Abstract
Mitosis is a process involving a complex series of events that require careful coordination. Protein phosphorylation by a small number of kinases, in particular Aurora A, Aurora B, the cyclin-dependent kinase–cyclin complex Cdk1/cyclinB, and Polo-like kinase 1 (Plk1), orchestrates almost every step of cell division, from entry into mitosis to cytokinesis. To discover more about the functions of Aurora A, Aurora B, and kinases of the Plk family, we mapped mitotic phosphorylation sites to these kinases through the combined use of quantitative phosphoproteomics and selective targeting of kinase activities by small-molecule inhibitors. Using this integrated approach, we connected 778 phosphorylation sites on 562 proteins with these enzymes in cells arrested in mitosis. By connecting the kinases to protein complexes, we associated these kinases with functional modules. In addition to predicting previously unknown functions, this work establishes additional substrate-recognition motifs for these kinases and provides an analytical template for further use in dissecting kinase signaling events in other areas of cellular signaling and systems biology.
INTRODUCTION
Mitosis is a crucial step of the cell cycle that is tightly regulated by the spatial and temporal interplay of a wide array of proteins. For cells to divide successfully, a series of complex processes must occur in a timely and accurate manner, including DNA replication, condensation of chromosomes, maturation and separation of centrosomes, nuclear envelope breakdown, formation of a microtubule-based spindle lattice, sister chromatid separation and segregation, and cytokinesis. Errors in these processes are often the underlying cause of developmental defects and cancerous transformation and can be fatal for a cell. Although these processes are diverse and executed by a cadre of functional classes of proteins, posttranslational protein phosphorylation by a small group of serine-threonine kinases orchestrates many aspects of most steps from mitotic entry to exit.
The families of cyclin-dependent (Cdk), Aurora, and Polo-like (Plk) kinases have emerged as the primary regulators of cell division. Cdk1/cyclinB is widely regarded as the master regulator of mitosis and is responsible for entry into and progression through mitosis as well as mitotic exit (1, 2). In mammals, the family of Aurora kinases consists of three paralogs, Aurora A, B, and C (3), which share a high degree of sequence conservation in their kinase domains. Activation of the Aurora kinases occurs by multiple distinct mechanisms, including autophosphorylation of their T loops and protein binding (4). The interaction with certain proteins not only contributes to kinase activation but also governs the spatially and temporally distinct subcellular localization of the three family members. The localization of Aurora A to centrosomes and proximal microtubules is distinct from that of Aurora B and C, which bind to centromeres early in mitosis before translocating to the central spindle in anaphase and the midbody during cytokinesis. Aurora A has been implicated in the G2-M transition (5–9), centrosome maturation and separation, as well as the formation of a bipolar spindle (4). Aurora B promotes chromosome bi-orientation (chromosome attachment to opposite poles of a mitotic spindle), correction of syntelic (sister kinetochores attached to the same spindle pole) and merotelic (one kinetochore attached to both spindle poles) microtubule-kinetochore attachments, and is required for tension-dependent activation of the spindle assembly checkpoint (10–13). Furthermore, Aurora B promotes chromosome condensation, sister chromatid cohesion, and abscission at the end of cytokinesis (4). Besides a function in spermatogenesis (14, 15), Aurora C does not seem to have an essential role in somatic cells, and its function in cancer cells is still unclear.
The Plk family consists of four members in mammals (Plk1 to 4), of which Plk1 is the most widely studied with regard to its function in mitosis (16). Plk4 is involved in centriole duplication (17), whereas Plk2 and 3 seem to have interphase functions (18). All four Plk enzymes share a common structure, with an N-terminal kinase domain and a C-terminal Polo-box domain (PBD). Activation of Plks occurs by phosphorylation of their T loops (19). Aurora A is the primary upstream kinase responsible for phosphorylating the T loop at Thr210 of Plk1 in early mitosis (7, 9); Plk1 phosphorylation outside of the T loop may also be important for its activation in later stages of mitosis (20, 21). Plk1 contributes to mitotic entry by promoting Cdk1/cyclinB activation (22). Early in mitosis, Plk1 localizes to centrosomes; at later mitotic stages, Plk1 spreads along the spindle; in anaphase, Plk1 resides at the central spindle; during cytokinesis, Plk1 is present at the midbody. In prometaphase and metaphase, Plk1 can be found at kinetochores (16). Plk1 promotes centrosome maturation and spindle assembly and is required for the timely removal of cohesion and sister chromatid separation. By targeting substrates to the kinetochores, Plk1 is involved in the spindle assembly checkpoint and promotes stable microtubule-kinetochore attachments. Finally, Plk1’s role in mitotic exit includes activation of the anaphase-promoting complex/cyclosome (APC/C) and promotion of cytokinesis by the recruitment of proteins to the central spindle and midbody (23).
The abundance of these kinases is often increased in human cancers through various mechanisms, including gene amplification and degradation- resistant mutations, and these kinases promote malignant transformation in vivo and in vitro and are therefore attractive targets for cancer treatment (4, 24–26). Small-molecule inhibitor screens have discovered a panel of kinase inhibitors, many of which are undergoing evaluation in clinical trials, as well as their use as chemical probes to explore the biology of their respective targets. Hesperadin and ZM447439 are among the earliest examples of Aurora kinase inhibitors, which have proven to be invaluable tools in elucidating Aurora B biology (27, 28). Efforts to develop molecules with improved selectivity for Aurora A or B have led to the next-generation inhibitors MLN8054 (Aurora A) and AZD1152 (Aurora B) (29, 30). BI2536 is a Plk inhibitor, which has been shown in vitro to inhibit Plk1, 2, and 3, but reproduces cellular mitotic phenotypes consistent with Plk1 depletion by RNA interference (RNAi) (31).
Although extensive research on the Aurora and Polo kinases has revealed many biological processes in which they are involved, their precise molecular targets and sites of phosphorylation often remain elusive. We combined the specific nature of small-molecule kinase inhibitors with large-scale quantitative phosphoproteomics to identify 778 phosphorylation loci on 562 proteins from mitotic cells that are candidate targets of these kinases. Using this comprehensive approach, we observed cross-regulation between kinases, identified kinase-specific as well as common targets, interrogated the evolutionary conservation of these phosphorylation events, and established a broad-scale understanding of the regulatory functions of these kinases during cell division. We validated a subset of these sites in vitro and in HeLa cells and uncovered a potential mechanism by which Aurora A promotes spindle bipolarity through the site-specific phosphorylation of the protein NuMA.
RESULTS
Inhibition of Aurora A, Aurora B, and Plk1 by small-molecule inhibitors
We used a panel of small-molecule inhibitors to target specific kinases in mitotic HeLa cells and characterized their effects. We used the inhibitor MLN8054, which at lower concentrations is selective for Aurora A, but at higher concentrations also inhibits Aurora B. For Aurora B, we used two inhibitors, ZM447439 and AZD1152, and compared their specificities for Aurora B versus Aurora A. We used BI2536, which is selective for Plk1, Plk2, and Plk3 but cannot be used to distinguish among them. To determine the inhibitor concentration for maximal selective inhibition of the target kinase, we synchronized HeLa cells in S phase with a double-thymidine block, arrested the cells in metaphase with Taxol, and then added the inhibitors for 45 min in the presence of the proteasome-blocking agent MG132. We included MG132 to prevent protein degradation, which also prevents mitotic exit. Inhibition of protein degradation was essential in distinguishing decreases in protein phosphorylation due to inhibition of kinase activity from potential protein turnover. Control cells were synchronized with thymidine, arrested with Taxol, and treated with MG132, but were not exposed to inhibitors.
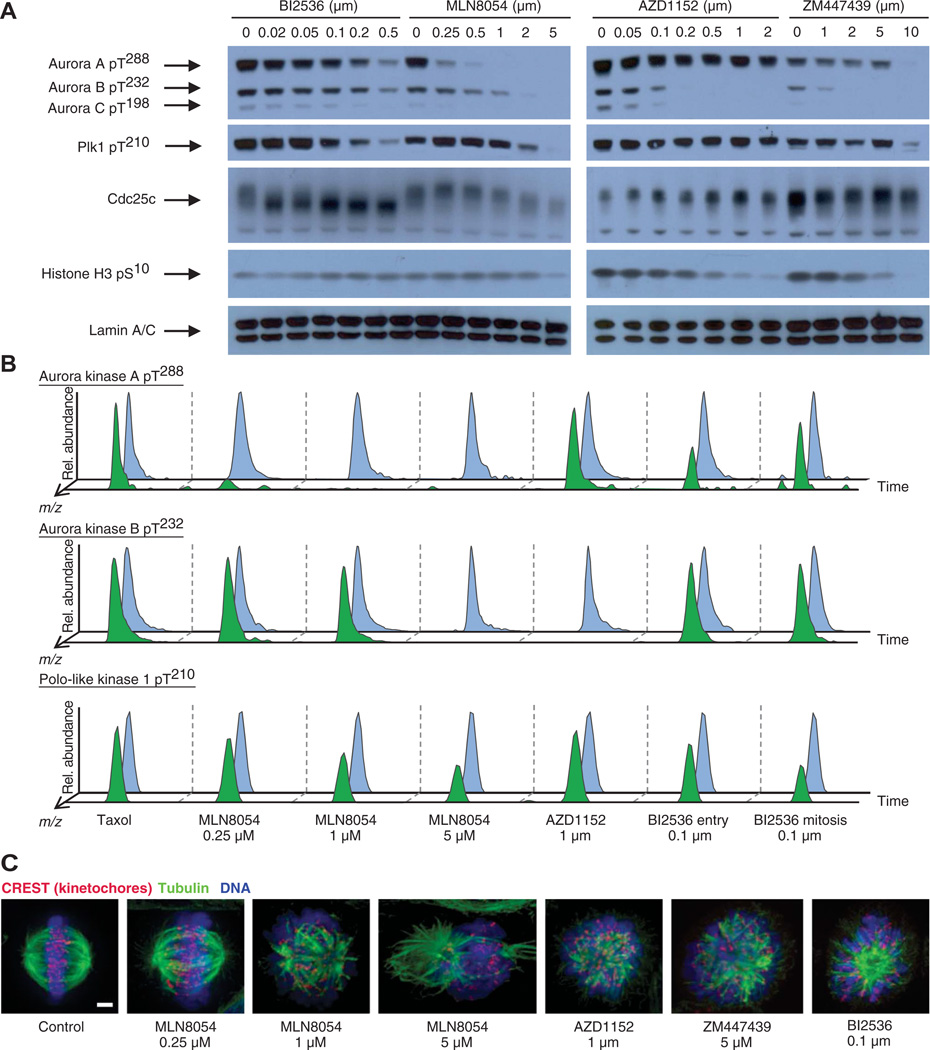
We monitored the phosphorylation status of the T loops of Aurora A, B, and C and Plk1 (an Aurora A substrate), as well as phosphorylation of the downstream targets Cdc25c (a known Plk1 substrate) and Ser10 on histone H3 (an Aurora B substrate), by Western blot analysis (Fig. 1A). On the basis of reduced electrophoretic mobility of Cdc25, a concentration of 100 nM BI2536 inhibited Plk activity. At higher concentrations, Plk1 T loop phosphorylation was reduced, along with reduced activity of Aurora A and B. This is consistent with the role of Plk1 in promoting the activation of Aurora A through phosphorylation of binding partners, and then Aurora A promotes the activation of Plk1 through phosphorylation at Thr210 (7, 9). On the basis of T loop phosphorylation of Aurora A and B, 1 µM MLN8054 inhibited Aurora A activity to a greater extent than it inhibited Aurora B activity. At an MLN8054 concentration of 5 µM, T loop phosphorylation of all three Aurora kinases, as well as phosphorylation of the T loop of Plk1, was undetectable. AZD1152 inhibited Aurora B autophosphorylation at concentrations as low as 100 nM, whereas Aurora A autophosphorylation was unchanged even in the presence of 2 µM AZD1152. At a concentration of 2 µM, ZM447439 appeared to completely inhibit autophosphorylation of Aurora B and C. Effective inhibition of phosphorylation of Ser10 on histone H3, an Aurora B substrate, was evident only with 5 µM ZM447439 or 2 µM AZD1152, despite the apparent complete inhibition of Aurora B autophosphorylation at these concentrations of the inhibitors, which may be the result of the relative inaccessibility of phosphatases to this site or other kinase activities that contribute to Ser10 histone H3 phosphorylation, such as Msk1 (32). However, even at these high concentrations of the inhibitors, Aurora A autophosphorylation appeared largely unchanged.
Fig. 1.
Characterization of small-molecule inhibitors of Aurora A, Aurora B, and Plks. (A) Western blot analysis of BI2536, MLN8054, AZD1152, and ZM447439 inhibitor titration in Taxol-arrested HeLa cells using antibodies against the phosphorylated T loop of all three Aurora kinases, the phosphorylated T loop of Plk1 (pT210), Cdc25c, and phosphorylated Ser10 of histone H3 (pS10). Anti–lamin A/C blots are shown as loading controls. Plk1 Thr210 is phosphorylated by Aurora A, Cdc25c is phosphorylated by Plk1, and his-tone H3 Ser10 is phosphorylated by Aurora B. Blot shown is representative of four independent experiments. (B) Raw LC-MS/MS data extracted for mass/charge ratios (m/z) corresponding to the Aurora A pT288 and Aurora B pT232 autophosphorylated activation loop phosphopeptides and the Plk1 pT210 activation loop phosphopeptide under each inhibitor condition. Blue trace: peptide from light, control cells; green trace: peptide from heavy, inhibitor-treated cells. All traces are extracted to ±2 parts per million (ppm) mass deviation from theoretical. Y axis is relative abundance on an arbitrary scale. (C) Immuno-fluorescence micrographs of spindles of control cells or cells treated with the indicated inhibitors. CREST (red) is a marker of kinetochores, and tubulin (green) marks microtubules. Proteins were labeled with antibodies. DNA (blue) was labeled with PicoGreen. Scale bar, 3 µm.
To confirm the inhibition observed by Western blotting, we quantified by mass spectrometry the phosphorylation status of peptides containing Aurora A Thr288, Aurora B Thr232, and Plk1 Thr210 from cells arrested in Taxol and exposed to the inhibitors (Fig. 1B). In cells exposed to 250 nM MLN8054, phosphorylation of the Aurora A T loop Thr288 was reduced by 7-fold relative to uninhibited cells and was nearly undetectable in the presence of an MLN8054 concentration of 1 µM (28-fold reduction). Similarly, autophosphorylation of Aurora B Thr232 was nearly undetectable in cells exposed to 1 µM AZD1152 relative to uninhibited cells. Consistent with reports that establish Aurora A–mediated transactivation of Plk1 in early mitosis (7, 9), phosphorylation of the T loop of Plk1 was reduced by a factor of 2 in cells treated with 5 µM MLN8054 relative to uninhibited cells, although at this concentration of inhibitor, Aurora A was essentially inactive. We noticed a similar persistence in Plk1 T loop phosphorylation by Western blotting even under conditions in which Aurora A appeared completely inhibited (Fig. 1A). We speculate that under the conditions of our experiments, either the phosphatases responsible for dephosphorylating this site may not be maximally active or Aurora A is one of multiple mitotic kinases that contribute to phosphorylation of Plk1 at Thr210. We also examined the phosphorylation of these peptides in the presence of BI2536 at two different points in the cell cycle, at the entry into mitosis and during mitosis. The “mitotic entry” condition was accomplished by releasing cells from a double-thymidine block for 3 hours, adding Taxol for 5 hours, and then adding MG132 and inhibitor for 45 min, whereas the mitotic population was generated by releasing cells from a double-thymidine block for 3 hours and then adding Taxol for 10 hours, followed by treatment with MG132 and inhibitor for 45 min. During mitosis, the presence of BI2536 reduced phosphorylation of Plk1 at Thr210 by a factor of 2 relative to uninhibited cells.
Examination of cellular phenotypes induced by inhibitor treatment confirmed previous reports of defects in spindle morphology and chromosome alignment (Fig. 1C and fig. S1) (27–31). Whereas control cells formed a mitotic spindle and aligned their chromosomes at the metaphase plate, low concentrations of MLN8054 induced minor spindle defects with few misaligned chromosomes. With increasing amounts of MLN8054, more severe spindle defects, multipolar spindles, and chromosome alignment defects were observed. HeLa cells treated with 1 µM AZD1152 or 5 µM ZM447439 showed severe chromosome misalignment and aberrant spindles. Cells treated with 100 nM BI2536 arrested in mitosis with monopolar spindle defects.
BI2536 inhibits Plk1, Plk2, and Plk3 in vitro and generates phenotypes resembling Plk1 knockdown in cells (33). To determine the effect of BI2536 on Plk4, we performed in vitro kinase assays with Plk1, Plk4, a peptide substrate, and BI2536 at concentrations ranging from 1 nM to 10 µM and then evaluated peptide phosphorylation by mass spectrometry (fig. S2). Whereas Plk1 was inhibited at nanomolar concentrations, Plk4 activity was unaffected even at the highest concentration tested (10 µM), suggesting that Plk4 is likely not inhibited at 100 nM, the concentration we selected for our studies.
Quantitative phosphoproteomics of mitotic HeLa cells treated with kinase inhibitors
To scan mitotic cells for substrates of the Aurora A, Aurora B, and Plk enzymes, we quantitatively determined differences in the phosphorylation status of proteins from mitotically arrested cells treated with the respective kinase inhibitors by the SILAC (stable isotope labeling with amino acids in cell culture) method (34). HeLa cells were labeled with either isotopically “heavy” ([13C6,15N2]lysine and [13C6, 15N4]arginine) or natural amino acids in tissue culture (fig. S3A ). Both populations of cells were arrested in metaphase of mitosis, and heavy-labeled cells were treated with kinase inhibitor for 45 min in the presence of MG132, whereas unlabeled cells were treated with MG132 but not with inhibitors (fig. S3, B and C). For some experiments with BI2536-mediated inhibition of Plks, we also examined phosphorylation status changes for cells arrested at metaphase (BI mitosis) and cells entering mitosis (BI entry). We used a single, optimum concentration of BI2536 (0.1 µM), AZD1152 (1 µM), and ZM447439 (5 µM) to inhibit Plks and Aurora B (fig. S3C). To differentiate between Plk targets during mitotic entry from those specific to mitosis, we conducted two sets of experiments: one in which BI2536 was added to cells arrested in metaphase and one in which BI2536 was added as cells were entering mitosis (fig. S3B). To dissect Aurora A from Aurora B targets, we performed a titration series of MLN8054 at 0.25, 1, and 5 µM. Aliquots of control and inhibitor-treated cells were analyzed by flow cytometry to confirm the efficiency of cell cycle synchronization by staining with propidium iodide and the monoclonal antibody MPM-2 (35), a mitotic marker that recognizes phosphoepitopes on mitotic proteins (fig. S4). Whereas most populations of Taxol-arrested cells across the different conditions had 4N DNA populations that were enriched in MPM-2–positive cells (89.3 to 92.6%), those in the BI2536 mitotic entry condition were somewhat reduced (72.2%) in MPM-2 immunoreactivity, indicating that some late G2 cells are likely present in this population. These inhibitor-treated, heavy cells were mixed with light control cells and lysed, and the resulting protein lysate was digested to peptides. These peptides were separated by strong-cation exchange (SCX) chromatography, and phosphopeptides were isolated from separate SCX fractions with titanium dioxide microspheres and analyzed by high-performance liquid chromatography–tandem mass spectrometry (LC-MS/MS).
A total of 457,480 singly, 258,217 doubly, and 36,640 triply phosphorylated peptides were sequenced across 542 LC-MS/MS runs representing all eight conditions, with an associated overall peptide false discovery rate (FDR) of 0.2% (table S1). Phosphorylation loci were assigned by creating specific identifiers, called ModSites. We developed this ModSite method because Aurora kinases phosphorylate a consensus sequence that is also the site for trypsin cleavage, which produces many N-terminally phosphorylated peptides and for which the assignment of a specific phosphorylation locus was often difficult. Therefore, we created identifiers that allow potential ambiguity in assigning phosphorylation to any specific locus when the following criteria were met: (i) Another acceptor residue occurred within two amino acids of a potential phosphoacceptor residue (Ser, Thr, or Tyr). (ii) A different locus assignment was ascribed to any of these adjacent residues in any other phosphopeptide identified across all experiments. (iii) The difference between the most extreme N-terminal and the C-terminal phosphoacceptor residue in a collection of many occurrences was less than six amino acids. For example, the autophosphorylated Aurora A T loop was observed throughout our experiments as both RTpTLCGTLDYLPPEMIEGR and RpTTLCGTLDYLPPEMIEGR. Rather than report both results with separate ratios, we classified them as a single, ambiguous identifier called a “ModSite,” in this case ModSite [O14965_(T287,T288)], representing the averaged quantitative results from each of the individual observations, and calculated the SD. Those without ambiguity had only a single phosphorylated site included in the averaged data tabulated from each observation (for example, NuMA Ser2062, Q14980_(S2062); KLGNpSLLR). The collection of phosphopeptides mapped to 33,017 unique ModSite identifiers on 6061 proteins (2.5% protein-level FDR) that were observed in two or more experiments.
Although mock-treated control cells were present in each SILAC experiment, we chose to further assess the baseline quantitative precision of our analytical strategy by comparing heavy- and light-labeled cells, both arrested in metaphase with Taxol and exposed to MG132, but not treated with any kinase inhibitor. Changes in phosphorylation status were expressed as the log2 ratio of phosphopeptide LC-MS peak areas from heavy-labeled to light-labeled cells. In this control sample, only 0.66% (increased phosphorylation) and 0.55% (decreased phosphorylation) ModSites exhibited a change of more than 2.5-fold when comparing the heavy-labeled cells with the light-labeled cells (fig. S5 and Table 1). We also examined the change in the proportion of sites exhibiting increased or decreased phosphorylation in response to exposure of the cells to the various inhibitors. We found that the AZD1152 and ZM447439 ratios were largely similar for each ModSite (fig. S6) and were therefore combined into one category (AZDZM). Exposure of the cells to any of the inhibitors resulted in a greater increase in the proportion of sites exhibiting decreased phosphorylation compared to those exhibiting increased phosphorylation (Table 1). The largest changes in the percentage of sites exhibiting an increase or a decrease occurred in cells exposed to BI2536, with inhibition of Plk in cells entering mitosis having the greatest percentage of change in phosphorylation site status. For each of these experiments, the fold change in phosphorylation status was calculated as the log2 ratio of the phosphorylation status detected in the heavy-labeled cells exposed to the inhibitor versus the light-labeled Taxol-arrested and MG132-treated control cells.
Table 1.
Net change in overall phosphorylation in response to kinase inhibitors.
| 2.5-Fold down-regulation (%) |
2.5-Fold up-regulation (%) |
|
|---|---|---|
| Taxol | 0.55 | 0.66 |
| MLN8054, 0.25 µM | 1.03 | 0.90 |
| MLN8054, 1 µM | 2.02 | 0.88 |
| MLN8054, 5 µM | 4.13 | 1.14 |
| AZDZM | 2.36 | 0.92 |
| BI2536 mitosis | 5.48 | 1.34 |
| BI2536 entry | 7.81 | 2.76 |
Aurora kinase A versus Aurora kinase B targets
On the basis of the inhibitor validation experiments (Fig. 1), we expected an increasing reduction in phosphorylation of direct targets of Aurora A with increasing concentrations of MLN8054, whereas phosphorylation of Aurora B targets should be largely unaffected at MLN8054 concentrations of 0.25 and 1 µM, but should exhibit decreased phosphorylation in cells treated with 5 µM MLN8054 or with AZD1152 or ZM447439. Aurora A substrates would remain mostly unchanged at single, optimum concentrations of the more selective Aurora B inhibitors AZD1152 and ZM447439.
To visualize trends in our data sets, we collected 434 ModSite ratios that were identified in each MLN8054 0.25 mM (MLN0.25), 1 µM (MLN1), 5 µM (MLN5), and AZDZM condition and that also met the conservative cutoff requirement of at least a 2.5-fold reduction (log2 of phosphorylation status of heavy-labeled cells in the presence of inhibitor versus light-labeled control cells ≤−1.4) in either MLN5 or AZDZM relative to uninhibited cells (table S2). This array of ModSites was then analyzed by agglomerative hierarchical clustering methods (fig. S7), which produced three general classes of ModSites.
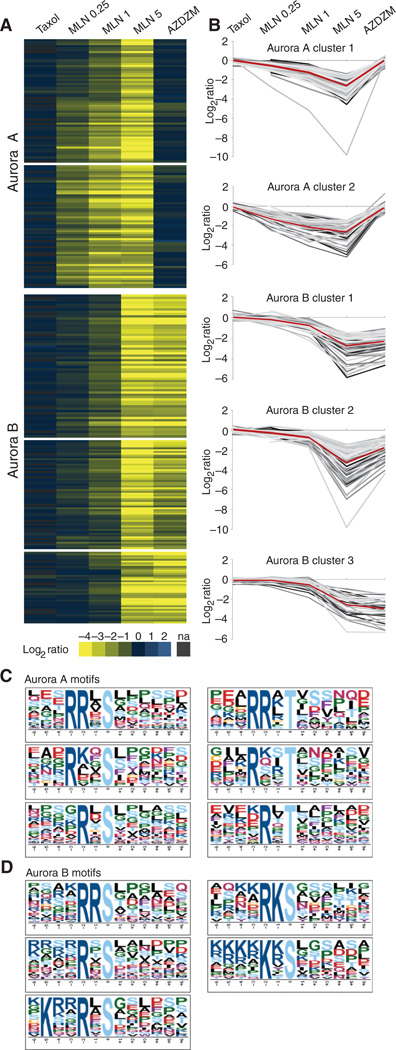
ModSites in the first class displayed a ratio profile expected for potential Aurora A targets (Fig. 2A); that is, they exhibited an increasing reduction in phosphorylation as the concentration of MLN8054 increased and little reduction in response to AZD1152 or ZM447439. For example, the autophosphorylation site Thr288 on Aurora A itself was unchanged in Taxol-arrested cells that were not exposed to an inhibitor (0.1), was reduced (−2.7) in MLN0.25, and continued to decrease with increasing concentration of MLN8054 inhibitor (−4.9 at MLN1 and −9.8 at MLN5), but was essentially unchanged (0.02) in the AZDZM condition. In general, ModSites in this first class followed this trend with slight deviations in the degree of inhibition at the different MLN8054 concentrations (Fig. 2B). Motif enrichment of the amino acid sequences surrounding phosphorylated residues in this class revealed a refinement of the canonical Aurora A motif (RRXp[S/T]), with an absolute requirement for arginines in the −2 and −3 position and an additional preference for leucines in the −1 and +1 position: RR[L/X]p[S/T][L/X]; 29% of all ModSites in these clusters exhibited this motif (Fig. 2C). Less stringent motifs with an arginine required in the −2 (RXp[S/T]) or −3 (RXXp[S/T]) position were also observed in 38 and 65% of all ModSites in this cluster. An expanded motif analysis of these potential Aurora A targets established that 91% of the ModSites in this class conformed to an Aurora A–like motif with a basic amino acid in either the −2 or the −3 position.
Fig. 2.
Cluster and motif analysis of putative Aurora A and B substrates. (A) Candidate Aurora A and B substrates identified by hierarchical clustering of ModSites with ratios reduced in either the MLN5 or the AZDZM conditions by 2.5-fold or more relative to uninhibited cells (na, not available). (B) Line graphs of log2 ratios of the ModSites of the corresponding Aurora A and B subclusters from panel A. The red line represents the average. (C) Motif analysis of both Aurora A–subclustered ModSites. (D). Motif analysis of all three Aurora B–subclustered ModSites.
The second class of ModSites exhibited ratio profiles predictive for Aurora B targets, with nearly unchanged values in the Taxol-arrested cells without inhibitor treatment, MLN0.25, and MLN1 conditions and strongly reduced values in the MLN5 and AZDZM conditions (Fig. 2A). The autophosphorylation site of Aurora B at Thr232 followed this trend, with nearly unchanged ratios in Taxol-arrested control cells (0.15), MLN0.25 (0.04), and MLN1 (−0.38), and strongly reduced values in MLN5 (−4.1) and AZDZM (−4.3). Within this class, three distinct subclasses could be distinguished that differed in the magnitude of inhibition in the MLN5 and AZDZM conditions (Fig. 2B). Motif enrichment for these ModSites showed a strong preference (55%) for basic amino acids immediately upstream and adjacent to the phosphorylation site ([R/K]p[S/T]). This is in contrast to potential Aurora A substrates that infrequently (9%) displayed a basic residue in this −1 position. Furthermore, the strong preference for arginine seen in the Aurora A motifs was not observed in this cluster: Only 8% of the ModSites in this cluster conformed to the classical Aurora A consensus sequence RRXp[S/T]. Additional basic residues, and more frequently lysine than arginine, were found even further upstream (positions −4 through −6) in the motifs present in this Aurora B cluster than were present in the motifs in the cluster for Aurora A. In general, glycine or a hydrophobic residue such as leucine occupied the +1 position (Fig. 2D). Of the ModSites in this cluster, 94% displayed a phosphorylation site with one of these motifs.
The third class of ModSites in this array displayed ambiguous behavior (fig. S8A). For example, some of them had similar relative reductions in phosphorylation in MLN1 and AZDZM conditions (subclass 1) (fig. S7B), whereas others had only reduced phosphorylation in response to the MLN5 (subclass 2) or the AZDZM (subclass 3) condition (fig. S8B). Although ModSites in the first subclass are likely real Aurora A or B targets, or targets of both kinases, the second and third subclasses are more difficult to interpret. Motif analysis of ModSites in these three ambiguous subclusters generated motifs intermediate to the two Aurora kinases: one with arginine in the −2 position and preferences for leucine in the −1 and +1 position (as observed for Aurora A targets), and another one with additional basic residues in the −1 to −6 position, reminiscent of Aurora B targets (fig. S8C).
We inspected the ModSites in the Aurora A and B clusters for proteins described in the literature as substrates of either kinase and found that some of these proteins were already known as substrates. In some cases, the phosphorylation sites identified in these clusters were also previously reported (fig. S9, A and B). These findings serve to further strengthen our confidence in the overall approach and the assignment of these ModSites to their respective candidate kinases.
Identification of Plk targets
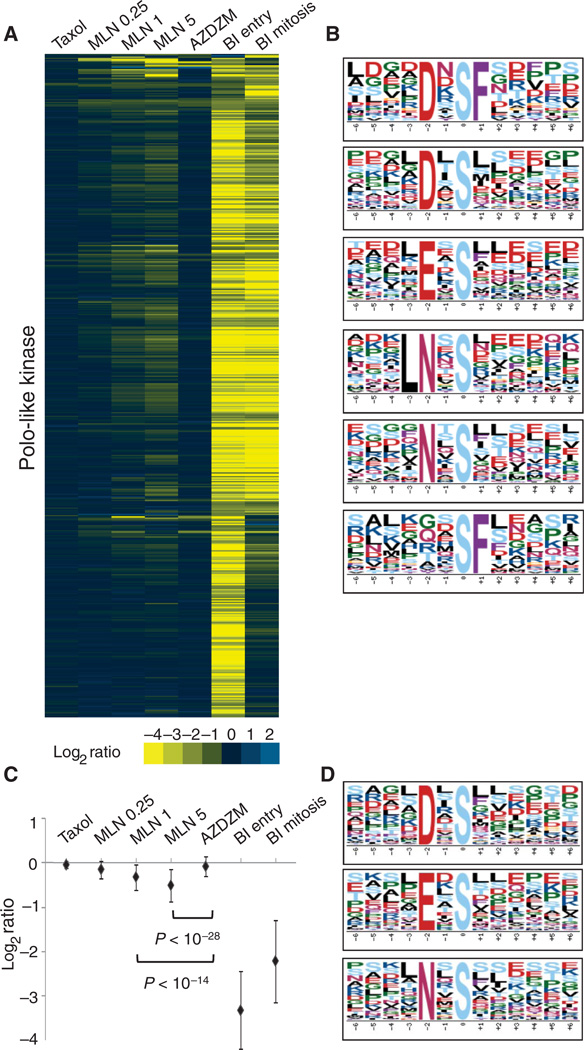
We began our analysis for Plk targets by extracting ModSites from our data collection that were present in each control (Taxol-arrested but not exposed to inhibitor), MLN0.25, MLN1, MLN5, and AZDZM condition and had a ≥2.5-fold reduction (log2 ratio ≥−1.4) in ratio in either BI2536 treatment of cells entering mitosis (BI entry) or BI2536 treatment of cells arrested in mitosis (BI mitosis) relative to uninhibited cells (table S3). We clustered this collection of 528 ModSites (Fig. 3A and fig. S10), revealing many ModSites with reduced ratios in both the BI mitosis and the BI entry samples (58.71%). A subset of the 528 ModSites had reduced ratios only in the BI entry samples (36.55%), and a small group exhibited reduced ratios only in the BI mitosis samples (4.73%). Motif analysis revealed not only the canonical Plk1 motif [D/E]Xp[S/T][FLIYWVM] (36) but also several variations of this motif with an absolute requirement for an aspar-agine in the −2 position (NXp[S/T] and LNXp[S/T]) instead of an aspartic or glutamic acid (Fig. 3B). We also identified another distinct potential Plk recognition motif with only phenylalanine in the +1 position (p[S/T]F) that appeared to be sufficient for Plk targeting without any additional preference upstream of the phosphorylation site, as well as a more specific variant of this phenylalanine-based motif (DXp[S/T]F). Of the ModSites in the Plk cluster, 74% conform to any of these motifs. A subset of these candidate Plk substrates was previously annotated as substrates of Plk1 (fig. S9C), confirming the validity of our analysis.
Fig. 3.
Cluster and motif analysis of putative Plk substrates and regulation of Plk by Aurora A. (A) Candidate Plk substrates identified by hierarchical clustering of ModSites with ratios reduced by 2.5-fold or more in either the BI entry or the BI mitosis condition relative to uninhibited cells. (B) Motif analysis of all Plk phosphorylation sites from the array in panel A. (C) Averaged ratios of all Plk substrates shown in panel A. Error bars indicate 1 SD. The difference between MLN1 or MLN5 and AZDZM is significant as indicated by P values. (D) Motif analysis of ModSites in the −0.5 MLN5 ratio space. Plk consensus motifs are shown.
We developed a mass spectrometry–based in vitro kinase assay with naturally occurring peptides as substrates to verify that these new Plk motif elements could be attributed to Plk1 activity. In brief, peptides derived from HeLa cells were exhaustively dephosphorylated with a cocktail of phosphatases, fractionated by SCX, and used as substrate “libraries” in an in vitro kinase reaction with recombinant Plk1. The phosphopeptides were isolated and analyzed by LC-MS/MS, and statistically significant motifs were extracted from the collection of individually sequenced phosphopeptide results (fig. S11A). With this in vitro assay using purified Plk1, we identified the same motifs as those observed in the SILAC assays with cells, supporting the notion that LNXp[S/T], NXp[S/T], and p[S/T]F could be additional Plk1 substrate motifs (fig. S11B). Although several small hydrophobic amino acids in the +1 position alone were sufficient as Plk1 recognition motifs in vitro (fig. S11B), we observed a general preference for these hydrophobic amino acids in the +1 position in cells only when they were identified as part of a motif that also contained asparagine, aspartic, or glutamic acid upstream of the phosphorylated serine (Fig. 3B).
We also performed the mass spectrometry–based in vitro kinase assay with recombinant Aurora A (co-purified with TPX2) (fig. S11C) or Aurora B [co-purified with INCENP (inner centromere protein)] (fig. S11D) and found the same consensus motifs as those that we identified in cells exposed to Aurora inhibitors (Fig. 2, C and D).
Kinase cross-regulation
Visual inspection of our clustered data array of BI2536-inhibited ModSites (Fig. 3A and fig. S10) suggested that many sites showed reduced ratios in the MLN1 and MLN5, but not AZDZM, conditions. These ModSites could be categorized into two groups. The first was composed of only 21 ModSites that were >2.5-fold reduced in both the MLN5 and either of the BI2536 conditions and contained many multiply phosphorylated peptides with sites consistent with both Plk1 and Aurora motifs, suggestive of direct co-regulation by both kinases at adjacent sites. The second group contained the remaining ModSites that displayed an average inhibition ratio of −0.3 in the MLN1 sample and −0.5 in the MLN5 sample, both of which were statistically significant when compared to their reduction in AZDZM samples (Welch’s t test, P < 10−14 and P < 10−28; Fig. 3C). Because MLN inhibits Aurora A, this subset of sites that exhibit reduced phosphorylation in either BI condition and are also significantly reduced in the MLN samples compared to the AZDZM (Aurora B–inhibited) samples suggests that, in addition to direct co-regulation of proteins by both Plk and Aurora A, a broader cross-regulation of Plk may occur either directly or indirectly through Aurora A. We hypothesized that if Plk activity depends on Aurora A, then phosphopeptides in the MLN5 data set with ratios ~−0.5 should contain a Plk-like motif. Indeed, inspection of peptide sequences in this ratio space in the MLN5 condition revealed not only Aurora consensus motifs but also those previously observed for Plk1 (Fig. 3D). No enrichment for Plk1 motifs was observed at the identical ratio space in either the AZDZM or the Taxol (control) data set.
To evaluate whether Aurora A or Aurora B substrates were also likely regulated by Plk, we analyzed whether Aurora A and B ModSites that exhibited >2.5-fold reduction in phosphorylation also exhibited reduced phosphorylation when Plk was inhibited in either BI2536 condition. Many ModSites in the Aurora A cluster, but not those in the Aurora B cluster, had reduced ratios when Plk was inhibited (fig. S12, A and B). Although the average of all ModSite ratios in both the BI entry and the BI mitosis samples was reduced for ModSites in the Aurora clusters, for the BI mitosis data set this reduction was due to a large number of strongly reduced ratios rather than an overall ratio reduction. In contrast, ModSites from the BI entry samples had an overall ratio reduction of −0.4 (fig. S12C) and were enriched in the Aurora A cluster relative to the Aurora B cluster (Welch’s t test, P = 3 × 10−7). As observed in the analysis for the Plk cluster, many of the ModSites in the Aurora A cluster that exhibited reduced ratios in BI entry and BI mitosis samples belonged to multiply phosphorylated peptides and contained sites with both Aurora and Plk motifs, again suggestive of direct co-regulation of those proteins at neighboring sites on the same tryptic peptide (fig. S12D). Motif analysis of ModSites in the −0.4 ratio space in the BI entry data set showed the presence of an Aurora-like motif (fig. S12E) that is intermediate between that of Aurora A and that of Aurora B, although more similar to that of Aurora A, indicating that phosphorylation by Aurora A may also be broadly regulated by Plk activity.
Evolutionary conservation of phosphorylation sites and motifs
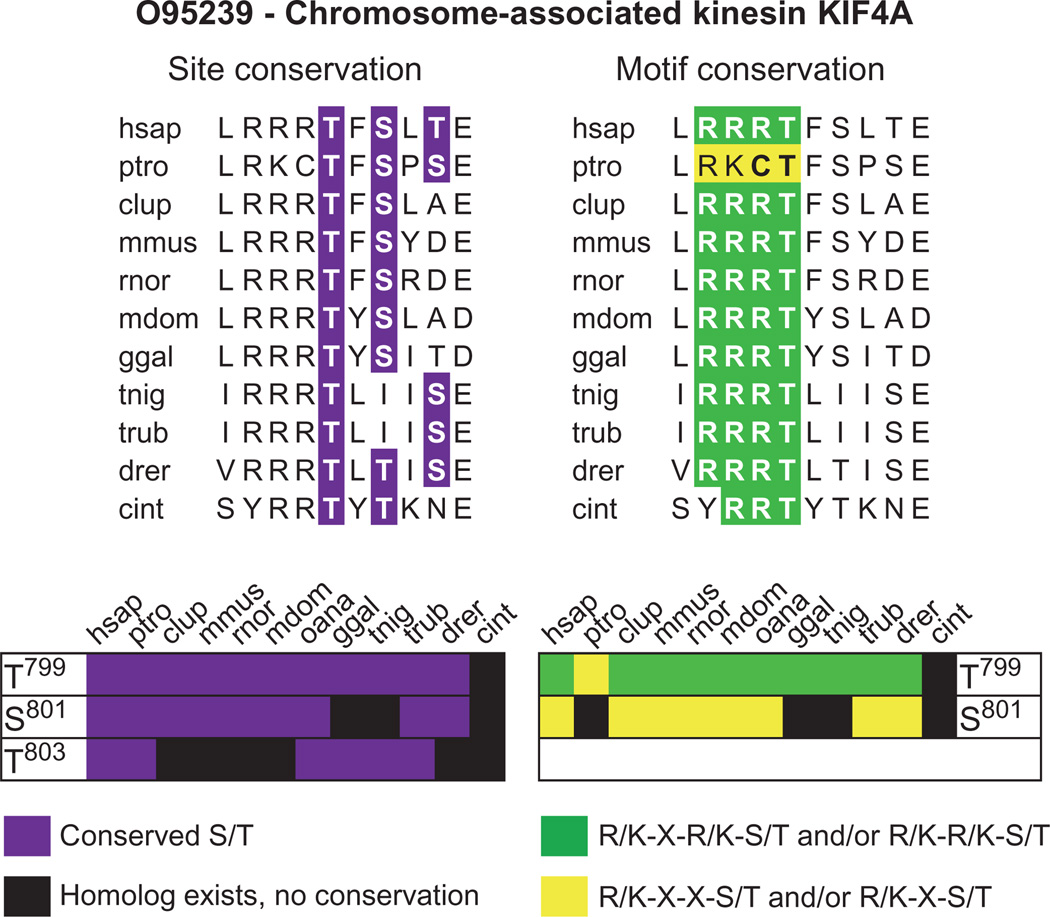
On the basis of our observations of the enrichment of specific motif-containing ModSites in our data sets, we assessed the structural environment and evolutionary conservation of these ModSites to determine whether there was evidence for selection pressure for motif preservation. Using algorithms to predict secondary structures as well as ordered versus unordered regions in proteins, we found that only 10% of all identified ModSites were located in secondary structures containing protein domains or in ordered regions, whereas roughly 90% were found in loops and unordered regions (fig. S13), which is consistent with the reported location of phosphorylation sites in Cdk1 substrates (37). We also determined the evolutionary conservation of these ModSites. Orthologs were identified for all ModSite-containing proteins and aligned, and each candidate phosphoacceptor residue in a ModSite was investigated independently for its conservation, as well as the conservation of surrounding amino acids consistent with one of our kinase motifs (table S4 and fig. S14). Hierarchical clustering revealed that most sites are conserved in vertebrates, but only a few sites were conserved across all species (for instance, Ser62 on Arf1, Ser500 on MCM7, Ser987 on NAT10, and Thr292 on UBP5). Motif-containing sites displayed a strong conservation of specific motif elements such as a basic residue in the −1 position for Aurora B or D/E/N in the −2 position and phenylalanine in the +1 for Plk across all species in which the respective sites were preserved (fig. S14), supporting the notion of modular conservation of the kinase-substrate relationship. We noted that when ambiguity occurred in our ModSite descriptors, this could sometimes be resolved by distinguishing between motif and site conservation, as in the case of the chromosome-associated kinesin KIF4A [O95239_(T799,S801,S803)] (Fig. 4). We also observed strong conservation for the T loop phosphorylation sites and motifs of Aurora A, Aurora B, and Plk1 (fig. S15), suggesting an early establishment of their mode of activation and conservation of this mechanism during evolution in eukaryotes.
Fig. 4.
Evolutionary site versus motif conservation. Representative sequence alignments of one ModSite for the chromosome-associated kinesin KIF4A [O95239_(T799, S801, T803)] with indicated site (purple) and motif conservation (yellow and green). This ModSite is a predicted substrate of Aurora B. The combination of site and motif conservation of T799 indicates this as the site most likely phosphorylated by Aurora B relative to the other ambiguous sites in this identifier. Abbreviations for the amino acids are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; I, Ile; K, Lys; L, Leu; N, Asn; P, Pro; R, Arg; S, Ser; T, Thr; and Y, Tyr.
Connectivity, functional enrichment, and subcellular localization of candidate substrates
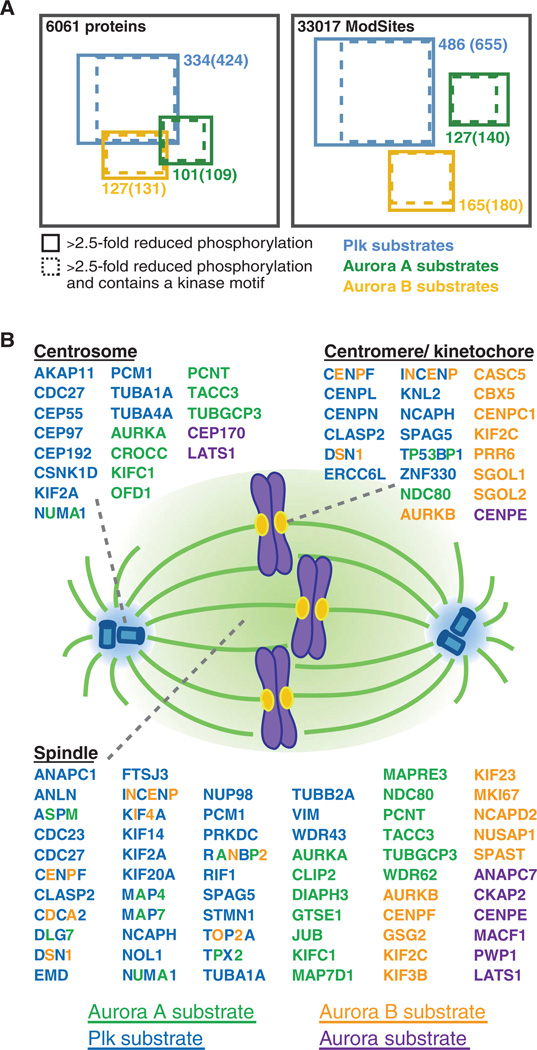
Even the most selective of small molecules may exhibit off-target effects. To safeguard against assigning a substrate to a kinase incorrectly, we imposed the additional requirement that ModSites ascribed to a particular kinase class must adhere to one of the motifs generated for that class by subsequent bioinformatic analyses. In addition, given that only Taxol and the two BI2536 treatments were necessary to identify candidate Plk substrates, we added ModSites to this group that had ratios that fit the strict >2.5-fold inhibition requirement in either or both BI2536 conditions and adhered to a motif for that cluster (table S5). The application of these requirements resulted in 127, 165, and 486 ModSites in each of the Aurora A, Aurora B, and Plk clusters, respectively (Fig. 5A).
Fig. 5.
Candidate Aurora A, Aurora B, and Plk substrates and their spindle localization. (A) Diagram of proteins and unique ModSites identified in HeLa cells by quantitative chemical phosphoproteomics. Total number of identifications is indicated in black. Blue squares indicate candidate Plk substrates; green squares, candidate Aurora A substrates; and orange squares, candidate Aurora B substrates. Number of proteins and ModSites in each group that displayed >2.5-fold reduction in phosphorylation after inhibitor treatment, as well as those that contained a minimal kinase motif (in parentheses) is given. (B) Depiction of candidate Aurora A (green), Aurora B (orange), Aurora ambiguous (purple), and Plk (blue) substrates based on their subcellular localization to centromeres or kinetochores, centrosomes, and the spindle. Proteins with more than one colored letter are substrates of more than one of these kinases. The number of colored letters is not an indication of the number of phosphorylation sites matched to a particular kinase.
To assess the overall connectivity of the collection of candidate substrates from all three kinases, we analyzed our data using STRING (38) (fig. S16). The resulting protein interaction map produced several clusters of high connectivity that contained proteins involved in cell cycle regulation, RNA processing and splicing, centrosome assembly, nuclear transport, and DNA damage repair. Aurora A was implicated in splicing processes (39), and many of the functional hits found in that screen are also present as Aurora A substrates in our data set.
Because of the important roles of Aurora A, Aurora B, and Plk1 in the cell cycle regulation of the mitotic spindle, we more closely examined our data set for proteins involved in (i) the assembly and dynamics of the mitotic spindle and its anchoring points, (ii) the assembly of centrosomes at the minus ends of microtubules, and (iii) the structure and function of centromeres and kinetochores at microtubule plus ends (Fig. 5B). We found 64 spindle-associated proteins to be candidate substrates of Aurora A, Aurora B, Plk, or some combination thereof. Five of these proteins were phosphorylated by Aurora A and Plk (ASPM, DLG5A, MAP4, NuMA1, and TPX2), three by Aurora B and Plk (INCENP, KIF4A, and TOP2A), and one by all three kinases (RANBP2). In addition to the cross-regulation of Plk and Aurora A themselves described earlier, we observed co-phosphorylation of binding partners and coactivators, such as TPX2 and INCENP. Our analysis revealed phosphorylation by these mitotic kinases of proteins that associate with spindle microtubules (CLIP2, HAUS6, and NUSAP1) and promote microtubule assembly (MAP4, MAP7, and MAP7D1) and disassembly (STMN1), nucleation (CLASP2 and TUBGCP3), and general organization of the spindle poles (DLGAP5, NuMA1, PCM1, and PCNT) as well as the spindle itself (SPAG5, TACC3, and ASPM). The family of kinesin proteins was also represented in our candidate Aurora B and Plk substrates. The only kinesin that we found predicted to be phosphorylated by Aurora A, KIFC1, is also the only minus end–directed kinesin that we identified as a candidate substrate in our data set. Among the plus end–directed kinesins, KIF2A, KIF14, and KIF20A were candidate Plk targets, whereas KIF2C, KIF3A, KIF3B, and KIF23 were candidate Aurora B targets. In addition, we observed potential coregulation of the plus end–directed kinesin KIF4A by both Aurora B and Plk.
Consistent with the localization patterns of these kinases, we observed phosphorylation of candidate substrates at the centrosome predominately by Aurora A and Plk, whereas targets that localized to the kinetochore or centromere were most likely phosphorylated by Aurora B and Plk. Proteins at the kinetochore that were phosphorylated by Aurora A alone (NDC80 and Nup133), or additionally by one of the other two kinases (TP53BP1), also localized to the spindle or exchanged with a soluble, cyto-plasmic protein pool. We found that proteins essential for proper spindle checkpoint function (NDC80, ERCCL6, CENPE, CENPF, SGOL1, SGOL2, and SPAG5) were candidates for phosphorylation, as well as proteins that constitute microtubule attachment sites at the kinetochore (KNL2, NDC80, CASC5, and DSN1) and its interacting proteins (CBX5). In addition, phosphorylation of chromosomal passenger complex proteins (AURKB and INCENP), which correct erroneous microtubule-kinetochore attachments, was also responsive to kinase inhibitor treatments. Although many proteins localize to the kinetochore only during mitosis, a subset of proteins that belong to the constitutive centromere-associated network (CCAN) are found at the centromere throughout the cell cycle and were identified as possibly phosphorylated by Aurora B, Plk, or both (CENPC1, CENPL, CENPN). We also identified members of the non–structural maintenance of chromosomes (non-SMC) condensin complex (NCAPD2, NCAPG, and NCAPH) as likely substrates of Aurora B and Plk.
We identified proteins that are integral components of the centrosome (CROCC, CEP192, CEP55, ODF1, PCM1, and PCNT) or are essential for centrosome assembly, cohesion, and function as potentially phosphorylated by Aurora A or Plk. Others are involved in anchoring and assembling of the mitotic spindle (FGFR1OP, HAUS6, CEP97, CEP170, and TUBGCP3) and microtubule dynamics at the centrosome (KIF2A, KIFC1, and HAUS8).
Verification of candidate kinase targets
To verify that our candidate substrates were not specific to Taxol-dependent checkpoint activation, we determined whether they were present under different conditions of spindle checkpoint activation and phases of the cell cycle. We analyzed (i) cells arrested in mitosis with nocodazole, (ii) cells collected by mitotic shake-off from release from a double-thymidine block in the absence of any microtubule targeting agents (unperturbed mitosis), and (iii) cells from an asynchronous population and compared each of these using the SILAC technique to Taxol-arrested cells. This allowed us to determine the relative abundance difference of our candidate substrate phospholoci among these three conditions in which they were established versus each other.
Most candidate substrates initially observed under Taxol arrest conditions were also observed during nocodazole arrest (98% exhibited a nocodazole/Taxol SILAC ratio of >0.3) (table S5). Similarly, most were also found during an unperturbed mitosis (93% with unperturbed mitosis/Taxol ratio greater than 0.3). The median ratio of a candidate target from any of the three kinases was 0.04 in asynchronous cells versus Taxol arrest, which is roughly equivalent to the percentage of those cells in mitosis, and 91% of them exhibit an asynchronous/Taxol SILAC ratio of <0.15. Western blot of the activating T loops for all three kinases and flow cytometry analysis of these different cell cycle conditions supported these SILAC data (fig. S17). Thus, it appears that most of these substrates are generally phosphorylated during an unperturbed mitosis as well as multiple conditions of checkpoint activation, and most of these phosphorylations appear to be specific to mitosis versus other phases of the cell cycle.
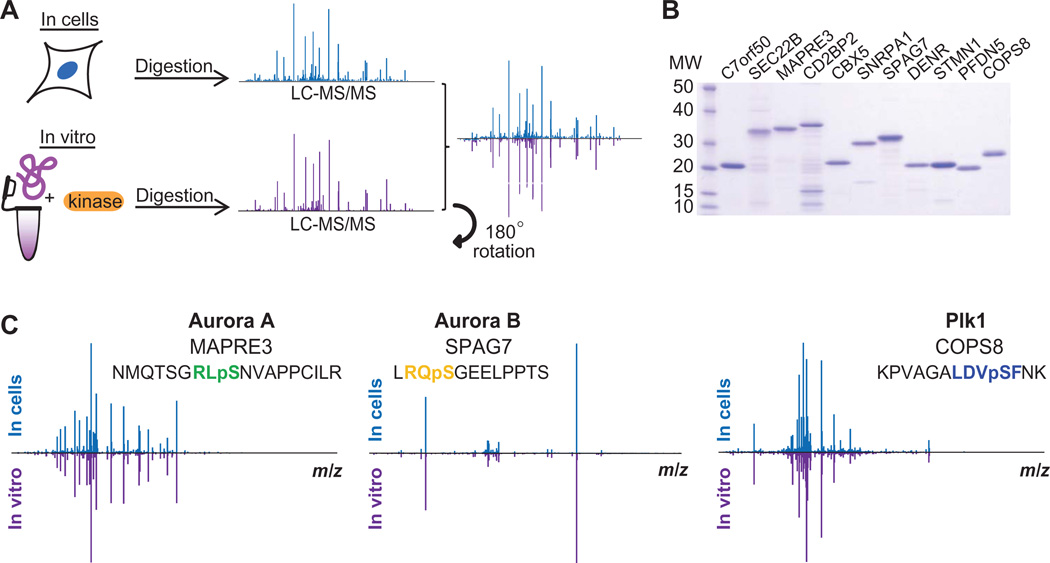
To verify that the ModSites we identified by this method were phosphorylated by Aurora A, Aurora B, or Plk1, we expressed a subset of the target proteins with these sites in bacteria, purified them, subjected each to in vitro phosphorylation reactions with the respective kinase, and determined their phosphorylation status by mass spectrometry with and without kinase treatment (Fig. 6 and fig. S18). Whereas we did not observe phosphorylation in any control reaction, we identified the same ModSite found in cells in the large-scale experiment in these in vitro kinase reactions. Furthermore, direct comparison of ion spectra of phosphopeptides identified in cells and in vitro exhibited a high degree of overlap of MS2 fragment ions and highly similar distributions of these ion intensities (Fig. 6C). Although fragment ions were readily identified in many spectra, some were dominated by a strong neutral loss peak originating from the loss of phosphoric acid from the precursor ion during fragmentation. However, even in these cases, spectra derived from cellular and in vitro sources were clearly from the same peptide (for example, see DENR in fig. S18). Collectively, these experiments increase our confidence that the proteins that we identified are phosphorylated in cells and can be real, cellular targets of these kinases.
Fig. 6.
Verification of kinase substrates by in vitro kinase reactions (A). Scheme of phosphorylation site identification in cells and in vitro. (B) Coomassie gel of purified substrates for in vitro kinase reactions. (C) Reciprocal plots of MS2 peptide fragmentation spectra identified as part of the large-scale proteomics analysis and in the in vitro kinase reactions for the Aurora A substrate MAPRE3, the Aurora B substrate SPAG7, and the Plk1 substrate COPS8.
Validation of Plk targets by Plk1 immunoprecipitation
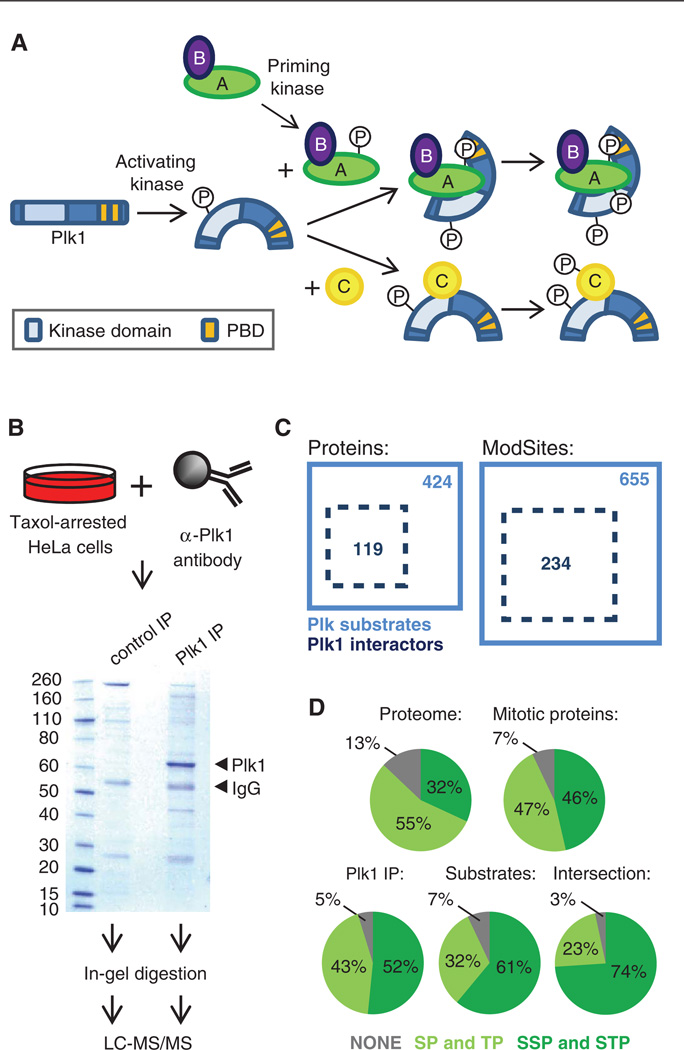
Although the cellular effects observed with BI2536 resemble phenotypes seen upon depletion of Plk1 by RNAi (33), BI2536 inhibits three Plk family members (Plk1, Plk2, and Plk3) to a similar degree in vitro. One mechanism by which Plk1 selects its substrates is by binding of its PBD to previously phosphorylated recognition sequences on these substrates or on adjacent adaptor proteins (Fig. 7A). All four Plk enzymes contain PBDs with different affinities for their respective recognition sequences, a mechanism thought to contribute to their substrate specificity (40). To determine whether the BI2536-sensitive ModSites that we identified could be phosphorylated by Plk1, we immunoprecipitated Plk1 from Taxol-arrested HeLa cells and identified interacting proteins by SDS–polyacrylamide gel electrophoresis (SDS-PAGE)–LC-MS/MS (Fig. 7B). Using this approach, we mapped 35.7% of the BI2536-sensitive phosphorylation sites (table S5) to proteins identified in a Plk1 immunoprecipitation with at least sixfold more total peptides than in a control immunoglobulin G (IgG) immunoprecipitation (table S6), suggesting that at least for this subset of candidate targets, Plk1 is the most likely Plk family member responsible for their phosphorylation (Fig. 7C). Many proteins identified in the Plk1 immunoprecipitation experiment were not found as BI2536-sensitive target proteins, and we speculate that because Plk1 may interact with and phosphorylate a single member of a multiprotein complex, not all proteins identified in the Plk1 immunoprecipitation would be expected to be BI2536-sensitive.
Fig. 7.
Identification of Plk1-interacting proteins. (A) Schematic of the mechanism of Plk1 activation and substrate recognition. Phosphorylation of Plk1 at Thr210 by kinases, such as Aurora A, leads to its activation. Through its Polo-box domain (PBD), Plk1 recognizes and interacts with some of its substrates, which have previously been phosphorylated by a priming kinase at an Sp[S/T]P sequence, leading to additional phosphorylation of these substrates by Plk1. PBD-independent interactions of Plk1 and substrates, which subsequently lead to phosphorylation of these substrates by Plk1, are also possible. (B) Scheme and Coomassie gel of Plk1 immunoprecipitation (IP). Plk1 and IgG bands are indicated by arrows. Substrates of Plk1 that interact directly with Plk1 through the PDB (labeled as A in panel A) or through other sites (labeled as C in panel A), as well as proteins bound to PBD-binding partners (secondary interactions, labeled as B in panel A) and direct Plk1-interacting proteins that are not substrates (not shown in panel A) are expected to coprecipitate. (C) Diagram of overlap of proteins and unique ModSites identified as Plk1 interactors by Plk1 IP (dotted black box) and Plk substrates identified in the large-scale phosphoproteomics experiments with BI2526 (solid blue box). (D) Distribution of [S/T]P and S[S/T]P sites in the whole proteome, in the 6061 proteins identified across all experiments (mitotic proteins), in all Plk1-interacting proteins identified in the Plk1 IP, in the 424 BI2536-sensitive candidate substrates, and in the 119 protein intersection of BI2536-sensitive substrates that were also in the Plk1 IP.
Plk1 binds to Sp[S/T]P and p[S/T]P sequences in candidate substrate proteins. We compared the occurrence of these motifs in whole proteome, in the 6061 proteins identified throughout our analyses (“mitotic proteins”), in all Plk1-interacting proteins identified in the Plk1 immunoprecipitation, in the 424 BI2536-sensitive substrates, and in the 119 protein intersection of BI2536-sensitive substrates that were also in the Plk1 immunoprecipitation and found an increase in Sp[S/T]P-containing proteins across this enrichment series (Fig. 7D).
Functional analysis of NuMA phosphorylation
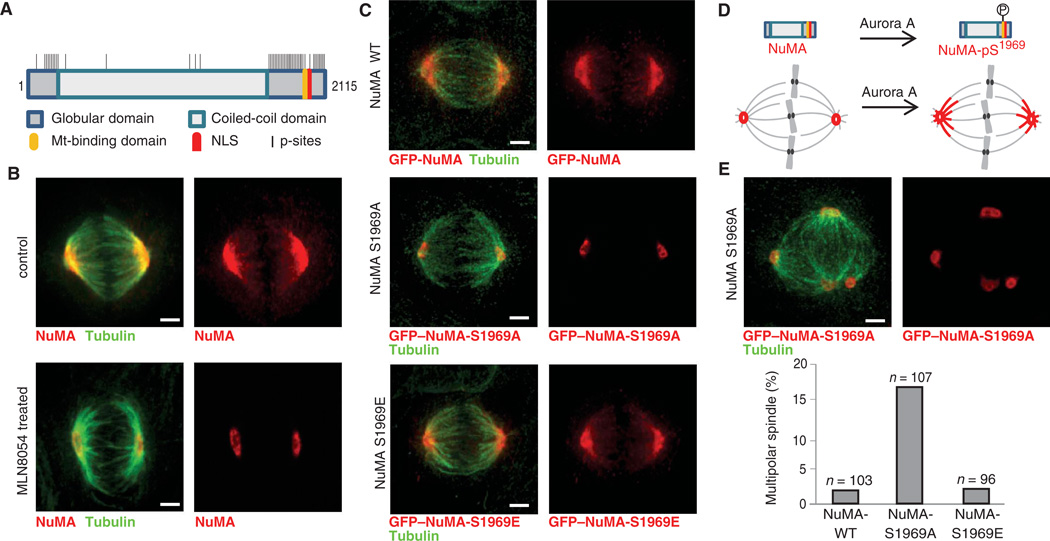
We identified 89 ModSites on the large coiled-coil protein NuMA (Fig. 8A). During interphase, NuMA is localized to the nuclear matrix (41). Upon the onset of mitosis, NuMA is phosphorylated and re- leased into the cytoplasm, where it binds to the dynein-dynactin complex and is transported along microtubules to the spindle poles. At the spindle pole, NuMA forms an insoluble matrix, which acts as a microtubule organization center. Dephosphorylation of NuMA during anaphase attenuates its interaction with the dynein-dynactin complex and thereby its localization at spindle poles. It was previously proposed that phosphorylation of NuMA by Cdk1 is necessary for NuMA’s association with dynein and thereby its spindle localization (42). In our analysis, six of the phosphorylation sites identified on NuMA displayed inhibition ratios and phosphorylation motifs characteristic of Aurora A targets, and four other sites had reduced ratios when treated with BI2536 and contain Plk motifs. To determine whether either of these kinases was important in NuMA sub-cellular targeting, we imaged HeLa cells in the presence and absence of MLN8054 and BI2536. In untreated mitotic cells, NuMA localized to spindle poles and along spindle microtubules (Fig. 8B). Due to the difficulty in comparing bipolar spindles in untreated cells to monopolar spindles in BI2536-treated cells, we did not score NuMA localization behavior with this inhibitor. However, in MLN8054-treated cells, the localization of NuMA in mitotic cells was restricted to the pericentriolar region and was not observed at spindle microtubules proximal to the poles (Fig. 8B), suggesting that Aurora A activity is involved in promoting transport of NuMA along the spindle. To establish whether this mislocalization was the result of direct phosphorylation of NuMA, we individually mutated several phosphorylated serine or threonine residues to alanine and transiently expressed green fluorescent protein (GFP)–NuMA, as well as GFP-NuMA mutants, in HeLa cells. Wild-type GFP-NuMA and most of these single-site GFP-NuMA mutants exhibited a localization pattern consistent with that of endogenous NuMA (Fig. 8C and fig. S19A). Although NuMA-S1969A localized to the nucleus during interphase, in mitosis this mutant exhibited a localization pattern indistinguishable from that of endogenous NuMA in MLN8054-treated cells (Fig. 8C and fig. S19A). A phosphomimetic S1969E mutant exhibited wild-type localization behavior (Fig. 8C and fig. S19A). Ser1969 is located at the end of the microtubule-binding domain of NuMA, which is important for the binding to microtubules during mitosis (43). Mutation of Ser1991 to alanine (NuMA-S1991A) resulted in disruption of the nuclear localization sequence (NLS) and mislocalization of NuMA to the pericentriolar region outside the nucleus, as well as the formation of cytoplasmic filaments in interphase (fig. S19B). During mitosis, however, NuMA-S1991A localized correctly to spindle poles and microtubules. This phenotype closely recapitulates those observed in cells expressing a NuMA variant lacking a functional NLS (44). We also tested whether any combination of two or more Aurora A or Plk1 sites would lead to additional phenotypes, or possibly alter the NuMA-S1969A phe-notype, but found that only combinations of mutated sites that included S1969A displayed the same abrogation of NuMA localization to spindle microtubules (fig. S19C). Together, this suggests that phosphorylation of NuMA on Ser1969 by Aurora A is essential for its transport along the spindle in mitosis (Fig. 8D). Closer examination of the spindle morphology in HeLa cells transfected with NuMA-WT (wild-type), NuMA-S1969A, and NuMA-S1969E revealed a substantial increase (fivefold) in the number of cells with multipolar spindles in NuMA-S1969A compared to cells expressing NuMA-WT or NuMA-S1969E (Fig. 8E), suggesting that phosphorylation of NuMA on this site is important for its function in spindle maintenance.
Fig. 8.
Analysis of NuMA phosphorylation. (A) Scheme of NuMA protein structure with domains, nuclear localization sequence (NLS), and identified phosphorylation sites. Mt-binding, microtubule binding. (B) Micrographs of endogenous NuMA (red) and tubulin (green) immunofluorescence in meta-phase in control and MLN8054-treated HeLa cells. (C) Micrographs of GFP–NuMA-WT, of GFP–NuMA-S1969A, and of GFP–NuMA-S1969E (red) and tubulin (green) immunofluorescence. (D) Depiction of Aurora A phosphorylation of NuMA on Ser1969 in the Mt-binding domain and its effect on NuMA spindle localization. (E). Micrographs of GFP–NuMA-S1969A (red) and tubulin (green) immunofluorescence in a multipolar spindle. Quantification of percentage of multipolar spindles in HeLa cells transfected with GFP–NuMA-WT, GFP–NuMA-S1969A, or GFP–NuMA-S1969E. The number of cells evaluated is indicated. Images in panels B, C, and E are maximum z projections of selected spinning disc confocal optical sections. Scale bar, 3 µm.
DISCUSSION
Recent improvements in mass spectrometry instrumentation and procedures for phosphopeptide isolation and analysis have resulted in a rapid expansion of catalogs of phosphorylation loci in the literature, many without any association to a specific cellular signal, phenotype, or kinase. We have presented here the design and deployment of experiments that assign the mitotic kinases Aurora A, Aurora B, and Plk to respective substrate loci by combining quantitative phosphoproteomics and efficient kinase inhibition with small-molecule chemical inhibitors. Using this approach, we determined 778 sites of phosphorylation on 562 proteins that could be ascribed to these enzymes (table S5). Many of the kinase substrates we identified have known functions in mitotic processes but have not been connected to Aurora or Plk kinase activity. Establishing these kinase-substrate relationships enhances our knowledge about the molecular mechanisms by which these kinases regulate mitotic progression. Although the design of our experiments and the nature of the Aurora A and B inhibitors allowed us to distinguish between candidate substrates of Aurora A and B, we recognize that either Plk1, 2, or 3 could be responsible for the phosphorylation sites observed in the Plk data sets. Inhibition studies with Plk4 and BI2536 indicated that it was unlikely that Plk4 is the upstream kinase for these sites (fig. S2); however, the similar inhibition profiles of Plk1, 2, and 3 with BI2536 in vitro do not allow for distinction between these three enzymes when this inhibitor is used. Nevertheless, the dominant role of Plk1 in mitosis and the cellular phenotypes observed with BI2536 (25, 33), as well as the results from our in vitro motif analysis (fig. S11), our substrate verification (Fig. 6 and fig. S18), and the Plk1 immunoprecipitation (Fig. 7), allow us to speculate that many of the substrates we identified might be phosphorylated by Plk1.
Using bioinformatic techniques, we dissected, associated, and classified our lists of candidate target proteins. An analysis of protein interactions of candidate kinase substrates revealed clusters of high connectivity for proteins involved in cell cycle regulation and centrosome assembly (fig. S16). A closer look at the proteins involved in these processes showed a strong connection between specific kinase activities and the subcellu-lar localization of their substrates (Fig. 5). For instance, most candidate substrates that localized to the centrosome were connected with Plk or Aurora A activities or both, whereas most centromeric proteins are candidate substrates of Plk or Aurora B activity or both. We identified many proteins involved in centromere assembly and cohesion, in kinetochore-microtubule attachment, and in correction of erroneous microtubule attachment as targets of Plk and Aurora B. Similarly, we found Aurora A and Plk substrates that constitute the centromere and anchor the mitotic spindle at microtubule minus ends.
Our data suggest that Plk activity specifically influenced the kinase activity of Aurora A during mitotic entry, but not in cells arrested at metaphase. Consistent with previous reports (7, 9), we observed that Aurora A regulated Plk1 activity directly by phosphorylation of its T loop. Using our approach, we identified the cross-regulation of Aurora A and Plk1 by monitoring the phosphorylation state not only of their respective T loops, but also on a system-wide scale on a diverse array of substrates.
Our analysis provides a versatile approach for deciphering complex signaling events on a system-wide scale. With this method, we identified a large number of cellular Aurora A, Aurora B, and Plk substrates and connected them to biological processes essential for mitotic progression and successful completion. The scale of the analysis provided statistical power in refining and extending the known motifs for these kinases, in addition to discovering previously unknown motifs. Moreover, the phosphorylation sites discovered in this analysis present an invaluable resource for understanding signaling mechanisms that control mitosis. For instance, we found a specific Aurora A phosphorylation site on the mitotic spindle apparatus protein NuMA and determined that phosphorylation of this site was essential for the migration of NuMA along the mitotic spindle and its function in spindle maintenance.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal bovine serum (Hyclone) and penicillin-streptomycin (100 U/ml and 100 mg/ml, respectively; Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. For SILAC experiments, HeLa cells were grown in arginine- and lysine-free DMEM with 10% dialyzed fetal bovine serum supplemented with either [13C6,15N2]lysine (100 mg/liter) or [13C6,15N4]arginine (100 mg/liter) (Cambridge Isotope Laboratories Inc.) (heavy population) or identical concentrations of iso-topically normal lysine and arginine (light population) for at least six cell doublings.
Immunofluorescence
HeLa cells were fixed with 3% formaldehyde, permeabilized with phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (PBST), and incubated with primary and secondary antibodies at room temperature. DNA was labeled with Hoechst 33342 (Sigma) or PicoGreen (Molecular Probes, Invitrogen). Images in Fig. 1C were collected as Z stacks with 0.3-mm spacing using a 100×, 1.35 numerical aperture (NA) objective on a Nikon TE2000U inverted microscope with a spinning disk confocal system (Perkin Elmer) and processed with MetaMorph software. Images in fig. S1 were collected with a 63×, 1.4 NA objective on an Axioplan2 Zeiss microscope with OpenLAB software.
Small-molecule inhibitors and antibodies
AZD1152 and BI2536 were synthesized in-house. ZM447439 was purchased (Tocris), and MLN8054 (Millennium) was obtained through a materials transfer agreement. Stock solutions of AZD1152, BI2536, and ZM447439 were prepared in dimethyl sulfoxide (DMSO) (Sigma), and MLN8054 in water. For a given inhibitor titration, the respective kinase inhibitor at the indicated concentration along with MG132 (1 mM, Sigma) was added to Taxol-arrested cells and analyzed by Western blotting four times. The following antibodies were used: antibody recognizing Plk1 pThr210 (BioLegend), antibody recognizing both Aurora A and Aurora B kinases phosphorylated at their T loops (Cell Signaling), antibody recognizing Aurora A (Cell Signaling), antibody recognizing histone H3 pS10 (Upstate), antibody recognizing NuMA1 (gift from D. Compton, Dartmouth), antibody recognizing TPX2 (gift from D. Compton, Dartmouth), antibody recognizing phosphorylated and nonphosphorylated Cdc25c (BioLegend), antibody recognizing lamin A/C (gift from F. McKeon, Harvard), CREST autoimmune serum (ImmunoVision), and antibody recognizing α -tubulin (Sigma). Secondary antibodies were obtained from Molecular Probes (Invitrogen) and Jackson ImmunoResearch.
Cellular phenotypes of kinase inhibitor–treated cells
For the analysis of cellular phenotypes, asynchronous HeLa cells were treated with kinase inhibitors and MG132 at the indicated concentrations for 45 min, fixed, and stained with CREST and α-tubulin antibodies and with Hoechst 33342. For each condition, at least 100 cells were inspected for their spindle and chromosome morphology. Cells were scored in five categories for spindle morphology based on the number of spindle poles as observed by tubulin staining (one, monopolar; two, bipolar; more than two, multipolar; not determinable, aberrant). Cells were categorized as pseudo-bipolar if they appeared to have two poles but displayed a compressed spindle with an increased number of astral microtubules emanating from the poles. Monopolar, multipolar, pseudo-bipolar, and aberrant spindles were scored as abnormal, whereas bipolar spindles were scored as normal. For chromosome phenotypes, cells were categorized as aligned, misaligned, rosette-like, halo-like, and monopolar. Alignment of chromosomes was based on both Hoechst and CREST staining. Cells displaying a monopolar spindle were scored as monopolar in chromosome phenotype. Misaligned, rosette-like, halo-like, and monopolar chromosome morphology were scored as abnormal, whereas aligned was scored as normal.
Flow cytometry analysis of kinase inhibitor–treated cells
Cell cycle synchronization of kinase inhibitor–treated HeLa cells was determined by flow cytometry analysis. HeLa cells were synchronized by a double-thymidine block (1 mM, Sigma) for 16 hours each time with 8 hours release in between. For mitotic arrest, 3 hours after the washout of the second thymidine block, Taxol (100 nM, Sigma) was added to the medium for 10 hours. For the 1 µM AZD1152 and 0.1 µM BI2536 mitosis and 5 µM ZM447430, 0.25 µM MLN8054, 1 µM MLN8054, and 5 µM MLN8054 conditions, the respective inhibitor at the indicated concentration along with MG132 (1 µM, Sigma) was added to the Taxol-arrested cells for 45 min. For mock treatment, Taxol-arrested cells were incubated with DMSO and MG132. For the 0.1 µM BI2536 entry condition, heavy and light HeLa cells were synchronized by a double-thymidine block as described above. Three hours after final release from thymidine block, 100 nM Taxol was added to heavy and light cells; 5 hours after addition of Taxol, heavy cells were treated with 0.1 mM BI2536 and 1 µM MG132 for 45 min, and light cells were treated with DMSO and 1 µM MG132 for 45 min. After inhibitor treatment, HeLa cells were collected by mitotic shake-off, fixed with 70% ice-cold ethanol, and stored overnight at 4°C. The next day, cells were washed twice with PBS; re-suspended in PBS containing 0.05% Tween 20, 5% bovine serum albumin, and either fluorescein isothiocyanate–conjugated MPM-2 antibody (Millipore) or Alexa Fluor 488–conjugated pS10 histone H3 antibody (Millipore); and incubated at 4°C for 1 hour with rotation. Cells were washed three times with PBS containing 0.05% Tween 20 and resuspended in PBS/Triton X-100/deoxyribonuclease-free ribonuclease/propidium iodide solution. Cells were incubated at room temperature for 30 min protected from light and analyzed on a FACSCalibur (BD Biosciences) flow cytometer. Cell doublets were removed by gating in software after acquisition.
Small-molecule inhibitor treatment of SILAC HeLa cells
To identify targets of kinase activity, we synchronized heavy- and light-labeled HeLa cells by a double-thymidine block as described above. Three hours after the washout of the second thymidine block, 100 nM Taxol was added to the cells for 10 hours. For the 1 µM AZD1152 and 0.1 µM BI2536 mitosis and 5 µM ZM447430, 0.25 µM MLN8054, 1 µM MLN8054, and 5 µM MLN8054 conditions, heavy-labeled HeLa cells were incubated with the respective inhibitor at the indicated concentration along with MG132 (1 µM, Sigma) for 45 min. For mock treatment, light cells were incubated with 1 µM MG132 and DMSO or water, depending on the solvent the respective kinase inhibitor was resuspended in. For the 0.1 µM BI2536 entry condition, heavy and light HeLa cells were synchronized by a double-thymidine block as described above. Three hours after final release from thymidine block, 100 nM Taxol was added to heavy and light cells; 5 hours after addition of Taxol, heavy cells were treated with 0.1 µM BI2536 and 1 µM MG132 for 45 min, and light cells were treated with DMSO and 1 µM MG132 for 45 min. After inhibitor treatment, HeLa cells were collected by mitotic shake-off and counted. Equal numbers of heavy and light HeLa cells were mixed, washed twice in PBS, snap-frozen, and stored at −80°C until lysis.
For nocodazole arrest, HeLa cells were synchronized by a double-thymidine block; 3 hours after washout of the second thymidine block, nocodazole (100 ng/ml) was added to the cells for 10 hours, and mitoti-cally arrested cells were collected by shake-off. For unperturbed mitotic cells, HeLa cells were synchronized by a double-thymidine block and released into growth media after the second thymidine block. Mitotic cells were collected by mitotic shake-off 9 hours after release. Mitotic shake-off was repeated after 10 and 11 hours to collect a sufficient number of cells, and all cells were pooled together.
Sample preparation, trypsin digestion, and SCX fractionation
Lysis and SCX chromatography were carried out as previously described (45). Twenty-four fractions were collected, lyophilized, and desalted on a 96-well Oasis MicroElution C18 SPE plate (Waters).
Phosphopeptide purification and analysis by mass spectrometry
For phosphopeptide purification, peptides were dissolved in 50% acetoni-trile (Honeywell Burdick & Jackson), 0.1% trifluoroacetic acid (TFA) (Honeywell Burdick & Jackson), and 2 M lactic acid (Sigma) and incubated with ~350 mg of TiO2 microspheres (Glycan Biosciences) for 45 min with agitation. After binding, the TiO2 microspheres were washed with 50% acetonitrile/0.1% TFA, and phosphopeptides were eluted with 50 mM di-sodium phosphate (Sigma) adjusted to pH 10 with ammonia (Sigma), dried, and desalted. Each phosphopeptide purification was analyzed in duplicate by nanoscale microcapillary LC-MS/MS essentially as described (46, 47) on an LTQ-Orbitrap (Thermo Electron).
Data analysis and phosphorylation-site assignments (ModSite)
The collected tandem mass spectra were data-searched with the SEQUEST algorithm (48), filtered to a <1% FDR with the target-decoy strategy (49), and reported. SILAC quantification was performed with a highly in-house modified version of the Xpress algorithm [http://tools.proteomecenter.org (50)]. All heavy-labeled sample/light-labeled sample ratios were log2-transformed and fit to a Gaussian distribution with Sigma Plot software. Log2-transformed ratios were adjusted to the calculated, experiment-specific distribution offset. For most of these phosphopeptides, sufficient site-determining ions were obtained in the MS2 fragmentation spectra to determine the localization of the phosphorylation site by SEQUEST. How-ever, peptides phosphorylated at their extreme termini exhibit fewer available site-determining ions during ion trap MS/MS owing to the instability of low-m/z (mass/charge ratio) species generated during collision-induced dissociation. Aurora kinases preferentially phosphorylate a consensus motif that contains basic residues [R/K]XXp[S/T] (51), which are also the sites of trypsin cleavage, leading to a large number of N-terminally phosphorylated peptides that are inherently problematic when assigning a phosphorylation locus with certainty. Algorithms that assign a score to all possible phosphorylation acceptor residues result in low scores for these phospho-peptides, leading to their exclusion from further analysis. To address this problem, we created identifiers that admit potential ambiguity in assigning phosphorylation to any specific locus when (i) another acceptor residue occurs within two amino acids of a database search result, (ii) a different locus assignment is ascribed to any of these adjacent residues in any other phosphopeptide across all experiments, and (iii) the difference between the most extreme N-terminal and C-terminal phosphoacceptor residues in a collection of many occurrences is less than six amino acids. Rather than report all possible results with separate ratios or discard these results entirely, we classified them as a single, ambiguous identifier, a ModSite, and averaged the quantitative results from each of the individual observations together and calculated the SD of the averaged value. All subsequent calculations were performed on these offset-corrected, averaged values for each site of phosphorylation (called ModSites).
All ratios were arrayed by ModSites in rows, and inhibitor conditions in columns. Agglomerative hierarchical clustering was carried out with the centered correlation similarity metric and centroid linkage in Cluster 3.0 (52). The output was visualized by Java TreeView 1.1.1 (53). Significant differences between ratios were determined with a two-tailed t test function assuming unequal variance in Excel. For pathway and evolutionary analysis, we expanded our arrays by eliminating the Taxol/Taxol ratio requirement used for clustering for the Aurora kinase substrates, and by eliminating the MLN as well as AZDZM conditions for Plk1 substrate analysis.
Motif analysis
Motif enrichment analysis of peptides in the different clusters was performed with Motif-X (54). For the overall percentage of motif-containing ModSites, the number of peptides containing a representative motif ([R/K]XXp[S/T] or [R/K]Xp[S/T] for potential Aurora A substrates; [R/K]XXp[S/T], [R/K]Xp[S/T], or [R/K]p[S/T] for potential Aurora B substrates; and [D/E/N]Xp[S/T] or p[S/T]F for potential Plk substrates) was determined.
Protein expression
For expression in bacteria, genes were amplified from a complementary DNA (cDNA) library and cloned into a modified version of the pet16b vector (Novagen/EMD Chemicals) containing a 10-His tag and a modified multiple cloning site. For protein expression, plasmids were transformed into Rosetta (DE3)pLysS (Novagen). Protein expression was carried out for 6 hours at 20°C. Ten-His–tagged proteins were purified with Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. Purified proteins were dialyzed overnight against 10 mM Hepes (pH 7.7), 0.1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% glycerol and stored at −80°C. For expression in insect cells, genes were amplified from a sequenced cDNA clone and cloned into a modified version of the pFastBac vector (Invitrogen) containing a 10-His tag (Plk1, TPX2, and INCENP) or pFastBac vector without tag (Aurora A and B). For bacmid generation, pFastBac constructs were transformed into DH10Bac Escherichia coli (Invitrogen). Recombinant bacmid DNA was purified, and recombination was confirmed by polymerase chain reaction (PCR). Recombinant bacmid DNA was transfected into Sf9 cells with Cellfectin (Invitrogen) according to the manufacturer’s instructions. Five days after transfection, P1 virus stock was isolated and further amplified. For protein expression, Sf9 cells were infected with amplified virus stocks, and cells were harvested 72 hours after infection. For Aurora A and TPX2 and Aurora B and INCENP co-expression, Sf9 cells were infected with both virus stocks. Three hours before harvesting, cells expressing Aurora A–TPX2, Aurora B-INCENP, Plk1, or Plk4 were treated with 100 nM okadaic acid (LC Labs). Ten-His–tagged proteins were purified with Ni-NTA agarose (Qiagen) according to the manufacturer’s instructions. Purified proteins were dialyzed overnight against 10 mM Hepes (pH 7.7), 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol and stored at −80°C.
In vitro kinase reactions
For Aurora A–TPX2 and Aurora B–INCENP in vitro protein kinase reactions, 1 µg of substrate was incubated with 75 ng of kinase in kinase buffer [20 mM Hepes (pH 7.7), 5 mM MgCl2, 0.1 mM EGTA, 0.1 mM DTT, 2.5 mM β-glycerophosphate, and 100 µM adenosine 5’-triphosphate (ATP)] at 30°C for 1 hour. For Plk1 in vitro kinase reactions, 1 µg of substrate was incubated with 75 ng of kinase in kinase buffer [20 mM Hepes (pH 7.7), 20 mM MgCl2, 0.1 mM EGTA, 0.1 mM DTT, 2.5 mM β-glycerophosphate, and 100 µM ATP] at 30°C for 1 hour. For control reactions, 1 µg of substrate was incubated with the respective kinase buffer at 30°C for 1 hour. Afterward, proteins were reduced with 5 mM DTT at 55°C, alkylated with 15 mM iodoacetamide (Sigma) at room temperature in the dark, and resolved by SDS-PAGE. Gel bands were excised destained trypsin-digested and analyzed by LC-MS/MS.
For in vitro peptide kinase assays, 3 µg of peptide substrates (New England Peptide) was incubated with 20 to 100 ng of kinase in the respective kinase buffer (see above) at 30°C for 30 min. For BI2536 inhibition studies, BI2536 was diluted in kinase buffer, and the kinase was added before peptide substrate was added. Afterward, the reactions were quenched with 0.1% TFA/3% methanol, desalted on an Oasis MicroElution C18 SPE plate (Waters), and diluted 1:100, and 1 µl was analyzed by Orbitrap LC-MS/MS.
Plk1 immunoprecipitation
For Plk1 immunoprecipitation, the Plk1 antibody (Sigma) was cross-linked with dimethyl pimelimidate to Protein G Agarose beads (Roche) according to the manufacturer’s instructions. Taxol-arrested HeLa cells were collected, washed with PBS, and lysed in lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM β-glycerophosphate, 1 mM sodium molybdate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors]. The lysate was clarified by centrifugation at 13,000g for 30 min at 4°C. The supernatant was transferred to a new tube and incubated with antibody-coupled beads for 2 hours at 4°C with rotation. Afterward, beads were washed and eluted with SDS-sample buffer at 70°C for 5 min, reduced with 5 mM DTT at 55°C, alkylated with 15 mM iodoacetamide (Sigma) at room temperature in the dark, resolved by SDS-PAGE, and visualized with Coomassie blue stain. Gel bands were excised, destained, trypsin-digested, and analyzed by LC-MS/MS.
Mutagenesis and transfection
Site-directed mutagenesis was performed with the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions and confirmed by DNA sequencing. HeLa cells were trans-fected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank D. Compton for providing reagents and for helpful discussions, D. Madden for help with baculovirus protein expression, J. Ruderman for providing active Aurora A kinase, F. McKeon for providing reagents, J. Gilmore and J. Milloy for technical discussions concerning data analysis, S. Cullati for proofreading the manuscript, and Millennium Pharmaceuticals for providing MLN8054.
Funding: This work was supported by NIH grant P20-RR018787 from the IDeA Program of the National Center for Research Resources, the American Cancer Society grant IRG-82-003-24 (to S.A.G.), and predoctoral fellowships from the National Institute of General Medical Sciences grant T32-GM008704 (to D.K.S. and B.K.F.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/4/179/rs5/DC1
Materials and Methods
Fig. S1. Spindle and chromosome morphology of inhibitor-treated HeLa cells.
Fig. S2. Plk1 and Plk4 inhibition by BI2536.
Fig. S3. Experimental setup and schematic of time line of HeLa cell synchronization.
Fig. S4. Flow cytometry analysis of inhibitor-treated HeLa cells.
Fig. S5. Ratio distributions.
Fig. S6. Comparison of the heavy-to-light ratios in AZD1152- and ZM447439-treated cells.
Fig. S7. Candidate Aurora kinase A versus B targets.
Fig. S8. Cluster and motif analysis of ambiguous Aurora substrates.
Fig. S9. Log2 ratio distribution of known Aurora kinase A, Aurora kinase B, and Plk1 substrates identified in this analysis.
Fig. S10. Candidate Plk targets.
Fig. S11. In vitro peptide kinase motif assay
Fig. S12. Regulation of Aurora A by Plk.
Fig. S13. Prediction of phosphorylation sites in structured and ordered regions of proteins.
Fig. S14. Evolutionary motif conservation.
Fig. S15. T loop sequence alignments.
Fig. S16. STRING and MCODE analysis of proteins predicted to be phosphorylated by Aurora A, Aurora B, or Plk.
Fig. S17. Western blot and flow cytometry analysis of nocodazole-arrested HeLa cells and HeLa cells collected by mitotic shake-off after thymidine release.
Fig. S18. Reciprocal plots of mass spectrometry results from in vitro kinase reactions and cellular proteomics analysis.
Fig. S19. The effect of NuMA phosphorylation on NuMA localization.
Details Regarding Data Availability
Description for Tables S1 to S6
References
Table S1. All ModSites and representative peptides.
Table S2. ModSites assigned to the Aurora kinase substrate cluster.
Table S3. ModSites assigned to the Plk substrate cluster.
Table S4. Analysis of site and motif conservation for candidate Aurora A, Aurora B, and Plk substrates across evolution.
Table S5. ModSite assignments to Aurora A, Aurora B, Aurora ambiguous, and Plk clusters.
Table S6. Plk1-interacting proteins.
Author contributions: A.N.K. coordinated the project, conducted most of the experimental work, analyzed most of the data, prepared the figures, and drafted the manuscript. D.K.S. conducted the Aurora A inhibitor study. B.K.F. established the informatics pipeline used for quantification and ModSite assignments. D.P. conducted the evolution alignments and data analysis. A.A.P. synthesized BI2536 and AZD1152. S.A.G. conceptualized the study, designed and performed the experiments, analyzed the data, and finalized the manuscript.
Competing interests: Compound MLN8054 requires a material transfer agreement (MTA) from Millennium Pharmaceuticals. S.A.G. has a paid consultancy with Millennium Pharmaceuticals relating to the clinical use of MLN8054 as an anticancer agent.
Data availability: Raw data files are available through https://proteomecommons.org/(see Supplementary Materials for codes).
REFERENCES AND NOTES
- 1.Dorée M, Hunt T. From Cdc2 to Cdk1: When did the cell cycle kinase join its cyclin partner? J. Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 3.Carmena M, Earnshaw WC. The cellular geography of Aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 4.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim. Biophys. Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouché JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, Ducommun B. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 6.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H. Aurora-A and interacting activator, the LIM protein Ajuba are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 7.Macůrek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 8.Satinover DL, Brautigan DL, Stukenberg PT. Aurora-A kinase and inhibitor-2 regulate the cyclin threshold for mitotic entry in Xenopus early embryonic cell cycles. Cell Cycle. 2006;5:2268–2274. doi: 10.4161/cc.5.19.3316. [DOI] [PubMed] [Google Scholar]
- 9.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 11.Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Philos. Trans. R. Soc Lond. B Biol. Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 13.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 14.Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sèle B, Jouk PS, Ohlmann T, Rousseaux S, Lunardi J, Ray PF. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat. Genet. 2007;39:661–665. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- 15.Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev. Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 16.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 17.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 18.Salaun P, Rannou Y, Prigent C. Cdk1 Plks and Auroras and Neks: The mitotic bodyguards. Adv. Exp. Med. Biol. 2008;617:41–56. doi: 10.1007/978-0-387-69080-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 2002;277:44115–44120. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 20.Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated ki-nase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–4908. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- 21.van de Weerdt BC, van Vugt MA, Lindon C, Kauw JJ, Rozendaal MJ, Klompmaker R, Wolthuis RM, Medema RH. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating Polo-like kinase 1. Mol. Cell. Biol. 2005;25:2031–2044. doi: 10.1128/MCB.25.5.2031-2044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 23.Petronczki M, Lénárt P, Peters JM. Polo on the rise—From mitotic entry to cytokinesis with Plk1. Dev. Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 2006;24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 25.Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf GR, Kraut N, Peters JM, Rettig WJ. BI 2536 a potent selective inhibitor of Polo-like kinase 1 inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 26.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora Bcouples chromosome alignment with anaphase by targeting BubR1 Mad2 and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann JJ, Warin N, Renaud F, De Savi C, Roberts NJ, Johnson T, Dousson CB, Hill GB, Perkins D, Hatter G, Wilkinson RW, Wedge SR, Heaton SP, Odedra R, Keen NJ, Crafter C, Brown E, Thompson K, Brightwell S, Khatri L, Brady MC, Kearney S, McKillop D, Rhead S, Parry T, Green S. Discovery synthesis and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of Aurora B kinase. J. Med. Chem. 2007;50:2213–2224. doi: 10.1021/jm061335f. [DOI] [PubMed] [Google Scholar]
- 30.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, Ray ET, Sells TB, Stringer B, Stroud SG, Vos TJ, Weatherhead GS, Wysong DR, Zhang M, Bolen JB, Claiborne CF. Antitumor activity of MLN8054 an orally active small-molecule inhibitor of Aurora A kinase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 32.Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J. Cell Sci. 2005;118:2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- 33.Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 34.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture SILAC as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 37.Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Weerdt BC, Littler DR, Klompmaker R, Huseinovic A, Fish A, Perrakis A, Medema RH. Polo-box domains confer target specificity to the Polo-like kinase family. Biochim. Biophys. Acta. 2008;1783:1015–1022. doi: 10.1016/j.bbamcr.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Sun QY, Schatten H. Role of NuMA in vertebrate cells: Review of an intriguing multifunctional protein. Front. Biosci. 2006;11:1137–1146. doi: 10.2741/1868. [DOI] [PubMed] [Google Scholar]
- 42.Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J. Cell Sci. 1995;108:621–633. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- 43.Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- 44.Saredi A, Howard L, Compton DA. NuMA assembles into an extensive filamentous structure when expressed in the cell cytoplasm. J. Cell Sci. 1996;109:619–630. doi: 10.1242/jcs.109.3.619. [DOI] [PubMed] [Google Scholar]
- 45.Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieguez-Acuna FJ, Gerber SA, Kodama S, Elias JE, Beausoleil SA, Faustman D, Gygi SP. Characterization of mouse spleen cells by subtractive proteomics. Mol. Cell. Proteomics. 2005;4:1459–1470. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villén J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 49.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 50.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohashi S, Sakashita G, Ban R, Nagasawa M, Matsuzaki H, Murata Y, Taniguchi H, Shima H, Furukawa K, Urano T. Phospho-regulation of human protein kinase Aurora-A: Analysis using anti-phospho-Thr288 monoclonal antibodies. Oncogene. 2006;25:7691–7702. doi: 10.1038/sj.onc.1209754. [DOI] [PubMed] [Google Scholar]
- 52.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 53.Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.