Abstract
Bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is an intracellular second messenger that regulates adaptation processes, including biofilm formation, motility, and virulence in Gram-negative bacteria. In this study, we have characterized the core components of a c-di-GMP signaling pathway in the model Gram-positive bacterium Bacillus subtilis. Specifically, we have directly identified and characterized three active diguanylate cyclases, DgcP, DgcK, and DgcW (formerly YtrP, YhcK, and YkoW, respectively), one active c-di-GMP phosphodiesterase, PdeH (formerly YuxH), and a cyclic-diguanylate (c-di-GMP) receptor, DgrA (formerly YpfA). Furthermore, elevation of c-di-GMP levels in B. subtilis led to inhibition of swarming motility, whereas biofilm formation was unaffected. Our work establishes paradigms for Gram-positive c-di-GMP signaling, and we have shown that the concise signaling system identified in B. subtilis serves as a powerful heterologous host for the study of c-di-GMP enzymes from bacteria predicted to possess larger, more-complex signaling systems.
INTRODUCTION
As a key second messenger in prokaryotes, bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) is of increasing interest due to its central roles in microbial adaptability and potential for clinical use as a stimulator of the innate immune response in humans (1–5). For example, c-di-GMP signaling pathways are shown in many bacteria to regulate the switch from planktonic to biofilm lifestyle, a state that increases the likelihood of persistent microbial infections and resistance to antibiotics. Conversely, dispersion of biofilms is often accompanied by a stimulation of virulence, and thus, c-di-GMP can also play an active role in pathogenesis. The paradigms for c-di-GMP signaling, derived almost exclusively from studies in Gram-negative bacteria, imply that increases in c-di-GMP levels lead to inhibited motility and enhanced biofilm formation, whereas decreased c-di-GMP levels may promote biofilm disassembly and lead to activation of virulence pathways (6–12).
The core components of c-di-GMP signaling pathways have been identified in Gram-negative bacteria, which allows for prediction of similar components in Gram-positive bacteria. For instance, c-di-GMP is synthesized by diguanylate cyclases (DGCs) (characterized as GGDEF domain proteins) and degraded by phosphodiesterases (PDEs) (characterized as EAL domain proteins and/or HD-GYP proteins) (13, 14). Receptors for c-di-GMP, including PilZ domain proteins, degenerate EAL domain proteins, and riboswitches, have also been identified and characterized (1, 14–16).
Despite the presence of putative components of c-di-GMP signaling pathways across the bacterial kingdom, c-di-GMP is difficult to detect in vivo, and roles for c-di-GMP in Gram-positive organisms are not well defined. The first diguanylate cyclase identified in Gram-positive bacteria was Mycobacterium smegmatis MSDGC-1, a protein with both DGC activity and PDE activity in vitro (17, 18). More recently, additional c-di-GMP signaling components have been characterized in M. smegmatis (19), Mycobacterium tuberculosis (20), Streptomyces coelicolor (21–23), and Clostridium difficile (15, 16, 24, 25). These studies establish c-di-GMP as a secondary messenger in Gram-positive bacteria, but until recently the possibility of c-di-GMP signaling in a widely studied Gram-positive model organism, Bacillus subtilis, had not been reported (26). To further examine c-di-GMP signaling roles in Gram-positive organisms, B. subtilis provides many advantages, including a diverse array of tools for genetic manipulation, lack of pathogenicity in humans, and a concise predicted c-di-GMP signaling network that can be examined without complications inherent to systems with high levels of functional redundancy.
Based on sequence analysis, the B. subtilis genome has the potential to encode four DGCs, two PDEs, a single bifunctional protein with GGDEF and EAL domains, and one PilZ domain c-di-GMP receptor. Recently, Chen et al. (26) performed genetic analyses that implicated a subset of these components, including YpfA, a putative c-di-GMP receptor, and YuxH, a putative PDE, in c-di-GMP signaling. In that study, deletion strains of genes encoding potential signaling factors coupled with mutagenesis of predicted c-di-GMP interaction sites provided evidence that YpfA and YuxH were involved in inhibition of B. subtilis motility (26).
To date, purified proteins possessing DGC, PDE, or c-di-GMP receptor characteristics from B. subtilis have not been characterized in vitro, and direct evidence for the presence of c-di-GMP in cells has not been demonstrated in vivo. In this work, we provide direct biochemical and molecular genetic evidence for a complete c-di-GMP signaling system in B. subtilis. Specifically, we demonstrate that three of the GGDEF domain-encoding enzymes possess diguanylate cyclase activity and one of the EAL-containing proteins harbors phosphodiesterase activity both in cells and as purified proteins in vitro. In addition, we show that the PilZ domain protein harbors specific c-di-GMP binding activity that readily distinguishes between GMP and c-di-GMP. We manipulate the intracellular concentrations of c-di-GMP by changing the expression levels of DGC and PDE activities, and we show that increased c-di-GMP correlates with decreased motility but with no effect on biofilm formation. On the basis of the work reported here, we rename the genes encoding the core components of c-di-GMP signaling accordingly and speculate as to the significance of this pathway in B. subtilis physiology.
MATERIALS AND METHODS
Cell extract preparation.
All B. subtilis strains were cultured in LB medium at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in cases where the Physpank promoter was induced. The cells were harvested (50-ml culture volume) at an optical density at 600 nm (OD600) of 1 by centrifugation at 4°C, and cell pellets were extracted immediately using the nucleotide extraction method reported by Spangler et al. (27). The extraction solvent was a mixture of acetonitrile-methanol-water (40:40:20 by volume). The cell pellet was resuspended in 3 ml ice-cold extraction solvent to quench metabolism and initiate the extraction process followed by a 15-min incubation step at 4°C. The cell suspension was then heated to 95°C for 10 min. After cooling, the suspension was centrifuged at 17,000 rpm for 5 min to separate insoluble material from the extracted molecules. The extraction of the resulting pellets was repeated twice with 2 ml extraction solvent at 4°C, and combined extraction volume (∼7 ml) was evaporated to dryness under vacuum. Residues were resuspended in 120 μl water under vigorous vortexing for subsequent analysis by reversed-phase LC resolution prior to electrospray ionization tandem mass spectrometry (LC–MS-MS).
LC–MS-MS analysis.
Detection of c-di-GMP was performed using the QTRAP 4000 triple quadrupole instrument (ABI Sciex, Foster City, CA). A Dionex UltiMate 3000 LC system (Dionex Corporation, Sunnyvale, CA) consisting of a binary pump, a temperature-controlled autosampler maintained at 5°C, and a column oven compartment maintained at 25°C, was interfaced to the electrospray ionization (ESI) Turbo V ion source of the 4000 QTRAP instrument. Twenty microliters of each sample dissolved in 100 μl of water was injected into a Nucleodur C18 pyramid column (50 mm by 3 mm; 3-μm particle size; Macherey-Nagel, Düren, Germany). Mobile phase A consisted of 10 mM ammonium acetate in 0.1% (vol/vol) acetic acid-water, while mobile phase B consisted of 100% methanol. The gradient conditions were 0 to 30% mobile phase B from 5 to 9 min, followed by holding at 30% mobile phase B for 1 min. The total run time was 15 min, and the flow rate employed was 400 μl/min (27).
The QTRAP instrument was operated in positive-ion multiple reaction monitoring (MRM) mode where both Q1 and Q3 were set to transmit different precursor/product ion pairs, while the collision energy (CE) in Q2 was varied depending on the precursor/product ion pair. The [M+H]+ precursor ion at m/z 691 was used for c-di-GMP, and the monitored product ions were m/z 248 (CE, 40 eV), m/z 540 (CE, 40 eV), and m/z 152 (CE, 61 eV). The product ions monitored for the uniformly 13C15N-labeled c-di-GMP ([M+H]+ at m/z 721) were m/z 263 (CE, 40 eV), m/z 560 (CE, 40 eV), and m/z 162 (CE, 61 eV), and the product ions monitored for cXMP ([M+H]+ at m/z 347) were m/z 153 (CE, 29 eV) and m/z 136 (CE, 59 eV). The selected precursor ion for pGpG was m/z 707, and the resulting product ions monitored were m/z 152 (CE, 55 eV) and m/z 248 (CE, 50 eV). The Turbo V ion source parameters were the following: the capillary was operated at 4,500 V, and the source temperature was set at 400°C. The curtain gas (N2) and collision gas (N2) settings were 10 lb/in2, the nebulization gas setting was 40 lb/in2, and the vaporization gas setting was 50 lb/in2. A dwell time of 50 ms and a declustering potential (DP) of 50 V were used for all the analytes.
The Analyst software version 1.4.1 (ABI Sciex) was used to acquire data. The software also has a built-in information-dependent acquisition (IDA) scan function that was used to obtain collision-induced dissociation (CID) spectra of both protonated c-di-GMP and of the “false” signal from 2′,3′-cyclic-GpGp. Using this function, the MRM scan of the precursor/product ion pair at m/z 691/152 was used as a survey scan to trigger CID on the precursor peak throughout the length of the run. The LC gradient and length of the IDA experiment were the same as described above for the MRM experiment.
Strain construction.
Each putative DGC gene was cloned into vector pDR111, which carries a spectinomycin resistance marker and a multiple cloning site (MCS) downstream of the IPTG-inducible promoter Physpank between two arms of the amyE gene (28, 29). Plasmids for GGDEF domain mutants were created by two-step PCR. To generate the ΔdgcW in-frame markerless deletion construct, the region upstream of dgcW was PCR amplified using the primer pair 2928/2929 and digested with EcoRI and XhoI, and the region downstream of fliW was PCR amplified using the primer pair 2930/2931 and digested with XhoI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of pMiniMAD2 that carries a temperature-sensitive origin of replication and an erythromycin resistance cassette to generate pDP391 (30). The pDP391 plasmid was introduced into strain PY79 by single-crossover integration by transformation at the restrictive temperature for plasmid replication (37°C) using mls resistance as a selecting factor. The integrated plasmid was then transduced into B. subtilis 3610. To evict the plasmid, the strain was incubated in 3 ml LB broth at a permissive temperature for plasmid replication (22°C) for 14 h, diluted 30-fold in fresh LB broth, and incubated at 22°C for an additional 8 h. The dilution and outgrowth procedures were performed three times. The cells were then serially diluted and plated on LB agar at 37°C. Individual colonies were patched on LB plates and LB plates containing mls to identify mls-sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR using primers 2928/2931 to determine which isolate contained the ΔdgcW allele. The ΔydaK construct was built similarly to that of ΔdgcW using the primer pairs 2932/2933 and 2934/2935 to generate pDP392. Primer pairs 2936/2937 and 2938/2939 were used for the ΔdgcK deletion construct pDP393.
The ΔdgcP::tet insertion-deletion allele was generated by two-step cloning using primer pairs 3037/3038 and 3039/3040. A plasmid containing a tetracycline drug resistance gene (pDG1515) was used as the vector backbone to obtain the insertion-deletion construct pSM43 (31, 32). Similarly, the ΔdgrA::erm insertion-deletion allele was generated by two-step cloning (primer pairs GXH489/GXH490 and GXH491/GXH492), and a plasmid containing an mls drug resistance gene (pAH52) was used as the vector backbone to obtain the insertion-deletion construct pDP407 (31, 32). The ΔpdeH::kan insertion-deletion mutation was generated using isothermal assembly of PCR amplicons (using primer pairs 3428/3429 and 3430/3431) and DNA containing a kanamycin drug resistance gene (pDG780) (31, 33). Assembled DNA was transformed into strain PY79 and transferred to the B. subtilis 3610 background using SPP1-mediated generalized transduction (34). To generate amyE::PpdeH-pdeH cat complementation construct pXG094, the sequence of the pdeH coding region with ∼500-bp upstream sequence was PCR amplified from B. subtilis 3610 chromosomal DNA with primers GXH514 and GXH515 and ligated into the EcoRI and BamHI sites of pDG364 (29, 35).
To generate the pKB149 vector containing the Pconstitutive (Pc) promoter, pDR90, which carries the Phyperspac promoter, a single lacO operator site, a polylinker, and a spectinomycin resistance gene cloned between two arms of the amyE gene, was used as a PCR template and amplified using primer pair 2006/2007. The resulting amplicon that contained the Phyperspac promoter but excluded the lacO operator site was digested with SphI and EcoRI and cloned into the SphI and EcoRI sites of pAH25 containing a polylinker and spectinomycin resistance marker between two arms of the amyE gene. pDR90 was a generous gift from David Rudner (Harvard Medical School), and pAH25 was a generous gift from Amy Camp (Mount Holyoke College).
To generate amyE::Pc-dgrA spec construct pXG095, a PCR product containing the dgrA coding region was PCR amplified from B. subtilis 3610 chromosomal DNA using primers GXH516 and GXH517, digested with SphI and BamHI, and ligated into the SphI and BamHI sites of pKB149. To examine the effect of c-di-GMP production on motility in our constitutive dgrA expression strain, the pXG100 construct (thrC::Physpank-dgcP mls) was generated via amplification of the dgcP gene cassette, including the leader sequence from Bacillus subtilis 3610 chromosomal DNA, using primers GXH532/GXH533. The amplicon was cloned via isothermal assembly into HindIII and SphI sites in pDP150, a plasmid containing an mls resistance gene, the Physpank inducible promoter, and coding sequence for the LacI repressor flanked by segments of the thrC gene for homologous recombination.
To analyze the function of heterologous GGDEF domain proteins, a parent construct, pXG101, was generated via PCR synthesis of the leader sequence from B. subtilis dgcP (nucleotides −60 to +3 relative to the translational start site) flanked by upstream HindIII and downstream NheI, SpeI, and SphI sites using six primers (GXH534 to GXH539) and cloned into HindIII and SphI sites of pDP150. pXG101 affords efficient expression of proteins from heterologous sources via a translational fusion with a leader sequence native to B. subtilis (35, 36).
To generate inducible translational fusion constructs in our amyE::Pc-dgrA spec strain, pXG102 (thrC::Physpank-Bs_dgcP-dcpA mls), pXG104 (thrC::Physpank-Bs_dgcP-cd1420 mls), and pXG105 (thrC::Physpank-Bs_dgcP-Atu1297 mls) were generated. Gene cassettes were amplified from Vibrio vulnificus, Clostridium difficile 630, and Agrobacterium tumefaciens C58 chromosomal DNAs, respectively, using primers GXH540/GXH521, GXH542/GXH543, and GXH544/GXH531. Amplicons were cloned into NheI/SphI sites of pXG101 via isothermal assembly, giving rise to translational fusion proteins with the following N-terminal sequences: MASIRY for V. vulnificus DcpA, MASFKE for C. difficile CD1420, and MASTAR for A. tumefaciens Atu1297 (nonnative sequence in italics) (32, 33, 35, 36).
SPP1 phage transduction.
To 0.2 ml of dense culture grown in TY broth (LB broth supplemented with 10 mM MgSO4 and 100 μM MnSO4 after autoclaving), serial dilutions of SPP1 phage stock were added and statically incubated for 15 min at 37°C. To each mixture, 3 ml TYSA (molten TY supplemented with 0.5% agar) was added, poured atop fresh TY plates, and incubated at 37°C overnight. Top agar from the plate that contained near confluent plaques was harvested by scraping into a 50-ml conical tube, vortexed, and centrifuged at 5,000 × g for 10 min. The supernatant was treated with 25 μg/ml DNase I before being passed through a 0.45-μm syringe filter and stored at 4°C.
Recipient cells were grown to stationary phase in 2 ml TY broth at 37°C. The cells (0.9 ml) were mixed with 5 μl SPP1 donor phage stock. Nine milliliters of TY broth was added to the mixture and allowed to stand at 37°C for 30 min. The transduction mixture was then centrifuged at 5,000 × g for 10 min, the supernatant was discarded, and the pellet was resuspended in the remaining volume. The cell suspension (100 μl) was then plated on TY fortified with 1.5% agar, the appropriate antibiotic, and 10 mM sodium citrate.
Expression and purification of candidate proteins in B. subtilis c-di-GMP signaling pathways.
pMBP-parallel (MBP stands for maltose-binding protein) and pHis-parallel vectors were used to generate clones for protein expression and purification (37). Coding sequences for all clones were amplified from B. subtilis strain 3610 genomic DNA using standard PCR techniques. Details of the resultant proteins from putative DGC expression plasmids are the following: pHis-YdaK-GGDEF, residues 111 to 283; pHis-DgcK-GGDEF, DgcK residues 176 to 359; pMBP-DgcW-GGDEF, residues 321 to 484; pHis-DgcW-PAS-GGDEF, residues 213 to 484; pMBP-DgcP-GGDEF, residues 423 to 576; pHis-DgcP-GAF-GGDEF, residues 117 to 578; full-length pHis-PdeH, residues 1 to 409; and full-length pHis-DgrA, residues 1 to 217. Expression vectors were transformed into Escherichia coli BL21(DE3) strains. One-liter cultures were inoculated with 15 ml of an overnight culture and grown at 37°C in LB medium containing 100 mg/ml ampicillin with or without 1 g/liter glucose for maltose-binding protein (MBP) or His tag fusion proteins, respectively. Protein expression was induced with 0.5 mM IPTG at an OD600 of 0.6. After induction, cells were grown for 16 h at 20°C, and cell pellets were harvested by centrifugation at 8,000 × g for 20 min prior to storage at −80°C.
Cell pellets for MBP fusion proteins were resuspended in equilibration buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1 mM sodium azide, and 1 mM Tris(2-carboxyethyl)phosphine (TCEP). After sonication, centrifugation, and filtration, clarified lysates were loaded onto a 10-ml amylose column (NEB). The columns were washed with 18 column volumes (CVs) of equilibration buffer followed by elution in 6 CVs of elution buffer (equilibration buffer containing 10 mM maltose). Similarly, cell pellets for His-tagged proteins were resuspended in buffer containing 25 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 1 mM TCEP. Clarified lysates were loaded onto an 8-ml nickel-nitrilotriacetic acid (Ni-NTA) column equilibrated in buffer containing 25 mM Tris-HCl (pH 8.0), 500 mM NaCl, 10% glycerol, and 10 mM imidazole, washed with 10 CVs of the same buffer, and eluted in a gradient from 10 to 250 mM imidazole over 8 CVs. After affinity purification, peak elution fractions were analyzed by SDS-PAGE, and relevant fractions were subjected to proteolysis with His-tagged tobacco etch virus (TEV) protease for 16 h at 4°C using a TEV/target protein mass ratio of 1:20. Putative DGCs were subsequently purified from the His-tagged TEV using Ni-NTA affinity chromatography, as the cleaved protein of interest does not bind to the affinity resin. Flowthrough fractions were pooled and subjected to ion-exchange chromatography. The DgcW-GGDEF, DgcW-PAS-GGDEF, and DgcP-GGDEF proteins were subjected to Source 15S cation-exchange chromatography (GE Healthcare) (equilibration buffer consisting of 20 mM HEPES [pH 7.0]; elution buffer consisting of 20 mM HEPES [pH 7.0] and 1 M NaCl), whereas the YdaK-GGDEF, DgcK-GGDEF, and DgcP-GAF-GGDEF proteins were purified via Source 15Q anion-exchange chromatography (GE Healthcare) (equilibration buffer consisting of 10 mM Tris [pH 8.0]; elution buffer consisting of 10 mM Tris [pH 8.0] and 1 M NaCl) using a gradient from 0 to 100% over 20 CVs in all cases. Final purification was performed via size exclusion chromatography on a HiLoad 16/60 Superdex 75 column (GE Healthcare) in 50 mM Tris (pH 8.0), 250 mM NaCl, and 0.3 mM TCEP.
In vitro diguanylate cyclase activity assays.
All diguanylate cyclase activity assays were incubated overnight at 37°C in 50 mM Tris (pH 7.5), 250 mM NaCl, 20 mM MgCl2, 1 mM TCEP, and 37 μM GTP with 10 μM enzyme. The reactions were terminated by incubation at 98°C for 10 min, and the samples were filtered and analyzed for the production of c-di-GMP by LC–MS-MS.
In vitro c-di-GMP phosphodiesterase activity assays.
All phosphodiesterase activity assays were incubated for 4 h at 37°C in 50 mM Tris (pH 7.5), 250 mM NaCl, 10 mM MgCl2, 0.3 mM TCEP, and 10 μM c-di-GMP with 10 μM enzyme. The negative-control groups do not contain MgCl2. The reactions were terminated by incubation at 98°C for 10 min, and the samples were filtered and analyzed for the production of pGpG product by LC–MS-MS (see below).
In vitro c-di-GMP receptor binding assays.
DgrA alone (100 μM) or mixed with either c-di-GMP (200 μM) or GMP (400 μM) was incubated on ice for 1 h and subjected to size exclusion chromatography on a HiLoad 10/300 Superdex 75 column (GE Healthcare) in buffer containing 30 mM sodium phosphate (pH 7.5) and 250 mM NaCl. For isothermal titration calorimetry (ITC) analyses, DgrA and YdaK samples were exchanged into 10 mM sodium citrate phosphate and 200 mM NaCl at pH 7.5 via size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare). c-di-GMP was dissolved into the same buffer, and titrations of proteins into c-di-GMP were conducted using a NanoITC low-volume calorimeter (TA Instruments). In a typical titration experiment, 300 μl of 4 to 6 μM c-di-GMP was placed in the sample cell of 174-μl working volume, while 50 μl of 30 to 150 μM protein was loaded in the titration syringe. Protein volume per injection varied from 0.36 to 1.49 μl with 150-s intervals between injections. After initial trials and parameter optimization, at least three titrations were carried out at 25°C for each protein-ligand pair (see Fig. S5 in the supplemental material).
Swarm expansion assay.
Cells were grown to mid-log phase at 37°C in LB broth and resuspended to an OD600 of 10 in phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4 [pH 8.0]) containing 0.5% India ink (Higgins). Freshly prepared LB containing 0.7% Bacto agar (25 ml/plate) was dried for 10 min in a laminar flow hood, centrally inoculated with 10 μl of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink demarks the origin of the colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate, and swarm radius measurements were taken along this transect. For experiments including IPTG, cells were propagated in broth in the presence of IPTG, and IPTG was included in the swarm agar plates (38).
RESULTS
Detection of c-di-GMP in vivo.
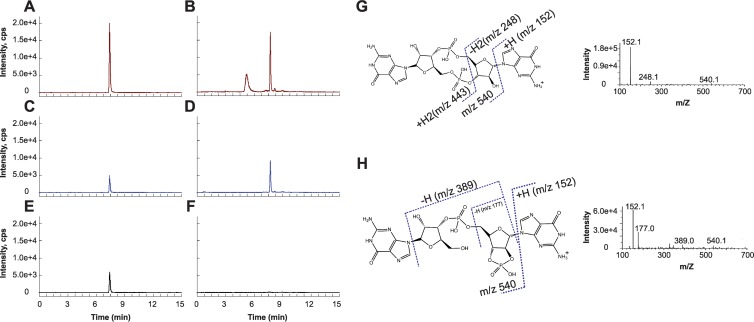
To determine whether c-di-GMP is produced in vivo in B. subtilis, we employed reversed-phase LC resolution prior to electrospray ionization tandem mass spectrometry (LC–MS-MS) to detect the presence of c-di-GMP in B. subtilis 3610 extracts. In the triple quadrupole mass spectrometer, ions matching the mass-to-charge ratio of the parent ion of interest were selected and introduced to a collision cell wherein collision-induced dissociation (CID) or “fragmentation” of the parent ion takes place. Based on a purified c-di-GMP standard, the mass-to-charge ratio of the parent ion was m/z 691, whereas the major CID fragment ions that were monitored in multiple reaction monitoring (MRM) mode were m/z 152, 248, and 540, listed in order of decreasing abundance, a property based on inherent parent ion fragmentation efficiencies at the employed collision energies. In this experiment, both the mass-to-charge ratio and the relative abundance of the fragment ions serve as a molecular fingerprint for the detected small molecule.
With LC–MS-MS, we identified a molecule in B. subtilis extracts with nearly identical high-performance liquid chromatography (HPLC) retention time and the same parent ion mass-to-charge ratio as c-di-GMP. Additionally, this molecule shared two MRM transitions due to fragment ions (m/z 540 and 152) with c-di-GMP (Fig. 1A to D). However, full CID analysis indicated that this molecule was not c-di-GMP due to the variance in its CID fragmentation spectrum from that of authentic c-di-GMP (Fig. 1G and H). In addition, the absence of the m/z 248 fragment ion from its CID spectrum gave us an MRM transition (m/z 691/248) that was distinct from that of c-di-GMP (Fig. 1E to H). By analyzing the CID data for the unknown molecule, we propose that the molecule is a guanosine dinucleotide with a 2′,3′-cyclic phosphate-2′,3′-c-GpGp (Fig. 1H). The origin or relevance of this molecule in B. subtilis is unknown, but the same molecular signature can also be seen in E. coli (27). We conclude that if wild-type B. subtilis synthesizes c-di-GMP under our growth conditions, it does so below the limit of detection of our method, which is 50 pg/μl.
Fig 1.
Development of an LC–MS-MS method to uniquely detect c-di-GMP. Three separate single reaction monitoring (SRM) transitions are shown for a synthesized c-di-GMP standard and a prevalent signal from B. subtilis lysates. (A to F) After collision-induced dissociation (CID), SRM transitions m/z 691 → 152 (A and B) and m/z 691 → 540 (C and D) are seen in both samples, whereas the m/z 691 → 248 transition (compare panels E and F) is unique to c-di-GMP. Intensity is shown as counts per second (cps). Two endogenous molecules in bacterial extracts have the same parent ion mass as c-di-GMP and elute at retention times of 5.2 min and 7.8 min, respectively. (G and H) Analysis of the CID spectra of c-di-GMP (G) and the latter peak from B. subtilis lysates (H) provides a molecular basis for the major transitions seen as indicated on the structures of c-di-GMP and the lysate molecule, predicted to be a guanine dinucleotide with a 2′,3′-cyclic phosphate, 2′,3′-c-GpGp.
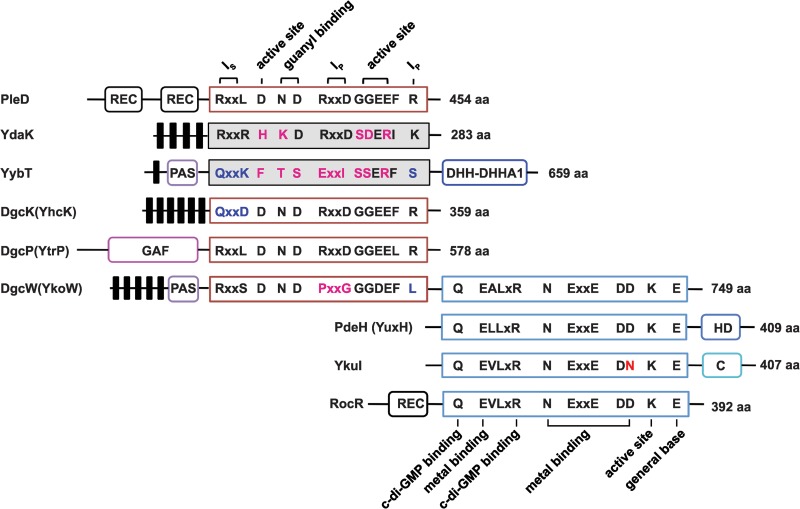
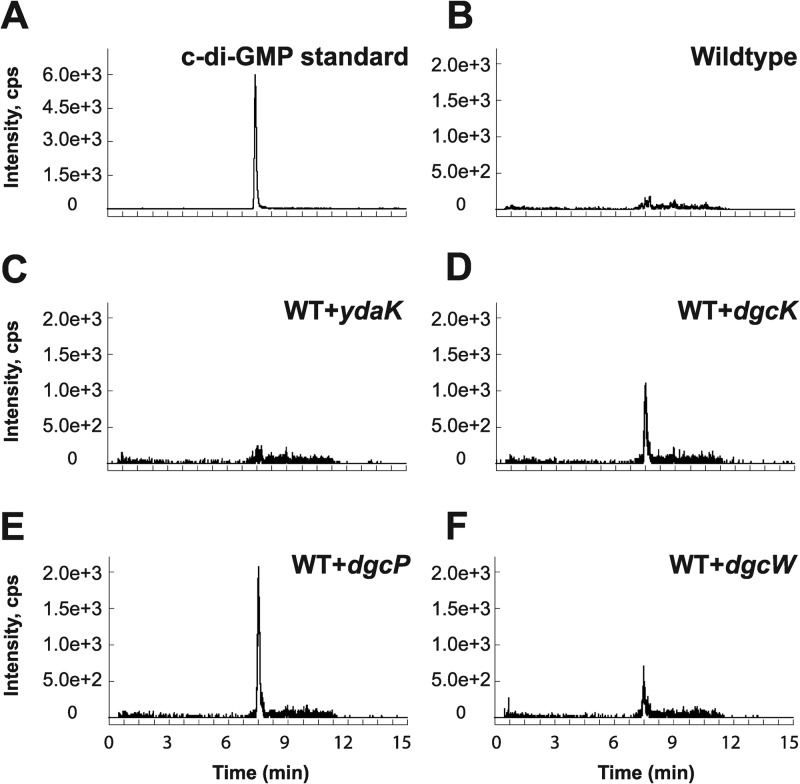
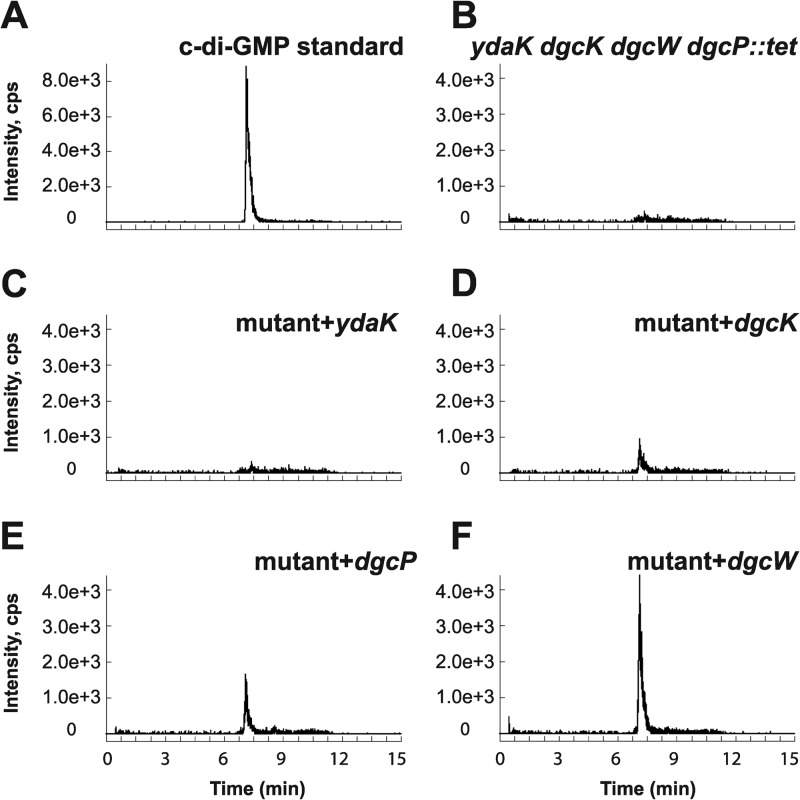
As we were unable to detect c-di-GMP in the wild-type 3610 strain during vegetative growth, we generated individual inducible expression strains for putative DGCs and analyzed these strains for the presence of c-di-GMP. Analysis of all putative open reading frames (ORFs) in the B. subtilis genome identified five proteins with predicted structural similarity to the GGDEF family of proteins: YybT, YdaK, YhcK, YtrP, and YkoW. The GGDEF domain of YybT has been characterized previously to exhibit ATP hydrolysis activity, but it is incapable of c-di-GMP synthesis, likely owing to degeneracy in the putative cyclase active site and was not considered further (Fig. 2) (39, 40). Overproduction from the IPTG-inducible Physpank promoter of three of the remaining putative DGC proteins, YhcK, YtrP, and YkoW, here renamed DgcK, DgcP, and DgcW, respectively, resulted in production of detectable levels of c-di-GMP from cell extracts (Fig. 3D to F; see Fig. S1 in the supplemental material), whereas expression of YdaK, a degenerate GGDEF protein, did not give rise to detectable levels of c-di-GMP (Fig. 3C). To confirm that one DGC did not indirectly stimulate c-di-GMP production through another, we overexpressed each candidate gene separately in a quadruple mutant that lacked all putative DGCs, ΔydaK ΔdgcK ΔdgcW ΔdgcP::tet mutant. Under these conditions, overexpression of DgcK, DgcP, and DgcW produced readily detectable levels of cytoplasmic c-di-GMP (Fig. 4 and Fig. S2). We conclude that DgcK, DgcP, and DgcW each possess diguanylate cyclase activity in vivo.
Fig 2.
Domain architectures of the putative diguanylate cyclases (DGCs) and c-di-GMP phosphodiesterases (PDEs) encoded by B. subtilis genes. PleD, a well-studied DGC from Caulobacter crescentus, is used as a reference for critical residues responsible for DGC activity and inhibition. RocR, a well-studied PDE from Pseudomonas aeruginosa, is used as a reference for critical residues responsible for PDE activity. The number of amino acids (aa) in each protein is shown. Putative DGCs with highly conserved active site GGDEF residues are outlined in red, whereas proteins with highly degenerate GGDEF sequences are shaded gray. Putative PDEs with highly conserved active site EAL residues are outlined in blue. Predicted transmembrane regions are shown as black bars. Ip is the primary inhibitory binding site for c-di-GMP, and Is is the secondary inhibitory binding site for c-di-GMP. Additional putative domains can be defined as follows: REC, receiver domain found in two-component signaling systems; PAS, a sensory domain of the PER/ARNT/SIM family known to respond to oxygen, redox potential, and light in other systems; GAF, a domain originally found in cGMP-specific phosphodiesterases, adenylyl and guanylyl cyclases and phytochromes, often serving as a cyclic nucleotide binding domain; DHH, a domain with possible phosphodiesterase function; DHHA1 domain, a DHH-associated domain and a member of DHH subfamily; HD domain, a domain with phosphohydrolase activity; C domain, C-terminal domain from YkuI of unknown function.
Fig 3.
In vivo analysis of c-di-GMP production by putative DGCs in inducible expression strains derived from wild-type (WT) B. subtilis. (A to F) Signals from the SRM transition m/z 691 → 248 obtained via LC–MS-MS are displayed for the c-di-GMP standard (A) and bacterial extracts of wild-type B. subtilis (B) and strains expressing the putative DGCs YdaK (C), DgcK (D), DgcP (E), and DgcW (F).
Fig 4.
In vivo analysis of c-di-GMP production by putative DGCs in a background lacking all endogenous DGCs. (A to F) Signals of SRM transition m/z 691 → 248 obtained via LC–MS-MS are displayed for the c-di-GMP standard (A) and bacterial extracts of B. subtilis ΔydaK dgcK dgcW dgcP::tet mutant strain (B) (labeled mutant in panels C to F) and strains expressing the putative DGCs YdaK (C), DgcK (D), DgcP (E), and DgcW (F).
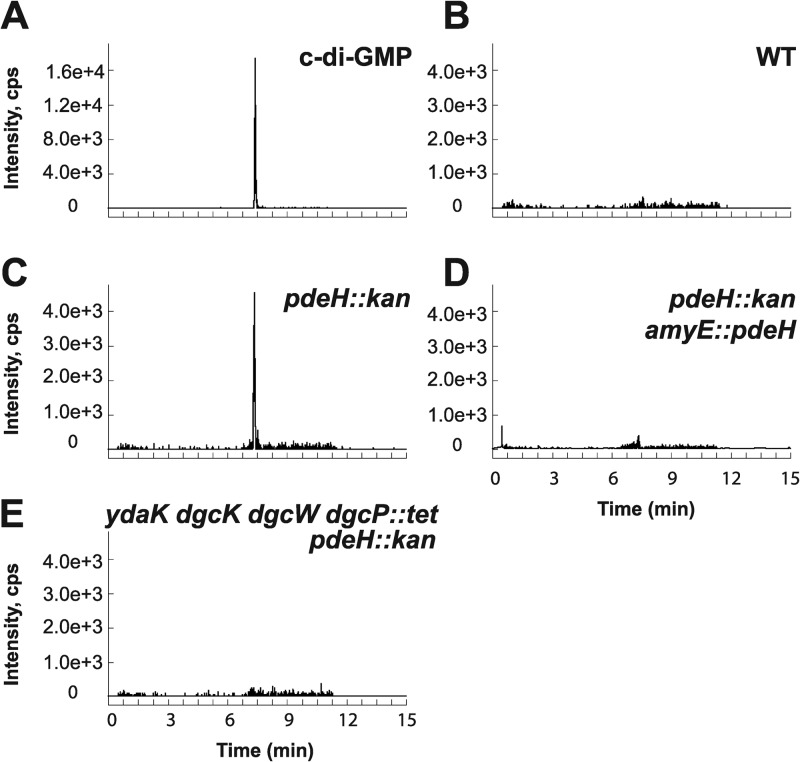
B. subtilis has three genes—dgcW, ykuI, and yuxH—that encode proteins with predicted EAL domains typically responsible for conversion of c-di-GMP to a linear pGpG dinucleotide. Previous studies showed that the ykuI gene product had no detectable PDE activity in vitro and was thus excluded from further analysis (41). Additionally, full-length DgcW (formerly YkoW) was shown to act primarily as a diguanylate cyclase as evidenced by the production of c-di-GMP in our recombinant strains (Fig. 3F and 4F), even though all residues reported to be involved in c-di-GMP phosphodiesterase activity are present in the EAL domain (Fig. 2). LC–MS-MS analysis of the ΔyuxH strain showed elevated c-di-GMP levels relative to the wild type, consistent with the proposal that YuxH is an active c-di-GMP phosphodiesterase (Pde), here renamed PdeH (Fig. 5; see Fig. S3 in the supplemental material). Complementation of the ΔpdeH mutant strain with a wild-type copy of pdeH expressed from the native PpdeH promoter region and inserted at an ectopic locus (amyE::PpdeH-pdeH) returned c-di-GMP to undetectable levels (Fig. 5D). The levels of c-di-GMP were also reduced to undetectable levels when cells were simultaneously mutated for PdeH and the three diguanylate cyclases DgcK, DgcW, and DgcP (Fig. 5E). We conclude that PdeH is a negative effector of c-di-GMP accumulation and that no other diguanylate cyclases besides DgcK, DgcW, and DgcP are sufficient for c-di-GMP synthesis in vegetative cells.
Fig 5.
In vivo detection of c-di-GMP in pdeH deletion strains (pdeH encodes a c-di-GMP phosphodiesterase). (A to E) LC–MS-MS was used to detect c-di-GMP via the SRM transition m/z 691 → 248 for c-di-GMP standard (A) and B. subtilis extracts from the wild-type (WT) strain (B), ΔpdeH::kan strain lacking a putative c-di-GMP phosphodiesterase (C), (D) ΔpdeH::kan amyE::pdeH strain showing active phosphodiesterase complementation, and ΔydaK dgcK dgcW dgcP::tet pdeH::kan strain lacking a putative c-di-GMP phosphodiesterase and all possible diguanylate cyclases (E).
Diguanylate cyclase, c-di-GMP phosphodiesterase, and c-di-GMP receptor activity in vitro.
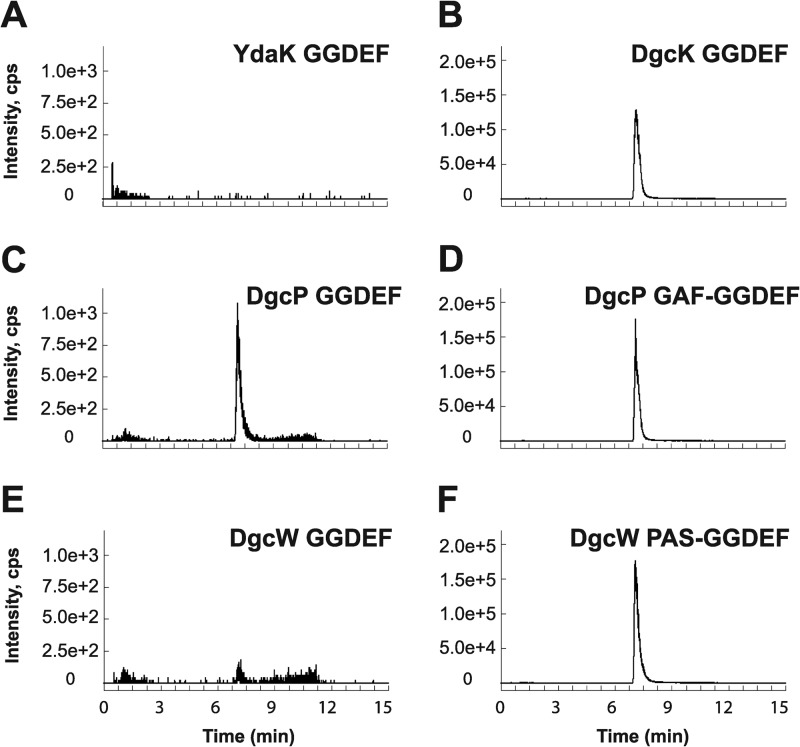
To directly measure diguanylate cyclase activity in vitro, we expressed and purified GGDEF domain fragments for YdaK, DgcK, DgcP, and DgcW from E. coli. In agreement with our in vivo overexpression data, only the three GGDEF proteins responsible for an increase in c-di-GMP levels in vivo, specifically, DgcK, DgcP, and DgcW, possessed diguanylate cyclase activity in vitro (Fig. 6; see Fig. S4 in the supplemental material). YdaK-GGDEF did not synthesize c-di-GMP from GTP under our experimental conditions, though we were able to measure c-di-GMP binding to this domain, likely at the conserved I-site, with a dissociation constant of ∼1.1 μM (Fig. S5). We conclude that DgcK, DgcP, and DgcW possess diguanylate cyclase activity both in vivo and in vitro, and YdaK may serve as a c-di-GMP receptor at high concentrations of c-di-GMP.
Fig 6.
In vitro detection of enzymatic activity of putative B. subtilis DGCs. To assess the activity of putative DGCs in vitro, LC–MS-MS was employed to detect enzymatic production of c-di-GMP from purified single- or dual-domain constructs containing GGDEF motifs. Ion peaks indicate production of c-di-GMP. (A to F) In total, four GGDEF single-domain proteins were analyzed, YdaK (A), DgcK (B), DgcP (C), and DgcW (E), as well as two dual-domain constructs containing GGDEF and either the GAF or PAS domain (D and F). As c-di-GMP copurifies with the DgcK GGDEF protein exogenously expressed and purified from E. coli, uniformly labeled [13C, 15N]GTP was used as a substrate for the DgcK GGDEF enzymatic reactions. For this isotope, the SRM transition m/z 721 → 263 was utilized (B), which corresponds to the m/z 691 → 248 transition used in all other cases.
DgcP and DgcW contain putative sensory domains, GAF and PAS, respectively, adjacent to their GGDEF domains. GAF domains have been shown to regulate enzymes, including histidine kinases and diguanylate cyclases, in response to cGMP, light, and oxygen levels (42–46). PAS domains similarly use cofactors to sense environmental cues including light or oxygen in the larger context of multidomain proteins (42, 47–52). Thus, we also examined dual-domain DgcP-GAF-GGDEF and DgcW-PAS-GGDEF proteins for DGC activity. The DgcP-GGDEF protein showed enzymatic activity that was significantly enhanced with the inclusion of the GAF domain (compare Fig. 6C and D). Similarly, DgcW-GGDEF was also weakly active as indicated by low levels of m/z 248 fragment ions, but the activity was dramatically increased with the inclusion of the adjacent PAS domain (compare Fig. 6E and F). We conclude that sensory inputs may well modulate the stimulatory activity of the GAF and PAS domains of DgcP and DgcW, respectively, in the cell (43–52).
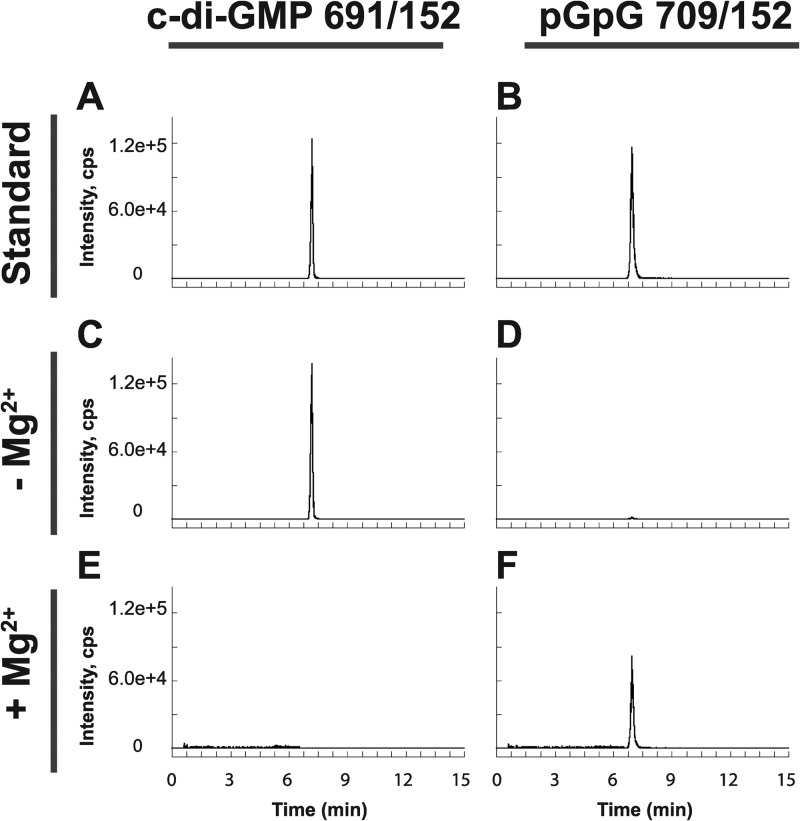
Having established both in vivo and in vitro that B. subtilis encodes three active diguanylate cyclases, we set out to analyze PdeH (formerly YuxH) similarly for its ability to hydrolyze c-di-GMP. On the basis of our in vivo data, we anticipated that PdeH would possess PDE activity. c-di-GMP was hydrolyzed to pGpG when incubated with PdeH as detected by using LC–MS-MS to monitor both substrate and product levels (Fig. 7). Furthermore, the activity of PdeH was Mg2+ dependent, in agreement with the published structural models showing divalent ions at the active site of the EAL proteins (53–55). We conclude that PdeH acts as a phosphodiesterase both in vivo and in vitro.
Fig 7.
In vitro characterization of PdeH demonstrates c-di-GMP phosphodiesterase activity. LC–MS-MS was performed to analyze the potential for conversion of c-di-GMP to the linear pGpG dinucleotide. SRM transitions m/z 691 → 152 and m/z 709 → 152 were monitored in each reaction for c-di-GMP consumption and pGpG production, respectively. (A to F) Standards of c-di-GMP (A) and pGpG (B) were analyzed and compared to reaction mixtures containing PdeH in the presence (C and D) or absence (E and F) of Mg2+.
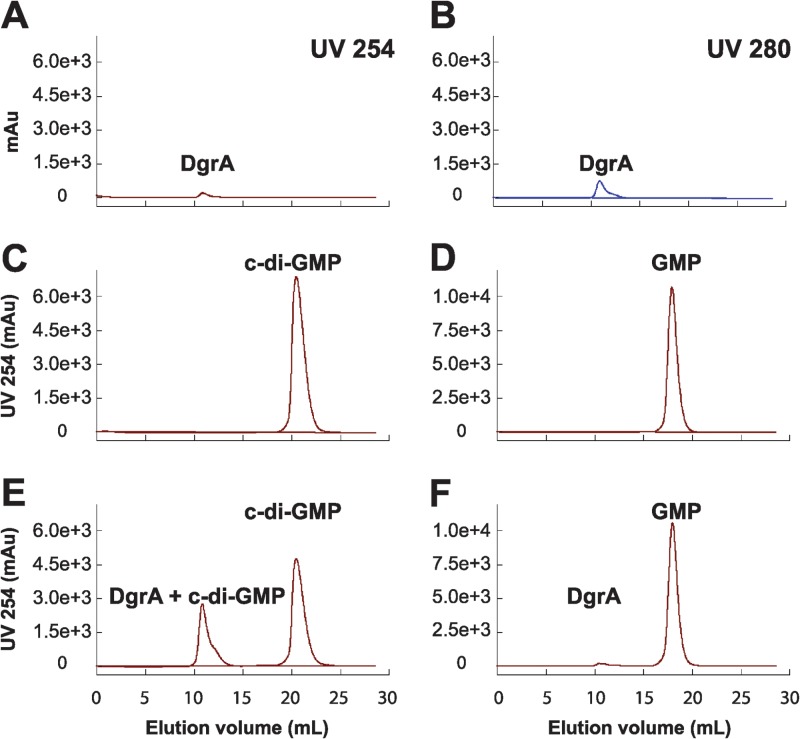
As we have demonstrated the presence of functional diguanylate cyclases and c-di-GMP phosphodiesterases in B. subtilis, we next focused on functional identification of a c-di-GMP receptor. The most likely candidate, YpfA, contains a PilZ domain common to c-di-GMP receptors, is the only predicted PilZ protein in B. subtilis, and is a homolog of YcgR—a c-di-GMP receptor from E. coli (56–60). To detect interaction, YpfA, c-di-GMP, or GMP was resolved individually or in combination by size exclusion chromatography (Fig. 8). Upon complex formation, bound nucleotides should elute at lower retention volumes owing to their association with the larger YpfA protein. In these experiments, the retention volume of c-di-GMP alone was 18 ml and shifted to approximately 11 ml in the presence of YpfA, a shift not seen for GMP. To confirm the size exclusion data, the dissociation constant of YpfA for c-di-GMP was measured by isothermal titration calorimetry (Kd [dissociation constant] = 11 nM; see Fig. S5 in the supplemental material). Thus, YpfA directly interacts with c-di-GMP, and we propose that the protein be renamed diguanylate receptor A (DgrA) to reflect its described function.
Fig 8.
In vitro analysis of the interaction of DgrA with c-di-GMP. Size exclusion chromatography was used to analyze the binding of purified DgrA to either c-di-GMP or GMP. (A to F) Absorbance of DgrA at 254 nm (A) and 280 nm (B) was monitored as a reference for comparison with the absorbance at 254 nm of individual nucleotides (C and D) or a mixture of DgrA and either c-di-GMP (E) or GMP (F). Elution of c-di-GMP at earlier retention volumes in the presence of DgrA is an indicator of complex formation. Equal quantities of 100 μM DgrA and 400 μM c-di-GMP or GMP were loaded in all experiments. Absorbance is shown in milli absorbance units (mAu).
Cytoplasmic accumulation of c-di-GMP inhibits motility through DgrA.
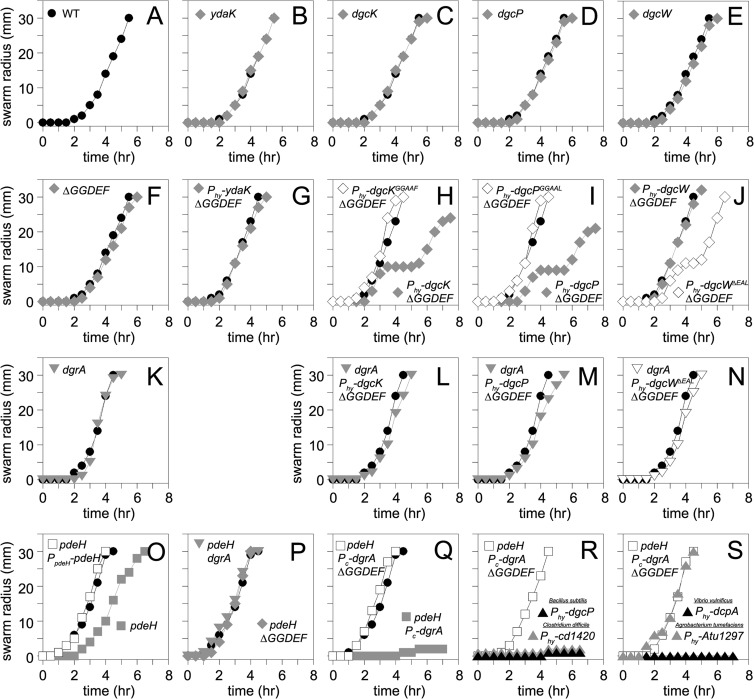
To investigate the biological roles of the four putative B. subtilis DGC proteins in motility and biofilm formation, we generated mutants with single deletions of dgcK, dgcP, dgcW, and ydaK, all combinations of double and triple deletions, the aforementioned quadruple mutation, and artificial overexpression constructs in which each gene was cloned downstream of the IPTG-inducible Physpank promoter and integrated at the ectopic amyE locus. None of the dgc single or combinatorial mutants displayed a defect in swarming motility (Fig. 9A to F). Cells that overexpressed DgcK and DgcP initiated swarming motility like the wild type but exhibited a transient cessation of motility (Fig. 9H and I). Cells overexpressing full-length DgcW swarmed like the wild type, but elimination of the EAL domain of the hybrid protein caused premature swarming cessation like the DgcK and DgcP constructs (Fig. 9J), hinting that the EAL domain might be either an active phosphodiesterase or a negative regulator of the DGC activity of DgcW. No biofilm defect was observed for any DGC mutant or overexpression construct (see Fig. S6 in the supplemental material). We conclude that overexpression of active DGCs result in a modest inhibition of motility under our conditions, and we infer that motility inhibition is the consequence of c-di-GMP accumulation.
Fig 9.
Swarm expansion assays of the indicated genotypes. (A) Swarming motility of the wild type (strain 3610). Each point is an average of three replicates. In all panels, lines indicated by filled circles represent the average behavior of the wild-type 3610 strain used as a control on the same day that a strain with the indicated genotype was tested. Note that some of the wild-type lines may be duplicated as multiple strains were tested in each assay. (B to F) Swarming motility of strains mutated in the indicated putative diguanylate cyclase genes (strains DS9289, DS9305, DS9537, and DS9883) or a ΔGGDEF quadruple mutant lacking all putative diguanylate cyclases (DS9884). (G to J) Swarming motility of ΔGGDEF strains expressing the indicated gene from a Physpank (Phy) promoter in the presence of 1 mM IPTG (gray diamonds) (strains NPS79, NPS80, NPS81, and NPS82). (K to N) Swarming motility of strains mutated for dgrA in an otherwise wild-type strain (NPS217) or in ΔGGDEF mutant backgrounds overexpressing GGDEF proteins that conferred a motility phenotype from panels H to J (NPS219, NPS220, and NPS221). Each assay was conducted in the presence of 1 mM IPTG. (O) Swarming motility of strains mutated for pdeH (gray closed squares; DK391) or mutated for pdeH in the presence of an ectopic PpdeH-pdeH complementation construct (open squares; NPS233). (P) Swarming motility of strains mutated for pdeH and dgrA (DK393) or mutated for pdeH and ΔGGDEF (DK392). (Q) Swarming motility of strains driving artificial constitutive expression of dgrA (Pc-dgrA) in a background mutated for pdeH (NPS235) or background mutated for pdeH and ΔGGDEF (NPS236). (R and S) Swarming motility of strains driving artificial constitutive expression of dgrA (Pc-dgrA) in a background mutated for pdeH and ΔGGDEF (NPS236) that also expresses the indicated GGDEF protein from an IPTG-inducible Physpank promoter (Phy) in the presence of 1 mM IPTG (NPS247, NPS252, NPS254, and NPS255).
One way in which c-di-GMP accumulation could inhibit motility is via a c-di-GMP output effector protein. One candidate for the effector protein is DgrA, as it is homologous to the E. coli c-di-GMP-dependent motility inhibitor YcgR, it was previously shown to inhibit motility in B. subtilis, and it binds directly to c-di-GMP in vitro (Fig. 8; see Fig. S5 in the supplemental material). Whereas cells mutated for dgrA displayed wild-type swarming motility (Fig. 9K), mutation of dgrA restored wild-type motility to backgrounds in which c-di-GMP was detected in vivo (i.e., overexpression strains of DgcK, DgcP, or DgcW lacking the EAL domain; compare Fig. 9H to J and Fig. 9L to N). We conclude that DgrA is necessary for motility inhibition when cellular levels of c-di-GMP are elevated.
To further examine whether c-di-GMP accumulation was responsible for motility inhibition, we tested the swarming motility of the pdeH mutant that resulted in high levels of c-di-GMP in the cytoplasm. The pdeH mutant swarmed slightly slower than the wild type (Fig. 9O). While subtle, the swarming defect was reproducible and was complemented by ectopic integration of the PpdeH-pdeH complementation construct (Fig. 9O). Furthermore, the slight reduction in motility was abolished when pdeH was mutated either in a strain lacking all four of the GGDEF proteins (DgcK, DgcP, DgcW, and YdaK) or in a strain lacking DgrA (Fig. 9P). We conclude that cells lacking PdeH were modestly impaired for motility and that wild-type motility can be restored to cells lacking PdeH when either DgrA or c-di-GMP production was abolished.
We wondered whether the subtle inhibition of motility in the absence of PdeH was due to poor expression of dgrA. When dgrA was cloned downstream of a modified Physpank promoter rendered constitutive by deletion of LacI operator sites and inserted at an ectopic locus (amyE::Pc-dgrA) of a pdeH mutant, motility was strongly inhibited in a manner dependent on the endogenous GGDEF proteins (Fig. 9Q). For a specificity control for DgrA, we also generated constitutive expression strains for two other putative c-di-GMP receptors in B. subtilis YdaK, a degenerate GGDEF protein capable of binding c-di-GMP (see Fig. S5 in the supplemental material) and YkuI, an EAL domain protein shown to bind, but not hydrolyze, c-di-GMP, in the pdeH mutant (41). Constitutive expression of neither YdaK nor YkuI exhibited any enhanced inhibition of motility, and both strains swarmed in a fashion indistinguishable from the pdeH mutant control (Fig. S7). Thus, if YdaK and YkuI are output effectors of c-di-GMP, their function is unrelated to motility inhibition. We conclude that motility inhibition due to c-di-GMP accumulation is dependent on high cytoplasmic levels of c-di-GMP sensed by the c-di-GMP receptor protein DgrA alone.
We noted that the strain deleted for the known functional diguanylate cyclases and the known c-di-GMP phosphodiesterase while expressing a constitutive DgrA protein (pdeH ΔGGDEF Pc-dgrA [Fig. 9Q]) was poised to inhibit motility if a source of c-di-GMP was provided. As such, we wondered if the strain was well suited to screen putative diguanylate cyclase activity of proteins from other organisms. Four diguanylate cyclases previously shown or predicted to possess enzymatic activity, DgcP (as a control from B. subtilis), Cd1420 (Clostridium difficile), DcpA (Vibrio vulnificus), and Atu1297 (Agrobacterium tumefaciens) were separately expressed from a Physpank promoter and a Shine-Dalgarno sequence from B. subtilis to ensure proper translation initiation (24, 61–63). The resulting constructs were ectopically integrated within the pdeH ΔGGDEF Pc-dgrA background. Overexpression of B. subtilis DgcP, C. difficile Cd1420, or V. vulnificus DcpA resulted in an inhibition of motility, whereas production of A. tumefaciens Atu1297 did not (Fig. 9R and S). The lack of a motility inhibition from the Gram-negative A. tumefaciens Atu1297 protein may be due to a requirement for a specific histidine kinase to activate the cyclase, as it possesses domains consistent with a response regulator in a two-component signaling system. We conclude that the B. subtilis ectopic expression strain is a fast, effective, and powerful bioassay for detecting c-di-GMP synthesis from foreign GGDEF proteins.
DISCUSSION
The major findings of the current study are that B. subtilis, a model system for the study of Gram-positive bacteria, has a concise signaling system that regulates the production and degradation of c-di-GMP and that this system has a role in regulating motility but apparently not biofilm formation. Our conclusions are based on the biochemical characterization of three active diguanylate cyclases, one active phosphodiesterase, and a single c-di-GMP receptor. Furthermore, we show that steady-state levels of c-di-GMP in vivo are undetectable during vegetative growth due to the activity of the c-di-GMP phosphodiesterase PdeH but can be elevated via the artificial expression of any of the diguanylate cyclases, DgcK, DgcP, or DcgW, or deletion of PdeH. Elevation of c-di-GMP levels leads to inhibition of swarming motility that requires the c-di-GMP receptor DgrA. By constructing a strain in which DgrA is constitutively expressed in a c-di-GMP null background, we show that the introduction of an active diguanylate cyclase results in enhanced inhibition of swarming motility (Fig. 9Q to S). Collectively, our data define the core components of a c-di-GMP signaling pathway in B. subtilis and present a novel system to functionally characterize putative components of c-di-GMP signaling from other organisms.
Our findings are in general agreement with a previous report but significantly extend that previous work to include direct detection of c-di-GMP, functional characterization of proteins, and the study of ydaK (26). Our data indicate that ydaK is not an essential gene and is not associated with a motility phenotype indicative of an active diguanylate cyclase when overexpressed, and though it binds c-di-GMP with moderate affinity, YdaK does not serve as a receptor to modulate swarming motility (Fig. 9; see Fig. S5 and Fig. S7 in the supplemental material). We can find no obvious consequence on biofilm formation by changes in intracellular c-di-GMP as found previously, but we conclude that high c-di-GMP is inhibitory for motility. We note that the motility defect we observed in the absence of PdeH (YuxH) was much less severe than previously reported, but inhibition was nonetheless due to DgrA (YpfA). We infer that the different degrees of inhibition of motility reported herein versus the previous report of a pdeH (yuxH) mutant may be attributable to different expression levels of DgrA (YpfA) under different laboratory conditions, a possibility supported by our DgrA constitutive expression strain that exhibits severely inhibited motility when c-di-GMP is elevated.
Collectively, our work directly demonstrates that B. subtilis has a complete signaling system that regulates intracellular c-di-GMP levels, and we have provided both in vitro and in vitro evidence for the function, or lack thereof, of each putative component of this signaling pathway. In addition, the work provides a tractable model for studying the role of c-di-GMP in many Gram-positive adaptation processes beyond motility and biofilm formation. Finally, the reduced complexity of the B. subtilis system allowed us to generate a strain defective in all known cyclases and phosphodiesterases. We demonstrate that our “c-di-GMP null” strain, combined with constitutive expression of the c-di-GMP receptor DgrA, can be used for the heterologous expression and functional analysis of putative cyclases from diverse bacteria that carry genes that often encode functionally redundant c-di-GMP proteins that ordinarily preclude the study of each protein in isolation. We further note that our system can be potentially modified further to screen for EAL phosphodiesterase proteins and genetic regulators that govern GGDEF activity.
Supplementary Material
ACKNOWLEDGMENTS
All mass spectrometry data were collected in the Indiana University (IU) METACyt Biochemical Analysis Center and Laboratory for Biological Mass Spectrometry. We thank R. Tamayo, D. A. Rowe-Magnus, and C. Fuqua for providing genomic DNAs from C. difficile, V. vulnificus, and A. tumefaciens; Jonathan Karty (IU Mass Spectrometry Facility) and Randy Arnold (IU Proteomics Facility) for scientific and technical assistance; C. Troiano and A. Munchel for collection of preliminary data in the early stages of this work; members of D. B. Kearns' lab for technical advice and helpful discussions; and D. P. Giedroc for comments on the manuscript. c-di-GMP purification was carried out with the assistance of members of E. E. Carlson's and Z. D. Aron's laboratories at Indiana University.
This work was supported with funds provided by the Indiana University College of Arts and Science and NIH grant GM093030 to D.B.K.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00373-13.
REFERENCES
- 1.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 3.Yan HB, Chen WX. 2010. 3′,5′-Cyclic diguanylic acid: a small nucleotide that makes big impacts. Chem. Soc. Rev. 39:2914–2924 [DOI] [PubMed] [Google Scholar]
- 4.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray PM, Forrest G, Wisniewski T, Porter G, Freed DC, DeMartino JA, Zaller DM, Guo Z, Leone J, Fu TM, Vora KA. 2012. Evidence for cyclic diguanylate as a vaccine adjuvant with novel immunostimulatory activities. Cell. Immunol. 278:113–119 [DOI] [PubMed] [Google Scholar]
- 6.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 7.Landini P, Antoniani D, Burgess JG, Nijland R. 2010. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 86:813–823 [DOI] [PubMed] [Google Scholar]
- 8.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35:322–332 [DOI] [PubMed] [Google Scholar]
- 9.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 10.Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Barthel M, Peters V, Hardt WD, Romling U. 2011. Complex c-di-GMP signaling networks mediate transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One 6:e28351. 10.1371/journal.pone.0028351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozlova EV, Khajanchi BK, Sha J, Chopra AK. 2011. Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 50:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 14.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 15.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar M, Chatterji D. 2008. Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology 154:2942–2955 [DOI] [PubMed] [Google Scholar]
- 18.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427 [DOI] [PubMed] [Google Scholar]
- 19.Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 40:11292–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta K, Kumar P, Chatterji D. 2010. Identification, activity and disulfide connectivity of c-di-GMP regulating proteins in Mycobacterium tuberculosis. PLoS One 5:e15072. 10.1371/journal.pone.0015072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78:361–379 [DOI] [PubMed] [Google Scholar]
- 22.Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull TD, Ryu MH, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. 2012. Cyclic di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J. Bacteriol. 194:4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordeleau E, Fortier LC, Malouin F, Burrus V. 2011. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7:e1002039. 10.1371/journal.pgen.1002039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. 2012. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194:3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 194:5080–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81:226–231 [DOI] [PubMed] [Google Scholar]
- 28.Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 29.Guerout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61 [DOI] [PubMed] [Google Scholar]
- 30.Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166–1179 [DOI] [PubMed] [Google Scholar]
- 31.Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 32.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265 [DOI] [PubMed] [Google Scholar]
- 33.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- 34.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 14:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson DG, Smith HO, Hutchison CA, III, Venter JC, Merryman C. 2010. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods 7:901–903 [DOI] [PubMed] [Google Scholar]
- 36.Chen R, Guttenplan SB, Blair KM, Kearns DB. 2009. Role of the sigmaD-dependent autolysins in Bacillus subtilis population heterogeneity. J. Bacteriol. 191:5775–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheffield P, Garrard S, Derewenda Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34–39 [DOI] [PubMed] [Google Scholar]
- 38.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581–590 [DOI] [PubMed] [Google Scholar]
- 39.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao F, Ji Q, Soehano I, Liang ZX. 2011. Unusual heme-binding PAS domain from YybT family proteins. J. Bacteriol. 193:1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basle A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284:13174–13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savakis P, De Causmaecker S, Angerer V, Ruppert U, Anders K, Essen LO, Wilde A. 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol. Microbiol. 85:239–251 [DOI] [PubMed] [Google Scholar]
- 44.Biswas KH, Sopory S, Visweswariah SS. 2008. The GAF domain of the cGMP-binding, cGMP-specific phosphodiesterase (PDE5) is a sensor and a sink for cGMP. Biochemistry 47:3534–3543 [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki M, Li N, Bondarenko VA, Yamazaki RK, Baehr W, Yamazaki A. 2002. Binding of cGMP to GAF domains in amphibian rod photoreceptor cGMP phosphodiesterase (PDE). Identification of GAF domains in PDE alphabeta subunits and distinct domains in the PDE gamma subunit involved in stimulation of cGMP binding to GAF domains. J. Biol. Chem. 277:40675–40686 [DOI] [PubMed] [Google Scholar]
- 46.Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE. 2006. cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J. Biol. Chem. 281:2841–2846 [DOI] [PubMed] [Google Scholar]
- 47.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39:2685–2691 [DOI] [PubMed] [Google Scholar]
- 49.Cho HY, Cho HJ, Kim YM, Oh JI, Kang BS. 2009. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 284:13057–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi Y, Rao F, Luo Z, Liang ZX. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285 [DOI] [PubMed] [Google Scholar]
- 51.Henry JT, Crosson S. 2011. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65:261–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preu J, Panjikar S, Morth P, Jaiswal R, Karunakar P, Tucker PA. 2012. The sensor region of the ubiquitous cytosolic sensor kinase, PdtaS, contains PAS and GAF domain sensing modules. J. Struct. Biol. 177:498–505 [DOI] [PubMed] [Google Scholar]
- 53.Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190:3622–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed] [Google Scholar]
- 55.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. 2010. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402:524–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 57.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 58.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295–1305 [DOI] [PubMed] [Google Scholar]
- 60.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 61.Nakhamchik A, Wilde C, Rowe-Magnus DA. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74:4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328 [DOI] [PubMed] [Google Scholar]
- 63.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.