Abstract
The epidemiological and evolutionary dynamics of the two cocirculating lineages of influenza B virus, Victoria and Yamagata, are poorly understood, especially in tropical or subtropical areas of Southeast Asia. We performed a phylogenetic analysis of the hemagglutinin (HA) and neuraminidase (NA) sequences of influenza B viruses isolated in Guangzhou, a southern Chinese city, during 2009 to 2010 and compared the demographic and clinical features of infected patients. We identified multiple viral introductions of Victoria strains from both Chinese and international sources, which formed two phylogenetically and antigenically distinct clades (Victoria 1 and 2), some of which persisted between seasons. We identified one dominant Yamagata introduction from outside China during 2009. Our phylogenetic analysis reveals the occurrence of reassortment events among the Victoria and Yamagata lineages and also within the Victoria lineage. We found no significant difference in clinical severity by influenza B lineage, with the exceptions that (i) the Yamagata lineage infected older people than either Victoria lineage and (ii) fewer upper respiratory tract infections were caused by the Victoria 2 than the Victoria 1 clade. Overall, our study reveals the complex epidemiological dynamics of different influenza B lineages within a single geographic locality and has implications for vaccination policy in southern China.
INTRODUCTION
Influenza B viruses are members of the family Orthomyxoviridae which are known to infect only humans and seals and which were first isolated in 1940 (1, 2). In contrast to influenza A viruses, influenza B viruses are not divided into subtypes based on surface glycoproteins (hemagglutinin [HA] and neuraminidase [NA]) but rather are classified into two phylogenetically and antigenically distinct lineages: the B/Victoria/2/87-like (Victoria) lineage and the B/Yamagata/16/88-like (Yamagata) lineage (3–6). Both lineages currently circulate globally and evolve more slowly than human seasonal influenza A viruses, although reassortment between the Victoria and Yamagata lineages has frequently been documented (7–13).
The epidemiological and evolutionary characteristics of influenza B/Victoria and B/Yamagata lineages are complex and remain poorly understood, in part due to a lack of robust global sampling. The current understanding is that these two lineages have circulated or cocirculated during different time periods in different geographic regions. The Victoria and Yamagata lineages were first detected during the 1988-1989 influenza season, and they cocirculated worldwide from 1983 to 1990 (1, 4–6). In the 1990s, the Victoria lineage became less frequently isolated and was mainly restricted to East Asia (14, 15). The Victoria lineage is thought to have re-emerged in North America and Europe and spread worldwide around 2000 to 2002 (9, 14, 15). In China, both the Victoria and Yamagata lineages have been cocirculating for a substantial period of time (16–20), with more frequent dominance of the Yamagata lineage in the 1990s, especially in northern China (21, 22). In contrast, during 2004 to 2008, Victoria strains became widespread in China, including in Zhejiang province, in Hunan province, and in Beijing (17, 19, 23–25). In regions neighboring China, such as Hong Kong and Taiwan, both influenza B lineages were isolated during 2000 to 2010, with different lineages dominating in different years (13, 26, 27). Most studies of influenza B virus undertaken to date have concentrated on the surveillance of viral activity and have not fully addressed the persistence, patterns of regional migration, epidemiology, and clinical features of the different lineages of influenza B virus in a quantitative manner.
The disease burden of influenza B virus is significant, especially among children and young teenagers (26). For example, during 2003 to 2008, excessive mortality attributable to influenza B virus infections was estimated to be higher than that attributable to seasonal A/H3N2 or A/H1N1 virus in China (28). Influenza B virus has caused more hospitalizations than influenza A virus in postpandemic H1N1/09 seasons (20). Influenza B virus infections also caused around one-quarter of influenza-associated hospitalizations in Hong Kong during 2000 to 2010 and were responsible for the major proportion of influenza burden in Taiwan during 2004 to 2012 (26, 27).
To better understand the epidemiological and evolutionary dynamics of influenza B virus, we studied the phylogeny and clinical features of infections reported during March 2009 to August 2010 in Guangzhou City, a subtropical metropolis in Guangdong province in southern China. Located northwest of Hong Kong, Guangzhou is a key national transportation hub and the biggest financial center in southern China, with a population of more than 12 million and direct jurisdiction over 12 districts. We mapped the sequences of influenza B virus isolates sampled in Guangzhou to the global phylogeny of influenza B virus and explored the association between genetic variation and demographic and clinical features.
MATERIALS AND METHODS
Hospital-based acute respiratory infection surveillance in Guangzhou, southern China.
A hospital-based sentinel network was established in four districts in central Guangzhou (Baiyun, Yuexiu, Liwan, and Haizhu) to monitor acute respiratory diseases in 2008 (29). The participating sites for this study included seven clinics and departments in local hospitals (Fig. 1), which enrolled patients with acute respiratory infections or community-acquired pneumonia confirmed by chest X ray between March 2009 and August 2010. The clinical definition of acute respiratory infection was fever with a temperature greater than 37.3°C, with at least one respiratory symptom, such as cough, sore throat, coryza, or shortness of breath (29). At the time of enrollment, the demographic and clinical characteristics of each subject were recorded, including gender, age, and symptoms. Approval was obtained from the ethics committee of all participating hospitals, and informed consent was obtained for all patients.
Fig 1.
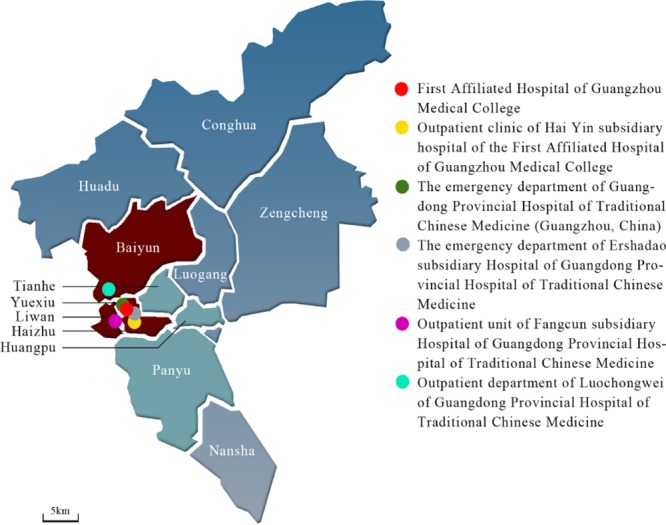
Map of Guangzhou City, China, and study sites. The Baiyun, Yuexiu, Liwan, and Haizhu districts in central Guangzhou City are in red. Other districts are in blue or green. Six study sites are shown as circles with different colors and described in the key.
Collection of clinical specimens, virus culture, and sequencing.
Nasopharyngeal and/or throat swab samples were collected from 205 subjects with acute respiratory infection or community-acquired pneumonia who were admitted to sentinel hospitals. Samples were stored at −80°C for further virus isolation and identification. Influenza B virus infection was identified and genotyped by real-time PCR following the protocol for influenza B virus RNA identification (PCR fluorescence probing diagnostic kits; Da An Gene, Guangzhou, China). Positive specimens were cultured in Madin-Darby canine kidney (MDCK) cells for 5 to 7 days. Viral genomic RNA was extracted from cultured supernatants followed by one-step RT-PCR to amplify the hemagglutinin (HA) and neuraminidase (NA) genes. The PCR products were gel purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) and sequenced with an ABI Prism 3130xl automated sequencer (Applied Biosystems, Foster City, CA). Primers were described previously (29) and supplied by Takato Odagiri (Laboratory of Influenza Virus Surveillance Center for Influenza Virus Research, National Institute of Infectious Diseases, Tokyo, Japan).
Phylogenetic analysis.
We analyzed the 205 newly acquired full-length HA and NA sequences of human influenza B viruses collected in Guangzhou from March 2009 to August 2010 (GenBank accession numbers are given below). We also analyzed global “background” human influenza B virus HA and NA sequences, which were downloaded from the Influenza Virus Resource at the National Center for Biotechnology Information (GenBank) website (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) (30). We obtained a total of 2,950 HA and 1,120 NA sequences, which were aligned using the MUSCLE program in MEGA 5 (31) with manual adjustment. Maximum likelihood (ML) phylogenies of the global data sets were inferred by using the rapid tree-searching approach implemented in the RAxML (version 7.04) program (32) and employing the general-time-reversal (GTR) substitution model with a gamma-distributed rate parameter. A total of 200 independent searches for ML phylogenies were performed with different random maximum parsimony starting trees in RAxML, and the phylogeny with the highest likelihood score was selected. A total of 500 pseudoreplicates were generated, and bootstrapping analyses were carried out by a similar ML method implemented in RAxML.
To provide an antigenic context for our study, we downloaded additional background human influenza B virus HA and NA sequences with available collection dates (between 1 January 2008 and 31 December 2011) from the EpiFlu database of the Global Initiative on Sharing All Influenza Data (GISAID) (33). We used GISAID genetic sequences because these could be paired with antigenic information provided in periodic WHO influenza reports (34). Together with the newly acquired HA and NA sequences from Guangzhou, we obtained 301 Victoria HA sequences, 294 Victoria NA sequences, 115 Yamagata HA sequences, and 114 Yamagata NA sequences in total. In this case, ML phylogenies were inferred in PhyML v3.0 (35) using the general-time-reversal substitution model with gamma-distributed among-site rate variation and a portion of invariable sites (GTR+Γ+I) and with bootstrap analyses of 1,000 pseudoreplicate data sets.
Correlating demographic and clinical features of influenza B virus patients with virus genetic variation.
Demographic and clinical information was available for 182 of the 205 influenza B virus cases from Guangzhou with available sequence data. Comparisons of demographic and clinical features between the Victoria and Yamagata lineages were performed by an analysis of variance (ANOVA) and nonparametric Kruskal-Wallis test for quantitative characteristics and chi-square and Fisher's exacts test for categorical variables, respectively. A P value of <0.05 was considered statistically significant except when a Bonferroni correction was used to adjust for multiple comparisons.
The strength of association between phenotypes (age, gender, and various clinical classes; see below) and the viral phylogeny was determined using two phylogeny-trait association statistics available within the Bayesian tip association significance testing (BaTS) program (36): the parsimony score (PS) and the association index (AI). BaTS provides a statistical test of the null hypothesis that phenotypes are associated randomly with the underlying phylogeny. For this analysis, phylogenetic trees of the HA were estimated using Bayesian Markov chain Monte Carlo methods available in the BEASTv1.6.1 package (37) and assuming a relaxed (uncorrelated lognormal) molecular clock (38) with a GTR+Γ+I substitution model. Bayesian analyses were run for 5 × 108 steps, sampling trees were done every 5 × 104 steps, and the first 1,000 trees were discarded as burn-in. To determine whether there was phylogenetic clustering by age and gender of the patient, we grouped viruses into three age classes (<15, 16 to 30, and >30 years old) and by gender, respectively. To determine whether there was phylogenetic clustering by clinical features, each patient was classified as positive or negative for each clinical symptom individually: upper respiratory tract infection, dizziness or headache, myalgia or bone pain, physical discomfort and hypodynamia, rhinitis, itching or sore throat, dry cough without phlegm, expectoration, and gastrointestinal symptoms. Body temperature was grouped into three classes: <38°C, 38.1 to 39.0°C, and >39.1°C.
Nucleotide sequence accession numbers.
All 205 HA sequences and 205 NA sequences generated here have been submitted to GenBank and assigned accession numbers KC986405 to KC986814.
RESULTS
Influenza virus activity in Guangzhou, 2009 to 2010.
During the study period, March 2009 to August 2010, notable waves of influenza B viruses were observed in Guangzhou City before and after the emergence of the influenza A H1N1/09 pandemic virus (Fig. 2A). Seasonal influenza A and influenza B viruses cocirculated in the spring and summer of 2009 before the pandemic H1N1/09 virus was introduced into the city. Pandemic H1N1/09 replaced seasonal influenza A viruses by September 2009 and dominated the epidemic for the rest of that year.
Fig 2.
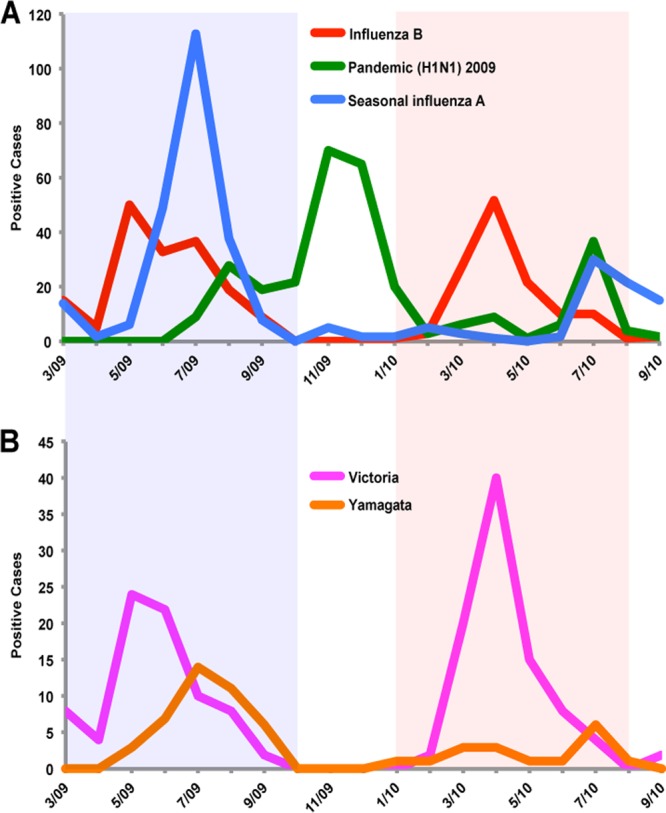
Influenza virus activity in Guangzhou City from April 2009 to September 2010. (A) Monthly number of laboratory confirmed influenza infection by type: influenza B virus in red, pandemic H1N1/2009 in green, and seasonal influenza A virus in blue. (B) Monthly number of influenza B virus infections by the Victoria (B/Victoria/2/87) and Yamagata (B/Yamagata/16/88) lineages. The 2009 and 2010 periods of influenza B virus epidemic activity are shaded in light blue and pink.
Based on epidemiological patterns (Fig. 2), we defined two separate influenza B virus epidemics, the first from March 2009 to October 2009 and the second from January 2010 to August 2010. Laboratory diagnosis showed that both the Victoria and Yamagata lineages cocirculated in the 2009 influenza B virus epidemic (Victoria represented 78 of 119 [66%] influenza B viruses), while Victoria viruses largely dominated the influenza B virus epidemic in 2010 (91 of 108 [84%] viruses) (Fig. 2B).
Distinct epidemiological dynamics of influenza B Victoria and Yamagata lineages.
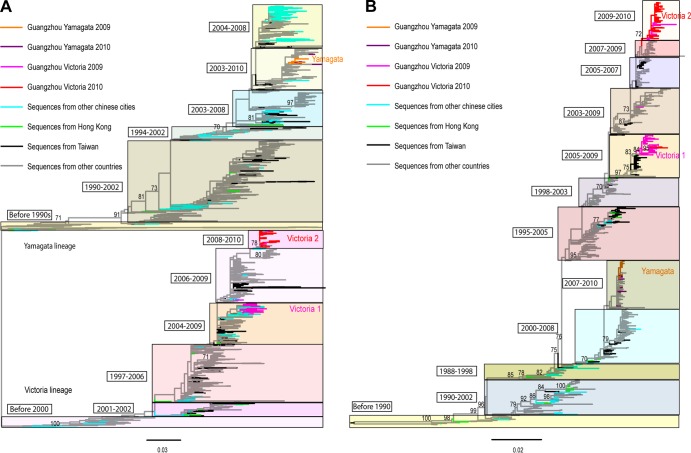
The phylogenetic analyses of influenza B virus HA (Fig. 3A) and NA (Fig. 3B) sequences revealed that distinct viral lineages circulate in Guangzhou, indicative of multiple introductions into the region. The number of independent introductions was estimated from the phylogenetic tree as Guangzhou strains separated by viruses from other locations, or by strains associated with particularly long branches, as well as singleton lineages (39–41).
Fig 3.
Global phylogenetic trees of the HA gene and NA gene of influenza B virus. (A) HA phylogeny, midpoint rooted; (B) NA phylogeny, rooted by the oldest sequence. The bootstrap support values for key branches in the trees are indicated. Sequences from different localities have their branches indicated by colors described in the key.
Yamagata lineage (HA phylogenetic tree).
Yamagata viruses isolated in 2009 in Guangzhou showed little phylogenetic distinction; they were closely related to each other and to sequences from the United States and Germany but more distant from sequences of Chinese and Taiwanese origins. This suggests a viral introduction into Guangzhou from outside China (although this picture may change with a greater sampling of Chinese sequences). In addition, one sequence was separated from other Guangzhou 2009 sequences by a sequence from Germany, indicating another independent introduction from outside China. Hence, there were at least two independent sources from outside China linked with the Yamagata epidemic in Guangzhou in 2009, one of which seemingly generated most of the infections.
Of the nine Yamagata viruses sampled from the 2010 epidemic, six belonged to the large cluster of Yamagata viruses isolated in 2009, suggesting that this lineage persisted between epidemics. The remaining three Yamagata 2010 viruses were phylogenetically distinct from the 2009 cluster and closely related to sequences from the United States, Germany, and Shanghai, China, separately. Hence, there have been at least three independent introductions of the Yamagata lineage from both international and Chinese sources into Guangzhou in 2010, but they did not cause a widespread epidemic, since the Victoria lineage dominated in 2010.
Victoria lineage (HA phylogenetic tree).
We observed two large phylogenetically distinct Victoria lineages circulating in Guangzhou in 2009 to 2010, defined as the Victoria 1 (V1) and Victoria 2 (V2) lineages, respectively (Fig. 3). Of the 78 Victoria viruses isolated in 2009, 52 (67%) fell into the V1 lineage. V1 was closely related to viruses sampled from other cities in the same province (Zhuhai and Shenzhen) and three other Chinese provinces further east (Shanghai, Ningbo, and Nanchang). Chinese sequences in the V1 lineage were phylogenetically distinct from sequences from outside China, indicating that the V1 epidemic in Guangzhou was linked with viruses locally circulating in southern China. In contrast, two Guangzhou V1 sequences clustered with sequences from Shenzhen (in southern China) and the United States, suggesting independent introductions into Guangzhou and (infrequent) virus movements between these places.
Twenty-six of the Victoria 2009 viruses belonged to a different lineage, V2, that included sequences from many countries, including China, the United States, South Korea, Lebanon, Japan, Guam, and Mongolia. In contrast to the cocirculating V1 and V2 lineages in 2009, all 2010 Victoria viruses fell into the V2 lineage and had a lower genetic diversity than viruses in the V1 lineage.
NA phylogenetic trees.
Although there were some topological differences between the HA and NA phylogenetic trees, they presented a broadly similar picture of Yamagata and Victoria lineages in 2009 and 2010 (Fig. 3B). Interestingly, in contrast to the HA analysis, three Victoria NA sequences (two V1 and one V2, all from 2009) clustered with the Yamagata lineage and one Yamagata NA sequence from 2009 clustered with the Victoria lineage, indicative of interlineage reassortments. Similarly, some of the NA sequences of the V2 clade clustered with those of the V1 clade and hence were compatible with intralineage reassortments.
Antigenic characteristics of influenza B lineages and reassortment.
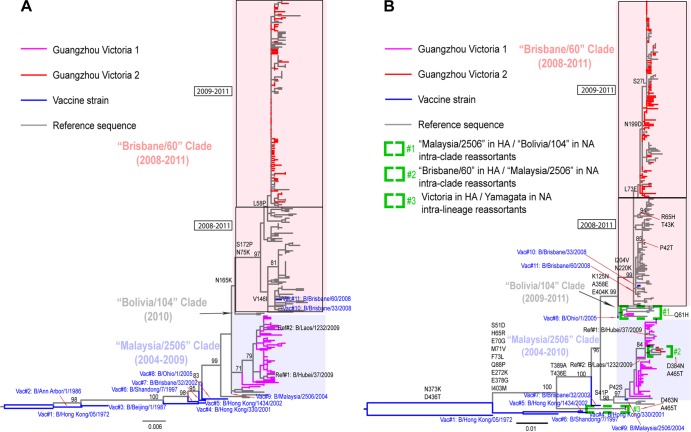
To clarify the antigenic difference between the V1 and V2 lineages, we explored the phylogenetic relationship between HA sequences from Guangzhou and sequences from reference strains that have been antigenically characterized (Fig. 4A). A phylogenetic analysis illustrated that the Guangzhou V2 viruses fell within the B/Brisbane/60/2008 (Brisbane/60) genetic clade, whereas the Guangzhou V1 viruses were more closely related to the older B/Malaysia/2506/2004 virus. According to the interim report from the WHO (34), the B/Brisbane/60/2008 clade is characterized by the HA amino acid substitutions N75K, N165K, and S172P, relative to the B/Malaysia/2506/2004 clade. Compared with the original B/Brisbane/60/2008 virus, the most recently collected viruses in the V1 clade, including all Guangzhou viruses, carry the additional amino acid substitutions I146V and L58P (Fig. 4A).
Fig 4.
Phylogenetic trees of the HA and NA genes of the Victoria lineage of influenza B virus. (A) HA phylogeny; (B) NA phylogeny. The trees are rooted by the oldest vaccine strain, B/Hong Kong/05/1972, and bootstrap support values and major amino acid substitutions are mapped to key branches. Intraclade and intralineage reassortants are indicated as described in the key. The Brisbane/60 (pink) and the Malaysia/2506-like (light blue) clades are indicated. Between the Brisbane/60 and Malaysia/2506 clades, two South American viruses—B/Bolivia/104/2010 and B/Bolivia/1526/2010—represent a new lineage, falling close to the recently emerged clade represented by B/Bolivia/104/2010 (Bolivia/104 clade).
In hemagglutination inhibition (HI) assays (34), all reference viruses in the V2 clade reacted well with antisera raised against B/Brisbane/60/2008 vaccine strain, or B/Brisbane/60/2008-like strains (B/Paris/1762/2009, B/Hong Kong/514/2009, and B/Odessa/3886/2010). Two reference viruses phylogenetically close to the Guangzhou V1 clade, B/Hubei/37/2009 and B/Laos/1232/2009, were antigenically close to the older vaccine strain B/Malaysia/2506/2004 and the B/Malaysia/2506/2004-like strain (B/Hong Kong/45/2005). Overall, these data suggest that Guangzhou V1 and V2 are antigenically different and represent variants from the B/Malaysia/2506/2004-like and B/Brisbane/60/2008-like lineages, respectively.
A phylogeny of the NA gene in the Victoria lineage is presented in Fig. 4B. The majority of NA sequences clustered in positions similar to those observed in the HA phylogeny. The exceptions are three reassortants: two interclade reassortants (one with the Malaysia/2506 HA and the Bolivia/104 NA and one with the Brisbane/60 HA and the Malaysia/2506 NA) and one interlineage reassortant (HA from the Victoria lineage and NA from the Yamagata lineage).
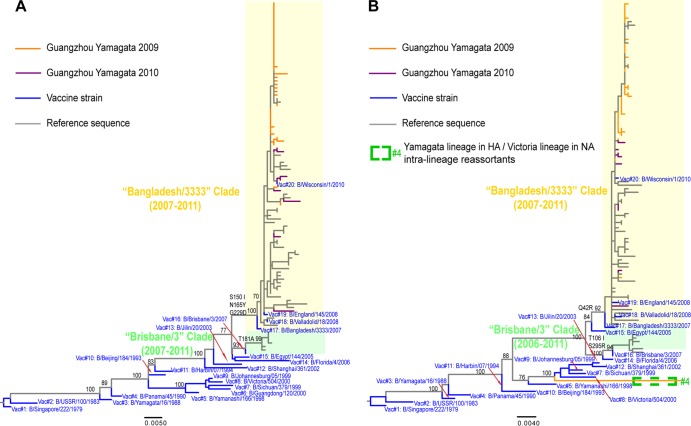
Phylogenetic analysis of the HA gene of the Yamagata lineage revealed that all of the Guangzhou viruses belonged to the B/Bangladesh/3333/2007 reference strain (Fig. 5A). According to HI assay results from the WHO interim report (34), all reference viruses in the B/Bangladesh/3333/2007 clade showed cross-reactivity with antisera raised against B/Bangladesh/3333/2007-like viruses (including B/Algeria/G-486 and the new vaccine strain B/Wisconsin/1/2010). Hence, these data suggest that the Guangzhou Yamagata lineage belongs to the B/Bangladesh/3333/2007-like antigenic group. Similar to viruses of the B/Victoria lineage, the Guangzhou B/Yamagata lineage retained the B/Yamagata NA segment, with the exception of one virus collected in 2010, which was a reassortant between a Yamagata lineage in the HA and a Victoria lineage in the NA (Fig. 5B).
Fig 5.
Phylogenetic trees of the HA and NA genes of the Yamagata lineage of influenza B virus. (A) HA phylogeny; (B) NA phylogeny. The trees are rooted by the oldest vaccine strain, B/Singapore/222/1979, and bootstrap support values and major amino acid substitutions are mapped to the key branches. Intralineage reassortant is indicated as described in the key. The Bangladesh/3333-like (light yellow) and Brisbane/3-like (light green) clades are indicated.
Correlation of demographic and clinical features with viral genetic variation.
The lineage-specific demographic and clinical features of influenza B virus patients are shown in Table 1. Strikingly, patients infected with Yamagata lineage viruses were significantly older than patients infected by either Victoria lineage viruses (median [interquartile range {IQR}], 32 [26 to 47] years for Yamagata versus 23 [17 to 29] for Victoria; P < 0.001, by a nonparametric Kruskal-Wallis test). Similarly, a higher proportion of adults over 31 years of age were infected by the Yamagata lineage than the Victoria lineage (56% versus 18 to 21%; P < 0.001, Fisher's exact test). In contrast, we found no significant difference in gender ratio, temperature, and clinical symptoms of patients infected by different viral lineages, except for fewer upper respiratory tract infections in V2 patients (P = 0.003, chi-square test). However, care must be taken in interpretation because of multiple testing issues. Furthermore, demographic and clinical features were not correlated with infection with the reassortant viruses (data not shown).
Table 1.
Clinical and demographic characteristics of patients with influenza B viruses in Guangzhou
| Characteristics | Value for lineage |
P | ||
|---|---|---|---|---|
| V1 (n = 45) | V2 (n = 94) | Yamagata (n = 43) | ||
| Demographic features | ||||
| Median age, in yrs (IQR)a | 23 (17–29) | 23 (17–29) | 32 (26–47) | <0.05 |
| No. (%) of patients <15 yrs oldb | 6 (13.3) | 14 (14.9) | 0 (0) | <0.001c |
| No. (%) of patients 16–30 yrs old | 31 (68.9) | 60 (63.8) | 19 (44.2) | |
| No. (%) of patients >31 yrs oldb | 8 (17.8) | 20 (21.3) | 24 (55.8) | |
| No. (%) of patients that were male | 19 (42.2) | 43 (45.7) | 22 (51.2) | >0.05 |
| Clinical symptoms | ||||
| Temp (°C),d mean ± SD | 38.60 ± 0.53 | 38.64 ± 0.56 | 38.38 ± 0.50 | >0.05 |
| No. (%) of patients <38.0°Ce | 12 (26.7) | 14 (14.9) | 12 (27.9) | >0.05 |
| No. (%) of patients 38.1–39.0°C | 28 (62.2) | 59 (62.8) | 26 (60.5) | |
| No. (%) of patients >39.1°C | 5 (11.1) | 21 (22.3) | 5 (11.6) | |
| No. (%) of patients with: | ||||
| Upper respiratory tract infection | 42 (93.3) | 64 (68.1) | 35 (81.4) | 0.003f |
| Dizziness or headache | 31 (68.9) | 68 (72.3) | 30 (69.8) | >0.05 |
| Myalgia or bone pain | 24 (53.3) | 55 (58.5) | 26 (60.5) | >0.05 |
| Physical discomfort and hypodynamia | 27 (60.0) | 65 (69.1) | 30 (69.8) | >0.05 |
| Rhinitis: nasal congestion, rhinorrhea, and sneezing | 19 (42.2) | 52 (55.3) | 30 (69.8) | 0.03f |
| Throat itching or sore throat | 32 (71.1) | 64 (68.1) | 34 (79.1) | >0.05 |
| Dry cough without phlegm | 17 (37.8) | 23 (24.5) | 19 (44.2) | 0.047f |
| Expectoration | 15 (33.3) | 48 (51.1) | 20 (46.5) | >0.05 |
| Gastrointestinal symptoms | 3 (6.7) | 10 (10.6) | 2 (4.7) | >0.05 |
Nonnormal distributed data are presented by median age and interquartile range (IQR). P value is calculated by the non-parametric Kruskal-Wallis test.
In the age group <15 years old, there was one child younger than 5 years old (3 months) infected with virus of the V2 lineage. In the age group >30 years old, four patients are older than 65; two were infected with virus of the V1 lineage, one had the V2 lineage, and one had the Yamagata lineage.
At least one of the expected values is smaller than 5. The P value was calculated by the Fisher exact test.
Temperature under the arm (axillary), normal distributed data. The P value was calculated by one-way ANOVA.
In the group that had temperatures less than 38°C, two patients had temperatures less than 37.3°C, one infected with the V2 lineage and one with the Yamagata lineage.
The Bonferroni correction was used for multiple comparisons (9 times). P values less than 0.0055 (0.05/9) are considered statistically significant.
To obtain a more quantitative measure of the possible association between demographic and clinical features and virus genetic variation, we used two phylogeny-trait association statistics, AI and PS. These analyses confirmed that Guangzhou influenza B viruses clustered by age and presence of upper respiratory tract symptoms of the patients they infected more strongly than by chance alone (P ≤ 0.007) (Table 2). In addition, this analysis revealed significant clustering by patient gender and gastrointestinal tract symptoms, based on both clustering statistics (P ≤ 0.007) (Table 2).
Table 2.
Results of the phylogeny-trait association test for different lineages of influenza B sampled in Guangzhou, southern Chinaa
| Comparison | Statisticb | Value (95% CIc) |
P | |
|---|---|---|---|---|
| Observed | Null | |||
| Gender of patient | AI | 7.99 (6.70–9.39) | 9.60 (8.41–10.71) | 0.012 |
| PS | 55.83 (51.0–60.0) | 60.50 (56.21–64.41) | 0.041 | |
| Age of patient | AI | 8.91 (7.74–10.12) | 10.50 (9.44–11.52) | 0.006 |
| PS | 55.14 (52.0–58.0) | 63.34 (60.10–66.08) | <0.001 | |
| Temp | AI | 10.88 (9.48–12.18) | 10.56 (9.53–11.54) | 0.694 |
| PS | 61.50 (59.0–64.9) | 62.70 (60.06–65.00) | 0.300 | |
| Upper respiratory tract infection | AI | 5.33 (4.23–6.40) | 6.68 (5.80–7.56) | 0.007 |
| PS | 32.51 (29.0–36.0) | 37.43 (35.26–39.17) | 0.001 | |
| Dizziness and headache | AI | 8.70 (7.46–9.87) | 7.93 (6.88–8.93) | 0.903 |
| PS | 47.49 (45.0–50.0) | 46.24 (43.12–48.66) | 0.859 | |
| Myalgia and bone pain | AI | 8.98 (7.60–10.37) | 9.32 (8.16–10.50) | 0.277 |
| PS | 57.63 (54.0–61.0) | 58.64 (54.53–62.43) | 0.372 | |
| Physical discomfort and hypodynamia | AI | 7.90 (6.61–9.13) | 8.51 (7.46–9.54) | 0.181 |
| PS | 49.51 (46.0–53.0) | 50.85 (47.61–53.53) | 0.299 | |
| Rhinitis: nasal congestion, rhinorrhea, and sneezing | AI | 8.98 (7.74–10.30) | 9.53 (8.41–10.64) | 0.196 |
| PS | 54.11 (51.0–58.0) | 59.79 (55.66–63.63) | 0.017 | |
| Itchy and sore throat | AI | 6.86 (5.76–8.09) | 7.78 (6.78–8.78) | 0.054 |
| PS | 42.87 (40.0–46.0) | 45.51 (42.98–47.93) | 0.052 | |
| Dry cough without phlegm | AI | 7.79 (6.55–8.96) | 8.47 (7.37–9.49) | 0.158 |
| PS | 47.57 (44.0–50.0) | 50.24 (46.95–52.83) | 0.120 | |
| Expectoration | AI | 8.81 (7.46–10.22) | 9.58 (8.40–10.74) | 0.147 |
| PS | 57.67 (54.0–62.0) | 60.34 (56.27–64.06) | 0.169 | |
| Gastrointestinal tract symptoms | AI | 2.11 (1.52–2.74) | 2.86 (2.30–3.40) | 0.007 |
| PS | 12.46 (12.0–13.0) | 14.61 (13.82–14.96) | 0.001 | |
Statistically significant results are in bold.
AI, association index; PS, parsimony score.
CI, confidence interval.
DISCUSSION
Our study of influenza B virus activity in Guangzhou, a subtropical city in southern China, revealed two distinct epidemic periods: March to October in 2009 (i.e., before the 2009 influenza A/H1N1 pandemic) and January to August 2010 (post-2009 pandemic). The influenza B virus epidemic of 2009 was characterized by the cocirculation of the Victoria and Yamagata lineages, with multiple viral introductions from both Chinese and international sources. The 2010 epidemic was largely dominated by the Victoria lineage, although there was evidence of independent introductions of the Yamagata viruses that did not lead to sustained activity. Notably, through comparisons of our Victoria virus sequences with those of vaccine and reference strains of known antigenic characteristics, we found that two antigenically and phylogenetically different lineages of the Victoria virus circulated in Guangzhou City. There was also evidence of intra- and interlineage reassortment events. Finally, phylogenetic clustering methods revealed notable differences in the age distribution of influenza B virus cases, with the Yamagata lineage infecting older people than either of the Victoria lineages and fewer upper respiration infections in the Victoria 2010 epidemic.
Multiple introductions of seasonal influenza A/H3N2, A/H1N1, 2009 pandemic H1N1, and influenza B viruses have previously been reported on the scale of single epidemics, and recent large-scale phylogenetic studies suggest that both temperate and tropical/subtropical regions could be the sources of the next global epidemic (20, 40–44). In line with these findings, our 2-year study in Guangzhou City reveals multiple introductions of influenza B viruses from other Chinese cities and from abroad. In addition, our phylogenetic analysis shows evidence of local persistence of influenza B viruses between two consecutive epidemics in Guangzhou in 2009 to 2010, as well as the disappearance of a predominant lineage at the end of the 2009 epidemic.
In line with our results from Guangzhou, short-term persistence of influenza B viruses was reported in another large subtropical city of southern China, Shenzhen (20). Like Shenzhen, Guangzhou hosts a large number of migrant workers who come to work in Guangzhou in spring and return to their hometowns in winter before the Chinese New Year. Guangzhou is also well connected with other regions of China and hosts the biannual Chinese Export Commodities Fair, which may also contribute to the complex patterns of multiple introductions and persistence of influenza B virus. However, there is no evidence in our short-term 2009–2010 influenza B virus data that Guangzhou is a preferred focus of global emergence of influenza viruses.
Although located in a subtropical region, Guangzhou adopts the Northern Hemisphere recommendations for influenza vaccines, in line with other cities in southern China and Hong Kong (26). Vaccine coverage is still relatively low in Guangzhou, with an estimated 13 vaccine doses used per 100 population (45), while a pediatric study reported a 6.5% vaccination rate for pandemic H1N1/09 in a boarding school (46). The vaccine is administered around October-December in anticipation of the Northern Hemisphere winter influenza season, peaking in January or February. However, considering that influenza B virus activity peaks in late spring in Guangzhou (April-May), we suggest that it may be optimal to postpone the vaccination campaigns in Guangzhou until the beginning of the year to improve the effectiveness of influenza vaccination. Since the vaccine also contains antigens for influenza A viruses, the optimal vaccine strategy also depends on the timing of influenza A virus activity. While we do not have any information on the seasonality of influenza A virus circulation in Guangzhou, other Southern Chinese cities experience influenza A virus activity in late spring, which would justify the suggested vaccine strategy (20, 47, 48). Although the emergence of the A/H1N1 pandemic virus in 2009 could have affected the seasonality of influenza B virus circulation in our Guangzhou study, past laboratory surveillance data suggest that seasonal influenza virus typically circulates in warmer months in this region (20, 47, 48).
In addition to a suboptimal timing of vaccination, Guangzhou experienced a partial mismatch between circulating influenza B virus strains and vaccine strains during our short study period. In 2009, when the Victoria and Yamagata lineages cocirculated in Guangzhou City, the recommended influenza B virus vaccine strain for use in the 2008–2009 Northern Hemisphere season was the B/Florida/4/2006-like Yamagata virus. Not only did the vaccine not cover the Victoria lineage (which represented 66% of the viruses isolated in Guangzhou in 2009), but also the vaccine was mismatched with respect to the Yamagata component. Indeed, our data suggest that the 2009 Yamagata viruses circulating in Guangzhou were antigenically close to the B/Wisconsin/1/2010 strain (B/Bangladesh/3333/2007-like virus), which was not included in the vaccine until 2013. In contrast, in the following year the Northern hemisphere 2009-2010 vaccine recommendations included a Victoria virus antigen, B/Brisbane/60/2008-like, which matched well with the predominant V2 lineage circulating in Guangzhou in 2010. To resolve the dilemma of cocirculating influenza B lineages, the WHO has recently suggested the use of quadrivalent vaccines containing two influenza B lineages, which have been approved for use in the 2013 Southern Hemisphere influenza season. Our study also supports the use of quadrivalent vaccines in Guangzhou and other areas in Southern China.
It is well known that influenza B virus predominantly infects children and young teenagers, in contrast to influenza A virus (especially A/H3N2) (28, 49, 50). We observed additional and notable age differences between influenza B lineages, with Yamagata strains infecting an older proportion of the population than Victoria strains. This difference could in part reflect greater background population immunity to Victoria strains among adults, consistent with the widespread circulation of these viruses since the 1980s in Asia. It may also be that the Victoria and Yamagata lineages differ in their intrinsic transmissibility, as the higher transmissibility of Victoria strains would be expected to result in a younger mean age at infection. Although influenza B viruses seem to be transmitted with a higher reproductive number (R0) in younger age populations (50), lineage-specific transmissibility estimates are not available. It has been reported that Yamagata-infected children were 2 to 3 years older than Victoria-infected children in Taiwan, but that study considered a very small data set, i.e., children with only 8 Victoria infections in total (51).
We did not find significant clinical differences by influenza B lineage, with the exception of fewer respiratory tract infections associated with the V2 lineage. Differences in the epidemiological characteristics and clinical features according to influenza type have been noted previously (52), but to our knowledge this is one of the few studies indicating further differences specific to the influenza B lineage. We note that the differences found here may not result in differences in severity and did not seem to be associated with specific point mutations (data not shown).
In Guangzhou, the pandemic A/H1N1 activity dominated during July 2009 to February 2010, with a peak in November 2009, between the two influenza B virus epidemics studied here. It is possible that cross-protective immunity from prior exposure to the pandemic virus may have reduced the severity of clinical symptoms in 2009, consistent with fewer upper respiratory infections in the 2010 epidemic, in which V2 was predominant. Furthermore, a well-matched influenza vaccine may have contributed to the mild clinical symptoms of the V2 epidemic in 2010.
We also observed a significant phylogenetic clustering by patient gender and presence of gastrointestinal symptoms. This needs careful interpretation. We have very limited data on gastrointestinal tract symptoms, and no association was reported between clinical symptoms and influenza genetic variation in previous influenza B virus and pandemic A/H1N1 studies (41, 53, 54). A better understanding of the association between demographic and clinical features and virus genetic variation will require additional data and long-term studies from other places.
In conclusion, this study highlights the complex epidemiological and evolutionary dynamics of influenza B virus. Although our findings rely on a relatively short surveillance period, they are indicative of interesting differences between lineages of influenza B viruses, potentially related to intrinsic differences in clinical presentation, transmissibility, or immunity of these viruses, which would be worthy of further investigation. More broadly, our data are also consistent with a meta-population model for the circulation of influenza B viruses, with multiple introductions into large and well-connected tropical Chinese cities and occasional local persistence between epidemics.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Combination Project of Guangdong Province from Guangdong Provincial Department of Science and Technology, China (grant no. 2010B091000018), The National Science and Technology Pillar Program during the 12th Five-year Plan Period from Ministry of Science and Technology, China (grant no. 2012BAI05B01), The Science Research Fund for Youth Scholar from First Affiliated Hospital of Guangzhou Medical School, China (grant no. 201220-gyfyy), and the Influenza Program of the Fogarty International Center, National Institutes of Health, which is funded by the Office of Global Affairs' International Influenza Unit, U.S. Department of Health and Human Services. E.C.H. is funded by an NHMRC Australia Fellowship.
We gratefully acknowledge the authors and the originating and submitting laboratories of the sequences from GISAID's EpiFlu Database on which this research is based. The sequences used are given in Table S1 in the supplemental material. All submitters of data may be contacted directly via the GISAID website (www.gisaid.org).
Yi Tan and Zifeng Yang carried out the study design; access: Yi Tan, Wenda Guan, and Zifeng Yang accessed data; Wenda Guan, Sihua Pan, Shiguan Wu, Yangqing Zhan, and Zifeng Yang carried out data collection and experiments; Yi Tan, Wenda Guan, and Tommy Tsan-Yuk Lam carried out data analysis; Yi Tan, Wenda Guan, Tommy Tsan-Yuk Lam, Cecile Viboud, Edward C. Holmes, and Zifeng Yang interpreted the data, and Yi Tan, Wenda Guan, Tommy Tsan-Yuk Lam, Cecile Viboud, Edward C. Holmes, and Zifeng Yang prepared the manuscript.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 11 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01039-13.
REFERENCES
- 1.Nerome R, Hiromoto Y, Sugita S, Tanabe N, Ishida M, Matsumoto M, Lindstrom SE, Takahashi T, Nerome K. 1998. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch. Virol. 143:1569–1583 [DOI] [PubMed] [Google Scholar]
- 2.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. 2000. Influenza B virus in seals. Science 288:1051–1053 [DOI] [PubMed] [Google Scholar]
- 3.Yamashita M, Krystal M, Fitch WM, Palese P. 1988. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 163:112–122 [DOI] [PubMed] [Google Scholar]
- 4.Kanegae Y, Sugita S, Endo A, Ishida M, Senya S, Osako K, Nerome K, Oya A. 1990. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 64:2860–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59–68 [DOI] [PubMed] [Google Scholar]
- 6.Rota PA, Hemphill ML, Whistler T, Regnery HL, Kendal AP. 1992. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J. Gen. Virol. 73:2737–2742 [DOI] [PubMed] [Google Scholar]
- 7.Krystal M, Young JF, Palese P, Wilson IA, Skehel JJ, Wiley DC. 1983. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc. Natl. Acad. Sci. U. S. A. 80:4527–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pechirra P, Nunes B, Coelho A, Ribeiro C, Gonçalves P, Pedro S, Castro LC, Rebelo-de-Andrade H. 2005. Molecular characterization of the HA gene of influenza type B viruses. J. Med. Virol. 77:541–549 [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Holmes EC. 2008. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 66:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo C, Morishita T, Satou K, Tateno Y, Nakajima K, Nobusawa E. 1999. Evolutionary pattern of influenza B viruses based on the HA and NS genes during 1940 to 1999: origin of the NS genes after 1997. Arch. Virol. 144:1881–1891 [DOI] [PubMed] [Google Scholar]
- 12.Matsuzaki Y, Sugawara K, Takashita E, Muraki Y, Hongo S, Katsushima N, Mizuta K, Nishimura H. 2004. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J. Med. Virol. 74:132–140 [DOI] [PubMed] [Google Scholar]
- 13.Chen GW, Shih SR, Hsiao MR, Chang SC, Lin SH, Sun CF, Tsao KC. 2007. Multiple genotypes of influenza B viruses cocirculated in Taiwan in 2004 and 2005. J. Clin. Microbiol. 45:1515–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw MW, Xu X, Li Y, Normand S, Ueki RT, Kunimoto GY, Hall H, Klimov A, Cox NJ, Subbarao K. 2002. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000–2001 and 2001–2002 seasons. Virology 303:1–8 [DOI] [PubMed] [Google Scholar]
- 15.Chi XS, Bolar TV, Zhao P, Rappaport R, Cheng SM. 2003. Cocirculation and evolution of two lineages of influenza B viruses in Europe and Israel in the 2001–2002 season. J. Clin. Microbiol. 41:5770–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi SX, Han GY, Liu YF. 2009. Virological surveillance and molecular characteristics of influenza B viruses in Hebei Province during 2004–2008. Zhongguo Yi Miao He Mian Yi 15:27–30 (In Chinese.) [PubMed] [Google Scholar]
- 17.Mao HY, Zhou M, Zhang YJ, Chen Y, Xu CP, Li Z, Yan JY, Lu YY. 2011. Genetic variation of the hemagglutinin and neuraminidase of influenza B viruses isolated in Zhejiang province during 1999–2010. Zhonghua Liu Xing Bing Xue Za Zhi 32:376–381 (In Chinese.) [PubMed] [Google Scholar]
- 18.Ren BZ, Wang NC, Feng JJ, Zhao R, Zhang FF. 2011. Analysis on the etiological surveillance of influenza/novel influenza A (H1N1) from 2009-2010 in Shanxi Province. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 25:20–22 (In Chinese.) [PubMed] [Google Scholar]
- 19.Liu YZ, Zhao X, Huang YW, Chen Z, Li FC, Gao LD, Li XY, Li WC, Hu SX, Tan MJ, Zhang HJ, Zhang H. 2012. Analysis of genetic features of influenza B virus in Hunan province from 2007 to 2010. Zhonghua Yu Fang Yi Xue Za Zhi 46:258–263 (In Chinese.) [PubMed] [Google Scholar]
- 20.Cheng X, Tan Y, He M, Lam TT, Lu X, Viboud C, He J, Zhang S, Lu J, Wu C, Fang S, Wang X, Xie X, Ma H, Nelson MI, Kung HF, Holmes EC, Cheng J. 2013. Epidemiological dynamics and phylogeography of influenza in southern China. J. Infect. Dis. 207:106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Guo Y, Guo J, Dong J. 2002. Characterization of HA1 gene of influenza B virus circulated in 1990 through 2000 in China. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 16:278–280 (In Chinese.) [PubMed] [Google Scholar]
- 22.Wu SH, Yu W, Zhang MM, Cui JQ, Fu RH, Zhao XG, He YH. 2006. Influenza surveillance from 1999 to 2005 in Liaoning regions. Zhonghua Liu Xing Bing Xue Za Zhi 27:238–240 (In Chinese.) [PubMed] [Google Scholar]
- 23.Mao HY, Lu YY, Yan JY. 2008. Molecular and antigenic characteristics of influenza B virus isolated in Zhejiang province in 2006. Zhonghua Liu Xing Bing Xue Za Zhi 29:413–414 (In Chinese.) [PubMed] [Google Scholar]
- 24.Zhang Y, Wen LY, Zhao X, Li Z, Guo JF, Xu CL, Wang M, Yu HJ, Yang WZ, Guo YJ, Shu YL. 2006. Antigenic and genetic study of influenza virus B circulated in China in 2004–2005. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 20:11–13 (In Chinese.) [PubMed] [Google Scholar]
- 25.Chen M, Peng XM, Wang XM. 2007. Analysis on the characteristics of HA1 gene of influenza virus B circulated in Beijing, 2005–2006. Zhonghua Liu Xing Bing Xue Za Zhi 28:308 (In Chinese.) [PubMed] [Google Scholar]
- 26.Chan PK, Chan MC, Cheung JL, Lee N, Leung TF, Yeung AC, Wong MC, Ngai KL, Nelson EA, Hui DS. 2013. Influenza B lineage circulation and hospitalization rates in a subtropical city, Hong Kong, 2000–2010. Clin. Infect. Dis. 56:677–684 [DOI] [PubMed] [Google Scholar]
- 27.Yang JR, Huang YP, Chang FY, Hsu LC, Lin YC, Huang HY, Wu FT, Wu HS, Liu MT. 2012. Phylogenetic and evolutionary history of influenza B viruses, which caused a large epidemic in 2011–2012, Taiwan. PLoS One 7:e47179. 10.1371/journal.pone.0047179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Shay DK, Jiang Y, Zhou H, Chen X, Zheng Y, Jiang L, Zhang Q, Lin H, Wang S, Ying Y, Xu Y, Wang N, Feng Z, Viboud C, Yang W, Yu H. 2012. Influenza-associated mortality in temperate and subtropical Chinese cities, 2003–2008. Bull. World Health Organ. 90:279–288B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang ZF, Zhan YQ, Chen RC, Zhou R, Wang YT, Luo Y, Jiang M, Li JQ, Qin S, Guan WD, Lai KF, Wen HL, Liang ZW, Li L, Zhong NS. 2012. A prospective comparison of the epidemiological and clinical characteristics of pandemic (H1N1) 2009 influenza A virus and seasonal influenza A viruses in Guangzhou, South China in 2009. Jpn. J. Infect. Dis. 65:208–214 [DOI] [PubMed] [Google Scholar]
- 30.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 33.GISAID Accessed 16 April 2013 EpiFluTM database. http://platform.gisaid.org/
- 34.MRC, National Institute for Medical Research Accessed 16 April 2013. Interim report from WHO Influenza Centre London, 2009–2011. WHO Influenza Centre, Medical Research Council (MRC), National Institutes for Medical Research, UK: http://www.nimr.mrc.ac.uk/who-influenza-centre/annual-and-interim-reports/ [Google Scholar]
- 35.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 36.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239–246 [DOI] [PubMed] [Google Scholar]
- 37.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Sengamalay NA, Spiro DJ, Volkov I, Grenfell BT, Lipman DJ, Taubenberger JK, Holmes EC. 2006. Stochastic processes are key determinants of short-term evolution in influenza A virus. PLoS Pathog. 2:e125. 10.1371/journal.ppat.0020125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson MI, Edelman L, Spiro DJ, Boyne AR, Bera J, Halpin R, Sengamalay N, Ghedin E, Miller MA, Simonsen L, Viboud C, Holmes EC. 2008. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 4:e1000133. 10.1371/journal.ppat.1000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes EC, Ghedin E, Halpin RA, Stockwell TB, Zhang XQ, Fleming R, Davey R, Benson CA, Mehta S, Taplitz R, Liu YT, Brouwer KC, Wentworth DE, Lin X, the INSIGHT FLU002 Study Group, Schooley RT. 2011. Extensive geographical mixing of 2009 human H1N1 influenza A virus in a single university community. J. Virol. 85:6923–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson MI, Tan Y, Ghedin E, Wentworth DE, St George K, Edelman L, Beck ET, Fan J, Lam TT, Kumar S, Spiro DJ, Simonsen L, Viboud C, Holmes EC, Henrickson KJ, Musser JM. 2011. Phylogeography of the spring and fall waves of the H1N1/09 pandemic influenza virus in the United States. J. Virol. 85:828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedford T, Cobey S, Beerli P, Pascual M. 2010. Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog. 6:e1000918. 10.1371/journal.ppat.1000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahl J, Nelson MI, Chan KH, Chen R, Vijaykrishna D, Halpin RA, Stockwell TB, Lin X, Wentworth DE, Ghedin E, Guan Y, Peiris JS, Riley S, Rambaut A, Holmes EC, Smith GJ. 2011. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc. Natl. Acad. Sci. U. S. A. 108:19359–19364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Ma S, Chen PY, He JF, Chan KP, Chow A, Ou CQ, Deng AP, Hedley AJ, Wong CM, Peiris JS. 2011. Influenza associated mortality in the subtropics and tropics: results from three Asian cities. Vaccine 29:8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Liu Y, Di B, Wang M, Shen J, Zhang Y, Chen X, Yuan J, Wu J, Li K, Lu E, Wu Y, Hao A, Chen X, Wang Y, Liu J, Pickerill S, Zheng B. 2011. Epidemiological investigation of an outbreak of pandemic influenza A (H1N1) 2009 in a boarding school: serological analysis of 1570 cases. J. Clin. Virol. 50:235–239 [DOI] [PubMed] [Google Scholar]
- 47.Shu YL, Fang LQ, de Vlas SJ, Gao Y, Richardus JH, Cao WC. 2010. Dual seasonal patterns for influenza, China. Emerg. Infect. Dis. 16:725–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du X, Dong L, Lan Y, Peng Y, Wu A, Zhang Y, Huang W, Wang D, Wang M, Guo Y, Shu Y, Jiang T. 2012. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat. Commun. 3:709. [DOI] [PubMed] [Google Scholar]
- 49.Olson DR, Heffernan RT, Paladini M, Konty K, Weiss D, Mostashari F. 2007. Monitoring the impact of influenza by age: emergency department fever and respiratory complaint surveillance in New York City. PLoS Med. 4:e247. 10.1371/journal.pmed.0040247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lunelli A, Rizzo C, Puzelli S, Bella A, Montomoli E, Rota MC, Donatelli I, Pugliese A. 2013. Understanding the dynamics of seasonal influenza in Italy: incidence, transmissibility and population susceptibility in a 9-year period. Influenza Other Respi. Viruses. 7:286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi CY, Wang SM, Lin CC, Wang HC, Wang JR, Su IJ, Liu CC. 2008. Clinical features of children infected with different strains of influenza B in southern Taiwan. Pediatr. Infect. Dis. J. 27:640–645 [DOI] [PubMed] [Google Scholar]
- 52.Yap J, Tan CH, Cook AR, Loh JP, Tambyah PA, Tan BH, Lee VJ. 2012. Differing clinical characteristics between influenza strains among young healthy adults in the tropics. BMC Infect. Dis. 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee RT, Santos CL, de Paiva TM, Cui L, Sirota FL, Eisenhaber F, Maurer-Stroh S. 2010. All that glitters is not gold—founder effects complicate associations of flu mutations to disease severity. Virol. J. 7:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li WC, Shih SR, Huang YC, Chen GW, Chang SC, Hsiao MJ, Tsao KC, Lin TY. 2008. Clinical and genetic characterization of severe influenza B-associated diseases during an outbreak in Taiwan. J. Clin. Virol. 42:45–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.