Abstract
The highly elongated single-celled cotton fibre consists of lint and fuzz, similar to the Arabidopsis trichome. Endoreduplication is an important determinant in Arabidopsis trichome initiation and morphogenesis. Fibre development is also controlled by functional homologues of Arabidopsis trichome patterning genes, although fibre cells do not have a branched shape like trichomes. The identification and characterization of the homologues of 10 key Arabidopsis trichome branching genes in Gossypium arboreum are reported here. Nuclear ploidy of fibres was determined, and gene function in cotton callus and fibre cells was investigated. The results revealed that the nuclear DNA content was constant in fuzz, whereas a limited and reversible change occurred in lint after initiation. Gossypeum arboreum BRANCHLESS TRICHOMES (GaBLT) was not transcribed in fibres. The homologue of STICHEL (STI), which is essential for trichome branching, was a pseudogene in Gossypium. Targeted expression of GaBLT, Arabidopsis STI, and the cytokinesis-repressing GaSIAMESE in G. hirsutum fibre cells cultured in vitro resulted in branching. The findings suggest that the distinctive developmental mechanism of cotton fibres does not depend on endoreduplication. This important component may be a relic function that can be activated in fibre cells.
Key words: Branch, endoreduplication, fuzz, Gossypium, lint, nuclear DNA content.
Introduction
Although the majority of plant trichomes are multicellular, those of Arabidopsis trichomes (leaf hairs) and Gossypium (cotton)—unique among crop plants—consist of extremely elongated single cells. The four commercially domesticated species of cotton (Gossypium hirsutum, G. herbaceum, G. arboreum, and G. barbadense) produce two types of seed fibres (lint and fuzz) in distinct waves. Lint production is usually initiated before or on the day of anthesis, whereas the development of fuzz fibres from rapidly enlarging spherical cells occurs in a second wave a few days later.
Arabidopsis trichomes are rarely formed adjacent to each other, and fate determination relies on a substrate depletion and lateral inhibition mechanism (Ishida et al., 2008; Pesch and Hulskamp, 2009). The activators of trichome fate in Arabidopsis are TRANSPARENT TESTAGLABRA1 (TTG1) and GLABRA1, 2, and 3 (GL1, GL2, and GL3). In cotton, the R2R3 MYB genes GhMYB109 and GaMYB2, which have high sequence similarities to GL1 and GaHOX1 (homologous to GL2), are specifically expressed in the early stages of fibre cell development. Ectopic expression of these cotton genes under control of the GL1 or GL2 promoter can restore formation of trichomes in their corresponding mutants (Suo et al., 2003; Wang et al., 2004). Functional homologues of Arabidopsis TTG1 and GL3 have also been cloned from G. hirsutum (Humphries et al., 2005; Shangguan et al., 2008). Anatomic and morphological observations reveal that cotton fibres seldom form clustering, like that observed for Arabidopsis trichomes, suggesting that a similar mechanism is involved in the regulation of fibre spacing. These findings suggest that the Arabidopsis genes involved in trichome formation pathways may also participate in cotton fibre patterning processes (Serna and Martin, 2006).
Upon Arabidopsis trichome initiation, a unique branched cellular architecture immediately appears, with mature Arabidopsis leaf trichomes typically having three branches borne on a stalk. Cells destined to become protrichomes exit the mitotic stage and enter into the endoreduplication progress. Protrichome cells form branches and then expand during endoreduplication. Endoreduplication is a modified cell cycle in which nuclear chromosomal DNA is repeatedly replicated without subsequent cell division (Kasili et al., 2011). Arabidopsis SIAMESE (SIM), a plant-specific cell cycle regulator, controls the onset of trichome endoreduplication (Churchman et al., 2006), and BRANCHLESS TRICHOMES (BLT) regulates endoreduplication levels (Bramsiepe et al., 2010; Kasili et al., 2011). Aside from the endoreduplication pathway, genetic and molecular data indicate that nine different loci influence branching via independent molecular pathways. KATANIN1 (KTN1), SPIKE1 (SPK1), ZWICHEL (ZWI), ETHYLENE RECEPTOR2 (ETR2), KIESEL (KIS), and PORCINO (POR) participate in microtubule biogenesis (Oppenheimer et al., 1997; Burk et al., 2001; G.T. Kim et al., 2002; Kirik et al., 2002; Qiu et al., 2002; Plett et al., 2009). ANGUSTIFOLIA (AN) localizes to punctate structures around the Golgi apparatus (Minamisawa et al., 2011) and is thought to regulate Golgi-related processes (Hülskamp, 2004), whereas STOMATAL CYTOKINESIS-DEFECTIVE 1 (SCD1) regulates cytokinesis (Falbel et al., 2003; Kang et al., 2003). In addition to these genes, STICHEL (STI) initiates trichome branch formation in a dose-dependent manner via an unknown mechanism (Ilgenfritz et al., 2003; Kasili et al., 2011). With the exception of sim, mutations in each of the above genes lead to trichome hypobranching. If cotton fibre cells are homologous to trichomes, then why are cotton fibres unbranched, and are they capable of branching? In this study, both of these long-standing questions are assessed.
Here, it is shown that endoreduplication was not concomitant with fibre outgrowth, corresponding to the BTL homologue being epistatic in fibre. Fibres could be induced to branch using targeted gene co-expression of (i) cotton BTL to elevate endoreduplication levels; (ii) Arabidopsis STI to replace the pseudogene in cotton; and (iii) cotton SIM to promote the mitosis to endoreduplication transition in developing fibre cells.
Materials and methods
Plant materials and growing conditions
Gossypium arboreum cv. DPL971, its natural fuzzless mutant DPL972, and G. hirsutum cv. MD51 ne were grown in field nurseries of a greenhouse at 25–32 °C with ~14h illumination. Bolls were selected from the same boll-bearing branches. Flowers were dated on the day they opened. This day was defined as 0 day post-anthesis (0 DPA). Roots, stems, and leaves were collected 20 d after germination, unless otherwise noted.
Genome DNA extraction and Southern blots
General procedures for DNA manipulation were as described by Sambrook and Russell (2001). Southern blots were performed under both low- and high-stringency conditions. Stringency is defined as the degree of complementarity. Depending on evolutionary distance, gene sequences in cotton and Arabidopsis may have diverged extensively. Low-stringency hybridization permitted the probe to bind to related but not necessarily identical genes in the same family or to locate a gene in G. arboreum that was similar to a known gene in Arabidopsis. With high-stringency hybridization, the probe ideally bound to a completely complementary sequence. Digoxigenin-labelled high-stringency hybridization was performed in accordance with the Cheunglab public protocol (http://www.dartmouth.edu/~staphy/protocol/southern_with_dig_probe.html). Low-stringency hybridization was carried out at 42 °C, and the membranes were washed with 0.2× SSC (0.3M sodium chloride, 0.03M sodium citrate, pH 7.0) and 0.1% (w/v) SDS.
RNA analysis
Total RNA was isolated using Column Plant RNAout 2.0 (Tiandz, Beijing, China). For RNA in situ hybridization, cotton ovules were collected and fixed in a solution of 4% (w/v) formaldehyde and 0.1% (v/v) Tween-20 in diethylpyrocarbonate-treated phosphate-buffered saline (PBS). The fixed ovules were processed according to the protocol described by Mayer et al. (1998). The samples were infiltrated with paraffin. Tissue sections were 8 µm in thickness. Digoxigenin-11 UTP (Roche) was incorporated into antisense or sense RNA probes. Preparation of radiolabelled probes and northern blot analysis were carried out as described by Sambrook and Russell (2001). Equal mixtures of 3, 5, 7, 10, 15, 20, and 25 DPA ovule or fibre RNA were used.
Data analysis
The whole-genome uncombined shotgun sequence of the wild diploid cotton species G. raimondii, which was sequenced and made available by Monsanto and Illumina, was downloaded from the GenBank database. A local Blast search was carried out using the related genes of Arabidopsis as the queries. Sequence information was extracted if the E-value was ≤10.
Nuclear isolation and DNA content measurement
Procedures for nuclear isolation and DNA microspectrophotometric measurements (using a CRAIC microspectrophotometer) of 20 and 25 DPA fibre cells were as previously described (Van’t Hof, 1999). The nuclear DNA content of G. arboreum fibre specimens was estimated by comparing fluorescence values with those of chicken erythrocytes, based on 2C DNA=2.4 pg as the standard (Clowes et al., 1983; Kapraun and Nguyen, 1994). Gossypium arboreum root tip cells were used as the 2C reference value standard. DPL971 root tips (3–4cm in length) were harvested and fixed in formaldehyde as described above. After rinsing in phosphate buffer for 10min, cell walls were enzymatically degraded by incubating root tips in 2.0% (w/v) cellulase R-10 (Sigma) and 0.5% (w/v) pectinase (Sigma) for 1.5h at 37 °C in a rotary shaker at 30rpm. Nuclear isolation and DNA measurements were performed as described above.
For determining early-stage ovule DNA content, the ovule fixation, subsequent sucrose infiltrations, embedding, sectioning at 50 µm, and dehydration were as previously described (Wu et al., 2007). The MMI CellCut laser microdissection system (Molecular Machines & Industries) was used for laser capture of epidermal cells. Cell wall enzymolysis, nuclear isolation, and DNA measurements of the collected cells were performed as described above.
For flow cytometric analysis, in vitro cultured calli were chopped with a razor blade in 0.1M citric acid and 0.5% (v/v) Tween-20, and the nuclear suspension was prepared using the two-step protocol described by Doležel et al. (2007). The nuclei were analysed using the BD FACSVerse flow cytometer.
Protein–protein interaction assays and western blot analysis
Entry clones containing full-length cDNA of GaBLT or a cDNA fragment of STI (encoding the N-terminal 454 amino acids) were transferred to the yeast two-hybrid vectors pGADT7-Rec and pGBKT7 (Clontech) and used to transform the yeast strain AH109 (Clontech).
Co-immunoprecipitation was performed similarly to as described by Staub et al. (1996) on 35S::STI+35S::GaBLT-GFP transgenic callus. Immunoprecipitation of the GaBLT–green fluorescent protein (GFP) protein was performed using an anti-GFP antibody (Santa Cruz Biotechnology). Protein G–agarose (Sigma) was used to precipitate the immunoprotein complexes. STI detection was performed with an anti-STI antibody. The anti-STI polyclonal antibody was raised against the N-terminal 454 amino acids of STI purified in recombinant yeast Pichia pastoris. Fibre protein extraction and western blot analysis were performed as described by Zhao et al. (2009). A 10 µg aliquot of proteins was separated in a 12% (w/v) SDS–polyacrylamide gel.
Bombardment of cultured cotton ovules, cotton transformation, and callus induction
Ovule culture and particle bombardment were conducted as previously described (Hof and Saha, 1997; H.J. Kim et al., 2002; Wang et al., 2010). Ovules of G. hirsutum MD51 ne at 2 DPA were bombarded, and then incubated in solid Beasley and Ting medium supplemented with 2.0 µM indole-3-acetic acid (IAA) and 2.0 µM gibberellic acid-3 (GA3) for 2 d. Thereafter, the cotton ovules were transferred onto liquid medium containing the same components.
The experimental procedures of G. arboreum DPL971 transformation and callus induction were mainly as described by Shang et al. (2009). The co-cultivation medium (pH 5.8) was as described in Smith et al. (1977). The subculture medium contained naphthaleneacetic acid (2.0mg l−1) and benzyladenine (0.5mg l−1). Vigorous calli were then selected and transferred onto fresh medium every 4 weeks until the transgenes were detected.
Cytology
Ovules were removed from the suspension medium and fixed in methanol:glacial acetic acid (3:1). Histochemical localization of β-glucuronidase (GUS) activity was performed according to the protocol reported by Jefferson et al. (1987). Small tufts of fibres were removed from the blue-stained GUS-expressing region. Fibre cells were separated using a microspectrophotometer based on the presence or absence of branching. In situ nuclear staining was as described by Hof and Saha (1997). The method for fibre nuclear segregation was as described above. Slide preparations and chromosome observation with 4’,6-diamidino-2-phenylindole (DAPI) staining were as previously described (Wang et al., 2007; Gan et al., 2011). The experimental procedures for callus histological observation (in 10 µm thick sections) was as described by Shang et al. (2009).
Sequence data in this article are presented with GenBank accession numbers, unless otherwise stated.
Results
Identification and cloning of homologous trichome-branching genes in G. arboreum
Based on known mechanisms in Arabidopsis, an attempt was made to identify homologues of SPK1, KTN1, ETR2, AN, ZWI, SCD1, KIS, POR, BLT, and STI in diploid G. arboreum. Putative full-length cDNAs of GaKTN1-1 (KC246031), GaKTN1-2 (KC246032), GaETR2-1 (KC246027), GaETR2-2 (KC246028), GaETR2-3 (KC246026), GaKIS (KC246029), and GaPOR (KC246030) were recovered based on Gossypium expressed sequence tags (ESTs; from GenBank) and the rapid amplification of cDNA ends (RACE) method. The homologues shared >50% overall sequence identity. All of them had conserved functional domains required for their particular biochemical reactions.
Because no highly homologous sequences of STI, SPK1, ZWI, AN, SCD1, and BLT for Gossypium were present in public databases, the Arabidopsis mRNA sequences were used to search against raw uncombined G. raimondii genome sequences. Primers were designed based on the most frequently identified combined short sequences (~100bp), and gene-containing fragments were amplified from G. arboreum genomic DNA or cDNA. Six potential genes were identified [designated GaSTI (JQ867271), GaAN (JQ711138), GaBLT (JQ711139), GaZWI (JQ711141), GaSPK1 (JQ711140), and GaSCD1 (KC246035)], and their deduced coded proteins all showed >70% identity to corresponding proteins in Arabidopsis. To ensure that other gene homologue sequences were not missed owing to the incomplete genome sequence or through base errors, the five sequences were evaluated using low- and high-stringency Southern blots. Only one hybridization band appeared in each lane under high-stringency conditions when using G. arboreum sequence probes (Supplementary Fig. S1 available at JXB online). These hybridization bands also appeared in the same location under low-stringency conditions when Arabidopsis-related sequences were used as a probe. This indicated that single copies of the five Arabidopsis homologous genes related to trichome branching had been identified in diploid G. arboretum (see the Materials and methods). However, these genes did not result in fibre branching in G. arboretum, and hence their expression patterns were investigated.
GaBLT is not expressed in fibres, and cotton STI is a pseudogene
The function of a gene is based upon its appropriate expression. The transcriptional levels of all the homologues of the 10 Arabidopsis genes were examined with northern blotting. The transcripts of GaKTN1-1, GaKTN-2, GaETR2-1, GaETR2-2, GaETR2-3, GaKIS, GaPOR, GaSPK1, GaZWI, GaAN, and GaBLT were detected in the 3–25 DPA ovule mixture (Supplementary Fig. S2A at JXB online). The expression of GaKIS, GaPOR, GaSPK1, GaZWI, and GaAN occurred at similar levels in fibres. GaKTN1, GaETR2, and GaSCD1 mRNA accumulated at significantly higher levels in fibres than in ovules. GaBLT transcripts were undetectable in fibres, and this was further confirmed with in situ hybridization (Supplementary Fig. S2B). GaSTI transcripts were not detected in the ovules or fibres. The presence of GaSTI transcripts in other tissues (cotyledon, hypocotyl; root, stem, leaf, at 5- and 15-leaf stages; petal and stamen mixture) was also tested. However, the transcripts were undetectable with northern blotting using seven different probes corresponding to its conceptual reading frame, or by quantitative RT–PCR (results not shown; see the Materials and methods and Supplementary Fig. S3). The above experiments were repeated using G. hirsutum RNA samples, and, again, no STI-like transcript was detected (data not shown). Thus, G. hirsutum STI and G. arboreum STI are considered pseudogenes (ψ). However, with the exception of ψGaSTI and GaBLT, the other identified genes were expressed in fibres, suggesting that fibre cells may have the potential for branching.
Nuclear DNA content of the fuzz fibre is constant, and a limited and reversible change occurs in lint
To test the possibility that the absence of GaBLT expression affects fibre endoreduplication levels, the nuclear DNA content of G. arboreum DPL971 fibres at 20 and 25 DPA was examined using the 2C criterion for a root tip nucleus. At 20 and 25 DPA, lint nuclear DNA averaged 3.3±0.28 pg (n=582) and 3.4±0.3 pg (n=547), whereas fuzz DNA content averaged 3.4±0.3 pg (n=510) and 3.5±0.25 pg (n=563), respectively. These results closely correspond to root tip measurements [3.4±0.25 pg, n=479; P > 0.05, Kruskal–Wallis one-way analysis of variance (ANOVA) on ranks and an all pairwise multiple comparison Dunn’s test].
The constant nuclear DNA content in mature fibre cells that had ceased elongation (~20 DPA) may prolong the early developmental stage that is amenable to division. In this case, the timing of endoreduplication may be altered in the early stages of fibre development, followed by degradation or other event(s) leading to the observed 2C level. Unfortunately, accurate measurement of DNA content from early developmental stages of fuzz fibres is difficult because the fuzz fibres are concealed amongst lint fibres. Consequently, as a proxy for detecting endoreduplication levels at the early fuzz developmental stages, the differences in fibre DNA content were compared between DPL971 (containing both lint and fuzz) and its fuzzless lint-normal mutant DPL972. Histological sections revealed that the fuzz of DPL971 was initiated at 3–4 DPA in our cultivation conditions (Wang et al., 2013). Nuclei from the fibres of both strains exhibited elevated DNA content (~3.4 pg, 2C to ~4.1 pg, 2.4C) of the same order of magnitude from 2 to 4 DPA (Supplementary Fig. S5A at JXB online). The statistical mean of the nuclear DNA content was slightly reduced from ~4.1 pg, 2.4C (4 DPA) to 3.8 pg, 2.2C (7 DPA) in DPL971. In contrast, 5 or 7 DPA DPL972 nuclear DNA content was equal to that at 4 DPA. These results demonstrate that the fuzz nuclei had a lower nuclear DNA content, which lowered the statistical mean nuclear DNA content of DPL971 samples that inevitably also contained fuzz nuclei. Furthermore, in the wild-type DPL971, the percentage of 5 and 7 DPA nuclei with the highest relative fluorescence units (RFU, 2.0) was lower than at 4 DPA, and the percentage of nuclei with 1.6 RFU (2C) was increased (Supplementary Fig. S5A). Thus, the increased proportion of 2C nuclei was attributed to fuzz because this profile did not appear in the nuclear determination of fuzzless DPL972 (Supplementary Fig. S5A). The above results indicated that fuzz DNA was constant (2C) at the fuzz fate determination and initiation stages, and elevated in lint.
The timing around lint initiation was also accessed. Swelling of the trichoblast cells that initiated lint growth occurred at –2 to –1 DPA in DPL971 (Supplementary Fig. S4 at JXB online). A laser microdissection system was used to collect adjacent epidermal cells from the DPL971 ovule at –4 to –5, –3, and –2 DPA) and from the lint and surrounding cells (at –1, 0, and 1 DPA). The DNA levels in epidermal cells did not differ from those of root tip cells before the occurrence of the swelling of the trichoblast. No detectable differences were found between the lint cells and the surrounding unexpanded cells (Supplementary Fig. S5B). This indicates that DPL971 lint fibre DNA content was constant before 1 DPA.
To summarize, the DNA content of fuzz cells was constant at the 2C level when endoreduplication did not occur. Upon initiation, an ephemeral DNA partial reduplication (note, it is unsure if it is an endoreduplication; see Discussion) was observed in lint, followed by a return to the 2C level.
Overexpression of GaBLT increases cotton cell endoreduplication levels, but co-expression with interactive Arabidopsis STI made no additional contribution to the DNA cycle
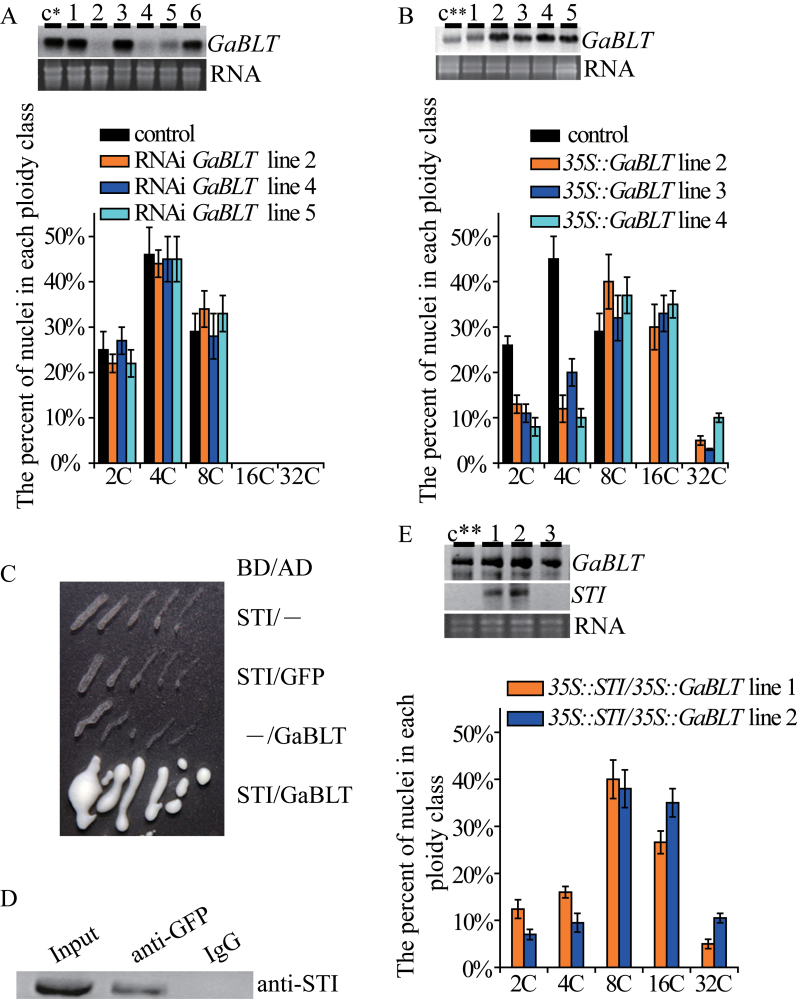
GaBLT was expressed in cotton callus cells (Fig. 1A, B). To test if GaBLT was essential for endoreduplication in cotton, RNAi (RNA interference) lines were generated in which the interfering RNA was targeted specifically to the 3’-coding region of GaBLT. Six independent transgenic cell lines were analysed with northern blotting for GaBLT transcripts. Three of these RNAi cell lines had significantly low levels of GaBLT, but no effect on nuclear DNA content (Fig. 1A), suggesting that GaBLT function was not required for endoreduplication. However, all transgenic cell lines overexpressing GaBLT driven by the 35S promoter showed a significant increase in DNA content relative to untransformed cell lines or pBI121 empty vector-transformed cell lines (Fig. 1B). Therefore, GaBLT was sufficient for endoreduplication in cotton callus cells.
Fig. 1.
The effects of GaBLT epistasis and overexpression on endoreduplication levels. (A) Northern blot with an antisense GaBLT probe showing GaBLT mRNA in independent GaBLT-RNAi cell lines (top panel). Flow cytometric DNA analysis of the effectual GaBLT-RNAi cell lines (lower panel). (B) Northern blot showing GaBLT mRNA in independent 35S::GaBLT transgenic cell lines (top panel). Flow cytometric DNA analysis of the effectual 35S::GaBLT cell lines (lower panel). (C) Interaction between STI and GaBLT in a −His +3-amino-1,2,4-triazole (30mM) plate. BD, GAL4-binding domain; AD, GAL4 activation domain. Note, STI and GFP do not interact. (D) Co-immunoprecipitation of GaBLT–GFP and STI. Transgenic callus extracts were immunoprecipitated with an anti-GFP antibody and detected with western blotting using an antibody raised against STI. IgG, pre-immune serum. (E) Northern blotting with an antisense GaBLT probe showing GaBLT mRNA in independent 35S::GaBLT and 35S::STI co-transformed cell lines (top panels). Flow cytometric DNA content analysis of GaBLT and STI co-expressing cell lines (lower panel). The control c* or c** lines were transformed with the pART27 or pBI121 empty vectors, respectively. DNA content of the non-transgenic cell lines was the same as the lines transformed with pART27 or pBI121. Error bars in (A), (B), and (E) indicate the SD for three independent experiments. The numbers in the top images of (A), (B), and (E) indicate cell lines.
In Arabidopsis, BLT interacts both genetically and physically with STI (Kasili et al., 2011). The hypothesis that GaBLT and STI proteins interact was tested using two methods. Yeast two-hybrid experiments demonstrated a clear interaction between full-length GaBLT and the N-terminal 472 amino acids of Arabidopsis STI (Fig. 1C). The interaction between STI and GaBLT in vivo was confirmed with an immunoprecipitation experiment in which the full-length GaBLT coding region was fused to GFP and co-expressed with STI in callus cells (Fig. 1D). The two protein products may act as part of a complex and induce endoreduplication. However, the nuclear DNA content of the cell lines co-expressing STI and GaBLT did not differ significantly from that of the 35S::GaBLT transgenic cell lines (Fig. 1B, E; P > 0.05, Kruskal–Wallis one-way ANOVA on ranks and Dunn’s test). The above results indicated that ectopic STI expression may have a specific function in conjunction with GaBLT in cotton, but that this role is not endoreduplication. Interestingly, the homologue of GaBLT in G. hirsutum was expressed in the cultured fibres (subsequent result in Supplementary Fig. S7 at JXB online), indicating that GaBLT alone was not enough to promote fibre branching and that an interactive STI homologue or an analogous factor may be necessary.
Overexpression of GaSIM promotes cotton cell transition from mitosis to endoreduplication
Mitosis occurs in differentiated fibre cells of cultivar MD51 ne ovules grown in in vitro culture (at 2 DPA, at a high temperature of 34 °C), producing multicellular, unbranched fibres harbouring individual nuclei in each cell (Hof and Saha, 1997, 1998). Trichomes of the Arabidopsis sim mutant are also multicellular and have normal morphology. SIM inhibits transition from the G2 phase to mitosis and contributes to endoreduplication (Churchman et al., 2006).
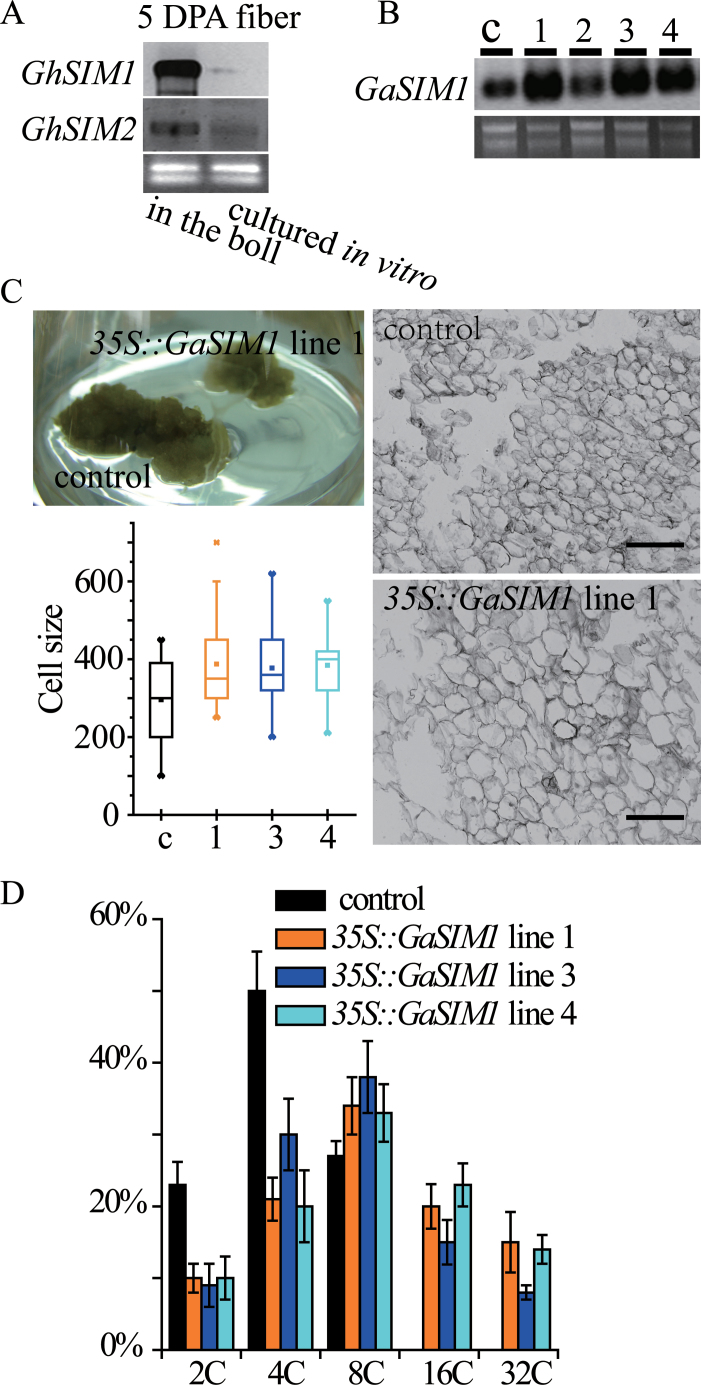
To assess the correlation between the multicellular phenotype and endoreduplication, two SIM homologous sequences in G. arboreum [GaSIM1 (KC246033) and GaSIM2 (KC246034)] were cloned based on the uncombined genome sequence of G. raimondii. Predicted GaSIM1 and GaSIM2 had all conserved motifs (Supplementary Fig. S6 at JXB online), suggesting a similar function to that of other members of the SIM family (Churchman et al., 2006). Next, an attempt was made to investigate the expression of GaSIM genes in cultured fibres, but producing elongated fibres from G. arboreum ovules was difficult. Hence, northern blotting was performed with probes corresponding to GaSIM1 and GaSIM2 using G. hirsutum fibre RNA samples. Transcription of both G. hirsutum SIM genes was significantly reduced in cultured fibres (Fig. 2A) which harboured multicellular fibres (see Supplementary Fig. S8; Hof and Saha, 1997, 1998).
Fig. 2.
Cotton SIM plays a role in repressing cytokinesis. (A) Northern blotting with antisense GaSIM1 and antisense GaSIM2 probes showing GhSIM1 and GhSIM2 mRNA levels in 5 DPA fibres of the G. hirsutum cultivar MD51 ne. The 5 DPA fibres came from in vivo (in the boll) or in vitro cultures (2 DPA ovules were cultured for 3 d). (B) Northern blot showing GaSIM1 mRNA levels in four independent 35S::GaSIM1 cell lines and the control (c). (C) Upper left panel: the phenotype of a cell line overexpressing GaSIM1 and a pBI121 empty vector (control) transformed cell line after two successive culture transfers. Lower left panel: cell size (µm2, area inside of the perimeter) of the callus. Calli were dispersed with water, and the cells were quantified with ImageJ software. The 35S::GaSIM1 lines differed significantly from the control (transformed with pBI121 empty vector) (P < 0.05, Kruskal–Wallis one-way ANOVA on ranks and Dunn’s test). Data are presented as box plots in which the box encompasses the 25th to the 75th percentile of the data, the line within the box is the median (50th percentile), and the error bars represent the 5th (lower bar) and the 95th (upper bar) percentiles. Right panels: representative micrographs of the transgenic and control cell lines. Bar=40 µm. (D) Nuclear DNA content of 35S::GaSIM1 cell lines and control (transformed with pBI121) as determined with flow cytometry. The percentage of nuclei in each ploidy class is indicated. Error bars indicate the SD for three independent experiments.
To investigate further the biological function of GaSIM genes, transgenic calli overexpressing GaSIM1 from the 35S promoter were created. The three positive transgenic 35S::GaSIM1 lines (Fig. 2B, lanes 1, 3, and 4) displayed a similar phenotype, showing a reduction in overall callus size and increased cell size compared with the control (transformed with empty pBI121) (Fig. 2C). These results suggest that cell division was reduced in lines overexpressing GaSIM1. Flow cytometric analyses confirmed that the 35S::GaSIM1 lines had extra rounds of endoreduplication, with increased levels of 16C and 32C cells detected (Fig. 2D). Based on the above results, it is inferred that GaSIM1 functions in repressing cotton cell cytokinesis, thus causing the transition from mitosis to endoreduplication.
Co-expression of GaBLT, GaSIM, and STI in in vitro cultured fibres causes branching
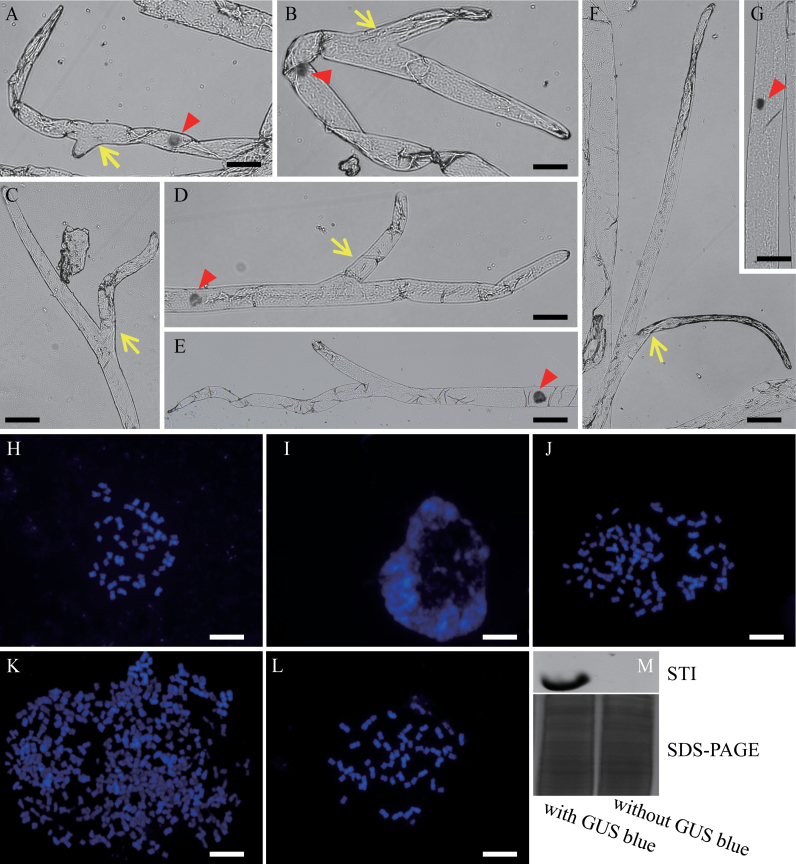
Cotton fibres are completely branchless in wild-type plants. Extensive scanning electron microscope observations of a large number of samples did not reveal any branching in fibres cultured in vitro. The transgenic fibres were then examined using the cotton ovule culture/biolistic transformation system, which is a quick and effective expression assay for detecting gene function and promoter specificity in fibres (H.J. Kim et al., 2002; Li et al., 2007; Wang et al., 2010). Northern blot analyses showed that the homologues of GaKTN1-1, GaKTN1-2, GaETR2-1, GaETR2-2, GaETR2-3, GaKIS, GaPOR, GaSPK1, GaAN, GaZWI, and GaSCD1 were expressed in cultured G. hirsutum fibres (data not shown). An attempt was made to take advantage of (i) fibre nuclear DNA amplification processes circumstantially in vitro; (ii) the specific GaSIM1 promoter of the mitosis to endoreduplication transition; and (iii) Arabidopsis STI functioning as a substitute for the cotton pseudogene. A GaBLT, GaSIM, and Arabidopsis STI expression construct was created, with the three genes being driven by the cotton fibre-specific promoter SCFP, which is activated at 0–25 DPA (Hou et al., 2008; Zhang et al., 2011). The construct also contained a SCFP::GUS reporter gene, which allowed visual identification of transformed fibre cells. GUS was expressed in some fibre cells (a tuft) 5 d after bombardment with the construct at 2 DPA, indicating successful transgene expression in these fibres (Supplementary Fig. S7A, B at JXB online). Interestingly, 12.3% (n=560) of the GUS-stained tufts had two-branched fibres that were shorter and thinner than non-transformed fibres (Fig. 3A–F; Table 1).
Fig. 3.
Co-expression of GaBLT, GaSIM, and STI can induce the fibre-branching phenotype. Ovules were collected at 2 DPA and bombarded with a construct to express GaBLT, GaSIM1, Arabidopsis STI, and GUS in fibres, followed by in vitro culture for 5 d. Branched fibres showing transgene expression as indicated by GUS staining were rinsed with water to reduce GUS staining, followed by nuclear staining with Schiff’s reagent (A–F) or separation of the nuclei and staining of the chromosomes with DAPI (J and K). (A– F) Branched fibres. (G) A bombarded fibre without GUS staining (branchless) as a negative control to see the increase in size of th enucleus in branching fibres. The fibre was also stained with Schiff’s reagent. (H) Metaphase chromosomes of a bombarded fibre without GUS staining. (I) Chromosome phenotype of a 7 DPA untreated fibre constantly growing in vivo (in the boll). (J) Chromosome phenotype of a bombarded but branchless fibre cell with GUS staining. (K) Chromosome phenotype of a bombarded and branched fibre cell with GUS staining. (L) Metaphase chromosomes of a G. hirsutum root tip cell. (M) Western blot showing Arabidopsis STI protein levels in fibres with or without GUS staining. Yellow arrows show branching sites; red arrowheads show cell nuclei. Bars=50 µM (A–G) or 10 µM (H–M).
Table 1.
Impact on cotton fibre branching following bombardment of cultured ovules with overexpression constructs containing the GUS reporter gene
| Ovule age when placed into culture | Bombarded genes overexpressed in fibres | No. of branched fibres | Total no. of GUS-stained fibre tufts |
|---|---|---|---|
| 2 DPA | STI+GUS | 0 | 93 |
| 2 DPA | GaSIM1+GUS | 0 | 89 |
| 2 DPA | GaBLT+GUS | 0 | 78 |
| 2 DPA | GaBLT+STI+GUS | 0 | 91 |
| 2 DPA | GaBLT+GaSIM1+GUS | 0 | 101 |
| 2 DPA | GaSIM1+STI+GUS | 3 | 483 |
| 0 DPA | GaSIM1+ STI+GUS | 0 | 494 |
| 2 DPA | GaBLT+GaSIM1+STI+GUS | 69 | 560 |
The branched and branchless fibres were captured using the laser microdissection system. Nuclear staining revealed that the GUS-stained fibres were all monocytes (Fig. 3A, B, D, E; n=252), with a larger nucleus (compared with a normal nucleus in Fig. 3G), in contrast to multicellular non-transgenic fibres (Supplementary Fig. S8 at JXB online), demonstrating the positive function of GaSIM. The impact of the transgene on branched and branchless fibres was then assessed by comparing the quantity of DNA with the chromosome number. The number of chromosome in the ovules supplier G. hirsutum is 52 (Wang et al., 2007). During metaphase, fibres lacking GUS staining had 52 chromosomes (Fig. 3H). Compared with the 5 DPA fibre cells grown in vivo, which had no separated chromosomes during interphase (Fig. 3I), >52 chromosomes were observed in the GUS-positive fibres growing in vitro (Fig. 3J, K), and the chromosome phenotype was similar to that of root tip cells (Fig. 3L). This indicated the occurrence of endoreduplication in the GUS-positive blue-stained fibres. Branchless fibres positively stained with GUS had no more than 104 chromosomes (4C, n=20), whereas branched fibres had up to 516 chromosomes (Fig. 3K; 16C, n=14). The above data showed that the DNA content of the transgenic fibres was elevated, attaining up to 16C when branching occurred. Additionally, ectopic expression of Arabidopsis STI protein in the GUS-stained fibre tufts was confirmed with western blotting (Fig. 3M).
To determine if all three genes (GaBLT, GaSIM1, and STI) functioned in fibre branching, different combinations of the genes and the GUS reporter were bombarded. Individually, none of the three genes promoted fibre branching (Table 1). Combinations of GaBLT+GaSIM1 or GaBLT+STI did not promote fibre branching (Table 1), successively indicating the necessity of STI and GaSIM1. However, a rare occurrence (0.6%) of branched fibres was observed in GaSIM1+STI construct-bombarded GUS-stained tufts (Table 1). This was a consequence of the autonomous expression of G. hirsutum BLT (GhBLT) in cultured fibres (Supplementary Fig. S9 at JXB online). All of the above data were obtained from fibres cultured beginning at 2 DPA. However, no GhBLT-expressing fibres were produced on 0 DPA ovules, which did not undergo fibre cell mitosis following cultivation for 5 d (Beasley and Ting medium with IAA and GA3) (Supplementary Fig. S9). When the 0 DPA ovules were bombarded with the GaBLT+STI construct, no branched fibres were observed (Table 1), indicating the necessity of exogenous GaBLT. Thus, the presence of all three genes was essential for branching in cultured fibres.
Discussion
Elevated endoreduplication levels are not sufficient for initiating cotton fibre branching
The nucleolus, which is a round or oval large body in the nucleus, consists of DNA (rDNA), RNA, and protein (Thiry and Lafontaine, 2005). The mean nucleolar volume increased rapidly, reached a maximum before 10 DPA, and was followed by a decline, according to varieties (De Langhe et al., 1978; Peeters et al., 1988). Whether the main contributors to enlargement are RNA, DNA, or protein is unclear. Thus, it cannot be confirmed if the lint DNA content elevation (Supplementary Fig. S5 at JXB online) was caused by endoreduplication, the nucleolar rDNA increasing [such as accumulation of extrachromosomal rDNA circles (Sinclair and Guarente, 1997)], or both. In addition, micronuclei were found in the fibres at or a little before 4 DPA, and were absent before 2 DPA; in the same variety and cultivation, until 4 DPA (at 2–3 DPA; the authors’ unpublished data), the lint DNA is elevated. So, the previous deduction that DNA content increasing in early developing fibres was due to an enlarged micronucleus (Taliercio et al., 2005) needs to be reconsidered.
In most genotypes, the degree of endoreduplication is highly correlated with that of Arabidopsis trichome branching (Hülskamp et al., 1994; Kasili et al., 2011). The Arabidopsis trichome undergoes progressive endoreduplication cycles during the early stages of cell morphogenesis, leading to increased nuclear DNA content. Mutations that affect trichome nuclear DNA content also alter trichome branching, with tetraploid lines producing overbranched trichomes compared with diploid lines (Perazza et al., 1999). Cotton fuzz cells were maintained at the G0 phase with a constant 2C nuclear DNA content (Supplementary Fig. S5A at JXB online), which may be correlated with their unbranched phenotype. Interestingly, no branching occurred while the DNA content of lint fibres increased from 2C to the maximum 2.4C over a 3 d period (2–5 DPA; Supplementary Fig. S5A), possibly because the endoreduplication (if it happened) levels did not attain a required threshold. Prior to the initiation of branching, the trichome nucleus undergoes three rounds of endoreplication (Hülskamp et al., 1994).
BLT has been conserved throughout angiosperm evolution, and a known link exists between the coordination of cell shape and nuclear DNA content in the Arabidopsis trichome. Both dicots and monocots have BLT homologues, which is indicative of conservation of their function throughout angiosperm evolution (Kasili et al., 2011). Gene performance and function are dependent on temporal and spatial expression patterns. GaBLT is unique in the genome (Supplementary Fig. S1 at JXB online); it is not widely expressed in the ovule and is absent in fibres. The overexpression of GaBLT in cotton callus cells significantly elevated DNA content (Fig. 1B), suggesting that it is potentially involved in endoreduplication. Differentiated fibres on cultured ovules expressing cotton BLT were branchless (Supplementary Fig. S9; Hof and Saha, 1997, 1998), even in the 35S::GaBLT fibres (Table 1), indicating that GaBLT alone was not sufficient to induce cultured fibre branching. Moreover, GaSIM1 repressed cotton cell cytokinesis (Fig. 2), and co-expression with GaBLT in cultured fibres significantly elevated the nuclear DNA content to 16C (~500 chromosomes; data not shown), but branched fibres were not induced (Table 1). Hence, a high endoreduplication level was not sufficient for initiation of cotton fibre branching.
Cotton fibres have branching potential
Arabidopsis stem trichomes are predominantly unbranched (Marks and Feldmann, 1989), with only 10–20% having two branches (Perazza et al., 1999). In contrast, nearly all of the stem trichomes on the triptychon mutant display two or three branches (Perazza et al., 1999). Additionally, stem trichomes subjected to STI overexpression exhibit two branching points (Ilgenfritz et al., 2003). These reports suggest that branching is controllable, as long as gene expression occurs at appropriate levels. Many Arabidopsis homologous branching genes were expressed in fibres (Supplementary Fig. S2A at JXB online), except GaBLT and ψGaSTI. The Arabidopsis trichome branching gene STI expressed with the endoreduplication inducers GaSIM and GaBLT in fibres induced branching (Fig. 3A–F). This demonstrated that trichome patterning genes could be used to produce homologous economically useful cotton fibres. The branched fibres may improve bulkiness, modifying batting, ease of hardening, and poor resilience. The branched-fibre changed the fibre physical structure which could improve bulkiness by increasing the gap between each fibre Thus, the branched fibres may be useful for non-woven applications, such as packing material used in quilts and cotton-padded coats.
Arabidopsis trichome branching involves a temporally defined sequence of events: endoreduplication, initiation of primary branching, continued endoreduplication, and secondary branching; primary and secondary branching are genetically distinct (Hülskamp et al., 1994; Folkers et al., 1997). The angle between the primary branches and the main stem is ~109 °, whereas it is ~85 ° between secondary branches, and the nucleus is typically positioned at the first branch point (Folkers et al., 1997). Hence, the cotton fibre branching architecture was more similar to the Arabidopsis trichome secondary branches based on two criteria: (i) the angle between the two branches was <90 °; and (ii) the nucleus was located below the branching point.
The genomic sequence of ψGaSTI showed an intact STI-like reading frame and codon bias, but additional and longer introns than STI (Supplementary Fig. S3 at JXB online). Whether ψGaSTI is a relic or if it is evolving to become functional is not known, but potential genes with the ability to regulate fibre branching in cotton may exist.
Cotton fibres have a unique identity compared with Arabidopsis trichomes and root hairs
Cotton fibres may be unique in that they are not a consequence of endoreduplication. In Arabidopsis, undifferentiated cells destined to become pro-trichome cells exit the mitotic cycle and enter into endoreduplication cycles, and the pro-trichome cells can form branches and expand during endoreduplication (Schnittger and Hülskamp, 2002; Ishida et al., 2008). Lint and fuzz showed no detectable endoreduplication before swelling (Supplementary Fig. S5 at JXB online), meaning that the endocycle was not an essential event for fibre initiation.
IAA (natural auxin) accumulates in cotton fibre initials. Genetic engineering to increase IAA levels in the ovule epidermis at the fibre initiation stage leads to a substantially increased number of lint fibres (Zhang et al., 2011). In addition, IAA is required for fibre production during in vitro culture of unfertilized ovules (Beasley, 1973; Beasley and Ting, 1973). These findings suggest that IAA is beneficial for fibre development. Arabidopsis root hairs, which are unbranched single cells, share a similar cell fate determination pathway (substrate depletion and lateral inhibition) with trichomes (Hülskamp, 2004; Ishida et al., 2008). IAA can induce root hair but not trichome production, and is not involved in the regulation of substrate depletion and the lateral inhibition pathway (Masucci and Schiefelbein, 1996; Rahman et al., 2002). This pathway acts upstream of, or independently from, the auxin pathway (Masucci and Schiefelbein, 1994, 1996; Ishida et al., 2008). High IAA levels have a negative effect on endoreduplication, and mutants in IAA signalling, biosynthesis, or transportation show an increased final DNA ploidy level (Ishida et al., 2010). Thus, two antagonistic pathways exist, and both are able to promote Arabidopsis root hair development. If substrate depletion and the lateral inhibition pathway play a role in fibre development, as mentioned previously, then we may reasonably conclude that fibres are similar to root hairs. However, hair cells require a lower active IAA supply than non-hair cells (Jones et al., 2008). Therefore, based on the present results, cotton fibres are not identical to Arabidopsis root hairs or trichomes, although they do share some homology. Fibres must have a regulatory mechanism that is not dependent on endoreduplication.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Southern blots with high stringency and low stringency.
Figure S2. Characteristics of the homologous genes controlling Arabidopsis trichome branching in G. arboreum.
Figure S3. Genomic sequence of ψGaSTI.
Figure S4. Histological observation of DPL971 seeds at different developmental stages.
Figure S5. Fibre nuclear DNA content at different developmental stages.
Figure S6. Alignment and evolutionary tree of GaSIM-related plant proteins.
Figure S7. Co-expression of GaBLT, GaSIM, and STI can induce fibre-branching phenotype.
Figure S8. Fibres with multicelled and unicelled phenotype.
Figure S9. Northern blots with antisense GaBLT probe showing GhBLT mRNA levels in cultured fibres.
Acknowledgements
We thank Professors Zhaosheng Kong and Yongmei Qin for their helpful suggestions and critical reading of the manuscript. We thank Professor Yan Pei for the SCFP promoter and Professor Peter M. Waterhouse for providing the pHANNIBAL/pART27 vector system. We also thank Professors Kunbo Wang and Fuguang Li for their technical assistance. This work was supported by a grant from the National Science and Technology Support Program of China [2013BAD01B03].
References
- Beasley CA. 1973. Hormonal regulation of growth in unfertilized cotton ovules. Science 179, 1003–1005 [DOI] [PubMed] [Google Scholar]
- Beasley CA, Ting IP. 1973. The effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. American Journal of Botany 60, 130–139 [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A. 2010. Endoreplication controls cell fate maintenance. PLoS Genetics 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. 2001. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. The Plant Cell 13, 807–827 [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG. 2006. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana . The Plant Cell 18, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes AW, Reidy MA, Clowes MM. 1983. Kinetics of cellular proliferation after arterial injury. I. Smooth muscle growth in the absence of endothelium. Laboratory Investigation 49, 327–333 [PubMed] [Google Scholar]
- De Langhe E, Kosmidou-Dimitropoulou S, Waterkeyn L. 1978. Effect of hormones on nucleolar growth and vacuolation in elongating cotton fibers. Planta 140, 269–273 [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2, 2233–2244 [DOI] [PubMed] [Google Scholar]
- Falbel TG, Koch LM, Nadeau JA, Segui-Simarro JM, Sack FD, Bednarek SY. 2003. SCD1 is required for cell cytokinesis and polarized cell expansion in Arabidopsis thaliana. Development 130, 4011–4024 [DOI] [PubMed] [Google Scholar]
- Folkers U, Berger J, Hulskamp M. 1997. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779. [DOI] [PubMed] [Google Scholar]
- Gan YM, Chen D, Liu F, Wang CY, Li SH, Zhang XD, Wang YH, Peng RH, Wang KB. 2011. Individual chromosome assignment and chromosomal collinearity in Gossypium thurberi, G. trilobum and D subgenome of G. barbadense revealed by BAC-FISH. Genes and Genetic Systems 86, 165–174 [DOI] [PubMed] [Google Scholar]
- Hülskamp M. 2004. Plant trichomes: a model for cell differentiation. Nature Reviews Molecular Cell Biology 5, 471–480 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Miséra S, Jürgens G. 1994. Genetic dissection of trichome cell development in Arabidopsis . Cell 76, 555–566 [DOI] [PubMed] [Google Scholar]
- Hof JV, Saha S. 1997. Cotton fibers can undergo cell division. American Journal of Botany 84, 1231–1231 [PubMed] [Google Scholar]
- Hof JV, Saha S. 1998. Growth and mitotic potential of multicelled fibers of cotton (Malvaceae). American Journal of Botany 85, 25–29 [PubMed] [Google Scholar]
- Hou L, Liu H, Li JB, Yang X, Xiao YH, Luo M, Song SQ, Yang GW, Pei Y. 2008. SCFP, a novel fiber-specific promoter in cotton. Chinese Science Bulletin 53, 2639–2645 [Google Scholar]
- Humphries JA, Walker AR, Timmis JN, Orford SJ. 2005. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Molecular Biology 57, 67–81 [DOI] [PubMed] [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua NH, Jürgens G, Hülskamp M. 2003. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiology 131, 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K, 2010. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis . Development 137, 63–71 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. 2008. A genetic regulatory network in the development of trichomes and root hairs. Annual Review of Plant Biology 59, 365–386 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HMO, Grierson CS. 2008. Auxin transport through non-hair cells sustains root-hair development. Nature Cell Biology 11, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY. 2003. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. The Plant Cell 15, 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF, Nguyen MN. 1994. Karyology, nuclear DNA quantification and nucleus–cytoplasmic domain variations in some multinucleate green algae (Siphonocladales, Chlorophyta). Phycologia 33, 42–52 [Google Scholar]
- Kasili R, Huang CC, Walker JD, Simmons LA, Zhou J, Faulk C, Hülskamp M, Larkin JC. 2011. BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development 138, 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. 2002. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO Journal 21, 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Williams MY, Triplett BA. 2002. A novel expression assay system for fiber-specific promoters in developing cotton fibers. Plant Molecular Biology Reporter 20, 7–18 [Google Scholar]
- Kirik V, Mathur J, Grini PE, Klinkhammer I, Adler K, Bechtold N, Herzog M, Bonneville JM, Hülskamp M. 2002. Functional analysis of the tubulin-folding cofactor C in Arabidopsis thaliana . Current Biology 12, 1519–1523 [DOI] [PubMed] [Google Scholar]
- Li L, Wang XL, Huang GQ, Li XB. 2007. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. Journal of Experimental Botany 58, 3227–3238 [DOI] [PubMed] [Google Scholar]
- Marks MD, Feldmann KA. 1989. Trichome development in Arabidopsis thaliana. I. T-DNA tagging of the GLABROUS1 gene. The Plant Cell 1, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1994. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiology 106, 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1996. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. The Plant Cell 8, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 [DOI] [PubMed] [Google Scholar]
- Minamisawa N, Sato M, Cho KH, Ueno H, Takechi K, Kajikawa M, Yamato KT, Ohyama K, Toyooka K, Kim GT. 2011. ANGUSTIFOLIA, a plant homolog of CtBP/BARS, functions outside the nucleus. The Plant Journal 68, 788–799 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. 1997. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proceedings of the National Academy of Sciences, USA 94, 6261–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters MC, Voets S, Wijsmans J, De Langhe E. 1988. Pattern of nucleolar growth in differentiating cotton fibres (Gossypium hirsutum L.). Annals of Botany 62, 377–382 [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne AM, Bonneville JM, 1999. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hulskamp M. 2009. One, two, three…models for trichome patterning in Arabidopsis? Current Opinion in Plant Biology 12, 587–592 [DOI] [PubMed] [Google Scholar]
- Plett JM, Mathur J, Regan S. 2009. Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana . Journal of Experimental Botany 60, 3923–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Jilk R, Marks MD, Szymanski DB. 2002. The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. The Plant Cell 14, 101–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. 2002. Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiology 130, 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Schnittger A, Hülskamp M. 2002. Trichome morphogenesis: a cell-cycle perspective. Philosophical Transactions of the Royal Society B: Biological Sciences 357, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Martin C. 2006. Trichomes: different regulatory networks lead to convergent structures. Trends in Plant Science 11, 274–280 [DOI] [PubMed] [Google Scholar]
- Shang HH, Liu CL, Zhang CJ, Li FL, Hong WD, Li FG. 2009. Histological and ultrastructural observation reveals significant cellular differences between Agrobacterium transformed embryogenic and non-embryogenic calli of cotton. Journal of Integrative Plant Biology 51, 456–465 [DOI] [PubMed] [Google Scholar]
- Shangguan XX, Xu B, Yu ZX, Wang LJ, Chen XY. 2008. Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. Journal of Experimental Botany 59, 3533–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91, 1033–1042 [DOI] [PubMed] [Google Scholar]
- Smith RH, James Price H, Thaxton JB. 1977. Defined conditions for the initiation of growth of cotton callus in vitro I. Gossypium arboreum . In Vitro Cellular and Developmental Biology-Plant 13, 329–334 [DOI] [PubMed] [Google Scholar]
- Staub JM, Wei N, Deng XW. 1996. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis . The Plant Cell 8, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo JF, Liang XE, Pu L, Zhang YS, Xue YB. 2003. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that is expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochimica et Biophysica Acta 1630, 25–34 [DOI] [PubMed] [Google Scholar]
- Taliercio E, Hendrix B, Stewart JM. 2005. DNA content and expression of genes related to cell cycling in developing Gossypium hirsutum (Malvaceae) fibers. American Journal of Botany 92, 1942–1947 [DOI] [PubMed] [Google Scholar]
- Thiry M, Lafontaine DL. 2005. Birth of a nucleolus: the evolution of nucleolar compartments. Trends in Cell Biology 15, 194–199 [DOI] [PubMed] [Google Scholar]
- Van’t Hof J. 1999. Increased nuclear DNA content in developing cotton fiber cells. American Journal of Botany 86, 776–779 [PubMed] [Google Scholar]
- Wang G, Zhao GH, Jia YH, Du XM. 2013. Identification and characterization of cotton genes involved in fuzz-fiber development. Journal of Integrative Plant Biology (in press). [DOI] [PubMed] [Google Scholar]
- Wang K, Guo WZ, Zhang TZ. 2007. Development of one set of chromosome-specific microsatellite-containing BACs and their physical mapping in Gossypium hirsutum L. Theoretical and Applied Genetics 115, 675–682 [DOI] [PubMed] [Google Scholar]
- Wang L, Li XR, Lian H, Ni DA, He Y, Chen XY, Ruan YL. 2010. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiology 154, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY. 2004. Control of plant trichome development by a cotton fiber MYB gene. The Plant Cell 16, 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Llewellyn DJ, White R, Ruggiero K, Al-Ghazi Y, Dennis ES, 2007. Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta 226, 1475–1490 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zheng XL, Song SQ, et al. 2011. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nature Biotechnology 29, 453–458 [DOI] [PubMed] [Google Scholar]
- Zhao PM, Wang LL, Han LB, Wang J, Yao Y, Wang HY, Du XM, Luo YM, Xia GX. 2009. Proteomic identification of differentially expressed proteins in the Ligon lintless mutant of upland cotton (Gossypium hirsutum L.). Journal of Proteome Research 9, 1076–1087 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.