Abstract
The role of a tetR transcriptional regulatory gene (SAV7471) in avermectin production in the Gram-positive soil bacterium Streptomyces avermitilis was investigated by gene deletion, complementation, and overexpression experiments. Gene deletion of the SAV7471 open reading frame resulted in avermectin overproduction. The deletion also resulted in overexpression of SAV7472-SAV7473 transcripts, which encode a protein of unknown function and a flavoprotein possibly involved in pantothenate and coenzyme A (CoA) metabolism. EMSAs and footprinting assays showed that SAV7471 can bind to two palindromic sequences with high similarity in the intergenic region between SAV7471 and SAV7472, a region that contains the apparent transcription start sites for each gene detected by rapid amplification of 5′ cDNA ends (5′-RACE). In addition to SAV7472-SAV7473, at least two genes (SAV1104 and SAV1258) involved in CoA metabolism are negatively controlled by SAV7471. By negatively regulating the transcription of the target genes SAV7472-SAV7473 and other genes involved in CoA metabolism, SAV7471 may affect cellular metabolic flux and may thereby indirectly regulate avermectin biosynthesis.
INTRODUCTION
Streptomyces spp. are Gram-positive, filamentous, soil-dwelling bacteria which are characterized by complex morphological differentiation and the ability to produce a wide variety of secondary metabolites, including antibacterial antibiotics, antitumor agents, immunosuppressants, and anthelmintic agents (1). The biosynthetic genes for a particular secondary metabolite are generally found in a gene cluster that includes a regulatory gene encoding a pathway-specific transcriptional activator. These specific regulators are controlled by various higher-level pleiotropic regulators, whose expression is typically affected by a variety of environmental and physiological cues (2).
Among the various secondary metabolites produced by Streptomyces, the avermectins (a series of 16-membered macrocyclic lactone derivatives generated by Streptomyces avermitilis) have potent anthelmintic and insecticidal properties and are therefore applied widely in agriculture, veterinary medicine, and human medicine (3). Because of their superior potency and commercial value, avermectins are considered one of the most important antibiotics on the market. The avermectin polyketide chain is derived from seven acetates and five propionates, together with a single 2-methylbutyric acid or isobutyric acid residue that forms the sec-butyl group (“a” components) or the isopropyl group (“b” components) attached to C-25 (4). The synthesis of initial aglycons (the first step in avermectin biosynthesis) requires four giant multifunctional polypeptides (AVES1, AVES2, AVES3, and AVES4) of the modular polyketide synthase (PKS), which are encoded by four large open reading frames (aveA1-aveA2 and aveA3-aveA4). The polyketide chain is subsequently cyclized, glycosylated at C-13 with the attachment of two oleandrose sugars, and methylated at the hydroxyl group of C-5 (5). The aveR gene in the avermectin biosynthetic gene cluster encodes a pathway-specific activator essential for transcription of all avermectin biosynthetic genes (6, 7).
The TetR family is a common class of transcriptional regulators that regulate a wide range of cellular activities and are widely distributed among bacteria. The regulated activities include antibiotic resistance, multidrug resistance, virulence, and development (8). The whole-genome sequencing project for S. avermitilis revealed 116 genes belonging to the TetR-family transcriptional regulators (9). This is far more than the 12 and 20 TetR-family genes in Escherichia coli and Bacillus subtilis. So far, a few TetR-family transcriptional regulators have been extensively studied in Streptomyces. ActR (ActII-ORF1) and AtrA are involved in regulation of actinorhodin biosynthesis (10–12), Pip and PqrA are repressors of drug resistance genes (13, 14), and ScbR is a repressor of γ-butyrolactone synthesis in Streptomyces coelicolor (15). SimR from Streptomyces antibioticus is a repressor of an efflux pump-encoding gene that confers resistance to the antibiotic simocyclinone (16). Recently, Ahn et al. predicted the target genes of TetR-family regulators based on genome context in Streptomyces (17). However, compared to the large numbers of this family, the transcriptional regulatory functions and regulatory mechanisms of most TetR-family genes are still unknown. Thus, investigation of these uncharacterized TetR-family regulators in Streptomyces should increase our understanding of the complex regulatory network involved in antibiotic biosynthesis.
SAV7471 encodes a 184-amino-acid putative TetR-family transcriptional regulator. In this study, we describe the regulatory role of SAV7471 in avermectin production in S. avermitilis ATCC 31267. We also show that SAV7471 serves as a transcriptional repressor that regulates SAV7472-SAV7473 operon expression by binding to two similar palindromic sequences in the intergenic region between SAV7471 and SAV7472.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. avermitilis strains used in the present study were ATCC 31267 (wild-type strain), D7471 (an SAV7471 deletion strain), and D7471-C (D7471 harboring SAV7471 overexpression plasmid pSEE7471). E. coli DH5α [supE44 ΔlacU169(ϕ80 lacZΔM15) hsdR17 recA endA1 gyrA96 thi-1 relA1] and BL21(DE3) [F− dcm ompT hsdS (rB− mB−) gal λ (DE3)] were used for plasmid construction and protein expression, respectively. E. coli strains were cultured at 37°C in LB medium. S. avermitilis strains were grown at 28°C on solid YMS medium for sporulation or in modified liquid YEME medium containing 25% sucrose for growth of mycelia for DNA extraction and protoplast preparation (22, 30). RM14 medium was used for regeneration of protoplasts (18). For avermectin production, the seed medium contained 30 g of soluble starch, 2 g of malt extract, 2 g of soy peptone, and 5 mg of CoCl2 · 6H2O per liter of H2O. The fermentation medium contained 70 g of soluble starch, 16 g of yeast extract, 0.5 g of K2HPO4 · 3H2O, 0.5 g of MgSO4 · 7H2O, 4 g of KCl, 5 mg of CoCl2 · 6H2O, and 2 g of CaCO3 per liter of H2O (19).
Gene deletion, complementation, and overexpression.
Targeted gene deletion mediated by homologous recombination was used to generate a SAV7471-null mutant. With ATCC 31267 genomic DNA as the template, a 316-bp fragment upstream of SAV7471 was amplified with the primers 7471 upstream-1 and 7471 upstream-2, and a 396-bp fragment downstream of SAV7471 was amplified with the primers 7471 downstream-1 and 7471 downstream-2 (see Table S1 in the supplemental material). The two fragments were recovered and used at a molar ratio of 1:1. A 712-bp fragment was amplified with the primers 7471 upstream-1 and 7471 downstream-2, digested by BglII/EcoRI, and cloned into BamHI/EcoRI-digested pKC1139 (20) to produce the SAV7471 gene deletion vector pKCD7471. pKCD7471 was introduced into S. avermitilis ATCC 31267 by protoplast transformation (18). Because S. avermitilis does not sporulate on RM14 medium, transformants regenerated on RM14 plates were transferred to solid YMS medium for sporulation. Double-crossover recombinant strains were selected as described previously (21). The transformants were spread on YMS agar plates containing apramycin. The plates were incubated initially at 28°C for 48 h and then at 39°C (a nonpermissive temperature for plasmid pKC1139 replication) for 7 days. Only mutants generated by a single crossover can grow on YMS containing apramycin in which pKCD7471 was integrated into the chromosome of S. avermitilis by homologous recombination. Insertion mutants were inoculated on YMS plates under nonselection conditions for two rounds of sporulation, and double crossover took place only in colonies sensitive to apramycin. The mutants were confirmed by PCR using one external primer (7471 V1 or 7471 V2) (see Table S1 in the supplemental material) located outside the homologous recombination regions coupled with one internal primer (7471 downstream-2 or 7471 upstream-1), and this was followed by DNA sequencing. The deletion strain was designated D7471.
A 973-bp DNA fragment carrying the complete SAV7471 gene and its possible promoter was amplified by PCR using the primers E7471 forward and E7471 reverse and then cloned into the EcoRI/BamHI-digested integrative vector pSET152 to produce the SAV7471 expression vector pSEE7471. For complementation analysis of the D7471 mutant, vector pSEE7471 was integrated into the attB site of the S. avermitilis chromosome after transformation. For SAV7471 overexpression, pSEE7471 together with pSET152 as a control was introduced into ATCC 31267 by protoplast transformation (22).
RNA extraction and quantitative RT-PCR analysis.
RNA was isolated as previously described (19). Mycelia of S. avermitilis grown in the fermentation medium for 2 days and 6 days were collected, frozen in liquid nitrogen, and ground to a fine powder. RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions and was treated with DNase I (TaKaRa, Shiga, Japan) to remove chromosomal DNA contamination. Quantitative real-time PCR (RT-PCR) was carried out in an ABI 7500 system using the SYBR Premix ExTaq (perfect real-time) kit (TaKaRa, Shiga, Japan). Primer pairs, which are listed in Table S1 in the supplemental material, were used to analyze transcript levels of aveR, aveA1, SAV7472, and SAV7473 in the various strains of S. avermitilis. The hrdB gene (SAV2444), which encodes the major σ factor in Streptomyces, was used as the internal control. The possibility of DNA contamination was ruled out by negative controls, which were conducted using PCR amplification without prior reverse transcription. PCR included a preincubation at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 45 s. The PCR products were loaded on 1.5% agarose gels and photographed after staining with ethidium bromide. Primer efficiencies were measured and calculated (23). The relative expression level was calculated using the comparative threshold cycle (CT) method. Results were normalized to the expression of hrdB. Each experiment was performed in triplicate.
Electrophoretic mobility gel shift assays (EMSAs).
The SAV7471 coding region was amplified using the primers His6-7471 forward and His6-7471 reverse and then digested with XhoI. pET28a(+) was digested with NcoI, blunted with Klenow fragment, and digested with XhoI. The purified fragments were ligated to generate the expression plasmid pET28a::SAV7471. The recombinant SAV7471 was tagged at the C terminus with a His6 oligopeptide. After confirmation by DNA sequencing, pET28a::SAV7471 was introduced into E. coli BL21(DE3) for protein expression. E. coli BL21(DE3) harboring pET28a::SAV7471 was grown at 37°C in 100 ml of LB with 50 μg ml−1 kanamycin to an optical density at 600 nm (OD600) of 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a final concentration of 0.2 mM, and the cultures were incubated for an additional 8 h at 16°C. The cells were harvested, washed twice with binding buffer (20 mM Tris base, 500 mM NaCl, 5 mM imidazole, 5% glycerol [pH 7.9]), and then resuspended in 10 ml of the same buffer. The cell suspension was sonicated on ice. After centrifugation, the supernatant was recovered, and His6-SAV7471 was purified from the whole-cell lysate using nickel-nitrilotriacetic acid (Ni-NTA) agarose chromatography (Novagen) according to the manufacturer's protocol.
EMSAs were performed according to the manufacturer's instructions (digoxigenin [DIG] gel shift kit, second generation; Roche). DNA fragments (probe 1 to probe 5) used as DIG-labeled probes were amplified by PCR with the primers listed in Table S1 in the supplemental material. The fragments were labeled with digoxigenin-11-ddUTP using recombinant terminal transferase. During the EMSA, the DIG-labeled DNA probe (0.3 nM) was incubated individually with various quantities of His6-SAV7471 at 25°C for 30 min in a binding buffer [20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), 0.2% (wt/vol) Tween 20, 30 mM KCl] containing 1 μg of poly(dI-dC) and 0.1 μg of poly l-lysine in a total volume of 20 μl. After incubation, protein-bound and free DNAs were separated by electrophoresis on 5.0% native polyacrylamide gels with 0.5× TBE buffer (44.5 mM Tris-HCl, 44.5 mM boric acid, and 1 mM EDTA, pH 8.0) as running buffer at 100 V and 4°C. The DNA was electroblotted onto a positively charged nylon membrane (Amersham) for 40 min at 400 mA in 0.5× TBE buffer. The signals were detected by chemiluminescence and recorded on X-ray film.
DNase I footprinting assays.
DNase I footprinting assays were performed using a fluorescence labeling procedure (24). DNA fragments were prepared by PCR using the fluorescence-labeled primers Probe 6 forward and Probe 6 reverse (see Table S1 in the supplemental material) and were purified from the agarose gel. Labeled DNA fragments (400 ng) and different concentrations of His6-SAV7471 were added to a final reaction mixture volume of 50 μl and incubated at 25°C for 30 min. DNase I digestions were carried out for 40 s at 25°C and stopped with 20 μl of 0.2 M EDTA (pH 8.0). After phenol-chloroform extraction and ethanol precipitation, the samples were loaded in an Applied Biosystems 3730 DNA genetic analyzer together with the internal-lane size standard ROX-500 (Applied Biosystems). The dye primer-based sequencing kit (Thermo) was used to precisely determine the sequences after the capillary electrophoresis results of the reactions were aligned. Electropherograms were analyzed with Genemarker v1.8 (Applied Biosystems).
Determination of transcriptional start points.
5′-RACE (rapid amplification of cDNA ends) experiments (5′/3′ RACE kit, second generation; Roche) were conducted to map the transcriptional start points of SAV7471 and SAV7472 according to the manufacturer's instructions. cDNA was synthesized using a gene-specific primer (SP1), and the template RNA was degraded with RNase H. After the cDNA was purified with the PCR product recovery kit (Tiangen, Beijing, China), a homopolymeric A tail was added to the 3′ end using terminal transferase. The tailed cDNA was amplified through 35 cycles with the oligo(dT) anchor primer and a specific nested primer (SP2). One microliter of the PCR product was amplified in a second PCR through 35 cycles with the PCR anchor primer and a specific nested primer (SP3) (see Table S1 in the supplemental material). Then the PCR products were cloned into the pMD18-T vector (TaKaRa, Shiga, Japan). Ligation products were used to transform competent E. coli DH5α cells, and at least six different positive clones were selected for plasmid extraction and were sequenced.
Fermentation and HPLC analysis of avermectin production.
Spores from various S. avermitilis strains cultured on YMS plates for 7 days were added to 250-ml flasks containing 50 ml of seed medium; the cultures were incubated for 24 h at 28°C on a rotary shaker (180 rpm). A 2.5-ml volume of the culture was then transferred to three 250-ml flasks each containing 50 ml of fermentation medium, and the cultures were grown for 10 days at 28°C on a rotary shaker (220 rpm). The fermentation broth (1.0 ml) was extracted with 4.0 ml of methanol for 30 min and centrifuged at 4,000 × g for 10 min. The supernatant was directly applied to a high-performance liquid chromatography (HPLC) system (model 600; Waters, Milford, CT) with a C18 column (10 μm; 4.6 [internal diameter] by 250 mm) developed with methanol-water (85:15) at a flow rate of 1.0 ml min−1. Avermectins were detected by UV absorption at 246 nm, with authentic samples of avermectin B1 used as internal standards.
S. avermitilis CoA levels.
Mycelia of S. avermitilis grown in the fermentation broth for 6 days were collected, frozen in liquid nitrogen, and ground to a fine powder. To deproteinize the lysates, 200 μl of cold 1 N perchloric acid was added to each sample (50 mg), homogenized thoroughly, and centrifuged at 14,000 × g at 4°C for 2 min to pellet precipitated protein. The supernatant was neutralized with cold 3 M potassium bicarbonate. The concentrations of coenzyme A (CoA) were determined for each sample using the PicoProbe acetyl-CoA assay kit (BioVision) according to the manufacturer's protocol.
RESULTS
SAV7471 plays a negative regulatory role in avermectin biosynthesis.
SAV7471 encodes a 184-amino-acid (aa) protein with an N-terminal TetR-family helix-turn-helix (HTH) DNA binding domain. To assess the in vivo biological significance of SAV7471, we cloned a DNA fragment containing the coding region of SAV7471 and its putative promoter into the Streptomyces integrating vector pSET152 to produce the SAV7471 overexpression plasmid pSEE7471. Introduction of pSEE7471 into the S. avermitilis wild-type strain ATCC 31267 dramatically decreased avermectin production, by about 75% (Fig. 1).
Fig 1.
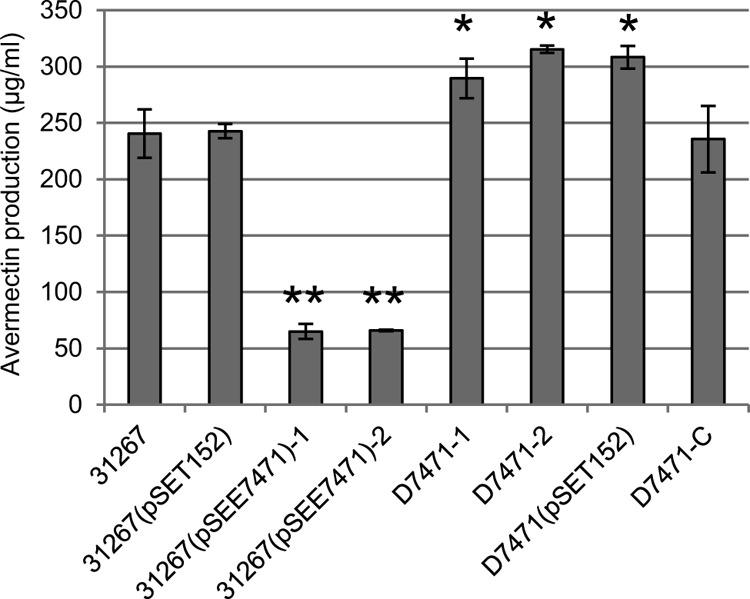
Effect of overexpression and deletion of the SAV7471 gene on avermectin production in S. avermitilis ATCC 31267. The strains left to right are wild-type ATCC 31267, the empty-plasmid-containing control ATCC 31267(pSET152), the SAV7471-overexpressing transformants 31267(pSEE7471)-1 and -2, D7471-1 and -2 (the SAV7471 deletion mutants), the D7471-2 empty plasmid-containing control D7471(pSET152), and the SAV7471 deletion complementation strain D7471-C (D7471-2 strain harboring pSEE7471). After incubation in fermentation broth for 10 days, cultures were extracted by methanol and were analyzed by high-performance liquid chromatography. Values are means ± standard deviations (SD) for three replicate flasks. Statistical significance was determined by comparing the mutant values to those of wild-type ATCC 31267. *, P < 0.05; **, P < 0.01.
We also constructed a SAV7471 deletion mutant in which the whole SAV7471 open reading frame (ORF) was deleted by a homologous recombination strategy. Deletion of SAV7471 did not affect S. avermitilis morphology (data not shown). Compared to S. avermitilis wild-type ATCC 31267, avermectin production was modestly increased in the SAV7471 deletion mutant D7471 (Fig. 1). To confirm that the SAV7471 deletion was the sole cause of the enhanced avermectin production, we reintroduced the SAV7471 gene into D7471-2 by integrating the pSET152-based complementation plasmid pSEE7471 into the chromosome. The avermectin yield of this complementation strain (termed D7471-C) was restored to that of the wild-type strain (Fig. 1). Thus, the absence of SAV7471 was the cause of the increased avermectin production in D7471. These findings, together with SAV7471 overexpression experiments, indicate that SAV7471 has a negative role in avermectin production.
SAV7471 represses the transcription of the adjacent genes SAV7472 and SAV7473.
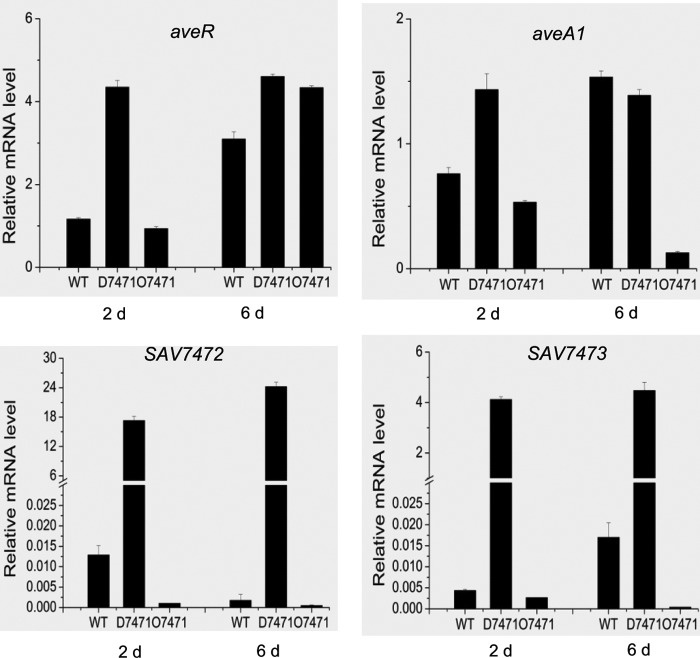
The aveR gene encodes a pathway-specific activator essential for transcription of all avermectin biosynthetic genes (6, 7). To test whether SAV7471 regulated avermectin production through a pathway-specific activator, we performed RT-PCR analysis in order to determine the expression levels of aveR and of the biosynthetic gene aveA1 in the SAV7471 deletion strain D7471, SAV7471-overexpression strain O7471, and in the wild-type strain ATCC 31267. RNA samples were isolated from cells grown in fermentation broth for 2 days and 6 days. The expression of the essential sigma factor gene hrdB (SAV2444) was used as an internal control. Compared to those of the wild-type strain, the transcript levels of aveR and aveA1 were increased in the SAV7471 deletion strain D7471 and were slightly decreased in the SAV7471 overexpression strain O7471 at 2 days when avermectin biosynthesis was initiated. But at 6 days, only the aveA1 transcript level was decreased in O7471, the transcript levels of aveR and aveA1 did not differ much in D7471 (Fig. 2). These results suggested that SAV7471 has a negative regulatory role in expression of the pathway-specific activator gene aveR.
Fig 2.
Detection of gene expression levels of aveR, aveA1, and SAV7472-SAV7473 in D7471 (the SAV7471 deletion mutant) and O7471 (SAV7471 overexpression transformant) by RT-PCR. RNA was prepared from cells grown in fermentation medium for 2 days and 6 days at 28°C with reciprocal shaking (220 rpm). hrdB (SAV2444), encoding the principal sigma factor, was used as the internal control. Control experiments with no reverse transcriptase confirmed that the RNA samples contained no chromosomal DNA.
RT-PCR analyses were also performed to test whether the deletion and overexpression of SAV7471 affected the expression of its adjacent genes, SAV7472 and SAV7473. SAV7472 and SAV7473 are separated by an intergenic region of only nine nucleotide base pairs, and RT-PCR analysis showed that they are in a transcriptional unit (data not shown). The transcription levels of SAV7472 and SAV7473 were very low in the wild-type strain ATCC 31267, were markedly increased in SAV7471 deletion strain D7471, and were decreased in SAV7471 overexpression strain O7471 at both times (Fig. 2), indicating that SAV7471 represses the expression of SAV7472 and SAV7473 either directly or indirectly. According to previous reports, TetR-family proteins usually repress their own expression (8), but because the entire ORF of SAV7471 was deleted in D7471, we could not analyze the expression level of SAV7471 in D7471.
SAV7471 interacts with the SAV7471-SAV7472 intergenic region.
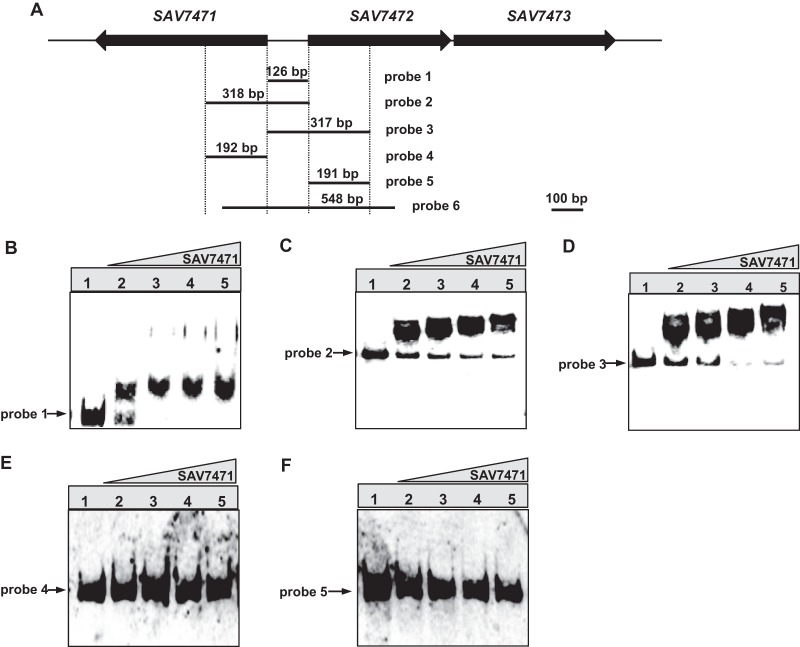
RT-PCR analysis showed that SAV7471 repressed the expression of SAV7472-SAV7473, and SAV7472 and SAV7473 were cotranscribed. To test whether SAV7471 protein interacted with the SAV7472-SAV7473 promoter region directly, we performed gel mobility shift assays (EMSAs). The recombinant C-terminal His-tagged SAV7471 was overexpressed in E. coli and purified by Ni affinity chromatography. Using this protein, we designed five DIG-labeled probes (probe 1 to 5) containing various regions for EMSAs (Fig. 3A). Of the five probes tested, probe 1 (126 bp of the SAV7471-SAV7472 intergenic region), probe 2 (318 bp of the upstream region of SAV7472 and covering a 126-bp overlap with probe 1), and probe 3 (317 bp of the upstream region of SAV7471 and covering a 126-bp overlap with probe 1), gave retarded signals (Fig. 3B to D), while retarded signals were not evident with probe 4 and probe 5, which do not include the SAV7471-SAV7472 intergenic region (Fig. 3E and F). Thus, the SAV7471-binding sites were located at the SAV7471-SAV7472 intergenic region. Because two retarded signals were detected with the probes, more than one SAV7471-binding site in the intergenic region was expected.
Fig 3.
Gel electrophoretic mobility shift assay (EMSA) for determination of SAV7471 binding sites in the upstream regions of SAV7472 and SAV7471. (A) Schematic representation of the probes used for EMSAs. The lengths and positions of the probes are shown. Probe 6 was used for DNase I protection experiments. (B to F) EMSA with probes 1 to 5. Each lane contained 0.3 nM probe. Lanes 1 to 5 contain 0, 50, 100, 200, and 400 ng of purified His6-SAV7471, respectively.
Determination of the SAV7471-binding sequences in the SAV7471-SAV7472 intergenic region.
EMSAs showed that SAV7471 interacts with the SAV7471-SAV7472 intergenic region. To determine the specific SAV7471-binding sequences, we performed DNase I footprinting experiments on a 548-bp DNA probe comprising the SAV7471-SAV7472 intergenic region in the presence or absence of the recombinant C-terminal His-tagged SAV7471. As shown in Fig. 4A, two protected regions were detected on the coding strand of the SAV7472 gene in the presence of 2 or 8 μM His6-SAV7471. The protected regions were named site 1 (AGAGAACCATCGTTCTCT, 18 nucleotides [nt]; underlining indicates inverted repeat sequences) and site 2 (GAGAACGTACGTTCTC, 16 nt), and binding specificity was greater for site 2 than for site 1. The two protected regions are palindromic sequences with high similarity. The consensus sequence of the SAV7471 binding site is a palindromic sequence: GAGAACSWWCGTTCTC (Fig. 4B). The result is consistent with previously reports that TetR-family proteins usually form symmetric TetR dimers and bind to palindromic operators.
Fig 4.

DNase I footprinting of the coding strand of the SAV7472 promoter region identified using His6-SAV7471. (A) The fluorograms correspond to the control DNA (with 10 μM bovine serum albumin [BSA]) and to the protection reactions (with 2 μM and 8 μM His6-SAV7471). Site 1 and site 2 indicate the corresponding sites protected by SAV7471. The arrows indicate the characteristic inverted repeat sequences. (B) Analyses of the consensus sequence of SAV7471 binding sites. S = G or C; W = A or T. The arrows indicate the characteristic inverted repeat sequences. (C) Nucleotide sequences of the promoter regions of SAV7471 and SAV7472. Sequences protected from DNase I digestion are indicated with shaded boxes (site 1 and site 2). The transcriptional start sites are indicated by dark bent arrows and boldface letters. The presumptive −35 and −10 elements of SAV7471 and SAV7472 promoters are boxed. The translational start codons are indicated by gray bent arrows and boldface letters.
To understand how SAV7471 represses the transcription of SAV7472-SAV7473, we determined the exact transcriptional start points of SAV7471 and SAV7472 by 5′-RACE. The RNA used for 5′-RACE was the same RNA that was prepared for RT-PCR analyses. The transcriptional start points are shown in Fig. 4C. In the coding strand of SAV7472, site 1 extends from positions −29 to −12 relative to the transcriptional start site, and is located between −35 region and −10 region. Site 2 ranges from positions +10 to +25 and is located downstream the transcriptional start site. In the case of SAV7471, site 1 extends from positions −7 to +11 with respect to the transcriptional start site and encompasses the transcriptional start site. Site 2 ranges from positions −43 to −28 and encompasses the −35 region. These observations suggested that, by binding to the palindromes in and downstream of the promoters of the two genes, SAV7471 blocks the attachment of RNA polymerase to the promoters and impedes RNA polymerase's progress along the strand, thus preventing transcription of SAV7472-SAV7473 and perhaps of itself.
Overexpression of SAV7472-SAV7473 leads to increased avermectin production.
The SAV7472 gene is predicted to encode a 153-amino-acid protein of unknown function (DUF1348), and SAV7473 encodes a 176-amino-acid protein annotated as a flavoprotein (PF02441) that is possibly involved in pantothenate metabolism. BLAST results showed that SAV7473 had a high similarity to phosphopantothenoylcysteine decarboxylase (PPC-DC) in Streptomyces venezuelae ATCC 10712 (identity, 65%; positives, 77%). Phosphopantothenoylcysteine decarboxylase catalyzes the decarboxylation of the cysteine moiety of 4′-phosphopantothenoylcysteine to form 4′-phosphopantetheine; this reaction forms part of the biosynthesis of coenzyme A (25). Overexpression of SAV7472-SAV7473 in wild-type ATCC 31267 led to a slight increase in avermectin production (Fig. 5). Avermectins are polyketide catalyzed by the type I polyketide synthase (PKS); acyl-CoA molecules are used as avermectin biosynthetic starter units and extender units during condensation steps. The increase in available starter units and extender units might lead to increased avermectin production.
Fig 5.
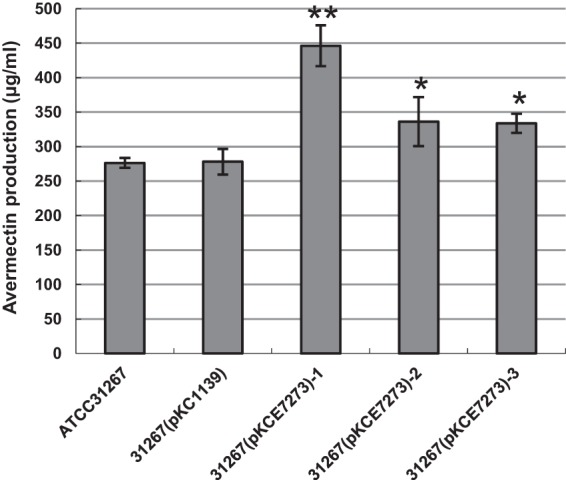
Effect of SAV7472-SAV7473 overexpression on avermectin production in S. avermitilis ATCC 31267. The strains (left to right) are wild-type ATCC 31267, plasmid control strain ATCC 31267(pKC1139), and SAV7472-SAV7473 overexpressing transformants 31267(pKCE7273)-1, -2, and -3. Values are means ± SD for three replicate flasks. Statistical significance was determined by comparing the mutant values to those of wild-type ATCC 31267. *, P < 0.05; **, P < 0.01.
Confirmation of putative SAV7471 target genes.
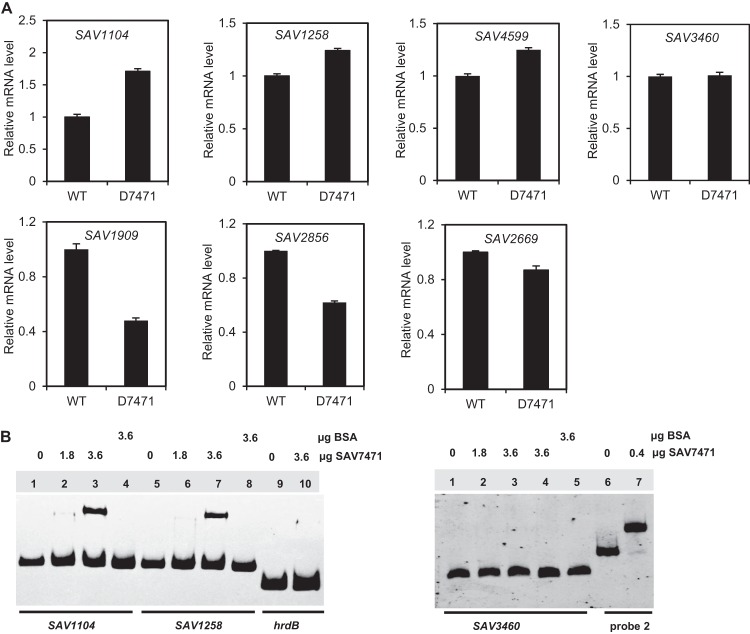
Interestingly, when the intergenic regions in the S. avermitilis genome were searched for the occurrence of SAV7471 binding sites using Virtual Footprint software (26), several genes involved in pantetheine and CoA metabolism were detected as putative SAV7471 targets (Table 1). Among them, coaD encodes a putative pantetheine-phosphate adenylyltransferase that is involved in pantetheine metabolism. The other genes encode a putative acyl-CoA synthetase, isobutyryl-CoA mutase, putative 3-hydroxyacyl-CoA dehydrogenase, and acetoacetyl-CoA thiolase, which are possibly involved in lipid metabolism. RT-PCR analysis showed that the expression of SAV1104, SAV1258, and SAV4599 was upregulated in the SAV7471 deletion strain, that the expression of SAV1909, SAV2669, and SAV2856 was downregulated in D7471, and that expression of SAV3460 was similar in D7471 and the wild-type strain (Fig. 6A). Like most TetR regulators, SAV7471 served as a repressor for SAV7472-SAV7473; we think it might repress other genes involved in CoA or pantetheine metabolism as well. Three of these genes (2 of them are upregulated) were further analyzed by EMSA to confirm whether they are SAV7471 target genes. Distinct retarded signals were observed in the presences of a high concentration (3.6 μg) of SAV7471 with probes from the upregulated genes SAV1104 and SAV1258, while no retarded signal was detected with probe SAV3460 (Fig. 6B). The results showed that SAV1104 and SAV1258 are also targets of SAV7471 and SAV7471 has a negative role in their transcription.
Table 1.
Putative SAV7471 target genes involved in pantetheine and CoA metabolism
| Gene | Function | Nucleotide positiona |
PWM scoreb | Sequence | ATG distancec | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| fadD3 (SAV1104) | Putative acyl-CoA synthetase, long-chain fatty acid-CoA ligase | 1388188 | 1388203 | 2 | GAGATCGACGGTGCTC | 43 |
| icmB (SAV3460) | Putative isobutyryl-CoA mutase, chain B | 4289699 | 4289714 | 2 | GATAACGTTCGTTCAC | 71 |
| coaD (SAV2669) | Putative pantetheine-phosphate adenylyltransferase | 3271874 | 3271889 | 3 | GAGAGCGAGGGACCTC | 3 |
| fadA1 (SAV2856) | Putative acetoacetyl-CoA thiolase | 3499879 | 3499894 | 3 | GTAAACCCTCGTTCAC | 38 |
| acsA1 (SAV4599) | Putative acetyl-CoA synthetase | 5615830 | 5615845 | 3 | GAGAACGTGTGGCCCC | 125 |
| fadD5 (SAV1258) | Putative acyl-CoA synthetase, long-chain fatty acid-CoA ligase | 1562606 | 1562621 | 4 | CAGTACGTGTGGTCCC | 107 |
| fadC1 (SAV1909) | Putative 3-hydroxyacyl-CoA dehydrogenase | 2335402 | 2335417 | 4 | GAGTGCGGGTGTTGCC | 84 |
| fadC1 (SAV1909) | Putative 3-hydroxyacyl-CoA dehydrogenase | 2335450 | 2335465 | 4 | GCGCACGCGGGTGCGC | 36 |
Genomic position. All sequences are minus strands.
Number of mismatches with respect to the consensus. PWM, positive weight matrix.
Values are distances (in nucleotides) to the predicted start codon of the downstream gene.
Fig 6.
EMSA to confirm binding of SAV7471 to upstream regions of putative SAV7471 target genes. (A) Detection of gene expression levels in D7471 and wild-type ATCC 31267 by RT-PCR. Quantifications were normalized to the expression of hrdB. Values are means and standard errors from three replicates. (B) EMSA determination of SAV7471 binding sites in the upstream regions of putative SAV7471 target genes. Each lane contained 0.3 nM probe. The amounts of His6-SAV7471 and BSA used are shown above each panel. The probes used are shown below the panels. As a positive control, probe 2 (the region upstream of SAV7472) was also analyzed. A region upstream of hrdB was used as a negative control. The addition of BSA rather than His6-SAV7471 was also used as a negative control.
S. avermitilis CoA levels.
CoA is made from pantothenate, cysteine, and adenosine. To determine the role of SAV7472-SAV7473 in pantothenate and CoA metabolism, we analyzed the concentration of CoA in wild-type S. avermitilis, the SAV7471 deletion mutant D7471, and the SAV7471-overexpressing transformant O7471. The CoA level of D7471 was slightly higher than that of the wild-type, and the CoA level of O7471 is lower than that of the wild type (Fig. 7). The results indicated that SAV7471 negatively regulated CoA biosynthesis.
Fig 7.
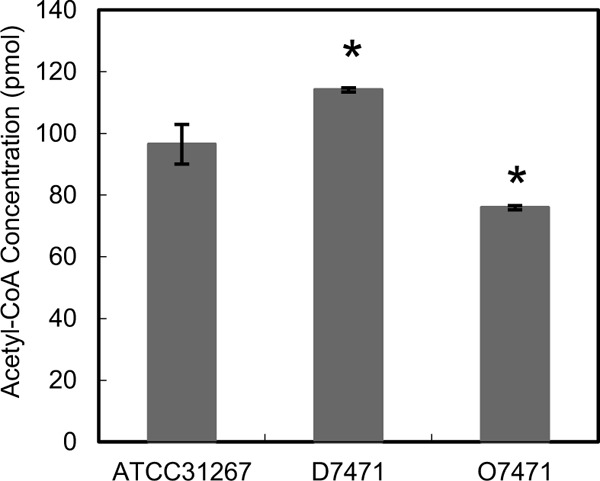
S. avermitilis CoA levels. CoA concentrations were measured from 50 mg of mycelia from wild-type S. avermitilis, D7471 (the SAV7471 deletion mutant), and O7471 (SAV7471 overexpression transformants). All strains were grown on fermentation medium for 6 days. The CoA levels are the means ± SD from three independent experiments. Statistical significance was determined by comparing the mutant values to those of wild-type ATCC 31267. *, P < 0.05.
DISCUSSION
The present work demonstrates that SAV7471 represses the transcriptional expression of the SAV7472-SAV7473 operon by binding to two similar palindromic sequences in the intergenic region between SAV7471 and SAV7472-SAV7473. In addition, SAV7471 was shown to have a negative role in avermectin biosynthesis. No similar palindrome sequence of the SAV7471 binding motif was found in the promoter regions of the avermectin biosynthesis regulatory gene aveR or of the avermectin biosynthesis structure genes. EMSA showed that SAV7471 cannot bind to the aveR promoter region in vitro (data not shown). Taken together, these results indicate that SAV7471 might regulate avermectin biosynthesis in an indirect manner.
The homologues of SAV7471 and the adjacent SAV7472-SAV7473 operon (SCO772 and SCO771-SCO770) are present in Streptomyces coelicolor (27), suggesting that this TetR-family transcriptional regulator may have conserved biological significance in vivo. BLAST results showed that SAV7473 has a high similarity to phosphopantothenoylcysteine decarboxylase (PPC-DC) in Streptomyces venezuelae ATCC 10712. Phosphopantothenoylcysteine decarboxylase catalyzes the decarboxylation of the cysteine moiety of 4′-phosphopantothenoylcysteine to form 4′-phosphopantetheine; this reaction forms part of the biosynthesis of coenzyme A (25). CoA-level analysis indicated that SAV7471 negatively regulated CoA biosynthesis in S. avermitilis. In addition to the SAV7473 gene, S. avermitilis has another gene, SAV6873 (dfp), which encodes a 405-amino-acid protein annotated as a putative phosphopantothenoylcysteine decarboxylase. Thus, the products of the SAV7472-SAV7473 operon together with the product of SAV6873 may be involved in pantothenate and CoA metabolism in S. avermitilis.
Avermectins are a group of polyketide antiparasitic macrolides. The biosynthesis of avermectin initial aglycons (6,8a-seco-6,8a-deoxy-5-oxoavermectin aglycons) requires a type I modular polyketide synthase (PKS) that carries out a total of 12 successive cycles of elongation that resemble the steps in long-chain fatty acid biosynthesis (5, 28). The loading domain of avermectin PKS utilizes either isobutyryl-CoA or 2-methylbutyryl-CoA as an avermectin biosynthetic starter unit, and malonyl-CoA or methylmalonyl-CoA is used as an extender unit during the following 12 rounds of condensation steps. Because SAV7471 negatively regulates some genes involved in pantothenate and CoA metabolism, we infer that deletion of SAV7471 may lead to the increase of the cellular CoA pool by stimulating the transcriptional level of target genes SAV7472-SAV7473 and other target genes involved in CoA metabolism; the subsequent increase in available starter units and extender units would lead to increased avermectin production. Though SAV7471 cannot regulate aveR directly, aveR expression was strongly affected by the loss of SAV7471 at 2 days. The results can be explained that the abundance of precursor for avermectin in D7471 might induce aveR expression. Xiong et al. showed that targeted disruption of bkdF in S. avermitilis inactivated avermectin's starter unit and additional precursor could stimulate the yield of oligomycin, another polyketide in this strain (29).
In summary, we have shown that the TetR-family protein SAV7471 represses the expression of SAV7472-SAV7473 and several other genes that are involved in pantothenate and CoA metabolism. By affecting the cellular pool of CoA pool in vivo, SAV7471 can have a regulatory role in avermectin production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation of China (grant no. 31170090) and the Ministry of Science and Technology of China (grant no. 2009CB118905).
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00716-13.
REFERENCES
- 1.Chater KF. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685–713 [DOI] [PubMed] [Google Scholar]
- 2.Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 [DOI] [PubMed] [Google Scholar]
- 3.Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S. 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15:361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cane DE, Liang TC, Kaplan LK, Nallin MK, Schulman MD, Hensens OD, Douglas AW, Albers-Schonberg G. 1983. Biosynthetic origin of the carbon skeleton and oxygen atoms of the avermectins. J. Am. Chem. Soc. 105:4110–4112 [Google Scholar]
- 5.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. 1999. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 96:9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. 2009. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 82:1089–1096 [DOI] [PubMed] [Google Scholar]
- 7.Guo J, Zhao JL, Li LL, Chen Z, Wen Y, Li JL. 2010. The pathway-specific regulator AveR from Streptomyces avermitilis positively regulates avermectin production while it negatively affects oligomycin biosynthesis. Mol. Gen. Genet. 283:123–133 [DOI] [PubMed] [Google Scholar]
- 8.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215–12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caballero JL, Malpartida F, Hopwood DA. 1991. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol. Gen. Genet. 228:372–380 [DOI] [PubMed] [Google Scholar]
- 11.Tahlan K, Ahn SK, Sing A, Bodnaruk TD, Willems AR, Davidson AR, Nodwell JR. 2007. Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63:951–961 [DOI] [PubMed] [Google Scholar]
- 12.Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58:131–150 [DOI] [PubMed] [Google Scholar]
- 13.Folcher M, Morris RP, Dale G, Salah-Bey-Hocini K, Viollier PH, Thompson CJ. 2001. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem. 276:1479–1485 [DOI] [PubMed] [Google Scholar]
- 14.Cho YH, Kim EJ, Chung HJ, Choi JH, Chater KF, Ahn BE, Shin JH, Roe JH. 2003. The pqrAB operon is responsible for paraquat resistance in Streptomyces coelicolor. J. Bacteriol. 185:6756–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. 2001. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015–1028 [DOI] [PubMed] [Google Scholar]
- 16.Le TB, Fiedler HP, den Hengst CD, Ahn SK, Maxwell A, Butter MJ. 2009. Coupling of the biosynthesis and export of the DNA gyrase inhibitor simocyclinone in Streptomyces antibioticus. Mol. Microbiol. 72:1462–1474 [DOI] [PubMed] [Google Scholar]
- 17.Ahn SK, Cuthbertson XL, Nodwell JR. 2012. Genome context as a predictive tool for identifying regulatory targets of the TetR family transcriptional regulators. PLoS One 7:e50562. 10.1371/journal.pone.0050562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacNeil DJ, Klapko LM. 1987. Transformation of Streptomyces avermitilis by plasmid DNA. J. Ind. Microbiol. 2:209–218 [Google Scholar]
- 19.Jiang LB, Liu YP, Wang P, Wen Y, Song Y, Chen Z, Li JL. 2011. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol. Lett. 33:1955–1961 [DOI] [PubMed] [Google Scholar]
- 20.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmids cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Chen Z, Zhao J, Song Y, Wen Y, Li J. 2004. Deletion analysis of oligomycin PKS genes (olmA) in Streptomyces avermitilis. Chin. Sci. Bull. 49:1–5 [Google Scholar]
- 22.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 23.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66 [DOI] [PubMed] [Google Scholar]
- 24.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103–113 [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss E, Zhai H, Brand LA, McLafferty FW, Begley TP. 2004. Mechanistic studies on phosphopantothenoylcysteine decarboxylase: trapping of an enethiolate intermediate with a mechanism-based inactivating agent. Biochemistry 43:15520–15533 [DOI] [PubMed] [Google Scholar]
- 26.Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189 [DOI] [PubMed] [Google Scholar]
- 27.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 28.Ikeda H, Nonomiya T, Omura S. 2001. Organization of biosynthetic gene cluster for avermectin in Streptomyces avermitilis: analysis of enzymatic domains in four polyketide synthases. J. Ind. Microbiol. Biotechnol. 27:170–176 [DOI] [PubMed] [Google Scholar]
- 29.Xiong W, Liang Y, Zheng Y. 2006. Enhancement and selective production of oligomycin through inactivation of avermectin's starter unit in Streptomyces avermitilis. Biotechnol. Lett. 28:911–916 [DOI] [PubMed] [Google Scholar]
- 30.Ikeda H, Kotaki H, Tanaka H, Omura S. 1988. Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob. Agents Chemother. 32:282–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.