Abstract
Objective
The gold standard for the diagnosis and evaluation of Crohn disease (CD) is endoscopy/colonoscopy, although this is invasive, costly, and associated with risks to the patient. Recently, circulating microRNAs (miRNAs) have emerged as promising noninvasive biomarkers. Here, we examined the utility of serum miRNAs as biomarkers of CD in children.
Patients and Methods
Studies were conducted using sera samples from patients with pediatric CD, healthy controls, and a comparison group of patients with pediatric celiac disease. Serum miRNA levels were explored initially using a microfluidic quantitative reverse transcription-polymerase chain reaction array platform. Findings were subsequently validated using quantitative reverse transcription-polymerase chain reaction in larger validation sample sets. The diagnostic utility of CD-associated serum miRNA was examined using receiver operating characteristic analysis.
Results
A survey of miRNA levels in the sera of control and patients with CD detected significant elevation of 24 miRNAs, 11 of which were chosen for further validation. All of the candidate biomarker miRNAs were confirmed in an independent CD sample set (n = 46). To explore the specificity of the CD-associated miRNAs, they were measured in the sera of patients with celiac disease (n = 12); none were changed compared with healthy controls. Receiver operating characteristic analyses revealed that serum miRNAs have promising diagnostic utility, with sensitivities for CD above 80%. Significant decreases in serum miRNAs were observed in 24 incident patients with pediatric CD after 6 months of treatment.
Conclusions
The present study identifies 11 CD-associated serum miRNA with encouraging diagnostic potential. Our findings suggest serum miRNAs may prove useful as noninvasive biomarkers in CD.
Keywords: biomarker, circulating miRNA, Crohn disease
The diagnosis of inflammatory bowel disease (IBD) is often achieved only months or years after the onset of symptoms. Several serological indicators of IBD have been identified; in general, they are antibodies directed against antigens expressed by organisms of the intestinal microbiome (1–4). For example, the anti-Saccharomyces cerevisiae antibody (ASCA) interacts with mannose epitopes of this yeast species and is present in 48% to 80% of patients with CD (5,6). In general, these markers are specific for IBD, but experience low sensitivity.
CD biomarkers can also be of value after the diagnosis is established, as measures of disease activity and predictors of outcome. The available serological markers have not proven useful in these contexts (7–13). Other serum and stool markers, such as C-reactive protein (CRP) and fecal calprotectin, are elevated in inflammatory and gastrointestinal diseases, but are not specific for IBD (14–19). The introduction of additional sensitive, specific, and noninvasive diagnostic markers may aid in the diagnosis of IBD, reduce patient risk and discomfort by reducing invasive testing, and accelerate the study of new treatments.
MicroRNAs (miRNAs) are short, noncoding RNAs that regulate target mRNAs via transcript degradation or translational repression. Cell- and tissue-specific miRNA expression profiles are altered in numerous disease states (20–30). The loss of all of the intestinal miRNA results in impaired barrier function and inflammation similar to IBD (31). With the exception of gastric and colorectal cancers (32,33), little is known regarding the function of miRNA in intestinal disease. Wu et al (34) profiled miRNA expression in colon biopsies in ulcerative colitis (UC), indeterminate colitis, infectious colitis, microscopic colitis, and irritable bowel syndrome. Significant changes were confirmed in 11 miRNAs in UC tissues when compared with normal controls, of which 5 were altered at least 2-fold. The authors focused on microRNA-192 (miR-192; 1.9-fold lower in active UC), showing that it localizes to colonic epithelia and is able to repress expression of the chemokine CXCL2 (MIP-2a) in a colonic epithelial cell line. They suggest that in UC, decreased miR-192 levels result in intestinal inflammation via increased CXCL2 secretion by epithelial cells. The same group has also investigated ileal and colonic miRNA expression, resulting in the identification of several miRNAs whose levels are altered in CD (35).
The recent discovery of circulating miRNAs possessing remarkable stability has prompted a number of studies investigating their potential merit as noninvasive biomarkers (36,37). Specific circulating miRNA profiles have now been described for various conditions, particularly cancer (36–41). In some cases, these circulating miRNA profiles are known to correlate with miRNA expression changes in the diseased tissue (41,42). Additionally, changes in circulating miRNA profiles may precede those of standard blood biomarkers (39,41), and several disease-specific profiles are known to possess both diagnostic and prognostic value (43,44). Taken together, these properties implicate miRNAs to be attractive, blood-based, noninvasive biomarkers.
To date, a single study has explored serum or plasma miRNAs within the scope of IBD. In an investigation of markers of colorectal cancer, Ng et al (45) described a significant decrease in plasma miR92 levels in a group of 20 elderly patients with IBD. Recently, Wu et al (46) identified peripheral blood cell miRNAs capable of distinguishing adult patients with CD and UC. Although a number of differentially expressed miRNAs were identified by this cell-based approach, the use of serum or plasma miRNA has the potential advantage of reflecting miRNA changes in the intestinal tissue itself, rather than blood cells.
Here, we have performed a study to test the hypothesis that the combination of epithelial damage and inflammation seen in CD results in altered circulating miRNA levels. We have identified a panel of serum miRNAs that are increased in patients with CD compared with healthy controls. These serum miRNAs show promise as clinically useful, noninvasive biomarkers for diagnosis and treatment response of pediatric CD.
PATIENTS AND METHODS
Serum Samples
Sera from patients with pediatric CD and healthy controls were obtained as part of institutional review board–approved studies at the Children’s Hospital of Philadelphia Research Institute (M.T. and M.B.L.); CD diagnosis was confirmed by standard parameters as previously described (47–49). Site of disease was defined according to the Montreal classification (50). CD activity was assessed using the Pediatric Crohn Disease Activity Index (PCDAI) (51). Controls were recruited for studies of growth and nutrition, had normal height and body mass index, and had no history of chronic diseases. Characteristics of patients with CD and controls are summarized in Table 1. Pediatric celiac disease serum samples and controls were obtained from Dr Alessio Fasano of the University of Maryland School of Medicine. Samples with visible evidence of hemolysis were excluded from the study. Serum was stored at −80°C until RNA isolation.
TABLE 1.
Baseline characteristics in patients with CD and controls
| CD (n = 46) | Controls (n = 32) |
|
|---|---|---|
| Age, y | 13.7 ± 3.0 | 13.1 ± 4.1 |
| Sex, male,% | 65.2 | 53.1 |
| Race, white,% | 84.8* | 65.6 |
| PCDAI | 34.7 ± 18.1 | |
| Median (range) | 32.5 (7.5, 80) | |
| No active disease (≤slO), % | 6.5 | |
| Mild disease (11–30), % | 39.1 | |
| Moderate/severe (> 30), % | 54.3 | |
| Months since diagnosis | 1.0 ± 1.9 | |
| Site of disease, n (%) | ||
| Isolated ileal disease | 0 | |
| Isolated colonic disease | 1 (2) | |
| Ileocolonic disease | 28 (61) | |
| Isolated upper GI tract disease | 1 (2) | |
| Perirectal involvement | 21 (46) | |
| Colonic involvement | 45 (98) | |
| Ileal involvement | 29 (63) | |
| Duodenal involvement | 13 (28) | |
| Gastric involvement | 40 (87) | |
| Esophageal involvement | 22 (48) |
Continuous variables presented as mean ± standard deviation unless otherwise indicated. Subjects may be counted in multiple disease site subgroups. CD = Crohn disease; GI = gastrointestinal; PCDAI = pediatric Crohn disease activity index.
P < 0.05 compared with controls.
RNA Isolation
Total RNA was isolated from 60mL of serum using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to manufacturer’s instructions. Exogenous Caenorhabditis elegans and human miRNAs were added as normalizing controls immediately following serum denaturation. RNA was eluted with 100mL elution solution (95°C) and stored at −80°C. Quality of total RNA preparations was confirmed using the Agilent 2100 Bioanalyzer with the RNA 6000 Pico kit (Agilent Technologies, Santa Clara, CA).
miRNA Analysis by Low-density Array
TaqMan Human MicroRNA Arrays (Applied Biosystems, Foster City, CA) were used to quantify serum miRNA content according to the manufacturer’s instructions. Reverse transcription (RT) products were preamplified and arrays were processed and analyzed by the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). miRNA levels were normalized against exogenous human embryonic-specific miRNAs (miR-302a and miR-372 for pool A arrays and miR-302d for pool B arrays) added in equal amounts during RNA isolation to control for assay variability (52).
qRT-PCR
Total RNA volumes of 1.33mL were reverse transcribed and amplified using the TaqMan MicroRNA Reverse Transcription and miRNA Assay Kits (Applied Biosystems) according to the manufacturer’s instructions. Reactions were performed in duplicate. miRNA levels were normalized to the levels of 2 exogenous C elegans miRNAs lacking homology to human sequences (cel-miR-54 and cel-miR-238) added during RNA isolation (36).
Enzyme-linked Immunosorbent Assay
Serum levels of CRP and ASCA IgG were determined in control and patients with CD using commercial enzyme-linked immunosorbent assay Kits (Calbiotech, Spring Valley, CA; ALPCO, Salem, NH) according to the manufacturer’s instructions.
Statistical Analysis
Significantly altered miRNAs from the low-density array (LDA) experiment were identified using Significance Analysis of Microarrays software (Stanford University, Stanford, CA). All other statistical calculations were performed using Stata 11.0 (StataCorp, College Station, TX). Fisher exact test, Mann-Whitney test, and Wilcoxon matched-pairs signed rank test were used to determine significance. Hierarchical cluster analysis (complete-linkage clustering with Euclidean (L2) distance) was performed using the 68 miRNAs detected in all 12 LDA samples. Receiver operating characteristic (ROC) curve analyses were used to determine diagnostic utility. Correlation between miRNAs was determined by Spearman correlation test. Heat maps were generated using Matrix2png (53). Expression data were presented as mean ± stanstandard error with 2-sided P values.
RESULTS
Discovery of CD-associated Circulating miRNA Using Low-density Array qRT-PCR
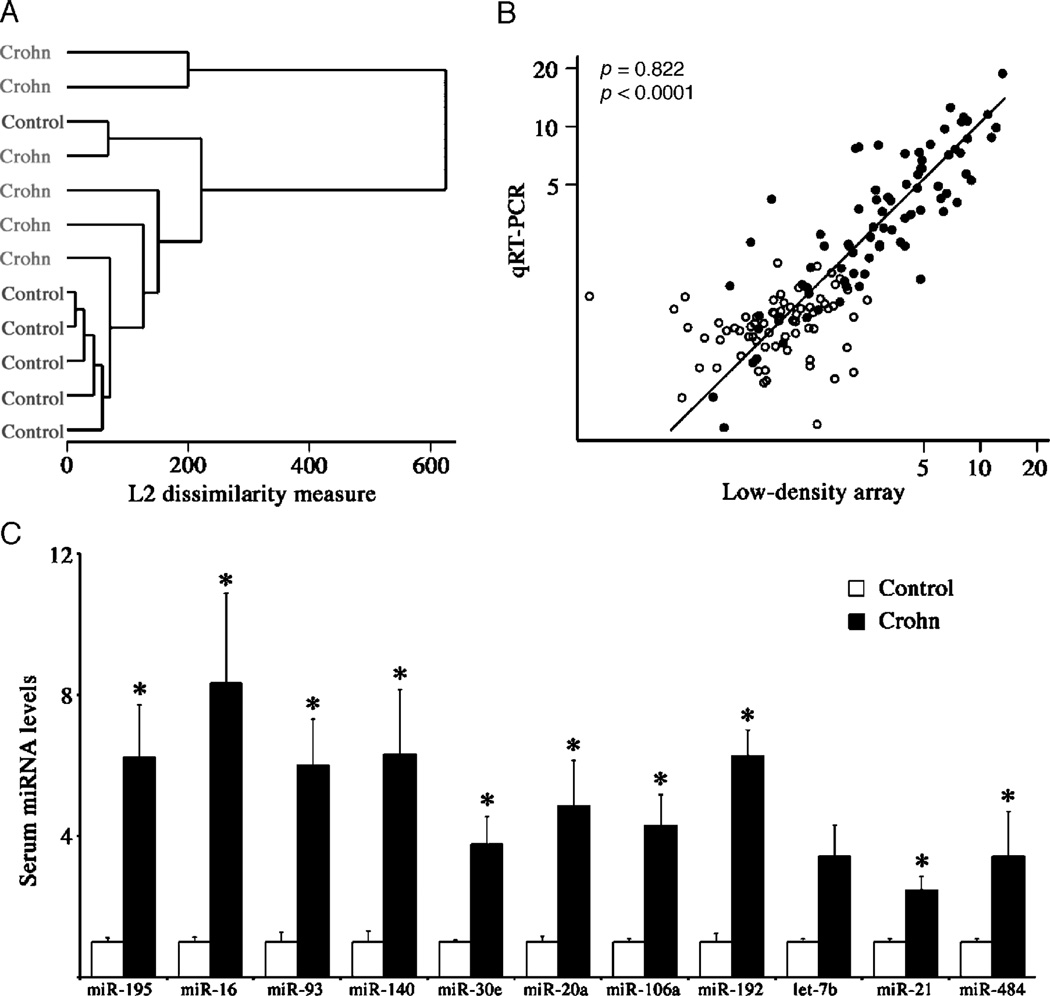
A microfluidic quantitative reverse transcription-polymerase chain reaction (qRT-PCR) LDA platform capable of detecting 667 mature human miRNAs was used to identify CD-associated miRNAs in an exploratory cohort of patients with CD with active disease (12.5 ± 1.4 years, 67% boys, PCDAI = 32.5 ± 8.4, n = 6), compared with age- and sex-matched healthy controls. Hierarchical clustering analysis revealed altered serum miRNA profiles in patients with CD compared with controls (Fig. 1A). Differences in individual serum miRNA levels between groups were identified using significance analysis of microarrays (54). A total of 24 miRNAs were increased at least 50% in patients with CD at a false discovery rate of 6% (Supplementary Table 1 [http://links.lww.com/MPG/A51] and Supplementary Fig. 1 [http://links.lww.com/MPG/A52]). Ten of the 24 miRNAs are members of 3 paralogous genomic miRNA clusters with oncogenic properties, namely miR-17–92, miR-106b-25, and miR-106a-363 (55–57). No circulating miRNAs were significantly decreased in patients with CD. A panel of 11 miRNAs was selected for direct confirmation by qRT-PCR; a single member (miR-20a, miR-93, and miR-106a) from each of the 3 paralogous miRNA clusters was included. Good agreement was observed between the array and individual qRT-PCR validation results (Fig. 1B). All but 1 of the selected miRNAs was significantly increased in the CD sera at least 2.5-fold (Fig. 1C).
FIGURE 1.
Low-density array (LDA) analysis of serum miRNA in patients with pediatric CD. A, Dendrogram showing hierarchical cluster analysis using 68 miRNAs detected in all samples by LDA. B, Scatterplot of relative serum miRNA levels of 11 CD-associated miRNAs and 2 unaltered control miRNAs as determined by LDA and individual qRT-PCR. Open circles, control samples; filled circles, CD samples; r, Spearman rank correlation coefficient. C, Comparison of relative levels of CD-associated miRNAs in control and CD samples determined by individual qRT-PCR. Data are presented as fold change in comparison with controls. *P <0.05. CD = Crohn disease; LDA = low-density array; miRNA = microRNA; qRT-PCR = quantitative reverse transcription-polymerase chain reaction.
Confirmation of CD-associated Circulating miRNA in an Independent Sample Set
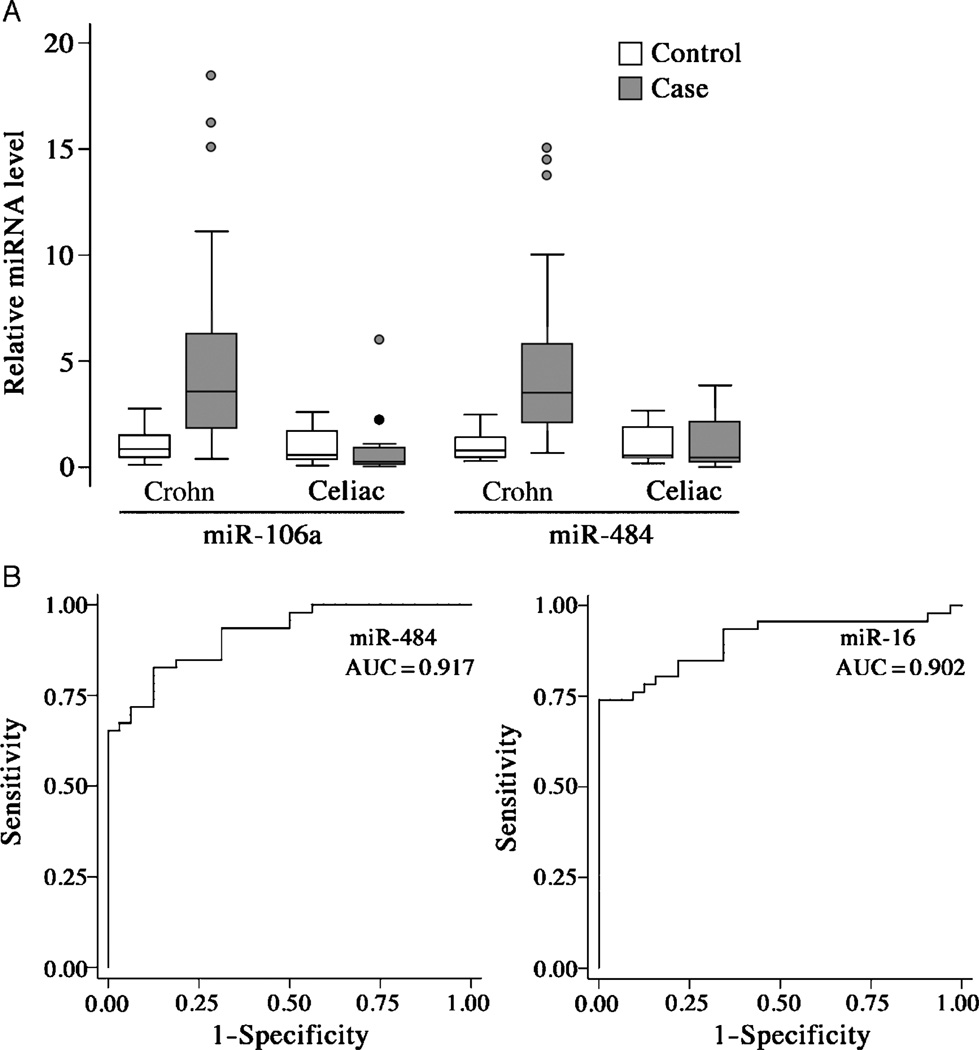
To verify the differences in serum miRNA levels between patients with CD and controls, qRT-PCR was performed on a larger, distinct set of incident cases of CD (n = 46), similar to the exploratory cohort in terms of demographic and disease characteristics, and control subjects (n = 32) (Table 1). Each of the selected 11 miRNAs was significantly elevated (range 2.7- to 8.7-fold) in the serum of patients with CD compared with controls at a P <0.0001 (Table 2). The results from miR-106a and miR-484 are illustrated in Figure 2A. No correlation was observed between miRNA levels or white or red blood cell count, indicating that the altered miRNA levels are not a simple reflection of hematologic changes. The miRNA levels also did not correlate with the albumin concentration, erythrocyte sedimentation rate, or CRP level (data not shown).
TABLE 2.
Serum miRNAs in patients with Crohn and celiac disease compared with controls
| Fold change |
||
|---|---|---|
| miRNA | Crohn (n = 46) | Celiac (n = 12) |
| miR-16 | 8.74 ± 1.54* | 1.04 ± 0.60 |
| let-7b | 7.49 ± 1.32* | 1.19 ± 0.73 |
| miR-195 | 5.67 ± 0.90* | 0.95 ± 0.53 |
| miR-106a | 4.79 ± 0.63* | 0.98 ± 0.49 |
| miR-20a | 4.63 ± 0.63* | 0.90 ± 0.42 |
| miR-30e | 4.60 ± 0.59* | 0.96 ± 0.34 |
| miR-140 | 4.51 ± 0.65* | 0.96 ± 0.37 |
| miR-484 | 4.50 ± 0.51* | 1.23 ± 0.44 |
| miR-93 | 4.48 ± 0.60* | 0.87 ± 0.49 |
| miR-192 | 4.24 ± 0.74* | 0.87 ± 0.27 |
| miR-21 | 2.72 ± 0.24* | 0.73 ± 0.27 |
Fold change is relative to controls and presented as mean ± SE.
P < 0.0001.
FIGURE 2.
Validation of CD-associated circulating miRNAs. A, Box-whisker plots of CD-associated serum miRNAs validated in an independent set of controls (n = 32) and CD cases (n = 46) as well as celiac cases (n = 12) and associated controls (n = 12). Box, 25% to 75%; whisker, upper, lower adjacent values; line, median; points, outside values. Data are presented as fold change in comparison with controls. B, Receiver operating characteristic curves of 2 CD-associated miRNAs in sera of patients with pediatric CD (n = 46) and healthy controls (n = 32). AUC = area under the curve; CD = Crohn disease; miRNAs = microRNAs.
Because CD sera were compared with sera of healthy control patients, it is possible that the observed elevations in panel miRNAs occur during enteritis of any kind. To address this possibility, miRNA levels were measured in the sera of children with active celiac disease and age-, race-, and sex-matched controls without celiac disease (Table 3). All 11 panel miRNAs elevated in CD serum were unaltered in the serum of celiac patients in comparison with healthy controls (Table 2 and Fig. 2A). The data suggest that elevated levels of the CD-associated circulating miRNAs are not a general result following intestinal tract inflammation and destruction, because active celiac disease involves inflammatory-mediated damage to the proximal small bowel mucosa.
TABLE 3.
Baseline characteristics in patients with celiac disease and controls
| Controls (n = 12) | Celiac disease (n = 12) | |
|---|---|---|
| Age, y | 14.0 ± 0.7 | 14.0 ± 0.8 |
| Sex, male,% | 25.0 | 25.0 |
| Race, white,% | 100 | 100 |
| EMA positive, % | 0 | 100* |
| tTG-IgA, U/mL | 0.8 ± 0.1 | 38.8 ± 8.5* |
| AGA-IgG, U/mL | 9.9 ± 2.0 | 19.5 ± 6.0 |
| AGA-IgA, U/mL | 3.9 ± 1.0 | 8.1 ± 2.4 |
Continuous variables presented as mean ± SE. AGA = anti-gliadin antibody; EMA = anti-endomysial antibody; tTG = anti-tissue transglutaminase antibody.
P < 0.05 compared with controls.
Spearman rank correlation revealed that each CD-associated miRNA was positively correlated (P <0.0001) with each of the other CD-associated miRNAs (Supplementary Fig. 2A [http://links.lww.com/MPG/A52]). Among the miRNAs, the strongest miRNA–miRNA correlation (r = 0.983) was between miR-20a and miR-106a, members of the paralogous clusters miR-17–92 and miR-106a-363, respectively, whereas the lowest correlation (r = 0.709) was between miR-192 and miR-484 (Supplementary Fig. 2B [http://links.lww.com/MPG/A52]). In contrast, levels of CD-associated miRNAs did not correlate with disease activity as determined by PCDAI score. Interestingly, esophageal disease involvement was significantly associated with higher serum levels of 8 CD-associated miRNAs (all except miR-16, miR-192, and let-7b; P <0.05).
Clinical Performance of Circulating miRNA as a Biomarker for CD
To assess the diagnostic utility of CD-associated miRNAs, we determined their receiver operating characteristics. ROC curves revealed that the CD-associated miRNAs have promising diagnostic properties, with area under the ROC curve (AUC) values of 0.82 to 0.92 (Table 4 and Fig. 2B), sensitivities of 70% to 83%, and specificities of 75% to 100% (Table 5). These values compared favorably to those of erythrocyte sedimentation rate and serum levels of CRP, ASCA IgG, and albumin, using standard diagnostic thresholds.
TABLE 4.
ROC analyses of panel miRNAs
| miRNA | AUC | 95% CI |
|---|---|---|
| miR-484 | 0.917 | 0.860–0.974 |
| miR-16 | 0.902 | 0.832–0.971 |
| miR-30e | 0.882 | 0.805–0.958 |
| miR-106a | 0.879 | 0.806–0.952 |
| miR-195 | 0.876 | 0.800–0.952 |
| miR-20a | 0.863 | 0.785–0.941 |
| let-7b | 0.860 | 0.781–0.939 |
| miR-21 | 0.853 | 0.770–0.936 |
| miR-93 | 0.852 | 0.769–0.935 |
| miR-192 | 0.834 | 0.744–0.923 |
| miR-140 | 0.821 | 0.728–0.915 |
AUC = area under the curve; CI = confidence interval; ROC = receiver operating characteristics.
TABLE 5.
Diagnostic properties of panel miRNAs and CD-related laboratory values
| miRNA | Sensitivity, % | Specificity, % | Correctly classified, % |
|---|---|---|---|
| miR-16 | 73.91 | 100 | 84.62 |
| miR-484 | 82.61 | 84.38 | 83.33 |
| miR-30e | 73.91 | 96.88 | 83.33 |
| miR-106a | 76.09 | 90.62 | 82.05 |
| miR-195 | 69.57 | 96.88 | 80.77 |
| miR-20a | 73.91 | 87.50 | 79.49 |
| miR-21 | 76.09 | 84.38 | 79.49 |
| miR-140 | 73.91 | 87.50 | 79.49 |
| let-7b | 82.61 | 75.00 | 79.49 |
| miR-192 | 78.26 | 78.12 | 78.21 |
| miR-93 | 71.74 | 84.38 | 76.92 |
| Test | Threshold | Sensitivity, % | Specificity, % | Correctly classified, % |
|---|---|---|---|---|
| CRP | 0.9 mg/dL | 63.64 | 93.55 | 76.00 |
| ASCA IgG | 10 U/mL | 62.22 | 80.65 | 69.74 |
| ESR | 20 mm/h | 52.17 | — | — |
| Alhnmin | 3.5 g/dL | 41.30 | — | — |
ASCA = anti-Saccharomyces cerevisiae antibody; CD = Crohn disease; CRP = C-reactive protein; ESR = erythrocyte sedimentation rate; miRNAs = microRNAs.
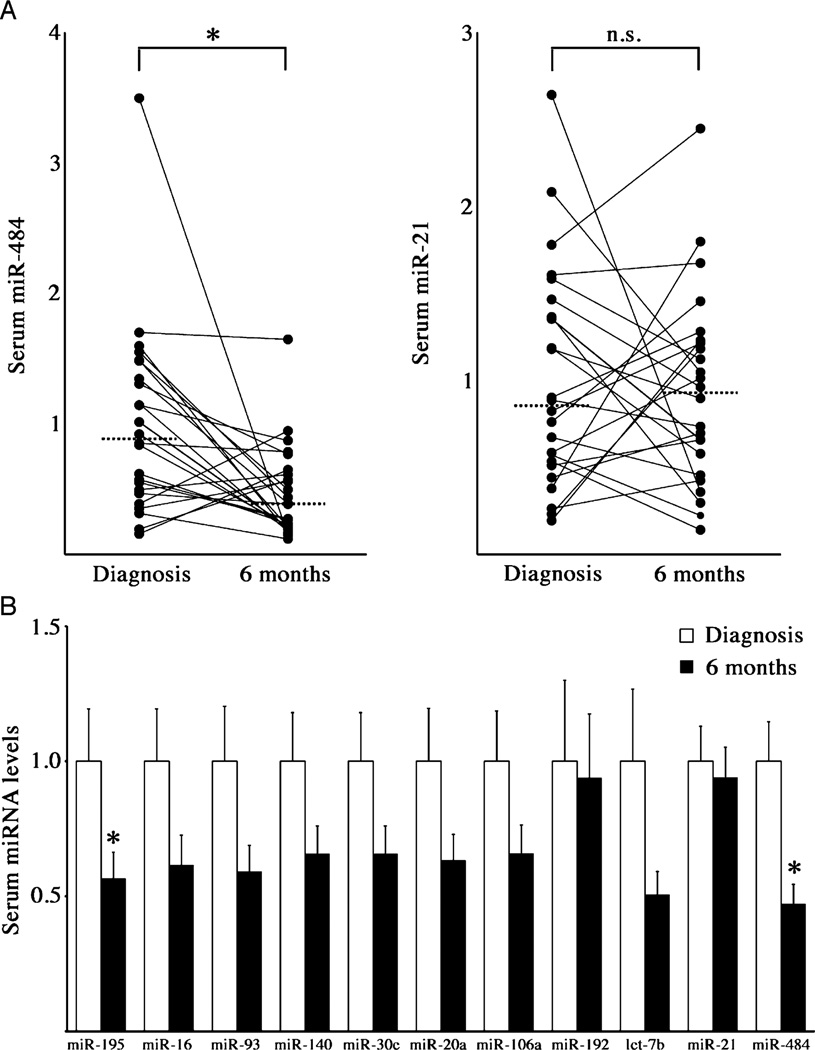
To determine whether CD-associated circulating miRNA levels change with treatment, we analyzed the sera of 24 patients with incident CD, similar to our larger cohort in terms of demographics and disease severity, at the time of diagnosis and 6 months later. Between baseline and 6 months, median PCDAI scores decreased significantly (P<0.001). At the 6-month study visit, the proportions of study subjects who had received the following medication during that interval were systemic steroids 16 (67%), methotrexate 1 (4%), 6-mercaptopurine or azathioprine 10 (42%), and infliximab 2 (8%). Significant reductions were observed in the levels of miR-484 and miR-195 (Fig. 3). All other panel miRNAs showed no significant change following treatment, although most trended downward (Fig. 3B). Changes in panel miRNA levels and PCDAI scores were not significantly correlated. These results suggest that CD-associated circulating miRNAs may be markers of response to therapeutic intervention.
FIGURE 3.
Response of CD-associated circulating miRNAs following treatment. A, Dot plots of 2 CD-associated miRNAs in sera of patients with pediatric CD at diagnosis and following 6 months of treatment (n = 24). Data are presented as fold change relative to level at diagnosis. Solid lines connect data points for each patient. Dashed line = median; *P = 0.003; n.s. = not significant, using the Wilcoxon matched-pairs signed rank test. B, Serum miRNA levels in patients with pediatric CD at diagnosis and following 6 months of treatment (n = 24). Data are presented as fold change relative to level at diagnosis. *P = 0.003 for miR-484 and P = 0.037 for miR-195 using the Wilcoxon matched-pairs signed rank test. CD = Crohn disease; miRNAs = microRNAs.
Analysis of CD-associated Circulating miRNAs Using Internal Reference miRNAs
One obstacle to the clinical use of circulating miRNA as a biomarker derives from its acellular nature. In assays of cellular miRNAs, a variety of “housekeeping” RNA species are used commonly to correct for differences in tissue mass, RNA yield, or quality. Because no such internal controls are present for assays of circulating miRNA, in our initial assays, 2 artificial C elegans miRNAs were added at the time of RNA purification and used as surrogate references (see Patients and Methods). However, it was noted in our LDA analyses that miR-150 and miR-342–3p were present at equivalent levels in patients with CD and controls (Supplementary Fig. 3A [http://links.lww.com/MPG/A52]). We therefore determined whether they could be used as internal reference miRNAs. The results obtained using the endogenous reference miRNAs were nearly identical to those obtained using exogenous reference miRNA (Supplementary Fig. 3B [http://links.lww.com/MPG/A52]). These results indicate that it may be possible to eliminate the use of exogenous miRNA in future analyses.
DISCUSSION
Circulating miRNAs have recently emerged as candidate biomarkers for disease, particularly cancer (36,37,39,58). The present study is the first to demonstrate the potential of circulating miRNAs as noninvasive biomarkers of pediatric CD. An initial screen of patients with CD by microfluidic qRT-PCR array identified a significantly altered serum miRNA profile in comparison with healthy controls. These findings were subsequently validated in a much larger set of cases and controls.
All 24 miRNAs significantly altered in CD sera were elevated. The pathogenesis of IBD is a complex process involving inflammatory signaling, lymphocyte infiltration of the gut, and epithelial cell damage. Each of these may result in increases in the levels of circulating miRNA. For example, exosomes secreted in the course of inflammatory signaling may carry specific miRNAs into the circulation. The intestine is a highly vascular organ, and thus activated lymphocytes in the lamina propria may contribute to circulating miRNA. Furthermore, injury to intestinal epithelia may result in increases in epithelium-specific miRNAs in the circulation, as has been observed for tissue miRNAs in heart or liver injury (39,59). For instance, we found that circulating miR-192 is elevated in CD; miR-192 is also the most greatly expressed miRNA in intestinal epithelia (31).
In a study of intestinal miRNA levels in CD, Wu et al (35) identified several miRNAs that are upregulated; these include 4 of the miRNAs we have described (miR-16, -20a, -21, and -106a). Let-7b, miR16, and miR-21 are greatly expressed in human dendritic cells, which likely contribute to the chronic inflammation of CD (60,61). In contrast, the internal control miRNAs (miR-150 and miR-342-3p) are not detected in intestinal epithelia (62). None of the panel miRNAs are restricted to a single cell type, yet they are greatly correlated with each other, suggesting that CD may be associated with a specific circulating miRNA signature that reflects both inflammation and enteritis.
Serological testing is frequently used in the diagnosis of children with suspected IBD, although evidence suggests current markers are suboptimal as screening tools for disease in this patient population, with reported sensitivities ranging from 55% to 71% (63–65). Thus, although the diagnosis of CD ultimately must be made on histopathologic grounds, the introduction of improved noninvasive testing may help to close the gap between the onset of symptoms and the final diagnosis, allowing for earlier treatment. Conversely, a negative screening test result may help reduce unnecessary endoscopy/colonoscopy.
The serum miRNAs examined here display encouraging diagnostic utility, performing favorably in comparison with some standard serological markers. MiR-484 and let-7b each possessed sensitivities > 80%, and 3 had specificities > 90% in comparison with healthy controls. In addition, each panel miRNA was unchanged in the serum of celiac patients compared with age-, race-and sex-matched controls, suggesting that these miRNAs may be specific for IBD or CD, rather than simply indicators of intestinal inflammation in general. This finding contrasts with current IBD serological markers, which are often present in non-IBD intestinal disease. For instance, ASCA is detected in a large proportion of patients with celiac disease, whereas perinuclear anti-neutrophil cytoplasmic antibodies can also be present in celiac disease or microscopic colitis (9,12,13).
Once the diagnosis of IBD is made, the current serological markers of CD are of limited use because they correlate poorly with disease activity or outcome in both adult and pediatric patients (66,67). Studies of circulating miRNA suggest that it may be a more dynamic biomarker; for example, levels of plasma miR-1, the most abundantly expressed miRNA in the heart (68), are elevated at the time of diagnosis in acute myocardial infarction and return to normal by the time of hospital discharge (38). Likewise, miRNA released from tumor cells can significantly alter circulating miRNAs levels, which normalize following tumor resection (42,69,70). We have found that after 6 months of treatment, serum miR-484 and miR-195 levels were significantly reduced from levels observed at the time of diagnosis. Reductions in panel miRNA levels did not significantly correlate with improved PCDAI scores. However, it remains possible that serum miRNAs accurately represent improvements at the mucosal level, as clinical scoring systems, as well as other surrogate markers, correlate poorly with mucosal healing (71,72).
A number of limitations to this preliminary analysis can be rectified with a larger cohort of cases and controls. The non-significant trend toward decreased circulating levels of several miRNAs in the longitudinal analysis may be because of lack of statistical power. In addition, it will be essential to obtain adequate numbers of CD cases reflecting different disease locations and types of complications. Likewise, it will be important to determine whether UC is characterized by a distinct set of circulating miRNAs, because these may be useful in distinguishing UC from colitis caused by CD. It will also be essential to analyze circulating miRNA levels in patients with gastrointestinal complaints not caused by CD (eg, infectious, allergic, or functional disease) because this is the group in whom noninvasive testing can be most useful. The analysis of other inflammatory conditions, such as rheumatoid arthritis, can also help establish the specificity of the CD-associated circulating miRNAs, although in practice, other clinical features can also serve to distinguish CD and nongastrointestinal disease.
In summary, this pilot study has identified a number of miRNAs significantly increased in the serum of patients with pediatric CD These CD-associated miRNAs display encouraging clinical utility that will require confirmation in large validation groups. Our findings suggest that large-scale investigations combining circulating miRNAs, laboratory, and genetic markers of CD may result in composite models with improved sensitivity and specificity for IBD in general and CD in particular.
Supplementary Material
Acknowledgments
The authors wish to thank Dr Alessio Fasano and Ms Debby Kryszak for providing sera from patients with celiac disease and associated controls.
We are grateful to all of the members of the Friedman laboratory; the Division of Gastroenterology, Hepatology, and Nutrition; those who have supported the Center for Pediatric IBD at Children’s Hospital of Philadelphia, and most of all, the patients who generously agreed to participate in research on IBD.
The study was supported by NIH R01DK079881, NIH K23DK082012, NIH R01KD60030, NIH K24 DK076808, and the Crohn’s and Colitis Foundation of America.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
The authors report no conflicts of interest.
REFERENCES
- 1.Dotan I. Serologic markers in inflammatory bowel disease: tools for better diagnosis and disease stratification. Exp Rev Gastroenterol Hepatol. 2007;1:265–274. doi: 10.1586/17474124.1.2.265. [DOI] [PubMed] [Google Scholar]
- 2.Koutroubakis IE, Petinaki E, Mouzas IA, et al. Anti-Saccharomyces cerevisiae mannan antibodies and antineutrophil cytoplasmic autoantibodies in Greek patients with inflammatory bowel disease. Am J Gastroenterol. 2001;96:449–454. doi: 10.1111/j.1572-0241.2001.03524.x. [DOI] [PubMed] [Google Scholar]
- 3.Peeters M, Joossens S, Vermeire S, et al. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. Am J Gastroenterol. 2001;96:730–734. doi: 10.1111/j.1572-0241.2001.03613.x. [DOI] [PubMed] [Google Scholar]
- 4.Quinton JF, Sendid B, Reumaux D, et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791. doi: 10.1136/gut.42.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubinsky M. What is the role of serological markers in IBD? Pediatric and adult data. Dig Dis. 2009;27:259–268. doi: 10.1159/000228559. [DOI] [PubMed] [Google Scholar]
- 6.Vermeire S, Joossens S, Peeters M, et al. Comparative study of ASCA (Anti-Saccharomyces cerevisiae antibody) assays in inflammatory bowel disease. Gastroenterology. 2001;120:827–833. doi: 10.1053/gast.2001.22546. [DOI] [PubMed] [Google Scholar]
- 7.Reumaux D, Sendid B, Poulain D, et al. Serological markers in inflammatory bowel diseases. Best Pract Res Clin Gastroenterol. 2003;17:19–35. doi: 10.1053/bega.2002.0347. [DOI] [PubMed] [Google Scholar]
- 8.Anand V, Russell AS, Tsuyuki R, et al. Perinuclear antineutrophil cytoplasmic autoantibodies and anti-Saccharomyces cerevisiae antibodies as serological markers are not specific in the identification of Crohn’s disease and ulcerative colitis. Can J Gastroenterol. 2008;22:33–36. doi: 10.1155/2008/974540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desplat-Jego S, Johanet C, Escande A, et al. Update on Anti-Saccharomyces cerevisiae antibodies, anti-nuclear associated anti-neutrophil antibodies and antibodies to exocrine pancreas detected by indirect immunofluorescence as biomarkers in chronic inflammatory bowel diseases: results of a multicenter study. World J Gastroenterol. 2007;13:2312–2318. doi: 10.3748/wjg.v13.i16.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olives JP, Breton A, Hugot JP, et al. Antineutrophil cytoplasmic antibodies in children with inflammatory bowel disease: prevalence and diagnostic value. J Pediatr Gastroenterol Nutr. 1997;25:142–148. doi: 10.1097/00005176-199708000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Proujansky R, Fawcett PT, Gibney KM, et al. Examination of antineutrophil cytoplasmic antibodies in childhood inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 1993;17:193–197. doi: 10.1097/00005176-199308000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Damoiseaux JG, Bouten B, Linders AM, et al. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic antibodies for inflammatory bowel disease: high prevalence in patients with celiac disease. J Clin Immunol. 2002;22:281–288. doi: 10.1023/a:1019926121972. [DOI] [PubMed] [Google Scholar]
- 13.Freeman HJ. Perinuclear antineutrophil cytoplasmic antibodies in collagenous or lymphocytic colitis with or without celiac disease. Can J Gastroenterol. 1997;11:417–420. doi: 10.1155/1997/210856. [DOI] [PubMed] [Google Scholar]
- 14.Beattie RM, Walker-Smith JA, Murch SH. Indications for investigation of chronic gastrointestinal symptoms. Arch Dis Child. 1995;73:354–355. doi: 10.1136/adc.73.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poullis AP, Zar S, Sundaram KK, et al. A new, highly sensitive assay for C-reactive protein can aid the differentiation of inflammatory bowel disorders from constipation-and diarrhoea-predominant functional bowel disorders. Eur J Gastroenterol Hepatol. 2002;14:409–412. doi: 10.1097/00042737-200204000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Walker TR, Land ML, Kartashov A, et al. Fecal lactoferrin is a sensitive and specific marker of disease activity in children and young adults with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2007;44:414–422. doi: 10.1097/MPG.0b013e3180308d8e. [DOI] [PubMed] [Google Scholar]
- 17.Pfefferkorn MD, Boone JH, Nguyen JT, et al. Utility of fecal lactoferrin in identifying Crohn disease activity in children. J Pediatr Gastroenterol Nutr. 2010;51:425–428. doi: 10.1097/MPG.0b013e3181d67e8f. [DOI] [PubMed] [Google Scholar]
- 18.Joishy M, Davies I, Ahmed M, et al. Fecal calprotectin and lactoferrin as noninvasivemarkers of pediatric inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2009;48:48–54. doi: 10.1097/MPG.0b013e31816533d3. [DOI] [PubMed] [Google Scholar]
- 19.Tibble JA, Sigthorsson G, Foster R, et al. Use of surrogate markers of inflammation and Rome criteria to distinguish organic from nonorganic intestinal disease. Gastroenterology. 2002;123:450–460. doi: 10.1053/gast.2002.34755. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Russell I, Chen C. MicroRNA and stem cell regulation. Curr Opin Mol Ther. 2009;11:292–298. [PubMed] [Google Scholar]
- 21.Vasilatou D Papageorgiou S, Pappa V, et al. The role of microRNAs in normal and malignant hematopoiesis. Eur J Haematol. 2010;84:1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 22.Cai B, Pan Z, Lu Y. The roles of microRNAs in heart diseases: a novel important regulator. Curr Med Chem. 2010;17:407–411. doi: 10.2174/092986710790226129. [DOI] [PubMed] [Google Scholar]
- 23.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Yi R, Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 2010;17:229–235. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey AK, Agarwal P, Kaur K, et al. MicroRNAs in diabetes: tiny players in big disease. Cell Physiol Biochem. 2009;23:221–232. doi: 10.1159/000218169. [DOI] [PubMed] [Google Scholar]
- 27.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 29.Kato M, Arce L, Natarajan R. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol. 2009;4:1255–1266. doi: 10.2215/CJN.00520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;39:654–1664. doi: 10.1053/j.gastro.2010.07.040. 1664 e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a micro-RNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slaby O, Svoboda M, Michalek J, et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;35:624–635. e1624. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 35.Wu F, Zhang S, Dassopoulos T, et al. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–77. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 40.Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong X, Du Y, Wang G, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 44.Silva J, García V, Zaballos A, et al. Vesicle-related microRNAs in plasma of NSCLC patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 45.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 46.Wu F, Guo NJ, Tian H, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thayu M, Denson LA, Shults J, et al. Determinants of changes in linear growth and body composition in incident pediatric Crohn’s disease. Gastroenterology. 2010;139:430–438. doi: 10.1053/j.gastro.2010.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thayu M, Shults J, Burnham JM, et al. Gender differences in body composition deficits at diagnosis in children and adolescents with Crohn’s disease. Inflamm Bowel Dis. 2007;13:1121–1128. doi: 10.1002/ibd.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubner SE, Shults J, Baldassano J, et al. Longitudinal assessment of bone density and structure in an incident cohort of children with Crohn’s disease. Gastroenterology. 2009;136:123–130. doi: 10.1053/j.gastro.2008.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. doi: 10.1136/gut.2005.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyams JS, Ferry GD, Mandel FS, et al. Development and validation of a pediatric Crohn’s disease activity index. J Pediatr Gastroenterol Nutr. 1991;12:439–447. [PubMed] [Google Scholar]
- 52.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 53.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 54.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kan T, Sato F, Ito T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uren AG, Kool J, Matentzoglu K, et al. Large-scale mutagenesis in p19(ARF)-and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan KS, Armugam A, Sepramaniam S, et al. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cekaite L, Clancy T, Sioud M. Increased miR-21 expression during human monocyte differentiation into DCs. Front Biosci (Elite Ed) 2010;2:818–828. doi: 10.2741/e143. [DOI] [PubMed] [Google Scholar]
- 61.Silva MA, Lopez CB, Riverin F, et al. Characterization and distribution of colonic dendritic cells in Crohn’s disease. Inflamm Bowel Dis. 2004;10:504–512. doi: 10.1097/00054725-200409000-00003. [DOI] [PubMed] [Google Scholar]
- 62.Davidson LA, Wang N, Shah MS, et al. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30:2077–2084. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruemmele FM, Targan SR, Levy G, et al. Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology. 1998;115:822–829. doi: 10.1016/s0016-5085(98)70252-5. [DOI] [PubMed] [Google Scholar]
- 64.Hoffenberg EJ, Fidanza S, Sauaia A. Serologic testing for inflammatory bowel disease. J Pediatr. 1999;134:447–452. doi: 10.1016/s0022-3476(99)70202-7. [DOI] [PubMed] [Google Scholar]
- 65.Sabery N, Bass D. Use of serologic markers as a screening tool in inflammatory bowel disease compared with elevated erythrocyte sedimentation rate and anemia. Pediatrics. 2007;19:e193–e199. doi: 10.1542/peds.2006-1361. [DOI] [PubMed] [Google Scholar]
- 66.Teml A, Kratzer V, Schneider B, et al. Anti-Saccharomyces cerevisiae antibodies: a stable marker for Crohn’s disease during steroid and 5-aminosalicylic acid treatment. Am J Gastroenterol. 2003;98:2226–2231. doi: 10.1111/j.1572-0241.2003.07673.x. [DOI] [PubMed] [Google Scholar]
- 67.Desir B, Amre DK, Lu SE, et al. Utility of serum antibodies in determining clinical course in pediatric Crohn’s disease. Clin Gastroenterol Hepatol. 2004;2:139–146. doi: 10.1016/s1542-3565(03)00321-5. [DOI] [PubMed] [Google Scholar]
- 68.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissuespecific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 69.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto Y, Kosaka N, Tanaka M, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–538. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 71.Afzal NA, Van Der Zaag-Loonen HJ, Arnaud-Battandier F, et al. Improvement in quality of life of children with acute Crohn’s disease does not parallel mucosal healing after treatment with exclusive enteral nutrition. Aliment Pharmacol Ther. 2004;20:167–172. doi: 10.1111/j.1365-2036.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- 72.Efthymiou A, Viazis N, Mantzaris G, et al. Does clinical response correlate with mucosal healing in patients with Crohn’s disease of the small bowel? A prospective, case-series study using wireless capsule endoscopy. Inflamm Bowel Dis. 2008;14:1542–1547. doi: 10.1002/ibd.20509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.