV-K113 gag and env genes with a splice acceptor site in env (Fig. 4A). Rec expression from these constructs was abolished by introducing four stop codons in its open reading frame. To discriminate between Rec-mediated RNA transport and nonspecific RNA leaks, we used two constructs containing or lacking the C-terminal RcRE, which was shown previously to be essential for specific Rec-mediated RNA transport (36, 37). The presence of functional splice sites upstream and downstream of the luciferase ORF permits only the transport of unspliced RNAs to be measured.
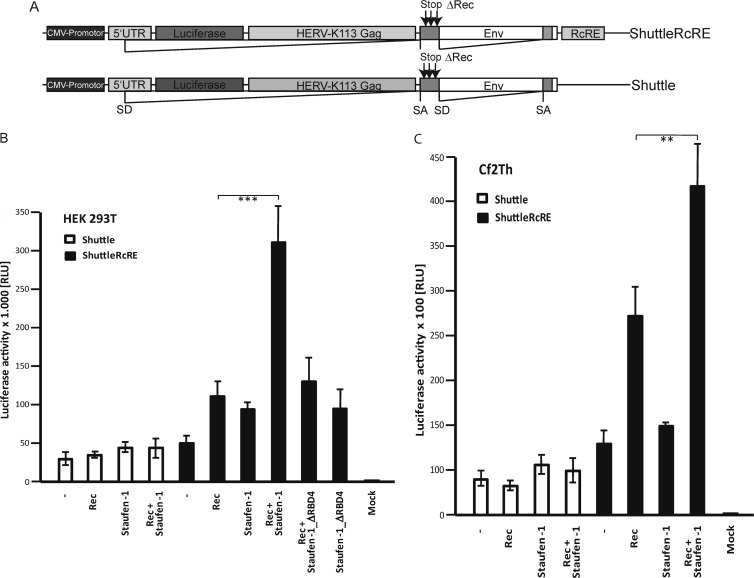
Fig 4.
Staufen-1 enhances Rec-mediated nuclear export and/or translation of viral RNA and leads to increased HERV-K(HML-2) particle production. (A) Schematic overview of the HERV-K(HML-2)-based reporter constructs used. Only ShuttleRcRE contains the RcRE site necessary for Rec binding. Intrinsic Rec expression was abolished by introduction of stop codons into the Rec ORF. CMV, cytomegalovirus; SA, splice acceptor; SD, splice donor. (B) Determination of Rec-mediated RNA transport efficiency and translation by a luciferase assay. HEK 293T cells were transfected with Rec and the indicated shuttle constructs. In addition, Staufen-1, Staufen-1_ΔRBD4 (a mutant with a deleted C-terminal part of the RBD4 region), or empty control vectors were cotransfected as indicated. The deletion mutant was expressed at a level similar to that of wild-type Staufen-1 (data not shown). Firefly luciferase values were normalized by a cotransfected Renilla luciferase expression construct. The signal without Staufen-1 was set at 100% for each shuttle construct. The effect mediated by cotransfection of Staufen-1 was evaluated relative to these measurements. ∗∗∗, P < 0.001. (C) The shuttle experiment was repeated with canine Cf2Th cells. Expression of human Staufen-1 results in an enhancement of luciferase expression in the presence of Rec, indicating increased nucleocytoplasmic transport and/or translation. ∗∗, P < 0.01. The experiment was repeated four times in HEK 293T cells and twice in Cf2Th cells, with similar results. Data from one representative experiment performed in quadruplicate for both cell types are shown with standard error bars. RLU, relative light units.
Coexpression of Staufen-1 with Rec and the shuttle construct lacking the RcRE did not result in a significant enhancement of luciferase activity in HEK 293T cells (Fig. 4B). In contrast, the reporter expression levels of the shuttle construct containing the RcRE increased by a factor of 5 when Staufen-1 was cotransfected with Rec. Rec expression alone enhanced transport and translation 2.5-fold in this system, while ectopic Staufen-1 overexpression doubled the luciferase expression level. Consistent with pulldown results, no increase was measured by the expression of a Staufen-1 protein lacking the C-terminal part of RBD4. Analogous results were obtained with canine Cf2Th cells not expressing endogenous Rec protein. In comparison to the results obtained with HEK 293T cells, transfection of human Staufen-1 had only an insignificant effect on the expression of the reporter gene (Fig. 4C). These results strongly indicate that Staufen-1 enhances Rec-mediated specific viral RNA shuttling from the nucleus into the cytoplasm and/or translation of the transcript at the ribosomes.
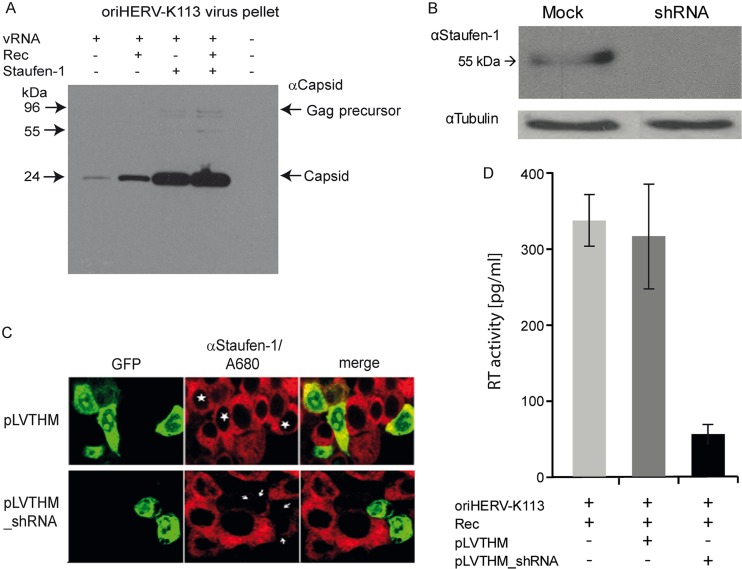
Staufen-1 has been shown to affect steps of the retroviral cycle beyond translation (30). We therefore extended our study to monitor the effects on particle production and release by comparing ultracentrifuge pellets from supernatants of HEK 293T cells transfected with a molecular clone of a reconstituted and presumably original HERV-K113 (oriHERV-K113) provirus encoding a functional Rec protein (10) in the presence and absence of cotransfected Staufen-1 and Rec. HEK 293T cells express only very low levels of endogenous Rec (5). A minority of the Rec transcripts expressed in this cell line has an open reading frame. A surplus of Rec alone was sufficient to cause a 5-fold increase in the level of HERV-K113 particles in the supernatant (Fig. 5A, compare first and second lanes). However, ectopic expression of Staufen-1 induced a 20-fold increase compared to the level in cells transfected with the virus construct alone (compare first and third lanes). At high Staufen-1 expression levels, a surplus of Rec had a very modest effect (an approximately 1.5-fold increase) (compare third and fourth lanes). Therefore, the Staufen-1-mediated increase depends on the Rec expression level, being more pronounced at lower levels. The same is true for Rec itself. If Staufen-1 is overexpressed, an increase in the Rec level results in a negligible enhancement of particle production.
Fig 5.
Effects of expression of Rec and Staufen-1 on HERV-K(HML-2) virus particle production. (A) HEK 293T cells were cotransfected with Rec-V5, Staufen-1, and oriHERV-K113, as indicated. Supernatants were filtered and pelleted by ultracentrifugation, and virus particles were detected by a HERV-K(HML-2) anticapsid polyclonal antibody. (B) Downregulation of Staufen-1 expression by a mix of Staufen-1-specific shRNAs. The top panel shows results of Western blotting with lysates of transfected HEK 293T cells. The blot was subsequently probed with an antitubulin antibody as a loading control. (C) Demonstration of the shRNA effect at the single-cell level. Cells transfected with pLVTHM_shRNAs (indicated by white arrows) show almost no staining with rat anti-Staufen-1 serum, while cells transfected with the empty vector pLVTHM (marked by stars) show staining similar to that of untransfected cells. The pLVTHM vector expresses GFP as a control. (D) In contrast to cells transfected with the empty pLVTHM vector, transfection with pLVTHM_shRNA significantly reduces the production of HERV-K113 virions. The experiment was repeated twice with duplicates. The means and standard errors are shown.
To determine whether a decrease in the Staufen-1 expression level affects particle production, we used a combination of five Staufen-1-specific shRNAs cloned into the pLVTHM vector that constitutively expresses GFP (as transfection control). Transfection of HEK 293T cells significantly reduced Staufen-1 protein expression (Fig. 5B). As shown in Fig. 5C, cotransfection of these shRNA vectors with HERV-K113 and Rec plasmids resulted in a 5-fold decrease in particle production, while no decrease was measured by cotransfection of the empty shRNA control vector pLVTHM. Therefore, a reduction in Staufen-1 expression has a profound influence on HERV-K(HML-2) virion production.
Staufen-1 interacts with the HERV-K(HML-2) Gag protein.
It was previously shown that Staufen-1 binds to the Gag protein of HIV-1 and plays a role in the encapsidation process of the viral RNA (30, 31). Taking into account this interaction, coimmunoprecipitation experiments were performed with lysates of HEK 293T cells transfected with expression vectors for FLAG–Staufen-1 and a V5-tagged version of a reconstituted HERV-K113 Gag protein (28). As shown in Fig. 6A, the HERV-K(HML-2) Gag protein coprecipitated Staufen-1, indicating an interaction between these proteins.
Fig 6.
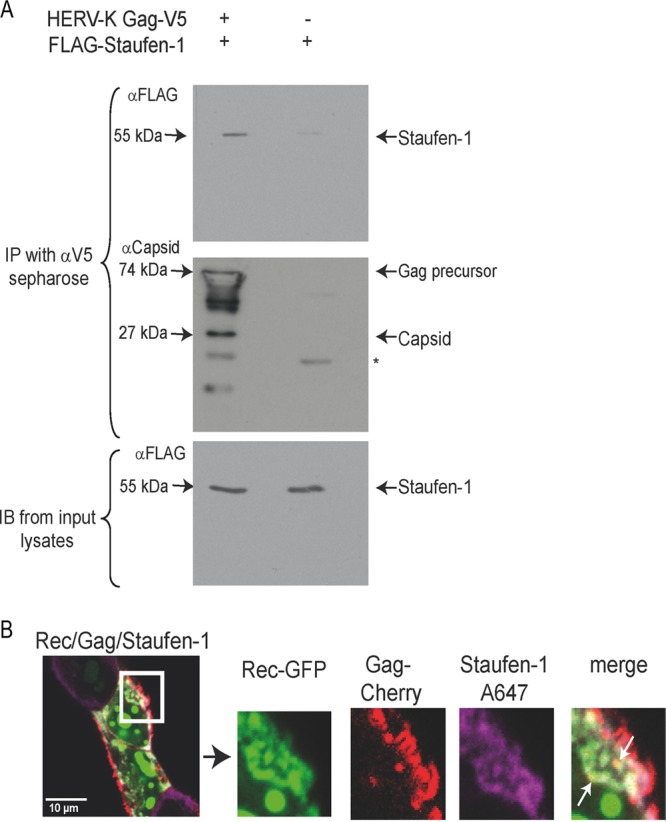
Staufen-1 is a binding partner of the HERV-K(HML-2) Gag protein. (A) Coimmunoprecipitation of FLAG–Staufen-1 with codon-optimized HERV-K113 Gag-V5 (28) using anti-V5-Sepharose. (Top) Coprecipitated FLAG–Staufen-1 detected by anti-FLAG. (Middle) Precipitated HERV-K(HML-2) Gag-V5. An anti-HERV-K(HML-2) capsid antiserum from rat was used for detection. The star marks the mouse light chain IgG band. (Bottom) The presence of FLAG–Staufen-1 in the lysates was verified. (B) Colocalization of Rec-GFP, HERV-K(HML-2) Gag-Cherry, and FLAG–Staufen-1 stained with anti-FLAG and anti-rabbit IgG-Alexa 647 in HEK 293T cells expressing oriHERV-K113. Small spots of colocalization are indicated by white arrows (see enlargement). No colocalization of HERV-K(HML-2) Gag with Staufen-1 is visible in the assumed budding structures at the cell membrane.
The subcellular colocalization of Cherry-labeled HERV-K113 Gag, Rec-green fluorescent protein (GFP), and Staufen-1–V5 was next analyzed in cells expressing all three proteins plus oriHERV-K113 as a provider of retroviral transcripts. Colocalization of labeled Rec, HERV-K(HML-2) Gag, and Staufen-1 appeared predominantly in spots in the cytoplasm close to the cell membrane (Fig. 6B). However, Rec and Staufen-1 were absent in Gag accumulations at the inner surface of the cell membrane. (Fig. 6B, enlargement). By electron microscopy, these areas were found to correspond to budding structures or clusters of viral particles (data not shown). These observations are consistent with results obtained for HIV-1. Rev is not present in viral particles, and Staufen-1 incorporation has been found to quantitatively correlate with vRNA and not with Gag (30, 39, 40). Therefore, the amount of Staufen-1 in budding structures or released HERV-K(HML-2) particles might simply be too small for detection using our approach. The presence of Rec or any Staufen-1 molecules in HERV-K(HML-2) virions has not yet been demonstrated.
Rec–Staufen-1–RNA complexes accumulate in stress granules.
Staufen-1 has many functions within the cell. One of its most striking tasks is the recruitment of RNAs into stress granules in stressed cells (41). During cellular stress situations, it is necessary to stop RNA translation for many of the proteins not required for stress control and reduction. Under cell stress conditions (e.g., arsenite-induced stress), the formation of stress granules and processing bodies (P-bodies) interrupts translation by diverting mRNAs from the polysomes (42). In contrast to stress granules, mRNAs in P-bodies are degraded under such conditions (42, 43).
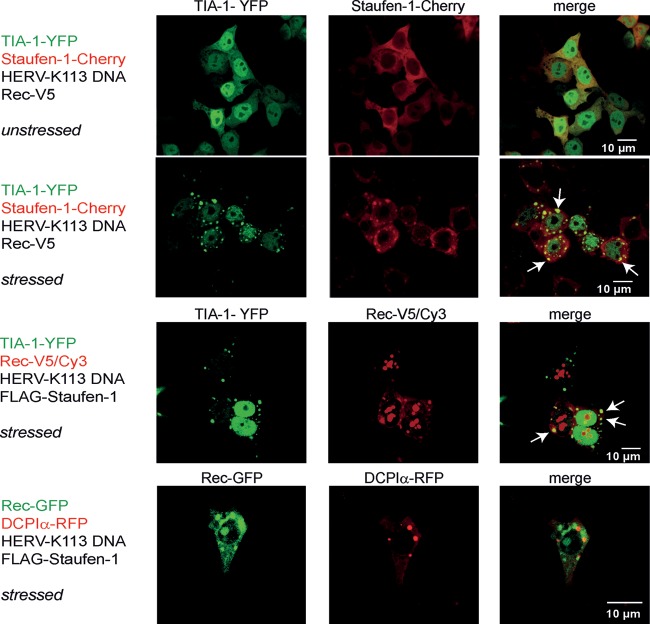
The interaction with Rec prompted us to investigate whether this viral protein colocalizes with Staufen-1 in the stress granules and P-bodies of cells expressing vRNA under cellular stress conditions. A fusion protein consisting of T-cell intracellular antigen 1 (TIA-1) and the yellow fluorescent protein (YFP) was coexpressed as a marker for stress granules (44), and while there was a diffuse pattern of TIA-1-YFP and Staufen-1–Cherry staining in unstressed HEK 293T cells, a clear dot-like colocalization pattern was seen in stressed cells (Fig. 7). In cells expressing Rec-V5, the viral protein was also found in TIA-1-containing stress granules but not in P-bodies visualized with a fusion protein between mRNA-decapping enzyme 1A (DCP1A) and the fluorescent protein (RFP) (43, 45, 46) (Fig. 7). These experiments demonstrated that Rec accumulates in stressed cells in stress granules but not in P-bodies, implying that translation of virus proteins is arrested but that vRNAs are not degraded.
Fig 7.
HERV-K(HML-2) Rec and Staufen-1 are present in stress granules. HEK 293T cells were transfected as indicated at the left of each panel. Fluorescence analysis was performed with untreated cells (top) or with cells stressed by 0.5 mM sodium arsenite. Colocalization is indicated by arrows.
DISCUSSION
The HERV-K Rec protein is a regulatory viral factor required for the efficient transport of intron-containing transcripts into the cytoplasm. This small viral protein comprises various functional domains, including a nuclear location signal, a nuclear export sequence, and presumably one or more regions for multimerization (36, 37, 47, 48).
To determine which host factors interact with Rec, we performed yeast two-hybrid analyses using the Rec variant encoded by the HERV-K113 element (17) as bait. In these screens, Staufen-1 was identified as a Rec binding partner. The interaction was confirmed by coimmunoprecipitations and was corroborated by immunofluorescence experiments using a reconstituted Rec variant that matches the consensus sequence of the youngest known elements of the HERV-K(HML-2) family. Moreover, Staufen-1 was shown to bind not only to Rec but also to the viral Gag protein. An interaction with the nucleocapsid region of the HIV-1 Gag protein has been reported previously (30).
Staufen-1 is an RNA transport protein that is expressed from the simplest to the most complex eukaryotes. It was first described in Drosophila melanogaster, where it plays an important role during “decision” processes and in the development of the anteroposterior axis during embryogenesis (49, 50). Furthermore, Drosophila Staufen influences the localization of various RNAs by direct binding to the target RNAs or as a component of ribonucleoprotein complexes (51, 52). Two counterparts of insect Staufen are present in humans: Staufen-1 and Staufen-2. Whereas Staufen-2 is synthesized exclusively in neuronal cells, Staufen-1 is expressed ubiquitously. It binds directly to RNAs at structured recognition motifs and is implicated in the transport processes of a variety of ribonucleoprotein complexes (53, 54). In neurons, for example, Staufen-1 is essential for the formation and distribution of RNA transport granules into dendrites (51, 55–60). The protein contains four double-strand RNA binding domains designated RBD2 to RBD5. RBD1 is not present in Staufen-1; it is found only in the Staufen-2 isoform and the insect homolog (61). We identified RBD4 as the Rec-interacting region. Similar to RBD3, this domain is likely to be involved in RNA binding and presumably also interacts with cellular proteins. The protein interacts with the cytoskeleton via its C-terminal tubulin binding domain (TBD) and enables Staufen-1-containing protein-RNA complexes (62) to arrive at their destinations, e.g., the rough endoplasmic reticulum or free polysomes (53, 54). There, Staufen-1 also has regulatory tasks during translational processes (32, 58). The protein stimulates translation within the 5′ UTR of repressed mRNAs carrying a special recognition motif (32) and is also an essential component of a recently described RNA decay mechanism (58, 63).
Our coimmunofluorescence and cell fractionation experiments indicate that the interactions of Staufen-1 with Rec take place in both the nucleus and the cytoplasm. It is known that Staufen-1 shuttles between these two compartments, and a significant nuclear or nucleolar localization has been described for cells expressing the NS1 protein of the influenza virus (64). These results and our findings make it likely that the expression of viral factors such as Rec and NS1 results in a retardation and subsequent accumulation of Staufen-1 in the nucleus.
Staufen-1 boosted the Rec-mediated nuclear export and/or translation of HERV-K(HML-2) RNA in our reporter shuttle assays. The particular step enhanced by Staufen-1 as well as the underlying mechanism remain to be determined. This effect was dependent on the presence of the RcRE. RNAs lacking this recognition motif did not benefit significantly from Staufen-1 overexpression, indicating that the relevant association of Staufen-1 with the vRNA for this process relies on Rec. It is currently unknown whether Staufen-1 binds directly to the HERV-K(HML-2) transcript, as was demonstrated previously for HIV-1 (30). We cannot exclude that Staufen-1 indeed interacts with structures in the RcRE or any other element on the vRNA, but a substantial effect on transport or translation was measured only if Rec was expressed. HERV-K(HML2) does not have a TAR element analogous to that used by Staufen-1 to attach to HIV-1 vRNA (32, 40). In the case of HIV, it is conceivable that Staufen-1 already binds directly to the TAR element in the nucleus via its RBD3.
The association with Rec appears to be critical for Staufen's involvement in the nuclear export of unspliced or partially spliced viral transcripts toward polysomes for translation or retroviral encapsidation of full-length HERV-K(HML-2) vRNAs into nascent viral particles (30, 60, 65, 66). Consistent effects were observed in experiments investigating the impact of Rec and Staufen-1 overexpression on particle production as well as in experiments in which Staufen-1 expression was downregulated by shRNAs. Staufen-1 and Rec overexpression resulted in a distinct enhancement of oriHERV-K113 production, while downregulation of endogenous Staufen-1 reduced particle production. The enhancing effect of Staufen-1 on virion production was greater than the effect in the shuttle experiments, pointing toward an involvement of Staufen-1 in replication steps subsequent to viral protein translation, in which the interaction with Gag plays a role. Particle production was especially augmented by Staufen-1 if the Rec expression level was low, and vice versa, indicating a mutual compensation between these two proteins in some aspects of their function. The promotion of virus production might be a cumulative effect of increased vRNA export, modulation of translation, and Gag assembly. A Staufen-1-mediated increase in retroviral particle production was described previously for HIV (39, 67). In those reports, the increase was attributed to a Staufen-1-mediated enhancement of HIV-1 Gag multimerization and assembly of immature capsids (39). RBD3 of Staufen-1 interacts with the nucleocapsid of the p55 Gag precursor of HIV-1, and an N-terminal region of the protein promotes Gag oligomerization (30). Interestingly, the increased HIV-1 particle assembly is associated with an increase in vRNA incorporation and a loss of infectivity (39, 40). The interaction of Staufen-1 with HERV-K(HML-2) Gag suggests a similar role for Staufen-1 in the multimerization and assembly of this endogenous virus.
The exact roles that Rev, Rex, Rec, and Staufen-1 play in the subcellular targeting and encapsidation process of genomic retroviral RNA and particle assembly are far from being understood. Several research groups reported for HIV-1 Rev that it “labels” unspliced and partially spliced viral RNAs and specifically targets these transcripts to their final destinations for RNA translation and packaging (2, 3, 68). At which step in the replication cycle the Rec–Staufen-1 complex dissociates from the vRNA destined for encapsidation or whether it is copackaged with the vRNA is not known. In HIV-1 assembly, Staufen-1 is packaged while bound to TAR, whereas Rev is not found in virions (30, 31, 40, 67).
Staufen-1 also has physiological functions during cell stress as a component of stress granules (41, 65, 69). These are dynamic aggregates containing mRNAs that are blocked during initiation of translation as well as 40S ribosomal subunits, translation initiation factors, and various RNA binding proteins (70). The RNA binding proteins TIA-1, TIA-R, and T-cell intracellular antigen 1 in particular self-aggregate under cell stress conditions and contribute to stress granule formation. Immunofluorescence experiments using TIA-1 as a stress marker and coexpression of vRNA revealed that Rec is present in stress granules induced by arsenite. Stress granules are an intermediate compartment between the translationally active polysomes and the RNA decay-linked P-bodies (43, 70). A highly dynamic exchange of mRNAs, including viral RNAs, occurs between these compartments. In contrast to stress granules, we did not see an association of Rec with P-bodies, which argues against P-bodies being a cellular complex with antiretroviral activity. This is in agreement with results of Abrahamyan and coworkers (71), who failed to find Staufen-1 and HIV-1 vRNA in P-bodies. Furthermore, in HIV-1-infected cells, Staufen-1 and HIV-1 Gag colocalized in ribonucleoprotein complexes distinct from typical stress granules, and the formation of regular arsenic-induced stress granules was inhibited by HIV-1 Gag.
In summary, based on our observations and all these considerations, Staufen-1 appears to associate with early nascent retroviral transcripts in the nucleus. For HERV-K(HML-2), this association is mediated by Rec, although an additional direct binding to the vRNA at this early stage cannot be excluded. However, only the Rec-mediated interaction significantly increases the nuclear export and/or translation of intron-containing transcripts and might link the ribonucleoprotein complex to the tubulin network through the C-terminal tubulin binding domain of Staufen-1 in the perinuclear region. This facilitates transport to polysomes, with a detour into storage granules under stress conditions. The association of Staufen-1 with Rec might also play a role in the transport of vRNA to rendezvous sites with the Gag protein, which themselves associate with Staufen-1. The exact roles that Rec and its new cofactor Staufen-1 play in vRNA transport processes, translation, encapsidation, and particle assembly remain to be deciphered.
ACKNOWLEDGMENTS
We thank Ewelina Caspers, Chris Daubitz, Maria Pack, and Kazimierz Madela for technical assistance and support and Steve Norley for critical reading of the manuscript. TIA-1–YFP and DCPIA-RFP were a generous gift from Nancy Kedersha (Brigham and Women's Hospital, Boston, MA).
Footnotes
Published ahead of print 7 August 2013
REFERENCES
- 1.Wodrich H, Krausslich HG. 2001. Nucleocytoplasmic RNA transport in retroviral replication. Results Probl. Cell Differ. 34:197–217. [DOI] [PubMed] [Google Scholar]
- 2.Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. 2010. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J. Virol. 84:6598–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. 2007. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 3:e54. 10.1371/journal.ppat.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Agostino DM, Felber BK, Harrison JE, Pavlakis GN. 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 12:1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beimforde N, Hanke K, Ammar I, Kurth R, Bannert N. 2008. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371:216–225. [DOI] [PubMed] [Google Scholar]
- 6.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 7:149–173. [DOI] [PubMed] [Google Scholar]
- 8.Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl 2):14572–14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanke K, Chudak C, Kurth R, Bannert N. 2013. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int. J. Cancer 132:556–567. [DOI] [PubMed] [Google Scholar]
- 11.Boller K, Schonfeld K, Lischer S, Fischer N, Hoffmann A, Kurth R, Tonjes RR. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89:567–572. [DOI] [PubMed] [Google Scholar]
- 12.Buscher K, Hahn S, Hofmann M, Trefzer U, Ozel M, Sterry W, Lower J, Lower R, Kurth R, Denner J. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 16:223–234. [DOI] [PubMed] [Google Scholar]
- 13.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65:4172–4180. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S, Ugurel S, Hanschmann KM, Strobel H, Tondera C, Schadendorf D, Lower J, Lower R. 2008. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res. Hum. Retroviruses 24:717–723. [DOI] [PubMed] [Google Scholar]
- 15.Herbst H, Kuhler-Obbarius C, Lauke H, Sauter M, Mueller-Lantzsch N, Harms D, Loning T. 1999. Human endogenous retrovirus (HERV)-K transcripts in gonadoblastomas and gonadoblastoma-derived germ cell tumours. Virchows Arch. 434:11–15. [DOI] [PubMed] [Google Scholar]
- 16.Lower R, Lower J, Frank H, Harzmann R, Kurth R. 1984. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J. Gen. Virol. 65(Part 5):887–898. [DOI] [PubMed] [Google Scholar]
- 17.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535. [DOI] [PubMed] [Google Scholar]
- 18.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 22:979–984. [DOI] [PubMed] [Google Scholar]
- 19.Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3:e10. 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79:12507–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22:814–817. [DOI] [PubMed] [Google Scholar]
- 22.Langner JS, Fuchs NV, Hoffmann J, Wittmann A, Brutschy B, Lower R, Suess B. 2012. Biochemical analysis of the complex between the tetrameric export adapter protein Rec of HERV-K/HML-2 and the responsive RNA element RcRE pck30. J. Virol. 86:9079–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suhasini M, Reddy TR. 2009. Cellular proteins and HIV-1 Rev function. Curr. HIV Res. 7:91–100. [DOI] [PubMed] [Google Scholar]
- 24.Younis I, Green PL. 2005. The human T-cell leukemia virus Rex protein. Front. Biosci. 10:431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. 2007. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 81:5607–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann S, Sauter M, Schmitt M, Baumert B, Best B, Boese A, Roemer K, Mueller-Lantzsch N. 2010. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J. Gen. Virol. 91:1494–1502. [DOI] [PubMed] [Google Scholar]
- 27.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228. [DOI] [PubMed] [Google Scholar]
- 28.George M, Schwecke T, Beimforde N, Hohn O, Chudak C, Zimmermann A, Kurth R, Naumann D, Bannert N. 2011. Identification of the protease cleavage sites in a reconstituted Gag polyprotein of an HERV-K(HML-2) element. Retrovirology 8:30. 10.1186/1742-4690-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanke K, Kramer P, Seeher S, Beimforde N, Kurth R, Bannert N. 2009. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus K and modulation of its functional activity by truncation of the cytoplasmic domain. J. Virol. 83:12790–12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5:41. 10.1186/1742-4690-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 24:2637–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 33:4797–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martel C, Macchi P, Furic L, Kiebler MA, Desgroseillers L. 2006. Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem. J. 393:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lower R, Boller K, Hasenmaier B, Korbmacher C, Muller-Lantzsch N, Lower J, Kurth R. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. U. S. A. 90:4480–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magin-Lachmann C, Hahn S, Strobel H, Held U, Lower J, Lower R. 2001. Rec (formerly Corf) function requires interaction with a complex, folded RNA structure within its responsive element rather than binding to a discrete specific binding site. J. Virol. 75:10359–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magin C, Lower R, Lower J. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takata H, Nishijima H, Ogura S, Sakaguchi T, Bubulya PA, Mochizuki T, Shibahara K. 2009. Proteome analysis of human nuclear insoluble fractions. Genes Cells 14:975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. 2007. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J. Virol. 81:6216–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74:5441–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 122:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. [DOI] [PubMed] [Google Scholar]
- 43.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22. [DOI] [PubMed] [Google Scholar]
- 44.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kedersha N, Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90:155–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kedersha N, Tisdale S, Hickman T, Anderson P. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552. [DOI] [PubMed] [Google Scholar]
- 47.Boese A, Galli U, Geyer M, Sauter M, Mueller-Lantzsch N. 2001. The Rev/Rex homolog HERV-K cORF multimerizes via a C-terminal domain. FEBS Lett. 493:117–121. [DOI] [PubMed] [Google Scholar]
- 48.Magin C, Hesse J, Lower J, Lower R. 2000. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology 274:11–16. [DOI] [PubMed] [Google Scholar]
- 49.Riechmann V, Ephrussi A. 2001. Axis formation during Drosophila oogenesis. Curr. Opin. Genet. Dev. 11(4):374–383. [DOI] [PubMed] [Google Scholar]
- 50.Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. 1998. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 12(12):1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. 1999. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10:2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. 2003. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. U. S. A. 100:2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. 1999. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duchaine TF, Hemraj I, Furic L, Deitinghoff A, Kiebler MA, DesGroseillers L. 2002. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 115:3285–3295. [DOI] [PubMed] [Google Scholar]
- 56.Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–525. [DOI] [PubMed] [Google Scholar]
- 57.Kiebler MA, DesGroseillers L. 2000. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron 25:19–28. [DOI] [PubMed] [Google Scholar]
- 58.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 26:2670–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krichevsky AM, Kosik KS. 2001. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696. [DOI] [PubMed] [Google Scholar]
- 60.Villace P, Marion RM, Ortin J. 2004. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32:2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miki T, Takano K, Yoneda Y. 2005. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct. Funct. 30:51–56. [DOI] [PubMed] [Google Scholar]
- 62.Milev MP, Ravichandran M, Khan MF, Schriemer DC, Mouland AJ. 2012. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Front. Microbiol. 3:367. 10.3389/fmicb.2012.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120:195–208. [DOI] [PubMed] [Google Scholar]
- 64.Falcon AM, Fortes P, Marion RM, Beloso A, Ortin J. 1999. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 27:2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brendel C, Rehbein M, Kreienkamp HJ, Buck F, Richter D, Kindler S. 2004. Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 384:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo M, Duchaine TF, DesGroseillers L. 2002. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 365:817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milev MP, Brown CM, Mouland AJ. 2010. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology 7:41. 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. 2005. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell 16:405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson P, Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 123:369–383. [DOI] [PubMed] [Google Scholar]