Abstract
The HIV-1 envelope glycoprotein (Env) mediates viral entry into host cells and is the sole target of neutralizing antibodies. Much of the sequence diversity in the HIV-1 genome is concentrated within Env, particularly within its gp120 surface subunit. While dramatic functional diversity exists among HIV-1 Env isolates—observable even in the context of monomeric gp120 proteins as differences in antigenicity and immunogenicity—we have little understanding of the structural features that distinguish Env isolates and lead to isolate-specific functional differences, as crystal structures of truncated gp120 “core” proteins from diverse isolates reveal a high level of structural conservation. Because gp120 proteins are used as prospective vaccine immunogens, it is critical to understand the structural factors that influence their reactivity with antibodies. Here, we studied four full-length, glycosylated gp120 monomers from diverse HIV-1 isolates by using small-angle X-ray scattering (SAXS) to probe the overall subunit morphology and hydrogen/deuterium-exchange with mass spectrometry (HDX-MS) to characterize the local structural order of each gp120. We observed that while the overall subunit architecture was similar among isolates by SAXS, dramatic isolate-specific differences in the conformational stability of gp120 were evident by HDX-MS. These differences persisted even with the CD4 receptor bound. Furthermore, surface plasmon resonance (SPR) and enzyme-linked immunosorbance assays (ELISAs) showed that disorder was associated with poorer recognition by antibodies targeting conserved conformational epitopes. These data provide additional insight into the structural determinants of gp120 antigenicity and suggest that conformational dynamics should be considered in the selection and design of optimized Env immunogens.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) facilitates viral entry into host cells through a series of receptor-mediated conformational changes that lead to fusion of the viral and host membranes. Env is a heavily glycosylated trimer of the gp120 surface subunit and gp41 transmembrane subunit heterodimers. As the primary target of the humoral immune response against HIV-1 (1–3), Env is the focus of intensive vaccine design efforts (4). HIV-1 escape from neutralizing antibodies generates exceptional diversity within the Env gene, which is particularly concentrated within the variable loops of gp120 (V1 to V5) (5–8). It is widely believed that an effective antibody-based HIV-1 vaccine would need to elicit antibodies capable of recognizing diverse Env isolates, ideally including “broadly” neutralizing antibodies (NAbs) as well as nonneutralizing antibodies with antibody-dependent cellular cytotoxicity (ADCC) effector functions, which appeared to correlate with protection in the RV144 HIV-1 vaccine trial (9). Indeed, the hopeful results of the RV144 trial, which provided evidence that vaccine-induced protection against HIV-1 may be possible (10), suggested that monomeric gp120 is a relevant HIV-1 vaccine immunogen and highlighted the importance of understanding the structural features that distinguish gp120 proteins and influence gp120 reactivity with neutralizing and ADCC-active antibodies (11–13).
Although the sequence and functional diversity of HIV-1 Env have been well-described (8, 14, 15), the degree of structural variability among global Env isolates, which must be overcome by broadly cross-reactive neutralizing and ADCC-active antibodies, is poorly understood. Similarly, it is unclear what structural features are associated with improved antibody recognition of Env immunogens and how these features vary among immunogens derived from distinct HIV-1 isolates. Cryo-electron microscopy studies have provided evidence that trimeric Env from distinct isolates can adopt different quaternary conformations on the virus surface (16, 17), but the detailed structural differences underlying these large-scale morphological rearrangements have not been resolved. Crystal structures of the HIV-1 gp120 core, with variable loops and glycosylation largely removed, have been determined for a number of Env isolates from multiple clades (18–26). The available structures indicate that the gp120 core is organized into a conserved inner domain composed of three layers (22), a heavily glycosylated outer domain, and a bridging sheet subdomain that forms upon CD4 binding (18). Furthermore, these truncated gp120 structures reveal a striking degree of structural conservation in the gp120 core across clades (26, 27). This conservation contrasts with the significant functional variability among diverse Env isolates, including differences in sensitivity to neutralizing antibodies (14), coreceptor usage (28, 29), CD4 reactivity (30), dependence upon CD4 (31, 32), antigenicity (15), and immunogenicity (33). This discrepancy between the functional diversity of Env and the apparent structural conservation of gp120 has often been attributed to quaternary structural differences in trimeric Env, which may not be apparent in monomeric gp120 (16, 21). However, differences in the antigenicity (15) and immunogenicity (33) of gp120 proteins from diverse isolates suggest that there is substantial conformational heterogeneity among isolates, even in the context of monomeric gp120.
The apparent homogeneity of the available gp120 core structures is rooted in the fact that the most divergent features of gp120, the variable loops and glycans (34), are truncated in order to facilitate crystallization. This is problematic for understanding the relationship between gp120 structure and antigenicity, because these elements are primary regulators of antibody binding to Env (35–37). Variable loop truncation has also been reported to allow the core to adopt the receptor-bound state, even in the absence of receptor, potentially favoring uniformity among the core structures (26). Additionally, most structures feature gp120 core in complex with a stabilizing ligand, such as soluble CD4 (sCD4) or antigen binding antibody fragments (Fabs), which may influence the observed core structure (21). The available crystal structures of truncated core constructs thus provide a detailed picture of how antibodies and receptors recognize the conserved elements of gp120, but the structural basis for isolate-specific antigenic differences among gp120 proteins remains poorly characterized.
To better understand the structure-function relationship that governs gp120 Env antigenicity, it is necessary to study constructs with intact variable loops (V1 to V5) and glycosylation under native conditions. The critical importance of characterizing gp120 in solution was supported by previous work that suggested that the global disorder of gp120 Env may disfavor antibody and receptor binding (38). Conformational flexibility is difficult to assess by X-ray crystallography, however, and requires complementary approaches, such as hydrogen-deuterium exchange with mass spectrometry (HDX-MS), which can elucidate the dynamic characters of proteins in solution. HDX-MS was used in two recent studies to examine disorder and structural flexibility in gp120 and to demonstrate that regions throughout the subunit, including the CD4 and coreceptor binding sites, are disordered in unliganded minimal core and full-length gp120 in solution (39, 40). Additionally, a recent study used molecular dynamics modeling to identify isolate-specific differences in residue communication networks within gp120 that contribute to differences in coupled motions among isolates. The study highlighted the role that molecular movements may play in generating conformational diversity among isolates, even within the gp120 core (41).
Here, we investigated the extent of isolate-specific structural differences in a panel of full-length gp120s in their unliganded and CD4-bound forms, and we examined whether site-specific differences in dynamics relate to differences in association with sCD4 and antibodies directed against a variety of linear and conformational epitopes. Four Env isolates with widely divergent phenotypes were compared (Table 1): SF162 (42), HXB2 (43), 1084i (44), and 1157ip (45). These isolates are derived from clades B and C, exhibit differences in coreceptor usage, vary in their sensitivity to neutralization by the CD4 binding site-directed antibody IgG1-b12, and exhibit a range of variable loop lengths and number of potential N-linked glycosylation sites (PNGS) (Table 1). Of these, HXB2 and 1084i are the most divergent isolates, with a sequence identity of 71.4%, based on 486 overlapping amino acids. Given the functional diversity of these Env isolates and the variability of their gp120 amino acid sequences, we hypothesized that there are measurable isolate-specific differences in solution structure and stability among the gp120s and that differences such as these might contribute to the antigenic variability of gp120 proteins.
Table 1.
Characteristics of the four Env isolates
| Isolate | Source | Clade | Coreceptor usage | b12 IC50a (μg/ml) | Loop length (no. of aa) |
No. of PNGS |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | C1 | V1/V2 | C2 | V3 | C3 | V4 | C4 | V5 | C5 | Total | |||||
| 1157ip | Plasma | C | CCR5b | 0.7c | 21 | 43 | 35 | 28 | 13 | 1 | 6 | 8 | 1 | 3 | 4 | 2 | 2 | 0 | 27 |
| 1084i | Plasma | C | CCR5d | 20e | 16 | 44 | 35 | 21 | 10 | 1 | 6 | 6 | 1 | 3 | 4 | 2 | 1 | 0 | 24 |
| HXB2 | TCLAf | B | CXCR4g | 0.01h | 26 | 40 | 36 | 34 | 11 | 1 | 5 | 8 | 1 | 3 | 4 | 1 | 1 | 0 | 24 |
| SF162 | CSFi | B | CCR5g | <0.01h | 25 | 40 | 35 | 28 | 12 | 1 | 3 | 6 | 1 | 3 | 5 | 1 | 1 | 0 | 21 |
The 50% inhibitory concentration of IgG-b12.
Data were obtained from reference 25.
Data were obtained from reference 102.
Data were obtained from reference 44.
Data were obtained from reference 103.
TCLA, T cell line adapted cells.
Data were obtained from reference 101.
Data were obtained from reference 104.
CSF, cerebrospinal fluid.
We used small-angle X-ray scattering (SAXS) and HDX-MS to characterize the full-length, glycosylated gp120 monomers in solution. SAXS provides a low-resolution structure for proteins in solution (46–48) and can identify the location of the large V1/V2 loops relative to the conserved core (40). HDX-MS provides complementary information on solvent accessibility and local structural dynamics by measuring the rate of deuterium incorporation into the amide groups of the protein backbone (49). Amides that are buried deep within a protein's hydrophobic core or that participate in stable hydrogen bond networks, such as those in protein secondary structures, take on deuterium slowly, while those in solvent-accessible, flexible, or dynamic regions of the native protein take on deuterium rapidly (50).
We observed that while the overall architecture of full-length, glycosylated gp120 is similar among diverse isolates, there are clear isolate-specific differences in the conformational dynamics of both unliganded and CD4-bound gp120, particularly within the gp120 inner domain. Our results provide insight into the structural factors that contribute to naturally occurring antigenic differences among full-length, glycosylated gp120 proteins and suggest that conformational dynamics should be considered in the selection and design of enhanced HIV-1 Env immunogens.
MATERIALS AND METHODS
Antibodies and CD4 constructs.
Antibodies IgG1-b12 (51–54), 17b (18, 35, 55–57), VRC01 (58), A32 (35, 59), and soluble two-domain CD4 (60) and CD4-IgG2 were obtained from the NIH AIDS Reagents Program. The antibodies N5i5, B18, and C4 were generously provided by Yongjun Guan and George Lewis (University of Maryland) (61, 62). Antibody M90 was purchased from Advanced BioScience Laboratories, Inc. (Rockville, MD) (63). Antibody CA13 (ARP3119) was obtained through the Programme EVA Center for AIDS Reagents of the National Institute for Biological Standards and Control (NIBSC; Hertfordshire, United Kingdom).
Expression and purification of gp120 glycoproteins.
Full-length gp120 glycoproteins HXB2, SF162, 1084i, and 1157ip K197N (referred to simply as 1157ip throughout the text) were expressed in African green monkey kidney cells (BSC40) infected with vaccinia virus encoding the corresponding gp120 sequence within the thymidine kinase locus as described previously (64). Plasmids carrying genes for Env sequences for HXB2 and SF162 were kindly provided by Leo Stamatatos, while those for 1157ip and 1084i were kindly provided by Ruth Ruprecht. All gp120 constructs possessed the same signal sequence, derived from HXB2. Briefly, cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen/Strep). Infection of BSC40 cells was carried out using a multiplicity of infection of 3, after which the cells were cultured in DMEM with 5% FBS and 1% Pen-Strep for 48 h. The culture supernatant was clarified by centrifugation, and Empigen BB was added to a final concentration of 0.25% to inactivate virus particles. After clarification, gp120 was purified by Galanthus nivalis lectin affinity, DEAE ion exchange, and size exclusion chromatography on a HighLoad 26/600 Superdex 200 column (GE Healthcare). Purified monomeric gp120s were homogenous, as determined by denaturing and reducing as well as blue native polyacrylamide gel electrophoresis (PAGE) (see Fig. S1 in the supplemental material).
Hydrogen-deuterium exchange with mass spectrometry.
HDX-MS was carried out as described previously (40) with a few modifications. Specifically, full-length, glycosylated gp120 proteins were diluted into a D2O-based phosphate buffer (10 mM phosphate, 144 mM sodium chloride, 85% D2O; pH 7.4) to a final gp120 concentration of 100 μg/ml. The sCD4 complexes of 1084i, 1157ip, and HXB2 were formed by incubating gp120 with a 3-fold molar excess of sCD4 for 1084i and HXB2 or both 3-fold and 5-fold molar excesses of sCD4 for 1157ip at room temperature for 1 h and at 4°C overnight. Soluble CD4-bound SF162 expressed in CHO cells was previously studied by using HDX-MS (40). CD4-bound gp120 samples were diluted to a final gp120 concentration of 100 μg/ml and were deuterated under conditions that were identical to those for the unliganded gp120 samples. Exchange reactions were quenched at 12 s, 1 min, 5 min, 30 min, and 4 h by adding an equal volume of ice-cold 1 M tris(2-carboxyethyl)phosphine (TCEP) in 0.2% formic acid titrated with sodium hydroxide such that the pH of the final solution was 2.5. The quenched exchange reaction was digested with pepsin (2:1 mass ratio) on ice for 5 min prior to flash-freezing in liquid nitrogen. In addition to these experimental samples, three control samples were prepared in parallel as previously described (40): an undeuterated sample, a “zero” exchange sample to correct for H-to-D exchange during pepsin digestion, and a fully deuterated sample to correct for D-to-H back-exchange during digestion and chromatography. Quenched and digested HDX samples were frozen in liquid nitrogen and stored at −80°C.
Deuterium uptake was monitored by liquid chromatography-MS (LC-MS) using a Waters Acquity ultraperformance LC (UPLC) system connected to a Waters Synapt high-definition MS quadrupole time of flight instrument. HDX samples were thawed on ice rapidly and loaded onto a 100-μl injection loop within a closed, insulated box with all lines and columns kept on melting ice to minimize back exchange. Peptides were trapped on an Acquity UPLC bridged ethylsiloxane/silica hybrid shield reverse-phase C18 guard column (2.1 by 5 mm; 1.7-μm particle size; Waters) in a running buffer of 0.1% trifluoroacetic acid (TFA). Peptides were eluted from the guard column and further resolved on a Hypersil gold reverse-phase C18 UPLC column (50 by 1 mm; 1.9-μm particle size; Thermo Scientific) by using a 15-min linear gradient from 5% solvent B to 100% solvent B (solvent A, 5% acetonitrile [ACN], 0.05% TFA; solvent B, 80% ACN, 0.05% TFA). To minimize sample carryover between runs, three “sawtooth gradients” from 5% to 80% acetonitrile over the course of 1 min were used to wash the main column, while the trap column was washed with successive injections of 10% formic acid, 80% methanol, 2:1 isopropanol-acetonitrile, and 80% acetonitrile between samples (65). Mass shifts were analyzed using HX Express (66), and the percent deuteration was calculated as shown in equation 1 (where m is the peptide mass centroid at a given time point, m0% is the zero time point peptide mass centroid, and m100% is the fully deuterated peptide mass centroid):
| (1) |
The estimated standard deviations for the percent deuteration values were calculated from independently prepared duplicate deuteration experiments using equation 2 as implemented in Microsoft Excel (where n is the number of replicates, xi is the measured value, and x̄ is the average value):
| (2) |
Peptic fragments were identified using a combination of tandem MS (MS/MS) data and the exact mass with the aid of the Protein Prospector program (http://prospector.ucsf.edu) as previously described (40). While glycan moieties on glycopeptides can retain deuterium, potentially affecting the observed rate of deuterium uptake (67), we observed that variability in site-specific glycosylation patterns (changes in the number of mannose, galactose, and fucose groups) had only a minor impact on the rate of deuterium exchange. Therefore, glycopeptide deuteration plots reflect the average deuterium uptake values for all available glycoforms. The heat map figures (for example, those in Fig. 2 and 5, below) were generated using Pymol (68).
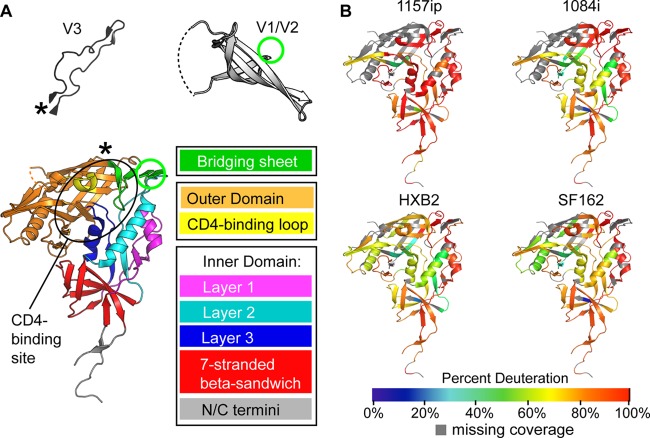
Fig 2.
Summary of isolate-specific differences in unliganded gp120 conformational dynamics. (A) The organization of monomeric gp120 into inner, outer, and bridging sheet domains is indicated by colors on the gp120 core with N- and C-terminal extensions (PDB ID 3JWD). The inner domain is further divided into mobile layers 1, 2, and 3 (magenta, cyan, and blue, respectively) that protrude from the 7-stranded β-sandwich (red) (22). Variable loops V1/V2 and V3 (from PDB 3U4E and PDB 2B4C, respectively) are included for reference. The positions of these elements in the context of the gp120 core are indicated by an asterisk for V3 and a green circle for V1/V2. The approximate location of the CD4 binding site is indicated with a black oval. The structures shown here and throughout the text reflect that of gp120 in a receptor-bound conformation, because no structure is currently available for full-length, unliganded gp120. These structures may not necessarily reflect the unliganded structure of full-length gp120 in solution. (B) Heat maps of HDX-MS data for unliganded gp120 reveal qualitative isolate-specific differences in structural dynamics. Colors mapped onto the gp120 core (PDB ID 3JWD) indicate the percent deuteration of peptides after 1 min of incubation in a deuterated buffer; warm colors correspond to high levels of deuterium uptake (dynamic regions), and cool colors correspond to low levels of deuteration (ordered or “protected” regions). Regions where peptide information is missing are indicated in gray.
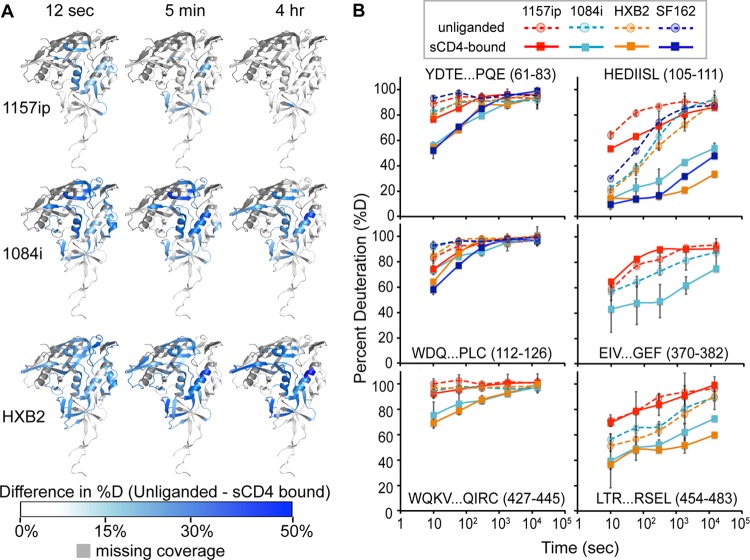
Fig 5.
Isolate-specific differences in sCD4-induced stabilization of gp120. (A) Difference heat maps show the spatial profile of sCD4-induced stabilization of gp120. The colors mapped onto the 3JWD structure indicate the quantitative differences in percent deuteration between unliganded and CD4-bound full-length gp120s. Darker blue areas correspond to greater CD4-induced stabilization. Missing peptide coverage is shown in gray. Difference heat maps for the 12-s, 5-min, and 4-h time points are shown. (B) Deuterium uptake plots (percent deuteration versus time) for identical or homologous peptides that are stabilized upon sCD4 binding. The peptide sequence and amino acid position (HXB2 numbering) are indicated for each peptide. The traces are color coded by isolates, as indicated in the symbol key. Stabilization as a result of sCD4 binding was observed as a downward shift in the deuterium uptake curve. Error bars reflect standard deviations, calculated as described in Materials and Methods.
Surface plasmon resonance.
All surface plasmon resonance (SPR) experiments were carried out on a Biacore T100 instrument (GE Healthcare) at 25°C in HBS-EP+ buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% P-20 at pH 7.4). Interactions between each gp120 and sCD4 were measured by immobilizing sCD4 (1 to 2 μg/ml in 10 mM sodium acetate [pH 5.5]) to densities of 85 response units (RU) and 185 RU on a CM5 chip by standard amine coupling with ethyl(dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide (EDC/NHS). Flow cell 1 was activated with EDC/NHS and capped with ethanolamine as the reference surface. After testing a variety of analyte flow rates to minimize potential mass transfer effects, gp120 injections were performed at 30 μl/min with a 4-min association phase and an 11-min dissociation phase. Duplicate gp120 samples were prepared independently with a 2-fold dilution series ranging from 1 μM to 15 nM. Regeneration of the sCD4 surface was performed with a 30-s injection of 10 mM glycine (pH 1.5) at 10 μl/min.
Interactions between gp120s and gp120-specific antibodies were measured using an antibody capture method. The four flow cells of a CM5 chip were immobilized with either goat anti-human or goat anti-mouse Fc-specific antibodies (Jackson ImmunoResearch Laboratories, Inc.) at 30 μg/ml in 10 mM sodium acetate buffer (pH 5.5) to a final density of ∼15,000 RU via amine coupling. Flow cell 1 was used as a reference surface, and flow cells 2, 3, and 4 were used to capture gp120-specific antibodies by injecting antibodies (diluted to 0.1 to 2 μg/ml in HBS-EP) at a flow rate of 10 μl/min with empirically determined contact times to achieve the desired surface density (typically around 60 RU). Independently prepared duplicate samples of gp120 were then injected over the captured antibody surface. The specific conditions for capture and gp120 injection, including flow rates, contact, and dissociation times and analyte concentrations are summarized in Table S1 in the supplemental material. Injections of sCD4-bound gp120 were performed using gp120 concentrations from 125 to 1.9 nM, all in the presence of 625 nM sCD4 (a 5-fold molar excess at a minimum). Blank injections contained the same concentration of sCD4. The goat anti-human or goat anti-mouse surface was regenerated with a 30- to 40-s injection of 10 mM glycine (pH 1.5) at 10 μl/min. All data were double reference subtracted, and curves were fit using a 1:1 Langmuir binding model within the BIAcore Evaluation software. The dissociation rates of gp120 from 17b and N5i5 were too slow to obtain reliable fits of the off-rates, even after more than an hour of dissociation. However, by fixing the dissociation rate to 0 in a custom binding model within the Biacore Evaluation software, it was possible to obtain estimates of the association rates. Fits of the experimental data using the 1:1 binding model and the fixed off-rate model produced comparable results, as judged by similar chi-square values (see Table S2 in the supplemental material). Estimated errors for on- and off-rates and dissociation constants were calculated from independently prepared duplicate concentration series for each analyte-ligand pair by using equation 2.
ELISAs.
ELISAs were performed by immobilizing 50 ng of gp120 per well of an Immulon 2HB plate (Thermo Scientific) in phosphate-buffered saline (PBS) at 4°C overnight. The following day, plates were blocked in a solution of PBS with 2% bovine serum albumin (BSA) and 0.01% Tween 20 for 2 h at room temperature and washed with Tris-buffered saline with 0.02% Tween 20. Primary antibodies were added at a starting concentration of 10 μg/ml for human antibodies or 5 μg/ml for mouse antibodies, diluted with a 4-fold dilution series into antibody dilution buffer (PBS with 1% BSA and 0.01% Tween 20), and allowed to incubate at room temperature for 1 h. Plates were washed, and secondary horseradish peroxidase (HRP)-conjugated goat anti-human or goat anti-mouse secondary antibodies (Jackson ImmunoResearch) were added to wells at 1:5,000 in antibody dilution buffer for 1 h at room temperature. After a final set of washes, the plates were developed with SureBlue Reserve trimethylbenzidine (KPL, Inc.) and stopped with 1 M HCl, and absorbance at 450 nm was measured on a Tecan Infinite M200 plate reader. Methods used in the peptide competition ELISA are described in the supplemental material (see Fig. S19).
Small angle X-ray scattering.
SAXS measurements were conducted on beam line 4-2 at the Stanford Synchrotron Radiation Laboratory (69). In-line size exclusion with SAXS was used to generate high-quality data sets for each gp120 protein. In these experiments, 100 μl of each sample at 3 to 5 mg/ml was injected onto a high-resolution Sepharose 200 column (GE Healthcare) with a flow rate of 50 μl/min in PBS. The focused 11-keV X-ray beam irradiated the eluted sample flowing through a thin-wall quartz capillary cell, placed at 1.7 m upstream of an MX 225HE detector (Rayonix, Evanston, IL). One-second exposures were collected every 5 s through the course of each run. The detector pixel numbers were converted to the momentum transfer (q = 4π × sinθ/λ, where 2 × θ is the scattering angle and λ is the X-ray wavelength of 1.127 Å, calibrated with silver behenate powder at the capillary position. Protein scattering data were processed by using MarParse, scaled for the transmitted beam intensity integrated for each exposure, and azimuthally averaged (69). A buffer blank was obtained from the average of the first 100 data points (before the void volume) and subtracted from the subsequent protein scattering data. The radii of gyration (Rg), the root-mean squared distance from the molecules' center of mass, was calculated from the slope of the Guinier plot and from the P(r) pairwise distance distribution histogram (77), Rg and I(0) for each frame were batch analyzed using the autoRg program (70), and frames with stable Rg values were merged using Primus (71). The one-dimensional (1D) SAXS curves were processed using GNOM to determine the Rg and Dmax values from the pairwise distance distribution functions (72–74). Low-resolution shape reconstructions were performed following the procedure described previously (40). In brief, DAMMIN (75) was used to generate a set of bead models, which were then aligned with each other. Positions of low occupancy were filtered using DAMAVER and DAMFILT to generate the final model (76).
RESULTS
Full-length, glycosylated gp120s exhibit similar morphologies based on SAXS.
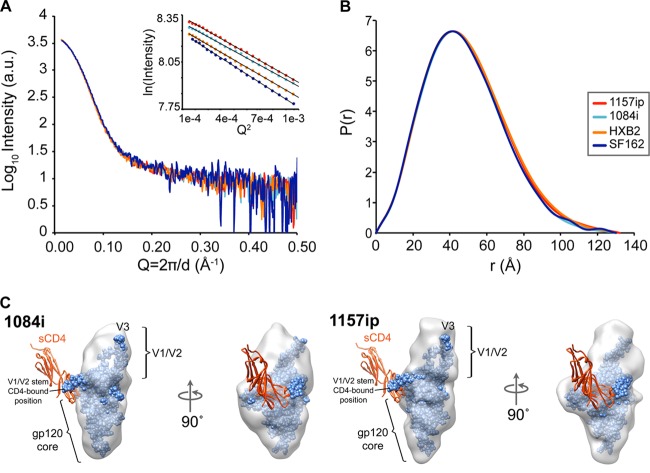
Small-angle X-ray scattering was used to study large-scale structural differences among the four isolates. Previous studies used this technique to observe changes in the position of V1/V2 loops upon sCD4 binding (40). If any of the isolates exhibited a distinct difference with regard to variable loop position, we would expect to observe this by SAXS. Because minor populations of dimeric contaminant can lead to large changes in the scattering profile, scattering data were collected with on-line size exclusion chromatography to ensure that only the homogenous monomeric population was sampled. The scattering patterns for the four gp120s are shown in Fig. 1A and are essentially superimposable. The inset for Fig. 1A shows that the Guinier plots for each gp120 sample were linear, confirming that the SAXS data from size exclusion chromatography-eluted samples were free of artifacts due to aggregation or interparticle effects and hence suitable for further analysis (77). Rg values for the four gp120s were in excellent agreement (Fig. 1B). The SAXS data suggested that only slight differences in the radii of gyration and maximal dimensions exist among the four gp120s examined, with general dimensional trends mirroring the expected increases in size based upon V1/V2 size and degree of glycosylation (Tables 1 and 2). An ab initio shape reconstruction was also performed for the gp120 monomers in their unliganded state. A comparison of reconstructions for 1157ip and 1084i gp120 is shown in Fig. 1C; all four gp120 reconstructions are compared in Fig. S2A in the supplemental material. Overall, the raw SAXS data and reconstructions indicated that the gp120 monomers possess similar morphologies. Additionally, the subunit organization observed for these gp120s matched previous observations with SF162 gp120 from Chinese hamster ovary (CHO) and 293E cell expression systems, with a prominent bulge positioned atop the gp120 core proximal to the likely V3 loop position. We inferred that this bulge corresponds to the position of V1/V2 in unliganded gp120 (40).
Fig 1.
Similar architecture of gp120 across isolates observed by SAXS. (A and B) SAXS curves (A) and pairwise distance distribution [P(r)] plots (B) show that the four full-length gp120s are similar in shape and size. Guinier plots (A, inset) indicate that the gp120 specimens did not exhibit aggregation or interparticle effects, which would have complicated the analysis. a.u., arbitrary units. (C) DAMMIN shape reconstructions of unliganded 1084i (left) and 1157ip (right) full-length gp120 (gray envelopes), with the gp120 extended core from 3JWD.pdb and containing the V3 loop from 2B4C.pdb (blue spheres) docked into the SAXS shape envelope. The V1/V2 stem in the CD4-bound conformation does not fit into the unliganded SAXS density. Instead, a lobe of mass atop the gp120 core is attributed to V1/V2 proximal to the likely V3 position in unliganded full-length gp120 (40). Two-domain sCD4 was not present in the SAXS experiment; however, it is shown as a red ribbon to aid in the interpretation of the SAXS envelopes.
Table 2.
SAXS-derived gp120 dimensions
| gp120 | Rg (Å) determined via: |
Dmax (Å) | |
|---|---|---|---|
| Guinier | GNOM | ||
| 1157ip | 37.4 ± 0.1 | 37.7 ± 0.1 | 132 |
| 1084i | 36.6 ± 0.1 | 36.8 ± 0.1 | 130 |
| HXB2 | 37.7 ± 0.1 | 37.9 ± 0.1 | 130 |
| SF162 | 36.6 ± 0.1 | 367. ± 0.1 | 130 |
The Porod-Debye region of the scattering curve along with Kratky plots were also compared (see Fig. S2B and C), as these can show differences in the degree of globular character and compactness (77, 78). These plots were nearly superimposable for the gp120s from the four isolates, indicating that they exhibited similar degrees of globular structure. Therefore, it appears that on a global scale, the gp120s were similar in their overall architecture, and the differences in deuterium uptake observed by HDX-MS (see below) largely reflect local differences in structural order and dynamics.
Isolate-specific differences in unliganded gp120 conformational dynamics measured by HDX-MS.
For the set of gp120s studied here, observable peptides provided deuteration information for 62 to 69% of the residues in the mature gp120 sequences: 62% for 1157ip, 67% for HXB2, and 69% coverage for both 1084i and SF162 (see Fig. S3 to S6 in the supplemental material). There was excellent peptide coverage of the gp120 inner domain sequence (∼98%, on average). In contrast, coverage of the outer domain and variable loops was lower and differed substantially among the isolates, due to greater sequence variability and glycosylation in these regions (see Fig. S7 in the supplemental material). For example, an L369P mutation between the clade C and B isolates likely disrupted a potential pepsin cleavage site (79) in the clade B isolates and yielded differential coverage of the CD4 binding loop across clades. Similarly, peptides spanning V1 and V4, the most densely glycosylated regions of gp120, were not observed for any isolates. Overall, differences in peptide coverage limited, but did not eliminate, our ability to characterize and compare the stability of certain regions of gp120 across isolates (see Fig. S8 in the supplemental material).
In order to measure the local dynamics of each full-length, glycosylated gp120, we quantified deuterium incorporation for every observable peptide in each isolate as a function of deuteration time (see Fig. S9 to S12 in the supplemental material). These data were summarized in the form of heat maps (Fig. 2; see also Fig. S13 in the supplemental material), in which the percent deuterium uptake for each peptide is indicated by the color on the gp120 core crystal structure with N- and C-terminal extensions (22). These heat maps make it possible to identify regions of order and disorder within full-length gp120 in solution. Notably, a number of peptides from layer 1 and the bridging sheet, which map to ordered α-helices and β-sheets in core crystal structures (Fig. 2A), were found to be rapidly deuterated based on HDX-MS analysis of unliganded, full-length gp120s in solution (Fig. 2B). Such discrepancies illustrate that secondary structures within the core regions of gp120 may be quite dynamic in the context of full-length gp120 in solution.
Heat maps enable qualitative comparison of differences in stability among the four gp120s. Figure 2B shows that there were many similarities among the isolates; for example, the CD4 binding loop, bridging sheets, and portions of the inner domain exhibited a relatively high degree of structural dynamics in all cases. Additionally, similar regions of gp120 were protected in all four instances (e.g., portions of the outer domain and the seven-stranded β-sandwich) (Fig. 2, 3, and 4). While there were general similarities, there were also abundant differences that could be observed among the four gp120s. Of the gp120s from the four isolates, 1157ip's stood out as the most dynamic; however, additional apparent differences in deuterium uptake over time (see Fig. S13 in the supplemental material) among the other gp120s suggest that each may have a unique stability profile.
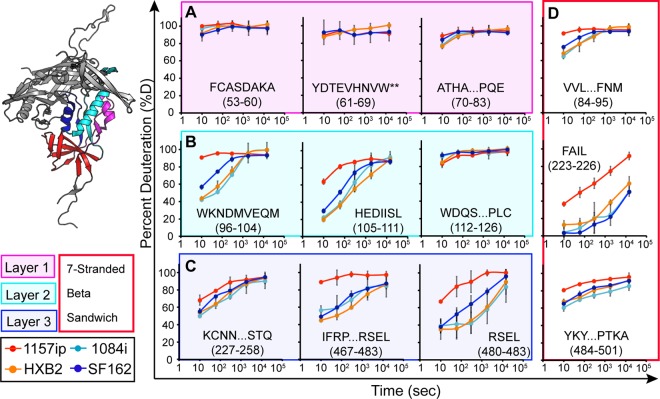
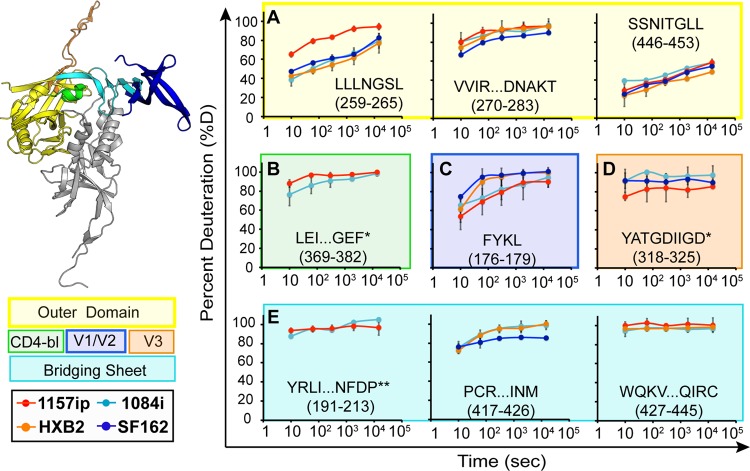
Fig 3.
Deuterium exchange profiles of homologous inner domain peptides. Each graph shows the deuterium uptake (percent deuteration) over time for peptides throughout the gp120 inner domain, which are either identical or homologous among the four gp120s. Each line in the graph reflects the deuterium uptake for that peptide in the context of a different isolate, as indicated in the figure legend. The peptide sequence and amino acid position (HXB2 numbering) are indicated for each graph. (A to C) Peptides from layers 1, 2, and 3 are color coded in the ribbon diagram, and deuteration uptake plots are grouped within the magenta, cyan, and blue boxes, respectively. (D) Peptides from the 7-stranded β-sandwich are bounded by a red box. A comparison of the deuterium uptake curves for a given peptide from multiple isolates revealed differences in stability, for example, the WKNDMVEQM peptide is more dynamic in 1157ip than in the other isolates. **, deuteration data obtained by subtraction of overlapping peptides. Error bars reflect standard deviations, calculated as described in Materials and Methods.
Fig 4.
Deuterium exchange profiles for the homologous outer domain, bridging sheet, and variable loop peptides. Each graph shows deuterium uptake (percent deuteration) over time for peptides in the gp120 outer domain (A), CD4-binding loop (CD4-bl) (B), variable loops V2 (C) and V3 (D), and bridging sheets (E). Each line on the graphs corresponds to deuterium uptake for that peptide in the context of a different isolate, as indicated. The peptide sequence and amino acid position (HXB2 numbering) are indicated for each peptide. Each graph is color coded according to the position of the peptide in the gp120 structure, as indicated in the ribbon diagram and symbol key to the left. *, the listed peptide sequence is from an isolate other than HXB2, because peptide coverage was missing for HXB2; **, deuteration data were obtained by subtraction of overlapping peptides. Error bars reflect standard deviations, calculated as described in Materials and Methods.
Comparisons of homologous peptides revealed significant variability, especially within the inner domains.
A more focused, direct comparison of individual peptides that were similar or identical among the isolates is shown in Fig. 3 and 4. Excluding glycoform variants, we identified 60, 49, 51, and 53 unique peptides for 1084i, 1157ip, HXB2, and SF162, respectively. Of these peptides, 29 were present in all four isolates, 9 were common to three isolates, and 10 were common to two isolates. Altogether, these peptides allowed us to compare deuterium uptake for 91% of residues within the inner domain and 42% of residues within the outer domain, bridging sheet, or variable loops across at least two isolates. However, it was only possible to compare deuterium uptake among four isolates for 81% of the inner domain residues and 17% of the residues in the outer domain, bridging sheet, or variable loops (see Fig. S8 in the supplemental material).
The stability of the gp120 inner domain varied substantially among the four isolates (Fig. 3). Layer 1 peptides were saturated with deuterium at the earliest time point in every isolate (Fig. 3A), indicating that this region is poorly ordered in the solution structure of full-length gp120. Because the deuterium uptake was so fast for these peptides, we could not detect differences among the isolates in this region. In contrast, layer 2 peptides (amino acids [aa] 96 to 104, 105 to 111, and 211 to 222) showed various degrees of protection from deuterium uptake at early time points in 1084i, HXB2, and SF162, but the corresponding peptides from 1157ip gp120 were nearly fully exchanged at matched time points (Fig. 3B; see also Fig. S14 in the supplemental material). Layer 3 peptides (aa 227 to 258 and aa 467 to 483) (Fig. 3C) and peptides from the 7-stranded β-sandwich (aa 84 to 95, 223 to 225, and 484 to 501) (Fig. 3D) showed a similar trend, in which 1157ip peptides were rapidly deuterated relative to those derived from other isolates. Therefore, it appears that inner domain secondary structures, which exhibit similar, though not identical, stabilization in the other three isolates, are substantially more dynamic in the context of 1157ip gp120.
Although few outer domain peptides could be directly compared across all four isolates, much of the outer domain sequence was comparable between at least two isolates (Fig. 4; see also S15 in the supplemental material). Most peptides from the outer domain were similarly stabilized among those isolates for which comparable peptides were available (e.g., aa 270 to 283 and aa 446 to 453). However, the LLLNGSL peptide (aa 259 to 265), which is directly linked to the inner domain, was deuterated more rapidly in 1157p than for the other isolates (Fig. 4A). The CD4 binding loop peptide (aa 369 to 382) was only slightly more dynamic in 1157ip than 1084i (Fig. 4B). Interestingly, the CD4 binding site-proximal peptide (aa 350 to 361) was significantly more stable in SF162 than in HXB2 (see Fig. S15 in the supplemental matrial). Overall, these results revealed conformational variations of the outer domain across isolates, although the extent of this variation appears limited relative to that observed for the inner domain.
Peptides from the variable loops and bridging sheet that were common to multiple isolates revealed few isolate-specific differences in deuterium uptake, partially due to the fact that these peptides were typically saturated with deuterium at the earliest time point of deuteration (Fig. 4C to E). The peptides FYKL (Fig. 4C) and YATGDIIGD (Fig. 4D) from V2 and V3, respectively, showed subtle differences in protection across isolates, but more complete coverage of these regions is needed to better characterize the extent of structural differences in the variable loops. Peptides within bridging-sheet beta strands (β-strands) β2, -3, and -21 (aa 112 to 127, 191 to 213, and 427 to 445, respectively) exhibited rapid deuterium uptake rates that were similar among all available isolates (Fig. 4E; see also Fig. S15). The PCRIKQIINM peptide (aa 417 to 426), which partly covers β-strands 19 and 20 of the bridging sheet, showed slight protection at early time points in all available isolates (Fig. 4E), perhaps due to hydrogen bonding with β-sheet 17, downstream of the CD4 binding loop, as observed in the core crystal structure (22).
In summary, significant isolate-specific differences in stability were observed throughout gp120. Most notably, peptides mapping to the gp120 inner domain were consistently more disordered in 1157ip than in matched peptides from other isolates. Many (but not all) peptides from the outer domain were similarly protected among the isolates that could be directly compared. Peptides mapping to the variable loops and bridging sheets were typically disordered, but poor peptide coverage and rapid saturation with deuterium made it difficult to discern differences in dynamics in these regions.
Isolate-specific differences persist in CD4-bound gp120.
Because CD4 binding has been shown to stabilize gp120 (38, 80) and induce ordering of the conserved CD4 and coreceptor binding sites (39, 40), we hypothesized that the CD4-bound forms of the gp120s are more structurally similar among isolates than unliganded gp120. To test this, we performed HDX-MS on sCD4-gp120 complexes for each isolate (Fig. 5). Overall, sCD4-bound gp120s were stabilized relative to their unliganded forms, taking on less deuterium than unliganded gp120s at matched time points (see Fig. S9 to S11 in the supplemental material). This was likely a result of multiple factors, including solvent occlusion, hydrogen bonding between sCD4 and the gp120 backbone, and a conformational change within gp120 that led to the formation of a more stable secondary structure. For regions of gp120 outside the CD4 binding site, for example, the bridging sheet and layers 1 and 2 of the inner domain, CD4-induced HDX protection is likely due to stabilization of secondary structures. The impact of sCD4 binding on deuterium uptake by gp120 is shown in the difference maps of Fig. 5A, in which blue areas indicate protection relative to unliganded gp120. Similar patterns of protection were observed among the four isolates, most notably in the CD4 binding loop, the bridging sheet, and layers 1, 2, and 3 of the inner domain (Fig. 5A; see also Fig. S16 in the supplemental material). However, the degree of protection as a result of sCD4 binding—the downward shift in deuterium uptake between unliganded and sCD4-bound gp120—varied among the isolates (Fig. 5B). Deuterium uptake plots for matched peptides from the sCD4-bound gp120s revealed that in contrast to 1084i and HXB2, 1157ip was poorly stabilized in the presence of sCD4 (Fig. 5B; see also Fig. S17 and S18). For example, 1084i and HXB2 peptides from the inner domain (e.g., aa 61 to 83, 105 to 111, and 454 to 483), bridging sheet (e.g., aa 112 to 126 and aa 427 to 445), and the CD4 binding loop for 1084i (aa 369 to 382) were dramatically stabilized in the presence of sCD4 relative to 1157ip. Notably, the sCD4-induced stabilization of 1084i and HXB2 was comparable to that observed for CHO cell-expressed SF162 gp120 (Fig. 5B, dark blue lines) demonstrated in previous work (40). Increasing sCD4 in the 1157ip-sCD4 mixture, from a 3-fold to a 5-fold molar ratio, did not increase the apparent protection of sCD4-stabilized peptides (data not shown), which suggests that sCD4 was present at saturating concentrations and all available gp120 was bound by sCD4. Thus, it appears that while similar regions of gp120 are stabilized as a result of sCD4 binding to diverse isolates, the sCD4-bound gp120s maintain different levels of structural dynamics and, as in the unliganded state, 1157ip gp120 remains relatively dynamic even with sCD4 bound.
Epitope stability and antigenicity: linear epitope-specific antibodies.
In order to determine whether differences in structural dynamics observed by HDX-MS affected ligand association with gp120, we used SPR to quantify the kinetics of sCD4 and antibody binding to the four gp120s and ELISAs to measure the levels of antibody stably bound to gp120. The determinants of antigenicity are complex and include, as minimal requirements, that (i) the residues that make up the antibody epitope be present in the primary amino acid sequence, and (ii) these residues, which form the antibody epitope, must be exposed or accessible for antibody recognition (81). Beyond these basic requirements, we hypothesized that the local conformational stability of an antigen might also impact the kinetics and/or the affinity of an antibody's interaction with its epitope, as has been previously proposed (81).
The antibodies B18, C4, and CA13 have been reported to recognize linear epitopes within helix 1 of gp120 (62, 82), and we confirmed their specificity in a peptide competition ELISA (see Fig. S19 in the supplemental material). The core epitopes of the three antibodies were absolutely conserved in all four isolates (Fig. 6B), yet exhibited significant differences in conformational dynamics based on HDX-MS (Fig. 3B). There was no detectable difference in linear epitope-specific antibody binding to the four gp120s by ELISA (Fig. 6A). We also investigated isolate-specific differences in the kinetics of gp120 binding to captured B18, CA13, and C4 by SPR (see Fig. S20 in the supplemental material). Although 1157ip stood out among the four isolates, with a more dynamic helix 1 based on HDX-MS, it was not distinguished by substantially different rates of association with or dissociation from these linear epitope-specific antibodies (Table 3). Furthermore, the affinities of C4 and CA13 for 1157ip were comparable to those observed for 1084i and HXB2, both of which featured a more ordered helix 1(Table 3 and Fig. 6C). In contrast, SF162, which was similar to 1084i and HXB2 in helix 1 stability, had affinities for CA13 and C4 that were at least 4-fold weaker than those of the other isolates. These findings suggest that the weaker recognition of SF162 by this panel of helix 1-specific antibodies may be due to factors other than the conformational stability of helix 1 (e.g., tertiary structural differences).
Fig 6.
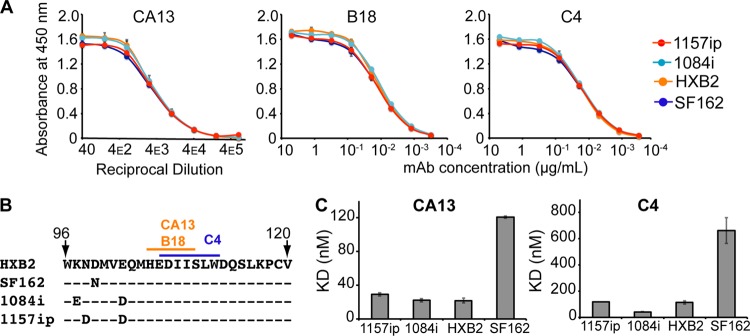
Linear epitope-specific antibody binding to a region of gp120 differentially stabilized across isolates. (A) Linear epitope-specific antibody (CA13, B18, or C4) binding to gp120 proteins measured by the ELISA. The curves are color coded by Env isolate as defined in the symbol key. Concentrations of monoclonal antibodies (mAb) are shown on the x axis for C4 and B18, and the reciprocal dilution of antibody supernatant is shown on the x axis for CA13. Curves are representative of at least two independent experiments; error bars indicate standard deviations from duplicate measurements. (B) The primary sequence recognized by antibodies CA13, B18, and C4 is absolutely conserved among the four gp120s. A dash indicates that the residue is conserved in a given isolate, and amino acid differences are indicated. This region is differentially stabilized in the four gp120s, based on HDX-MS (Fig. 3B) (C) Summary of SPR-derived binding constants for CA13 and C4 binding to the four gp120s. Raw and fitted SPR curves are shown in Fig. S20 in the supplemental material. The B18-gp120 SPR binding curves did not fit well to a 1:1 binding model; therefore, estimates of affinity are not available, although qualitative isolate-specific differences in binding to B18 were similar to those observed for CA13 and C4 (see Fig. S20). Error bars indicate standard deviations, calculated as described in Materials and Methods.
Table 3.
SPR binding parameters for linear epitope-specific antibodies
| Antibody | Parametera (units) | Affinity of antibody for the gp120 based on SPRb |
|||
|---|---|---|---|---|---|
| 1157ip | 1084i | HXB2 | SF162 | ||
| CA13 | ka (M−1 s−1) | 1.8 (1) × 104 | 1.96 (9) × 104 | 0.9 (1) × 104 | 0.36 (1) × 104 |
| kd (s−1) | 5.31 (4) × 10−4 | 4.3 (2) × 10−4 | 2.03 (1) × 10−4 | 4.32 (9) × 10−4 | |
| KD (nM) | 29 (2) | 22 (2) | 22 (3) | 121 (1) | |
| C4 | ka (M−1 s−1) | 2.6 (2) × 104 | 3.2 (1) × 104 | 1.1 (1) × 104 | 0.42 (5) × 104 |
| kd (s−1) | 3.1 (2) × 10−3 | 1.34 (1) × 10−3 | 1.29 (3) × 10−3 | 2.8 (1) × 10−3 | |
| KD (nM) | 118 (1) | 42 (2) | 110 (10) | 600 (100) | |
ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant.
Values are means ± standard deviations. Values in parentheses are errors for the last significant figure; 1.9 (1) ×104 is equivalent to 19,000 ± 1,000.
Dynamics and conformational epitope antigenicity: CD4 binding site.
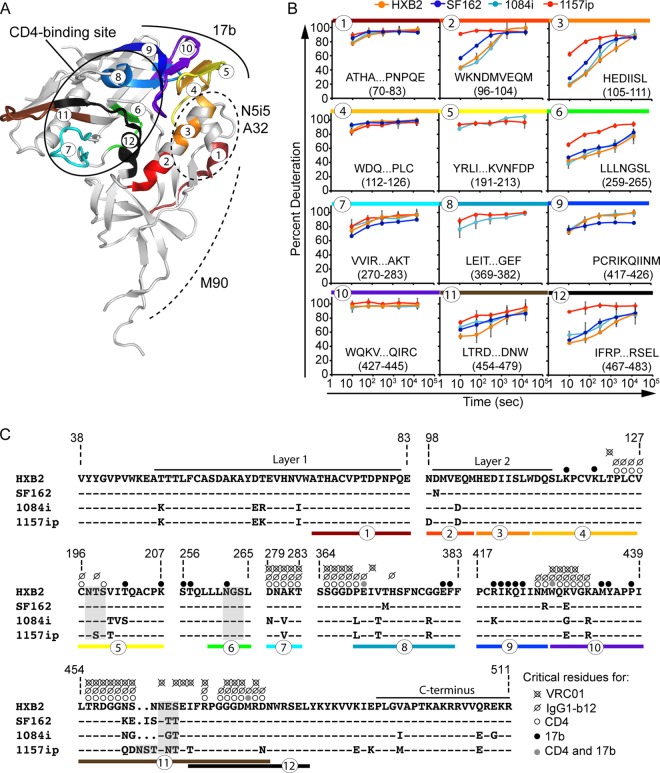
Because the majority of broadly cross-reactive HIV-1 NAbs and ADCC-active antibodies target discontinuous epitopes within gp120, we sought to determine whether differences in the stability and structural dynamics of gp120 correlated with differences in the antigenicity of conformational epitopes such as the CD4 binding site. A summary of the differences in the primary sequence and dynamics of peptide segments involved in conformational antibody epitopes is shown in Fig. 7. Many of the peptides that are directly involved in CD4 binding site-ligand interactions (Fig. 7A) are either saturated with deuterium at the earliest time point (peptides 4 and 10 in Fig. 7B) or are similarly disordered in the isolates for which data are available (e.g., peptides 7 and 8 in Fig. 7B). However, peptide 12, which contains a number of CD4 contact residues and helix 5, is significantly more dynamic in 1157ip (Fig. 7B). In an ELISA, the CD4 binding site ligands CD4-IgG2 and IgG1-b12 bound strongly to the two clade B isolates, SF162 and HXB2, and relatively weakly to the clade C isolates (Fig. 8). This pattern of differential binding across clades was particularly evident for IgG1-b12 and suggested that the observed differences in CD4 binding site ligand recognition of gp120 might be partly due to clade-specific primary sequence differences within the CD4 binding site (Fig. 7C). Therefore, we collected SPR data for sCD4, CD4-IgG2, and IgG1-b12 binding to all four gp120s (see Fig. S21 in the supplemental material), but we focused our comparison on the isolates 1084i and 1157ip, which are closely related in their primary amino acid sequences (Fig. 7C) yet exhibit dramatic differences in the stability of peptides both proximal (e.g., peptide 12 in Fig. 7B) and distal (e.g., inner domain peptides in Fig. 3) to the CD4 binding site. This pair of isolates presents an opportunity to study the impact of gp120 dynamics on recognition by CD4 binding site ligands while limiting confounding amino acid changes.
Fig 7.
Summary of isolate-specific differences in conformational dynamics and primary sequence for peptides involved in conformation-dependent antibody epitopes. (A) Colors and numbers mapped onto the 3JWD crystal structure indicate peptides within conformation-dependent antibody epitopes that can be tracked by HDX-MS. The structure is annotated to indicate the CD4 binding site (targeted by sCD4, IgG1-b12, and VRC01) as well as N5i5, A32, M90, and 17b epitopes. (B) Deuteration profiles (percent deuteration versus time) for peptides within conformation-dependent antibody epitopes. Plots are color coded and numbered as for panel A. (C) Amino acid alignment of the four isolates for regions of gp120 involved in conformation-dependent antibody binding. A dash indicates that the residue is conserved in a given isolate relative to HXB2; a period indicates an insertion in a separate sequence. N5i5 and A32 recognize a similar epitope involving layers 1 and 2 (61). Layer 1 and the C terminus are thought to be involved in the M90 epitope (63, 86). Critical residues for CD4, IgG1-b12, VRC01, and 17b binding are indicated with dots, as shown in the symbol key, and are based on references 21, 24, and 55. Peptides are indicated with colors and numbers as for panels A and B.
Fig 8.
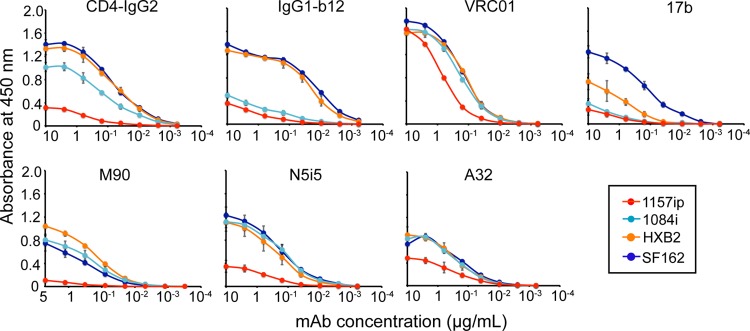
Conformation-dependent antibody binding to gp120s as determined by ELISA. Conformation-dependent antibodies specific for different regions of gp120 were tested: CD4 binding site ligands (CD4-IgG2, IgG1-b12, and VRC01), conformational inner domain antibodies (M90, N5i5, and A32), and the coreceptor binding site antibody (17b). Each graph corresponds to a different antibody binding to the four gp120s. The binding curves are color coded based on HIV isolate as shown in the symbol key. Antibody binding levels (measured as the absorbance at 450 nm) versus antibody concentrations are shown. Curves are representative of at least two independent experiments; error bars indicate standard deviations from duplicate measurements.
CD4-IgG2 bound poorly to 1157ip gp120 compared to 1084i in the ELISA (Fig. 8). These results were confirmed by SPR, which indicated that 1157ip had the weakest affinity for sCD4 and CD4-IgG2 of the four isolates, showing 5-fold- and 2-fold-weaker affinities for sCD4 and CD4-IgG2, respectively, than 1084i (Fig. 9C and Table 4). IgG1-b12 binding to 1084i fit poorly to a 1:1 binding model, which complicated a direct comparison of 1084i and 1157ip binding to this antibody based on SPR. The weak affinity of 1157ip for CD4 binding site ligands was largely due to lower rates of association (Fig. 9A), although 1157ip also showed a slightly higher rate of dissociation from sCD4 compared to 1084i (Fig. 9B). Thus, it appears that the increased conformational flexibility of 1157ip is associated with slower association rates and an overall weaker affinity for CD4 mimetics (Fig. 9C). This effect appears to be most prominent in CD4 binding site ligands that require contact with both the inner and outer domains of gp120, because while 1157ip bound to CD4-IgG2 at a very low level in the ELISA, the potent and broad CD4 binding site antibody VRC01, which primarily makes contact with the gp120 outer domain (24), bound to 1157ip at only a slightly reduced level relative to 1084i (Fig. 8).
Fig 9.
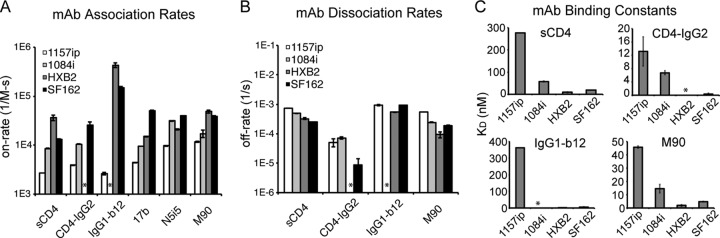
Summary of conformation-dependent antibody binding to gp120 as measured by SPR. (A) Rate of association of the four gp120 proteins with immobilized sCD4 or captured antibodies. Because of slow dissociation rates for 17b and N5i5, only on-rates could be derived for these antibodies. mAb, monoclonal antibody. (B) Rates of dissociation of the four gp120s from immobilized sCD4 or captured antibody ligands. Reliable off-rates could not be determined for 17b or N5i5. (C) Summary of binding constants for gp120 binding to immobilized sCD4 or captured antibody ligands. Large KD values reflect weaker binding interactions. Raw data and fitted curves for gp120 binding to conformation-dependent ligands are presented in the supplemental material. *, the experiment could not be reliably performed or the signal could not be fit with a 1:1 binding model. Error bars indicate standard deviations, calculated as described in Materials and Methods.
Table 4.
SPR binding parameters for conformation-dependent antibodies
| Antibody | Parametera (units) | SPR-derived binding values for gp120-ligand interactionb |
|||
|---|---|---|---|---|---|
| 1157ip | 1084i | HXB2 | SF162 | ||
| sCD4 | ka (M−1 s−1) | 2.67 (1) × 103 | 8.5 (2) × 103 | 37 (5) × 103 | 13.1 (4) × 103 |
| kd (s−1) | 7.41 (3) × 10−4 | 4.9 (1) × 10−4 | 3.3 (2) × 10−4 | 2.46 (6) × 10−4 | |
| KD (nM) | 277 (1) | 58 (2) | 9 (2) | 18.8 (2) | |
| CD4-IgG2 | ka (M−1 s−1) | 3.91 (8) × 103 | 10.5 (1) × 103 | NDc | 25 (4) × 103 |
| kd (s−1) | 5 (2) × 10−5 | 7.1 (6) × 10−5 | ND | 0.9 (5) × 10−5 | |
| KD (nM) | 13 (4) | 6.8 (6) | ND | 0.4 (3) | |
| IgG1-b12 | ka (M−1 s−1) | 2.6 (1) × 103 | —d | 430 (40) × 103 | 150 (10) × 103 |
| kd (s−1) | 9.4 (5) × 10−4 | — | 5.4 (1) × 10−4 | 9.2 (2) × 10−4 | |
| KD (nM) | 362 (1) | — | 1.3 (1) | 6.2 (6) | |
| M90 | ka (M−1 s−1) | 1.18 (4) × 104 | 1.7 (3) × 104 | 4.9 (4) × 104 | 3.89 (7) × 104 |
| kd (s−1) | 5.41 (7) × 10−4 | 2.4 (1) × 10−4 | 0.9 (2) × 10−4 | 1.88 (5) × 10−4 | |
| KD (nM) | 45.7 (8) | 14 (3) | 1.9 (6) | 4.8 (2) | |
ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant.
Values are means ± standard deviations. Values in parentheses are errors for the last significant figure; see Table 3.
ND, experiment not done.
—, data fit poorly to a 1:1 binding model.
The few observable differences in the stability of peptides involved in the CD4 binding site across isolates (Fig. 7) suggest that differences in conformational stability of regions outside the CD4 binding site (for example, the gp120 inner domain) likely also exert an influence on gp120's affinity for CD4 and binding site-targeted antibodies. This model is supported by previous work that showed that layers 1, 2, and 3 of the inner domain, regions that are dramatically destabilized in 1157ip relative to the other isolates, significantly impact gp120 interactions with CD4 and CD4 mimetics (83–85).
Dynamics and the antigenicity of conformational inner domain epitopes.
Because isolate-specific differences in gp120 stability were most apparent in the inner domain (Fig. 3 and 7B), we measured binding of the four gp120s to antibodies that have been reported to target conformational epitopes within this region: M90, A32, and N5i5 (Fig. 7A). M90 is thought to recognize a conformational epitope involving layer 1 and the N and C termini of gp120 (63, 86). N5i5 is a potent ADCC-active antibody that binds to a CD4-induced, discontinuous epitope similar to that of A32 (61) that includes layers 1 and 2 of the inner domain; its contact residues are absolutely conserved in the isolates tested here (George Lewis and Marzena Pazgier, personal communication). Because of the high level of sequence conservation within the epitopes recognized by these antibodies and in the N5i5 epitope, in particular, we were able to test the impact of gp120 dynamics on inner domain antigenicity with limited confounding effects from amino acid variability in the antibody epitopes (Fig. 7C).
Antibodies M90, A32, and N5i5 bound well to 1084i, HXB2, and SF162 but only weakly to 1157ip gp120 in the ELISA (Fig. 8). These results were supported by the SPR data, which showed that 1157ip bound to both N5i5 and M90 with the slowest on-rate of the four gp120s (Fig. 9A). Additionally, 1157ip had the fastest off-rate and weakest affinity for M90 of the four isolates (Fig. 9B and C and Table 4; see also Fig. S21 in the supplemental material). The rate of N5i5 dissociation was too slow to obtain reliable estimates of the off-rate or binding affinity, even when we used dissociation times of greater than 1 h (see Fig. S22 in the supplemental material). Overall, the relatively high degree of disorder within the 1157ip gp120 inner domain appeared to correlate with a lower rate of association with and a weaker affinity for inner domain-specific, conformation-dependent antibodies.
Antibodies recognizing CD4-induced epitopes bind slowly to 1157ip in both the unliganded and CD4-bound states.
HDX-MS analysis of sCD4-bound gp120 revealed that many regions of gp120 are stabilized as a result of sCD4 binding, including the bridging sheet microdomain and the gp120 inner domain (Fig. 5). Notably, these same regions are targeted by antibodies such as 17b and N5i5, which preferentially recognize CD4-bound gp120 (55, 61, 87). Because HDX-MS indicated that differences in stability are maintained in sCD4-bound gp120 (Fig. 5B), we sought to test the antigenicity of CD4-induced epitopes and the antigenic consequences of sCD4 binding across isolates by using SPR.
As with N5i5, 17b off-rates were slow for all isolates (with the exception of 1084i), and we were limited to estimates of the on-rate for 1157ip, HXB2, and SF162 (see Fig. S23 in the supplemental material). Notably, 1157ip had the lowest rate of association with 17b in its unliganded state and was poorly recognized by 17b in the ELISA (Fig. 8), despite having well-conserved 17b contact residues (Fig. 7C). Many peptides containing residues critical for 17b binding were saturated with deuterium at the earliest time point (peptides 4, 5, and 10 in Fig. 7B) or were similarly disordered in the isolates for which data were available (peptide 8 in Fig. 7B). SF162 exhibited stabilization in peptide 9 (Fig. 7B), which contains a number of residues critical for 17b binding (Fig. 7C), and bound strongly to 17b in the ELISA and SPR relative to the other isolates (Fig. 8 and 9). Conversely, peptide 6, which contains a residue critical for 17b binding at position 262 (HXB2 numbering) (55), was more disordered in 1157ip, as were peptides 2 and 3 (Fig. 7B), which are directly linked to β-strands 2 and 3 of the bridging sheet (highlighted as peptides 4 and 5, respectively, in Fig. 7A). Given the few differences in stability observed for peptides containing residues critical for 17b binding, it seems plausible that increased disorder observed in the inner domain of 1157ip is communicated to the bridging sheet, with implications for 17b binding to unliganded gp120.
Consistent with the stabilization of the 17b and N5i5 epitopes in sCD4-bound gp120 by HDX-MS (Fig. 5; see also Fig. S24 in the supplemental material), we observed a dramatic increase in 17b and N5i5 binding to all four gp120s upon addition of sCD4 (Fig. 10). It is worth noting that, in addition to epitope stabilization, tertiary structural changes, not readily detectable by HDX-MS, are also likely to contribute to the increase in 17b and N5i5 binding to sCD4-bound gp120. This may be especially true for 17b, as the lateral movement of V1/V2 upon sCD4 binding appears to coincide with ordering of the bridging sheet and formation of the 17b epitope (40).
Fig 10.
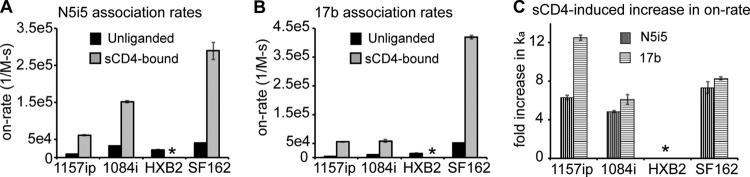
Changes in CD4i antibody binding to gp120 in the presence of sCD4 by SPR. (A) On-rates of N5i5 binding to the four gp120s in the presence and absence of sCD4. (B) On-rates of 17b binding to the four gp120s in the presence and absence of sCD4. (C) Change in rates of association (ka) of 17b and N5i5 with the gp120s as a result of sCD4 binding. Data are presented as the fold increase relative to the rate of association with unliganded gp120. *, the experiment could not be reliably performed. Error bars indicate standard deviations, calculated as described in Materials and Methods.
We investigated whether the differences in 17b and N5i5 reactivity across isolates observed in unliganded gp120 would be similarly apparent in the context of sCD4-bound gp120. SPR was used to measure the rate of gp120 binding to 17b and N5i5 in the presence of a 5-fold molar excess of sCD4. The rates of association with 17b and N5i5 increased for all isolates in the presence of sCD4, and the isolate rank order of highest (SF162) and lowest (1157ip) rates of association with the two antibodies was identical for unliganded and sCD4-bound gp120 (Fig. 10A and B and Table 5). Importantly, the increase in the rate of 1157ip binding to 17b and N5i5 upon addition of sCD4 was similar to that of the other isolates (Fig. 10C), indicating that although 1157ip gp120 wasn't as dramatically stabilized by sCD4 as the other gp120s, it remained capable of binding to sCD4 and undergoing a transition to a CD4-induced state. Overall, it appears that isolate-specific antigenic differences within CD4-induced epitopes are maintained in the sCD4-bound state, in agreement with the conformational stability profiles observed for the sCD4-bound gp120s with HDX-MS.
Table 5.
SPR binding parameters for CD4i epitope-specific antibodies
| Antibody | Parametera (units) | SPR-derived binding values for gp120-ligand interactionb |
|||
|---|---|---|---|---|---|
| 1157ip | 1084i | HXB2 | SF162 | ||
| N5i5 | ka (M−1 s−1) | 9.7 (2) × 103 | 31.2 (4) × 103 | 21.2 (6) × 103 | 39.4 (2) × 103 |
| kd (s−1) | —c | — | — | — | |
| KD (nM) | — | — | — | — | |
| N5i5 + sCD4 | ka (M−1 s−1) | 6.1 (2) × 104 | 15.1 (3) × 104 | NDd | 29 (2) × 104 |
| kd (s−1) | — | — | ND | — | |
| KD (nM) | — | — | ND | — | |
| 17b | ka (M−1 s−1) | 4.38 (3) × 103 | 9.5 (1) × 103 | 15.1 (3) × 103 | 50.6 (7) × 103 |
| kd (s−1) | — | 2.72 (1) × 10−5 | — | — | |
| KD (nM) | — | 2.86 (4 | — | — | |
| 17b + sCD4 | ka (M−1 s−1) | 5.5 (1) × 104 | 5.8 (5) × 104 | ND | 41.9 (6) × 104 |
| kd (s−1) | — | 1.3 (1) × 10−4 | ND | — | |
| KD (nM) | — | 2.2 (4) | ND | — | |
ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant.
Values are means ± standard deviations. Values in parentheses are errors for the last significant figure; see Table 3.
—, because of slow dissociation, it was not possible to reliably fit dissociation rates or binding constants for this interaction.
ND, could not be reliably performed.
Consideration of potential caveats of sCD4-induced stabilization of gp120 based on HDX-MS.
Two factors we considered that could potentially complicate interpretation of the HDX-MS results for sCD4-bound gp120 were isolate-specific differences in kinetics and affinities of binding to sCD4 and isolate-specific variability in the fraction of “active” gp120 within a purified gp120 preparation. Because 1157ip has the weakest affinity for sCD4, we considered the possibility that the greater levels of deuterium uptake by this isolate in the presence of sCD4 reflect a difference in the amount of time a molecule of 1157ip would spend in the sCD4-bound state or a lower level of sCD4 occupancy at equilibrium. However, given the affinity values calculated by SPR (Table 4) and the concentration of sCD4 used in complex formation, we estimate that only 0.01% more 1157ip would remain unbound at equilibrium compared to 1084i (0.28% versus 0.27% unbound gp120 for 1157ip and 1084i, respectively), an insignificant difference. Similarly, using a standard first-order rate equation to model the exponential decay of the gp120-sCD4 complex and the dissociation rates calculated by SPR (Table 4), after 12 s of deuteration, only 0.3% more dissociation would occur for 1157ip compared to 1084i, yet we observed dramatic differences in deuterium uptake at this time point (Fig. 5). Furthermore, we attempted to account for the weaker affinity of 1157ip for sCD4 by using a higher concentration of sCD4 in complex formation (5-fold molar excess) than with the other isolates (3-fold molar excess). The absence of additional stabilization in the 5-fold versus 3-fold molar excess samples suggests that sCD4 saturates the available 1157ip gp120 population. Thus, differences in CD4 occupancy at equilibrium or kinetic differences in sCD4 dissociation are unlikely to explain the dramatic differences in stability observed for sCD4-bound 1157ip compared to the other isolates.
Similarly, if purified 1157ip gp120 contained a relatively large fraction of inactive gp120, as a result of heterogeneity in disulfide bonding or glycosylation, for example, then addition of sCD4 would only weakly stabilize gp120 at the population level (although it may be capable of dramatically stabilizing individual molecules of active gp120). However, there is no indication that such a population exists in 1157ip; for example, Kratky plots of 1157ip indicate that the gp120s share a similar global structure (see Fig. S2). Furthermore, addition of sCD4 to 1157ip produces a similar increase in its rate of association with 17b and N5i5, as observed for the other gp120s (Fig. 10C). Because the rate of association is sensitive to the active concentration of analyte, this suggests that the fraction of gp120 that is active for sCD4 binding is similar among the four gp120s. Thus, we considered these caveats, but we concluded that they were unlikely to be responsible for the observed isolate-specific differences in the stability and antigenicity of sCD4-bound gp120. Instead, the differences appeared to reflect inherent structural features of each gp120.
DISCUSSION
Our understanding of the structural basis for antigenic and immunogenic differences among gp120 proteins from diverse isolates has been hindered by our inability to extract detailed structural information for intact Env glycoprotein constructs containing the variable loops and glycans, which are major determinants of isolate antigenicity and neutralization sensitivity. Crystal structures of truncated core constructs provide valuable information about the conserved elements of gp120 and their recognition by receptor and antibodies, but they do not elucidate how the conserved features are masked by the dominant variable elements or how these variable elements affect the conformation of distal, conserved epitopes in the context of full-length gp120 (88). Furthermore, X-ray crystallography does not probe protein dynamics in solution, which may be an important determinant of Env antigenicity (38).
Here, we used HDX-MS and SAXS in combination with antibody binding studies to investigate the structural and antigenic variability among full-length, glycosylated gp120 proteins from diverse HIV-1 isolates in order to understand the role of structural differences, such as variable loop position and conformational dynamics in modifying the antigenicity of gp120 proteins from diverse isolates. A comparison of the four full-length gp120s by SAXS (Fig. 1; see also Fig. S2) indicated that the overall morphologies of the gp120 monomers were similar and that they were comparable to SF162 gp120 expressed in 293E and CHO cells, with the large V1/V2 loops likely occupying a similar position atop the gp120 core (40). Radii of gyration differed by only ∼1 Å and appeared to be in good agreement with expected sizes related to variable loop length and glycosylation. Kratky plots of the SAXS data also indicated that the gp120s were similarly ordered, and none of the isolates appeared to be unfolded or noticeably different in global structure by this measure (see Fig. S2).
Implications of inner domain conformational variability for CD4 reactivity.
In contrast to the similar global organization observed for the gp120 monomers by SAXS, HDX-MS revealed significant differences in local dynamics among the four unliganded gp120 proteins (Fig. 2). This is the first report, to our knowledge, of experimentally measured, naturally occurring structural heterogeneity among full-length, glycosylated gp120 monomers from diverse isolates. Many of the differences were concentrated in the inner domain and at the interface of inner and outer domains (Fig. 3 and 4). This is consistent with previous computational (89) and structural studies, which suggested that the inner domain is capable of substantial structural variability. For example, layers 1 and 2 of the unliganded SIV core inner domain (90) adopt a conformation that is distinct from that observed in unliganded extended cores from multiple isolates (26) as well as CD4-bound HIV-1 isolates (18, 19). Similarly the inner domain exhibits substantial variability compared to the relatively inert outer domain in the crystal structures of gp120 bound to antibodies b12 and F105 (21). Our observation that diverse, full-length gp120s can exhibit significant differences in structural order contrasts with a recent structural study of unliganded extended core constructs that found that extended core proteins from diverse isolates were nearly superimposable (26). The results are not necessarily contradictory, however, given that variable loop truncation appears to favor a more uniform CD4-bound-like conformation (26). The crystal structures also lack glycosylation and are subject to packing constraints in the crystal, which could suppress isolate-specific differences in local dynamics and flexibility.
The gp120 inner domain is critical for maintaining gp120-gp41 interactions and mediating CD4 reactivity (22, 83, 85). In addition, recent work suggests that it is an important target for ADCC-active antibodies (61, 91). Site-directed mutagenesis studies have shown that residues in layers 1, 2, and 3 of the inner domain participate in a network of critical interactions in both soluble, monomeric gp120 and in the context of trimeric Env, and this network is required for stabilizing the transition to the CD4-bound state and maintaining noncovalent gp120-gp41 interactions in the Env trimer (83–85). Our results are consistent with these reports and provide a direct probe of the structural ordering in layers 1, 2, and 3 of the gp120 inner domain as a result of CD4 binding. The key residues that have been shown to mediate the transition to the CD4-bound conformation are absolutely conserved among the four isolates studied here, including H66, P212, and S375 (83). Although these residues are present in the primary sequence, our HDX-MS results suggested that the regions containing these residues are ordered to different degrees in the four gp120s and are notably disordered in 1157ip (Fig. 3). That disorder may preclude the formation of a stable bond network. Indeed, 1157ip had the weakest affinity for CD4-binding site ligands of the four isolates (Fig. 8 and 9), and HDX-MS analysis of the 1157ip gp120-sCD4 complex indicated that 1157ip was poorly stabilized in the presence of sCD4 relative to the other isolates (Fig. 5). While addition of sCD4 stabilized the gp120s to different degrees, the pattern of sCD4-induced ordering was found to be conserved among the four isolates. Specifically, the CD4 binding site, bridging sheet, and inner domain of gp120 were protected from deuterium uptake in all isolates in the presence of sCD4. CD4-induced ordering of the gp120 inner domain thus appears to be critical for viral entry, just as bridging sheet ordering is conserved and required for coreceptor binding (92).
Epitope dynamics and antibody binding.
In order to determine whether the differences in structural dynamics we observed by HDX-MS were related to antigenic differences among the gp120 proteins, we measured antibody binding to the panel of full-length gp120s. Only subtle differences in linear epitope-specific antibody binding were observed across isolates (Fig. 6), despite differences in the structural dynamics of the antibodies' epitopes among the four gp120s (Fig. 3B). This contrasts somewhat with early studies in non-HIV systems, which concluded that linear epitope flexibility correlates with improved recognition by antibodies. However, the conclusions of these previous studies were based on correlations between crystal structure temperature factors and the antigenicity of different regions within a single protein, where antibodies raised against peptides from highly mobile protein elements bound to the protein better than antibodies to regions of the same protein with low B factors (93, 94). There are a number of potential caveats associated with this type analysis, including differences in the immunogenicity of peptide sequences and differences in the tertiary structural context, which may alter antibody recognition. In contrast, our study looked at the binding of monoclonal antibodies to a common linear epitope differentially stabilized among the gp120 proteins. While relatively minor differences in linear epitope recognition were observed, one exception was the relatively poor recognition of SF162 gp120 by helix 1-specific antibodies. The stability of helix 1 as monitored by HDX-MS was similar in SF162, HXB2, and 1084i; thus, the difference in linear epitope-specific antibody recognition of SF162 may reflect a difference in the SF162 tertiary structure or possibly epitope occlusion by adjacent glycans.
Antibodies targeting discontinuous and conformational epitopes showed dramatic isolate-specific differences in gp120 binding (Fig. 8 and 9). Importantly, the isolate with the most dynamic inner domain, 1157ip, was poorly recognized by every conformation-dependent antibody tested with an epitope that either wholly (A32, N5i5, and M90) or partially (IgG1-b12, sCD4, CD4-IgG2, and 17b) involved the inner domain. The potential importance of inner domain disorder in limiting the antigenicity of the CD4 and coreceptor binding sites is further highlighted by the observation that many peptides in the CD4 and coreceptor binding sites were similarly disordered among the four isolates (Fig. 7B). Notably, VRC01 binding to 1157ip was only slightly reduced compared to other CD4 binding site ligands (Fig. 8). These data support a proposed model for the impressive breadth of VRC01 neutralization; because VRC01 predominantly makes contact with the gp120 outer domain, it is less sensitive to naturally occurring structural heterogeneity within the gp120 inner domain (24, 95).
Increased conformational dynamics appeared to correlate with reduced binding by multiple antibodies with distinct specificities; thus, it is unlikely that the effects we detected were simply due to amino acid changes within antibody contact sites. The contact residues of N5i5, in particular, were absolutely conserved among the four isolates (Lewis and Pazgier, personal communication), yet substantial differences in N5i5 association with gp120 were observed among the four isolates. Additional support for the proposal that structural stabilization leads to improved recognition by conformation-dependent antibodies is found in the observation that sCD4 binding induces ordering within the epitopes targeted by N5i5 and 17b (Fig. 5; see also Fig. S24 in the supplemental material), which correlates with increased N5i5 and 17b binding to sCD4-stabilized gp120 compared to the relatively dynamic unliganded gp120 (Fig. 10).
Overall, the data suggest that antibodies recognizing discontinuous epitopes are sensitive to differences in the structural dynamics of elements both proximal and distal to the antibody binding site. The data presented here, and those from previous studies (40, 83), suggest that conformational epitopes in the CD4 binding site, bridging sheet, and inner domain are coupled to one another. Thus, it is conceivable that enhanced conformational dynamics in specific regions of the HIV-1 gp120 Env due to distal amino acid changes could provide an additional pathway for escape from neutralizing and ADCC-active antibodies targeting conserved, conformational epitopes, allowing for structural heterogeneity in regions where amino acid changes are not tolerated due to functional requirements.
It will be valuable to extend this work to trimeric Env constructs in order to test the influence of conformational dynamics within the Env trimer on the neutralization phenotype of virion-associated Env. However, such a study would depend on the identification of homogenous, native Env trimers that are suitable for structural comparisons among isolates. Such constructs remain elusive despite substantial efforts (96). Soluble oligomeric gp140 ectodomain constructs, which are often thought to mimic the native Env trimer, appear to exhibit substantial heterogeneity from one isolate to another (82), and in many cases, they adopt nonnative conformations, especially within gp41 (M. Guttman et al., unpublished data). At present, full-length gp120s are the most tractable system for comparing structural order among isolates. In many respects, full-length gp120 appears to reflect the subunit organization of gp120 in the context of the trimer (40) and can provide valuable insight into the structure-function relationship of HIV-1 Env. Furthermore, because monomeric gp120 is likely to be immunogenic in the course of natural HIV-1 infection due to the labile interactions between gp120 and gp41, which allow for gp120 shedding (97), and because monomeric gp120 proteins are commonly used vaccine immunogens, including in the RV144 trial, it is important to understand the structural factors that influence antibody reactivity to monomeric gp120 proteins as well as trimeric Env constructs.
Significance for gp120 as a prospective vaccine immunogen.
The RV144 HIV-1 vaccine trial provided the first evidence that vaccine-induced protection against HIV-1 acquisition is possible and has rekindled interest in gp120 proteins as vaccine immunogens. Analysis of samples collected in the RV144 trial revealed that among other correlates of protection, ADCC activity was negatively associated with risk of infection in individuals with low levels of IgA antibodies (9). Notably, vaccinees with strong IgA responses to C-1 in the gp120 inner domain were at greater risk of infection compared to vaccinees without these antibodies, suggesting that IgA may inhibit ADCC-active antibody binding to this region. Thus, it was proposed that inner domain-specific, ADCC-active IgG antibodies, similar to the A32-like antibodies commonly elicited during HIV-1 infection (91), may play an important role in protection from HIV-1 acquisition (98). We observed that the stability of the C-1 region targeted by these antibodies varied among gp120 proteins from diverse isolates, and the differences in stability corresponded to differences in antibody binding to this region, including N5i5, which potently mediates ADCC (61). In light of previous studies, our results suggest that conformational dynamics may play a role in facilitating evasion of antibodies targeting this functionally conserved gp120 epitope and highlight the potential importance of characterizing the local conformational stability of future gp120 vaccine immunogens.
The relationship between gp120 conformational flexibility, antigenicity, and immunogenicity is likely to be complex. The data presented here, and in previous studies (38), suggest that flexibility modifies gp120 antigenicity; however, antigenicity is not equivalent to immunogenicity, and it remains to be determined what effect flexibility may have on the immunogenicity of Env constructs. Indeed, previous work revealed that the opposing influences of stability and dynamics are both likely to influence epitope immunogenicity. For example, Dey and colleagues demonstrated that the immunogenicity of the gp120 coreceptor binding site can be enhanced by using cavity-filled, disulfide-stabilized gp120 core proteins (99). Similarly, a single glycan deletion at the base of V2, which appears to shift the conformational equilibrium of Env toward a CD4-bound-like state (31), has been shown to elicit improved antibody titers with increased neutralization breadth in macaques (100). In contrast, a recent study by Liao et al. identified a conformationally variable linear epitope in V2 that was an important immunogenic target of antibodies isolated from vaccinees in the RV144 trial (13). In the design of improved antibody-based HIV-1 vaccines, the appropriate balance between immunogen stability and flexibility should be carefully evaluated in the context of both immunogenicity (the ability to elicit high titers of antibodies) and quality (the ability to elicit broad and potent antibodies). Our results support a role for HDX-MS in testing the relationship between the local stability of immunogens, their recognition by antibodies, and their immunogenicity, with the goal of improving antibody-based HIV-1 vaccine efficacy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by UAB CFAR grant P30-AI027767 through the CNIHR program for new HIV investigators (K.K.L.), NIH grants R00-GM080352 and R01-GM099989 (K.K.L.), P01-RR000166 (Washington National Primate Research Center), R01-AI076170, and Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) grant OPP1033102 (S.-L.H.). T.M.D. was supported by NIH training grants T32-AI7509 to -12. Data collection at SSRL was supported by grant number P41-RR001209 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health.
We acknowledge Tsutomu Matsui and Lester Carter at SSRL BL4-2 for valuable assistance with online FPLC SAXS data collection. We thank John Sumida and Mauro Acchione of the University of Washington Center for Intracellular Delivery of Biologics Analytical Biopharmacy core, supported by the Washington State Life Sciences Discovery Fund, for helpful discussions and use of the Biacore T-100 instrument. We acknowledge the contributors who generously provided materials that were important in carrying out these studies. We thank Yongjun Guan, Marzena Pazgier, and George Lewis at the University of Maryland for providing the antibodies N5i5, B18, and C4 and for helpful discussions. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 consensus subtype B Env (15-mer) peptides complete set, HIV-1 gp120 monoclonal antibody (IgG1 b12) from Dennis Burton and Carlos Barbas, HIV-1 gp120 monoclonal antibodies 17b and A32 (catalog number 11438) from James E. Robinson, HIV-1 gp120 monoclonal antibody (VRC01) from John Mascola, sCD4-183 from Pharmacia, Inc. (11780), and CD4-IgG2 from Progenics Pharmaceuticals. The reagent CA13 (ARP 3119) was obtained from the Centre for AIDS Reagents, NIBSC HPA UK, supported by the EC FP6/7 Europrise Network of Excellence, the NGIN Consortium, and the Bill and Melinda Gates GHRC-CAVD Project, and was donated by C. Arnold.
Footnotes
Published ahead of print 31 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01535-13.
REFERENCES
- 1.Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091–1094 [DOI] [PubMed] [Google Scholar]
- 2.Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. 1986. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233:209–212 [DOI] [PubMed] [Google Scholar]
- 3.Barin F, McLane MF, Allan JS, Lee TH, Groopman JE, Essex M. 1985. Virus envelope protein of HTLV-III represents major target antigen for antibodies in AIDS patients. Science 228:1094–1096 [DOI] [PubMed] [Google Scholar]
- 4.Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu. Rev. Med. 61:135–152 [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 6.Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch KA, Rainwater S, Jaoko W, Overbaugh J. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology 398:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19–42 [DOI] [PubMed] [Google Scholar]
- 9.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 11.Alam SM, Liao H-X, Tomaras GD, Bonsignori M, Tsao C-Y, Hwang K-K, Chen H, Lloyd KE, Bowman C, Sutherland L, Jeffries TL, Kozink DM, Stewart S, Anasti K, Jaeger FH, Parks R, Yates NL, Overman RG, Sinangil F, Berman PW, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Karasavva N, Rerks-Ngarm S, Kim JH, Michael NL, Zolla-Pazner S, Santra S, Letvin NL, Harrison SC, Haynes BF. 2013. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J. Virol. 87:1554–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong L, Sheppard NC, Stewart-Jones GBE, Robson CL, Chen H, Xu X, Krashias G, Bonomelli C, Scanlan CN, Kwong PD, Jeffs SA, Jones IM, Sattentau QJ. 2010. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 403:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao H-X, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang K-K, Chen X, Tsao C-Y, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang Z-Y, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore JP, McCutchan FE, Poon SW, Mascola J, Liu J, Cao Y, Ho DD. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350–8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JLS, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6(12):e1001249. 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Cell. Mol. Immunol. 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 20.Huang CC. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Do Kwon Y, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancera M, Majeed S, Ban YEA, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang S-H, Yang X, Zhang M-Y, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Cell. Mol. Immunol. 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diskin R, Marcovecchio PM, Bjorkman PJ. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17:608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merk A, Subramaniam S. 2013. HIV-1 envelope glycoprotein structure. Curr. Opin. Struct. Biol. 23:268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng HK, Malnati MS, Plebani A, Siccardi AG, Littman DR, Fenyö EM, Lusso P. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259–1265 [DOI] [PubMed] [Google Scholar]
- 29.Berger EA, Murphy PM, Farber JM. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657–700 [DOI] [PubMed] [Google Scholar]
- 30.Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, Gabuzda D, Smith AB, Sodroski J. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5(4):e1000360. 10.1371/journal.ppat.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harouse JM, Kunsch C, Hartle HT, Laughlin MA, Hoxie JA, Wigdahl B, Gonzalez-Scarano F. 1989. CD4-independent infection of human neural cells by human immunodeficiency virus type 1. J. Virol. 63:2527–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DH, Winters-Digiacinto P, Mitiku M, O'Rourke S, Sinangil F, Wrin T, Montefiori DC, Berman PW. 2010. Comparative immunogenicity of HIV-1 clade C envelope proteins for prime/boost studies. PLoS One 5(8):e12076. 10.1371/journal.pone.0012076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637–648 [DOI] [PubMed] [Google Scholar]
- 35.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PWIH, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 39.Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. 2010. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J. Virol. 84:10311–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, Kahn M, Garcia NK, Hu S-L, Lee KK. 2012. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J. Virol. 86:8750–8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sethi A, Tian J, Derdeyn CA, Korber B, Gnanakaran S. 2013. A mechanistic understanding of allosteric immune escape pathways in the HIV-1 envelope glycoprotein. PLoS Comput. Biol. 9(5):e1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng-Mayer C, Weiss C, Seto D, Levy JA. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. U. S. A. 86:8575–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, Wong-Staal F. 1984. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 226:1165–1171 [DOI] [PubMed] [Google Scholar]
- 44.Grisson RD, Chenine A-L, Yeh L-Y, He J, Wood C, Bhat GJ, Xu W, Kankasa C, Ruprecht RM. 2004. Infectious molecular clone of a recently transmitted pediatric human immunodeficiency virus clade C isolate from Africa: evidence of intraclade recombination. J. Virol. 78:14066–14069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J. Virol. 80:8729–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rambo RP, Tainer JA. 2010. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 20:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam CD, Hammel M, Hura GL, Tainer JA. 2007. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40:191–285 [DOI] [PubMed] [Google Scholar]
- 48.Svergun DI, Koch MHJ. 2002. Advances in structure analysis using small-angle scattering in solution. Curr. Opin. Struct. Biol. 12:654–660 [DOI] [PubMed] [Google Scholar]
- 49.Englander SW. 2006. Hydrogen exchange and mass spectrometry: a historical perspective. J. Am. Soc. Mass Spectrom. 17:1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan CR, Engen JR. 2009. Investigating solution-phase protein structure and dynamics by hydrogen exchange mass spectrometry. Curr. Protoc. Protein Sci. Chapter:Unit-17.617. 10.1002/0471140864.ps170s58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 88:10134–10137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbas CF, Björling E, Chiodi F, Dunlop N, Cababa D, Jones TM, Zebedee SL, Persson MA, Nara PL, Norrby E. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. U. S. A. 89:9339–9343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, Burton DR. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 55.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 57.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]
- 58.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moore JP, Thali M, Jameson BA, Vignaux F, Lewis GK, Poon SW, Charles M, Fung MS, Sun B, Durda PJ. 1993. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J. Virol. 67:4785–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. 1990. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res. Hum. Retroviruses 6:465–479 [DOI] [PubMed] [Google Scholar]
- 61.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. U. S. A. 110:E69–E78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retroviruses 10:371–381 [DOI] [PubMed] [Google Scholar]
- 63.di Marzo Veronese F, Rahman R, Pal R, Boyer C, Romano J, Kalyanaraman VS, Nair BC, Gallo RC, Sarngadharan MG. 1992. Delineation of immunoreactive, conserved regions in the external glycoprotein of the human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 8:1125–1132 [DOI] [PubMed] [Google Scholar]
- 64.Guo W, Cleveland B, Davenport TM, Lee KK, Hu S-L. 2013. Purification of recombinant vaccinia virus-expressed monomeric HIV-1 gp120 to apparent homogeneity. Protein Expr. Purif. 90:34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang J, Rand KD, Beuning PJ, Engen JR. 2011. False EX1 signatures caused by sample carryover during HX MS analyses. Int. J. Mass Spectrom. 302:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weis DD, Engen JR, Kass IJ. 2006. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 17:1700–1703 [DOI] [PubMed] [Google Scholar]
- 67.Guttman M, Scian M, Lee KK. 2011. Tracking hydrogen/deuterium exchange at glycan sites in glycoproteins by mass spectrometry. Anal. Chem. 83:7492–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA [Google Scholar]
- 69.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. 2007. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Crystallogr. 40:s453–s458 [Google Scholar]
- 70.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. 2007. ATSAS2.1: towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40:s223–s228 [Google Scholar]
- 71.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36:1277–1282 [Google Scholar]
- 72.Petoukhov MV, Svergun DI. 2007. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 17:562–571 [DOI] [PubMed] [Google Scholar]
- 73.Svergun DI. 1991. Mathematical methods in small-angle scattering data analysis. J. Appl. Crystallogr. 24:485–492 [Google Scholar]
- 74.Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25:495–503 [Google Scholar]
- 75.Svergun DI. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76:2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Volkov VV, Svergun DI. 2003. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36:860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mertens HDT, Svergun DI. 2010. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 172:128–141 [DOI] [PubMed] [Google Scholar]
- 78.Rambo RP, Tainer JA. 2011. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95:559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. 2008. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 22:1041–1046 [DOI] [PubMed] [Google Scholar]
- 80.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. U. S. A. 97:9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atassi MZ. 1975. Antigenic structure of myoglobin: the complete immunochemical anatomy of a protein and conclusions relating to antigenic structures of proteins. Immunochemistry 12:423–438 [DOI] [PubMed] [Google Scholar]
- 82.Jeffs SA, Goriup S, Kebble B, Crane D, Bolgiano B, Sattentau Q, Jones S, Holmes H. 2004. Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine 22:1032–1046 [DOI] [PubMed] [Google Scholar]
- 83.Finzi A, Xiang S-H, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Désormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang S-H, Mao Y, Sodroski J, Finzi A. 2013. The highly conserved layer-3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J. Virol. 87:2549–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kassa A, Madani N, Schön A, Haim H, Finzi A, Xiang S-H, Wang L, Princiotto A, Pancera M, Courter J, Smith AB, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J. Virol. 83:8364–8378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722–9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiang S-H, Doka N, Choudhary RK, Sodroski J, Robinson JE. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses 18:1207–1217 [DOI] [PubMed] [Google Scholar]
- 88.Drummer HE, Hill MK, Maerz AL, Wood S, Ramsland PA, Mak J, Poumbourios P. 2013. Allosteric modulation of the HIV-1 gp120-gp41 association site by adjacent gp120 variable region 1 (V1) N-glycans linked to neutralization sensitivity. PLoS Pathog. 9(4):e1003218. 10.1371/journal.ppat.1003218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korkut A, Hendrickson WA. 2012. Structural plasticity and conformational transitions of HIV envelope glycoprotein gp120. PLoS One 7(12):e52170. 10.1371/journal.pone.0052170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Cell. Mol. Immunol. 433:834–841 [DOI] [PubMed] [Google Scholar]
- 91.Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tainer JA, Getzoff ED, Alexander H, Houghten RA, Olson AJ, Lerner RA, Hendrickson WA. 1984. The reactivity of anti-peptide antibodies is a function of the atomic mobility of sites in a protein. Nature 312:127–134 [DOI] [PubMed] [Google Scholar]
- 94.Westhof E, Altschuh D, Moras D, Bloomer AC, Mondragon A, Klug A, Van Regenmortel MHV. 1984. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature 311:123–126 [DOI] [PubMed] [Google Scholar]
- 95.Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85:8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Forsell MN, Schief WR, Wyatt RT. 2009. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr. Opin. HIV AIDS 4:380–387 [DOI] [PubMed] [Google Scholar]
- 97.Moore JP, McKeating JA, Weiss RA, Sattentau QJ. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139–1142 [DOI] [PubMed] [Google Scholar]
- 98.Tomaras GD, Ferrari G, Shen X, Alam SM, Liao H-X, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, Poling B, Nicholson CO, Zhang R, Lu X, Parks R, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngarm S, Gilbert PB, Kim JH, Michael NL, Montefiori DC, Haynes BF. 2013. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl. Acad. Sci. U. S. A. 110:9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. 2009. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 5(5):e1000445. 10.1371/journal.ppat.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng-Mayer C, Liu R, Landau NR, Stamatatos L. 1997. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J. Virol. 71:1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siddappa NB, Watkins JD, Wassermann KJ, Song R, Wang W, Kramer VG, Lakhashe S, Santosuosso M, Poznansky MC, Novembre FJ, Villinger F, Else JG, Montefiori DC, Rasmussen RA, Ruprecht RM. 2010. R5 clade C SHIV strains with tier 1 or 2 neutralization sensitivity: tools to dissect Env evolution and to develop AIDS vaccines in primate models. PLoS One 5(7):e11689. 10.1371/journal.pone.0011689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang H, Rola M, West JT, Tully DC, Kubis P, He J, Kankasa C, Wood C. 2010. Functional properties of the HIV-1 subtype C envelope glycoprotein associated with mother-to-child transmission. Virology 400:164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ching LK, Vlachogiannis G, Bosch KA, Stamatatos L. 2008. The first hypervariable region of the gp120 Env glycoprotein defines the neutralizing susceptibility of heterologous human immunodeficiency virus type 1 isolates to neutralizing antibodies elicited by the SF162gp140 immunogen. J. Virol. 82:949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.