Abstract
SMC (Structural Maintenance of Chromosome) proteins act ubiquitously in chromosome processing. In Escherichia coli, the SMC complex, MukBEF, plays roles in chromosome segregation and organization. We used single-molecule millisecond multicolor fluorescence microscopy of live bacteria to reveal that a dimer of dimeric fluorescent MukBEF molecules act as the minimal functional unit. 8-10 of these complexes, on average, accumulated as ‘spots’ in 1-3 discrete chromosome-associated regions of the cell, where they formed higher-order structures. Functional MukBEF within spots exchanged with freely diffusing complexes at a rate of one complex every ~50 s in reactions requiring ATP hydrolysis. Thus, by functioning in pairs, MukBEF complexes may undergo multiple cycles of ATP-hydrolysis without being released from DNA, analogous to the behavior of well-characterized molecular motors.
SMC (Structural Maintenance of Chromosome) complexes share conserved architectures and function in chromosome maintenance in all domains of life, although the molecular mechanism by which they act in vivo is unknown (1-3). In eukaryotes, SMC heterodimers associate with a range of accessory proteins, acting in chromosome organization, sister chromosome cohesion and other chromosome biology functions, whereas in bacteria an SMC homodimer and associated accessory proteins act in chromosome maintenance (1). In E. coli and some other γ-proteobacteria, a distant SMC relative, MukB with accessory proteins MukE and MukF, replaces the typical SMC complex, but has similar functions (4, 5). Bacterial smc null mutants are frequently temperature-sensitive, produce anucleate cells and show disturbed chromosome organization at permissive temperature, indicating roles in SMC-mediated chromosome segregation and/or compaction (1, 6, 7). In E. coli undergoing non-overlapping replication cycles, MukBEF accumulates as ‘spots’ at ~1-3 discrete chromosome locations, typically at midcell and/or quarter cell, in the same regions as replication origins (6). Structural and biochemical MukBEF fragment studies report two subunit arrangements, 2:4:2 or 2:2:1, for MukB:E:F, dependent on whether ATP is absent or bound, respectively (8) (fig. S1A). Here our aim was to understand the molecular architecture of active SMC complexes in vivo, as well as the transformations undergone during ATP binding and hydrolysis, as complexes associate with, and dissociate from, the chromosome.
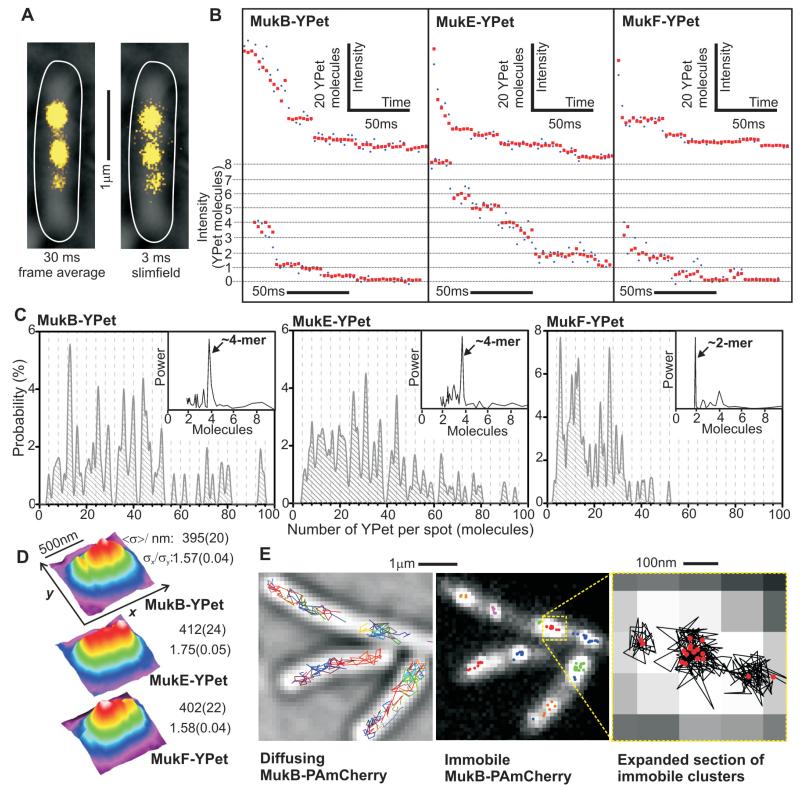
E. coli cells, in which endogenous MukBEF genes were replaced by functional YPet fluorescent fusions, were analyzed by slimfield microscopy, a strategy used previously for studying replisomes (9) (Fig. 1A, fig. S1-2, tables S1-3). Analysis of the numbers of MukB, E or F molecules in individual fluorescent spots showed broad stoichiometry distributions, even between spots in the same cell, with mean values 36 ± 3, 36 ± 4 and 19 ± 1 molecules (±s.e.m.) for MukB, E and F, respectively (9, 10) (Fig. 1B and C). Fourier analysis showed periodicities in stoichiometry of 4:4:2 molecules, respectively for MukB:E:F (Fig. 1C, insets). Spots were elongated parallel to the cell long-axis (Fig. 1D), suggesting that MukBEF complexes spanned several tens of nm, with a decrease of ~20% in measured spot width with increasing stoichiometry across the range measured, consistent with increasing compaction of MukBEF structures as more molecules are added (fig. S3, table S4).
Fig. 1.
MukBEF imaging. (A) Representative frame-average and slimfield MukB-YPet cell images (yellow), brightfield and cell outline overlaid (white). (B) Photobleaching of MukBEF-YPet spots, high (upper row) and low stoichiometry data (expanded sections, lower row), raw (blue) and filtered (red). (C) Stoichiometry distributions, N=51-84 cells. 4-mer interval grid-lines, power spectra (arbitrary units) inset. (D) False-color plots for mean 2D spatial distributions for slimfield images with a 3 ms integration time, N=197-237 spots. Estimates for FWHM <σ> and σx/σy for Gaussian fits parallel to x and y axes (s.d. error). (E) Live-cell PALM, diffusing (gray brightfield, tracks colored) and immobile MukB-PAmCherry (different clusters colored), expanded indicating tracks (black) and clusters (red).
Higher resolution data were obtained using live-cell PALM (Photo-Activated Localization Microscopy) (11) with functional PAmCherry fusions to MukBEF. Rapidly diffusing and relatively immobile populations, forming ~1-3 immobile elongated spots per cell were observed, as in slimfield images (Fig. 1E), the latter resolvable into sub-clusters containing closely associated individual PAmCherry molecules in a diameter of less than 40 nm (fig. S4 and S5).
Slimfield analysis of diffusing cellular YPet fluorescence (9) (fig. S6) indicated ~300-400 molecules/cell for MukB and E, and ~200 molecules/cell for MukF (table S5), in broad agreement with ensemble western estimates (12), implying only ~20% of cellular MukBEF is integrated into spot complexes. PALM single-particle tracking gave similar apparent diffusion coefficients for diffusing MukB, E and F despite large differences in individual molecular weights, compatible with them being components of the same large complexes (fig. S5).
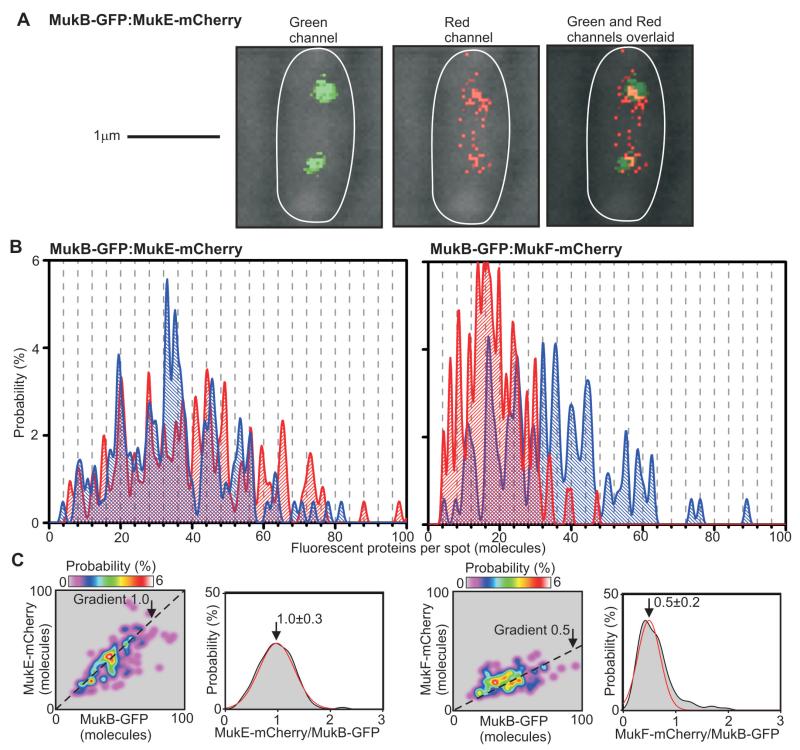
We confirmed the stoichiometry periodicity of 4:4:2 for MukB:E:F by measuring simultaneously the intensities of mCherry and GFP fusions to pairs of MukB, E and F in the same spots, (Fig. 2, fig. S1 and S7, table S6). A plot of the spot-by-spot stoichiometry gave mean ratio values of 1.0 ± 0.3 (±s.d.) and 0.5 ± 0.2 for relative content of MukE to MukB and of MukF to MukB, respectively. Thus the localized MukBEF spots contain ~8-10 dimer of dimer 4:4:2 complexes as minimal functional units.
Fig. 2.
Dual color single molecule millisecond imaging. (A) Brightfield (gray) and 3 ms fluorescence green (left panel) and red (middle panel) channels, overlaid (right panel) for dual label strain. (B) Unbiased kernel density stoichiometry estimation on mCherry (red) and GFP (blue) components for two dual label strains, 4-mer spaced grid-lines. (C) Stoichiometry of mCherry versus GFP component for each spot, dotted line gradients of 1.0 and 0.5; distribution of ratio of stoichiometry for mCherry and GFP components (gray) with Gaussian fit (red), mean ± s.d. indicated.
A 2:2:1 MukB:E:F ratio defines an ATP-bound state (8), resulting from displacement of one MukF and two MukE from a 2:4:2 putative ATP-free form. Given MukF forms stable homodimers (8, 13), MukF displacement may allow recruitment of a second 2:2:1 complex through MukF-mediated dimerization (8, 12, 14), generating the observed 4:4:2 periodicity. Indeed, ATP binding and MukB head engagement were essential for localized spot formation, because they were present in cells of a MukBEQ mutant that binds ATP but is hydrolysis-impaired (8, 15, 16), but not in cells carrying either nucleotide-binding (MukBDA) or engagement-deficient (MukBSR) mutations (17, 18) (fig. S8). The relative stoichiometries of MukBEQ:E:F in localized spots were similar to wild type, consistent with both being ATP-bound, as was the total number of MukBEQEF complexes/spot (fig. S9), and the cellular content of diffusing molecules (tables S4 and S5). Because MukBEQEF cells are Muk−, MukBEF complexes must hydrolyze ATP to be functional.
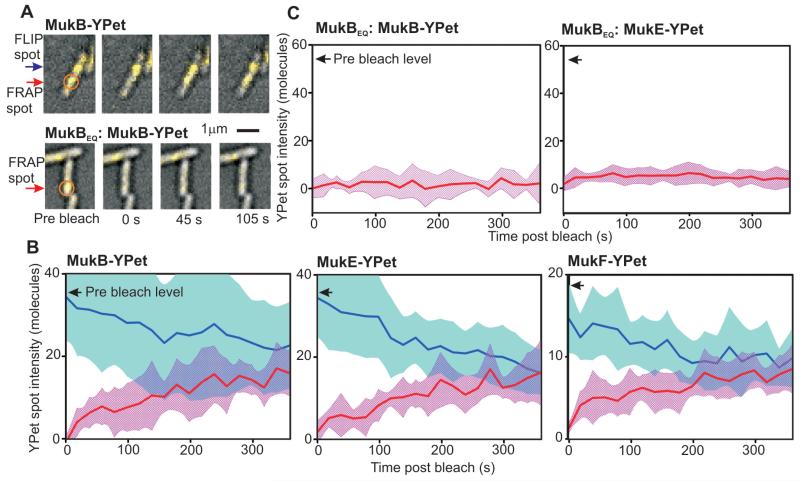
To investigate whether conformational changes during ATP hydrolysis are linked to MukBEF turnover, we compared fluorescence recovery after photobleaching (FRAP) on two MukBEQEF strains with wild type counterparts (Fig. 3A, fig. S10). Detection sensitivity was increased using longer cephalexin-treated cells in which the photoactive to bleached MukBEF-YPet content was higher; recovery up to 60% of pre-bleach levels over several minutes was observed (Fig. 3B). In comparison, steady-state cells gave up to 30% recovery from pre-bleach levels (fig. S10). Reaction-diffusion modeling indicated dwell times for single MukBEF 4:4:2 complexes of ~50 s, independent of cephalexin treatment, with no dependence on pre-bleach intensity. Localized spots outside the original bleach zone indicated fluorescence loss in photobleaching (FLIP) over a similar time scale, converging to similar steady-state intensities. Quantifying post bleach fluorescence for all localized spots indicated ~4-mer periodicity to MukB and MukE, and ~2-mer for MukF (fig. S11), consistent with integer units of 4:4:2 complexes turning over. In contrast to wild type MukBEF, MukBEQEF spots showed no recovery in fluorescence after photobleaching, showing that ATP hydrolysis promoted MukBEF dissociation from DNA (Fig. 3C, fig. S10).
Fig. 3.
Turnover of MukBEF complexes. (A) FRAP of MukB-YPet (upper panel) and ATP hydrolysis mutant MukBEQ-YPet (lower panel), laser focus (orange circle) and FRAP (red arrow) and FLIP (blue arrow) indicated; steady-state cells. (B and C) Mean FRAP (red) and FLIP (blue) traces for cephalexin-elongated cells, s.d. errorbounds (shaded), pre bleach levels shown (arrows), N=10-14 traces.
High-speed imaging allowed us to compare dim spots of rapidly diffusing wild type MukBEF complexes with those carrying either ATP-binding or -hydrolysis mutations. Fluorescence was converted to stoichiometries using single molecule YPet intensity (fig. S12). Wild type and hydrolysis mutants contained mixed populations of MukBEF complexes with ~30% in the 4:4:2 and ~70% in the 2:4:2 state, whereas complexes of the ATP-binding mutant were exclusively in the dimeric 2:4:2 state.
While ATP-hydrolysis is essential for the activity of SMC complexes, its mechanistic significance has been unclear. Our data indicate that the minimal functional MukBEF complex acting at discrete chromosome positions is an ATP-bound dimer of MukB dimers, with ATP binding and head engagement being necessary for stable chromosome association, and ATP hydrolysis required to release complexes from chromosomes. The observation that turnover of MukBEF complexes from chromosomes is slower than predicted from in vitro ATPase levels (8, 19) (fig. S1F), supports a model where ATP hydrolysis within each ATPase head pair is independent, with all four ATP molecules in the two closed dimer of dimer heads needing to be hydrolyzed almost simultaneously to completely release a single DNA–bound complex. A multimeric form of MukBEF would therefore allow release of one DNA segment and capture of a new segment without releasing the complex from the chromosome, a process akin to a rock climber making trial grabs to reach a hand hold, and one which could lead to ordered MukBEF movement within a chromosome, perhaps leading to DNA remodeling (fig. S1F). This is analogous to the processive ‘walking’ of the molecular motors kinesin and dynein along microtubules (20).The functional advantage of dimeric SMC complex oligomerization may be exploited by other SMC complexes, irrespective of the mechanism of multimerization. Like MukBEF, bacterial SMC-ScpAB forms relatively immobile complexes that accumulate at a few chromosome positions. Bacillus subtilis SMC-ScpAB can form multimeric complexes in vitro, with SMC and ScpB forming homodimers, and ScpA forming monomers or dimers (15, 21). Eukaryote SMC complexes also share similar characteristics to MukBEF in maintaining chromosomes, accumulating at discrete chromosome loci (22, 23) and turning over in ~seconds, as well as having the same distinctive architecture (24, 25). Although they capture DNA topologically in apparent heterodimeric complexes (26), higher order complexes might form, and exploit the type of rock climbing mechanism described here.
Supplementary Material
Acknowledgements
RR-L held the Todd-Bird Junior Research Fellowship of New College (Oxford University). SU was supported by a Mathworks scholarship. MCL was supported by a Royal Society University Research Fellowship. We thank R. Oliveira for assistance with FRAP experiments, and A. Kapanidis for discussions and provision of the PALM equipment. The experimental work was funded by an EPSRC grant to MCL (EP/G061009) and by a Wellcome Trust programme grant to DJS (WT083469MA).
Footnotes
All data are presented in the Supporting Online Material and bacterial strains are available on request.
The authors declare they have no competing financial interests.
References and Notes
- 1.Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Mol Microbiol. 2011;81:855. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 3.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niki H, et al. E.coli MukB protein involved in chromosome partition forms a homodimer with a rod-and-hinge structure having DNA binding and ATP/GTP binding activities. EMBO J. 1992;11:5101. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol Gen Genet. 1996;250:241. doi: 10.1007/BF02174381. [DOI] [PubMed] [Google Scholar]
- 6.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiraga S, et al. Mutants defective in chromosome partitioning in E. coli. Res Microbiol. 1991;142:189. doi: 10.1016/0923-2508(91)90029-a. [DOI] [PubMed] [Google Scholar]
- 8.Woo JS, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 9.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J Biol Chem. 2006;281:34208. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J. 2005;24:1921. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gloyd M, Ghirlando R, Guarne A. The role of MukE in assembling a functional MukBEF complex. J Mol Biol. 2011;412:578. doi: 10.1016/j.jmb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano M, Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 2004;23:2664. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B, et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam P, et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13:1941. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 18.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 19.Chen N, et al. ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli. J Bacteriol. 2008;190:3731. doi: 10.1128/JB.01863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kural C, et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 21.Mascarenhas J, et al. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 2005;6:28. doi: 10.1186/1471-2121-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Ambrosio C, Kelly G, Shirahige K, Uhlmann F. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol. 2008;18:1084. doi: 10.1016/j.cub.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 23.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J Cell Sci. 2011;124:685. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 24.Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol. 2006;16:333. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira RA, Heidmann S, Sunkel CE. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- 26.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 27.Bachmann BJ. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972;36:525. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazoe M, et al. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohsumi K, Yamazoe M, Hiraga S. Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol Microbiol. 2001;40:835. doi: 10.1046/j.1365-2958.2001.02447.x. [DOI] [PubMed] [Google Scholar]
- 32.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 33.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 35.English BP, et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A. 2011;108:E365. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden SJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J. 2010;99:3102. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. Journal of Collod and Interface Science. 1996;179:298. [Google Scholar]
- 39.Woldringh CL. In: Bacterial Chromatin. Dame RT, Dorman CJ, editors. Springer Biomedical/Life Science; The Netherlands: 2009. [Google Scholar]
- 40.Delalez NJ, et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acad Sci U S A. 2010;107:11347. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peccoud J, Ycard B. Markovian modelling of gene-product synthesis. Theoretical Population Biology. 1995;48:222. [Google Scholar]
- 42.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 43.Plank M, Wadhams GH, Leake MC. Millisecond timescale slimfield imaging and automated quantification of single fluorescent protein molecules for use in probing complex biological processes. Integr Biol (Camb) 2009;1:602. doi: 10.1039/b907837a. [DOI] [PubMed] [Google Scholar]
- 44.Chandrasekhar S. Stochastic problems in physics and astronomy. Reviews of Modern Physics. 1943;15:1. [Google Scholar]
- 45.Schütz GJ, Sonnleiter M, Schindler H. Ultrasensitive microscopy of the plasma membrane of living cells. Journal of Fluorescence. 2001;11:177. [Google Scholar]
- 46.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.