Abstract
In plants, sugars such as glucose act as signalling molecules that promote changes in gene expression programmes that impact on growth and development. Recent evidence has revealed the potential importance of controlling mRNA decay in some aspects of glucose-mediated regulatory responses suggesting a role of microRNAs (miRNAs) in these responses. In order to get a better understanding of glucose-mediated development modulation involving miRNA-related regulatory pathways, early seedling development of mutants impaired in miRNA biogenesis (hyl1-2 and dcl1-11) and miRNA activity (ago1-25) was evaluated. All mutants exhibited a glucose hyposensitive phenotype from germination up to seedling establishment, indicating that miRNA regulatory pathways are involved in the glucose-mediated delay of early seedling development. The expression profile of 200 miRNA primary transcripts (pri-miRs) was evaluated by large-scale quantitative real-time PCR profiling, which revealed that 38 pri-miRs were regulated by glucose. For several of them, the corresponding mature miRNAs are known to participate directly or indirectly in plant development, and their accumulation was shown to be co-regulated with the pri-miR by glucose. Furthermore, the expression of several miRNA target genes was found to be deregulated in response to glucose in the miRNA machinery mutants ago1-25, dcl1-11, and hyl1-2. Also, in these mutants, glucose promoted misexpression of genes for the three abscisic acid signalling elements ABI3, ABI4, and ABI5. Thus, miRNA regulatory pathways play a role in the adjustments of growth and development triggered by glucose signalling.
Key words: Abscisic acid, AGO1, DCL1, glucose, HYL1, miRNA, plant development, post-transcriptional control, seed germination.
Introduction
Photosynthesis-derived sugars are the main source of carbon skeletons and energy in plants. It is not surprising that glucose (Moore et al., 2003; Li et al., 2006), fructose (Cho and Yoo, 2011; Li et al., 2011), sucrose (Vaughn et al., 2002), and trehalose-6-phosphate (Delatte et al., 2011; Wahl et al., 2013) are important signalling molecules that impact on gene expression and several aspects of plant growth and development (e.g. germination, flowering, senescence; Smeekens et al., 2010; Eveland and Jackson, 2012; Stitt and Zeeman, 2012; Wahl et al., 2013). Integration of metabolic and circadian signals with environmental cues seems to coordinate starch degradation and sugar pools, thus impacting on carbon availability for growth (Stewart et al., 2011; Stitt and Zeeman, 2012). Sugar signalling also depends on the integration with other nutrient levels, such as phosphate, sulphate, and nitrogen, which are tightly related with circadian regulation and are required for proper plant development expression programmes (Rolland et al., 2006). In addition, sugars such as sucrose or glucose have been shown to interact with the hormones abscisic acid (ABA), ethylene (Yanagisawa et al., 2003; Karve et al., 2012), auxin, and cytokinin (Moore et al., 2003; Sairanen et al., 2012) in regulating plant development (Rolland et al, 2006) and stress responses (Price et al., 2003; Jossier et al., 2009).
The only sugar sensor that has been described unambiguously up to now is hexokinase 1 (HXK1), which senses glucose (Moore et al., 2003; Karve et al., 2012) and directly mediates the glucose-dependent transcriptional repression of the chlorophyll a/b-binding protein 2 gene (CAB2; Cho et al., 2006). Further evidence for transcriptional control mediating glucose responses comes from the observation that the APETALA2-type transcription factor ABI4, a component of the ABA signalling pathway, is involved in sugar-induced gene expression changes (Li et al., 2006; Bossi et al., 2009). In addition, post-transcriptional regulation is also involved in sugar signalling. For instance, evidence was found for sucrose affecting the translation of the mRNA for five Arabidopsis thaliana S-group bZIP-type transcription factors (Rahmani et al., 2009), and the degradation of the ethylene insensitive 3 (EIN3) transcription factor has been found to be glucose induced (Yanagisawa et al., 2003). Moreover, it was shown that glucose modulates the mRNA stability of the Arabidopsis transcription factor AtbZIP63 as part of a regulatory scheme integrating energetic and abiotic stress (Matiolli et al., 2011). Moreover, the expression of AtbZIP63 was found to be deregulated in the miRNA-processing mutant hyl1-2 (Matiolli et al., 2011). This latest observation begs the question of whether miRNA-related regulatory pathways participate in glucose responses. MIRNA genes are transcribed by RNA polymerase II, resulting in a primary miRNA transcript (pri-miR) that is first converted into a precursor miRNA (pre-miRNA) by the endonuclease DICER-like1 (DCL1). Subsequently, the C2H2-zinc finger protein SERRATE (SE), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and DCL1 interact to trim the pre-miRNA generating a 21-nucleotide double-stranded miRNA/miRNA* duplex that is exported to the cytoplasm through the action of the exportin HASTY and other factors (Voinnet, 2009). Methylation of the duplex fragment on the 3′-terminal nucleotides of each strand by HEN protects the fragment from degradation by exonucleases. The mature miRNA is recognized by ARGONAUTE1 (AGO1) which is in turn integrated into the RNA-induced silencing complex (RISC) through a process that might also require HYL1 activity (Eamens et al., 2009). miRNA complementarity to specific mRNA guides RISC-mediated target mRNA cleavage and/or translational repression (Mi et al., 2008; Voinnet, 2009).
Germination and seedling establishment are crucial initial steps for plant development. It is well established that high exogenous glucose delays these early plant developmental phases (Arenas-Huertero et al., 2000; Cheng et al., 2002; Gibson, 2005; Dekkers et al., 2008). In the current work, sensitivity to glucose during seedling development was examined to analyse the capacity of mutants hyl1-2 and dcl1-11, which are defective in pre-miRNA processing, and mutant ago1-25, which is impaired in miRNA activity, to modulate glucose responses. Young seedlings of these mutants showed a glucose hyposensitive phenotype, which was defined by the reduced capacity of glucose to delay early seedling development. This phenotype was correlated with the regulation of the expression of a set of 38 pri-miRs by 4% glucose, indicating that miRNA-related regulatory processes modulate the glucose-dependent delay of seedling development.
Materials and methods
Plant material and growth conditions
A. thaliana ecotype Columbia-0 (Col-0) was used as a wild-type control for the mutants ABA insensitive abi4-1 (Finkelstein, 1994), ABA-deficient aba2-1 (Schwartz et al., 1997), ago1-25 (Morel et al., 2002), dcl1-11 (Zhang et al., 2008), and hyl1-2 (Vazquez et al., 2004) and Landsberg erecta (Ler) was used as a wild-type control for the glucose-insensitive HXK1 knockout gin2-1 (Moore et al., 2003). Seeds were surface sterilized and sown in plates with half-strength Murashige and Skoog media (MS/2) containing 0.5% (w/v) agar (type E) with 1–6% glucose, 4% mannitol, or 0.5–5 µM ABA (all Sigma-Aldrich reagents). The plates were placed in the dark at 4 °C for 2 d for stratification and then incubated at 24 °C under continuous light (50 µmol m–2 s–1) for up to 14 d. For phenotypic assays, each plate contained at least 20 seeds of each specified genotype and the assays were carried out in duplicate or triplicate. Analysis of deviance (McCullagh and Nelder, 1989) was used for statistical evaluation of germination and development efficiencies. For gene expression analysis assays, 5mg of seeds of each evaluated genotype was plated in at least triplicate.
RNA isolation and cDNA synthesis
Total RNA was isolated following a LiCl-based protocol for seeds (Oñate-Sánchez and Vicente-Carbajosa, 2008), except for the addition of 3 µl of glycogen (20mg ml–1, Roche) in the last precipitation step. For pri-miR profiling, total RNA was treated with TURBO DNA-free (Ambion), according to the manufacturer’s instruction. For other quantitative real-time PCR (qRT-PCR) analyses, DNAse treatment was only carried out in the cases where the primers pairs could not be designed to span exon–exon junctions. Synthesis of cDNA was carried out with ImProm II Reverse Transcriptase (Promega), according to manufacturer instructions.
qRT-PCR analysis
Gene expression analysis by qRT-PCR was conducted as described by Czechowski et al. (2005) and Udvardi et al. (2008), evaluating gene expression with the Delta-Delta Ct method (Livak and Schmittgen, 2001). For the pri-miR profiling, cDNA was diluted four times before qRT-PCR analysis with SYBR Green (Applied Biosystems) on a 384-well 7900HT Real-Time PCR (Applied Biosystems), using PP2A (At1g13320), At2g283904; At5g15710, and UBQ10 (At4g05320) as reference genes (Czechowski et al., 2005). For the validation of pri-miR profiling results, qRT-PCR was performed with SYBR Green (Invitrogen) on a 96-well 7500 Fast Real-Time PCR (Applied Biosystems), using PP2A (At1g13320) as the reference gene. Differences in gene expression were considered significant for fold-changes ≥|1.5| between treated and control samples and for P < 0.05 according to two-tailed Student’s t-test. Mature miRNA quantification was conducted as described by Varkonyi-Gasic et al. (2007), using PP2A for normalization. The sequences of the qRT-PCR primers are given in Supplementary Table S1 (available at JXB online).
High-throughput analysis of short-term responses to glucose
Arabidopsis Col-0 growth conditions in liquid MS/2, treatments with 2% glucose and 2% mannitol (osmotic control) for 4h, and RNA extraction protocol for the analysis of short-term glucose responses were performed as described previously (Matiolli et al., 2011). Preparation and deep sequencing of small RNA (sRNA) libraries was conducted according to the manufacturer’s instructions by Ambry Genetics (Aliso Viejo, CA, USA) using Illumina’s Genome Analyzer IIx. The reads were analysed using CLC Genomics Workbench software (CLC Bio). All reads were matched against Arabidopsis miRNA mature sequences downloaded from miRBase (release 18; Kozomara and Griffiths-Jones, 2011). Mismatches were not allowed in the matching and maximum number of hits for a read was one. The read count values were estimated as the reporting expression value for each treatment. Differential expression of miRNAs was detected by test on proportions and Kal’s Z-test. Small RNA library data from this study were deposited in the GEO database under the accession number GSE43546.
Results
ago1-25, dcl1-11, and hyl1-2 mutants exhibit a glucose hyposensitive growth phenotype from germination to seedling establishment
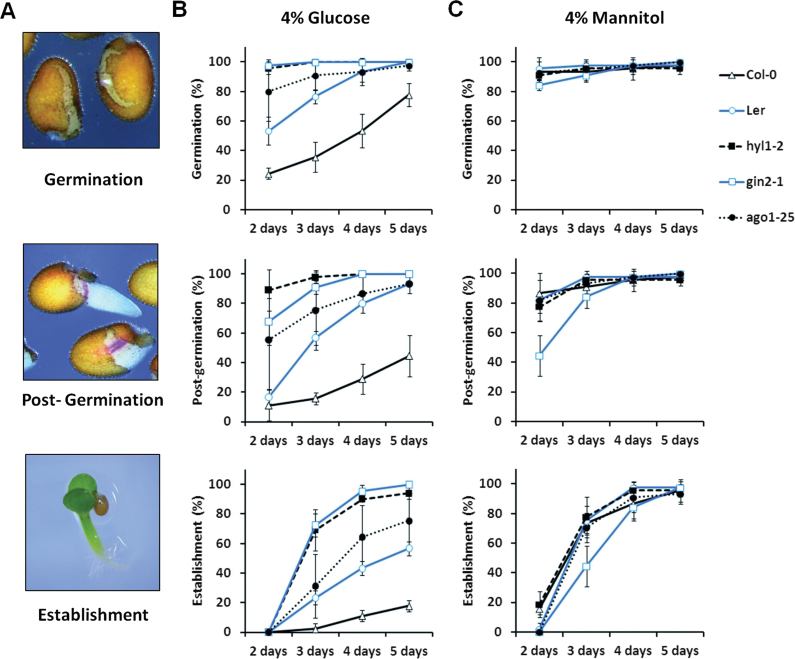
In order to evaluate the involvement of miRNA-related regulatory pathways in glucose-induced delay of Arabidopsis early seedling development (Nonogaki et al., 2010; Eveland and Jackson, 2012), the miRNA-biogenesis mutants dcl1-11 and hyl1-2, the miRNA-activity mutant ago1-25, and the corresponding wild type Col-0 were scored for germination (i.e. testa rupture), post-germination (i.e. radicle emergence and elongation), and establishment (i.e. cotyledon expansion and greening; Fig. 1A, Supplementary Fig. S1) in response to a range of glucose or mannitol (osmotic control) concentrations. The glucose-insensitive mutants gin2-1 (HXK1 glucose-sensor knockout mutant in Ler background; Moore et al., 2003) and abi4-1/gin6 (Apetala2-type transcription factor involved in ABA signalling in Col-0 background; León and Sheen, 2003) were included as controls.
Fig. 1.
Glucose-induced delay of early seedling development is dependent upon miRNA machinery activity. (A) Developmental phases that were monitored; germination (testa rupture), post-germination (radicle emergence and elongation), and establishment (cotyledon expansion and greening). (B) Effects of 4% glucose on germination and development of miRNA-deficient mutants ago1-25 and hyl1-2 was less severe than for wild-type Col-0 and Ler and was similar to the glucose-insensitive gin2-1. (C) Osmotic-control 4% mannitol could not reproduce the delay observed for glucose. In media not supplied with sugar, all seeds reached post-germination stage at 2 d and establishment within 3 d of light exposure (Supplementary Fig. S2A). In glucose-supplied media, growth arrest was not observed before seedling establishment stage was reached. Seeds of each genotype were sown in MS/2 plates supplied or not with the indicated sugar, kept for 2 d at 4 °C in the dark for stratification and transferred to continuous light (50 µmol m–2 s–1) at 24 °C. Germination, post-germination, and establishment were scored from 2–5 d of light exposure. Results presented are mean±SD of three independent experiments, each of which including 20 seeds (this figure is available in colour at JXB online).
A first set of experiments using a range of glucose concentrations between 0 and 6% revealed that from germination to seedling establishment, hyl1-2 and ago1-25 were significantly less sensitive to high concentrations of glucose than Col-0 wild-type seedlings. Their levels of sensitivity were comparable to the glucose-insensitive mutant abi4-1 (Supplementary Fig. S1). No differences in the rate of development between the genotypes were observed for seedlings grown on control media or media supplied with 1% glucose (Supplementary Fig. S1). Nevertheless, after 14 d of light exposure in the presence of 4% glucose, all seedlings of all genotypes reached the establishment phase (cotyledon expansion and greening; data not shown). Similar results were obtained with different seed batches (data not shown).
From these initial data, it was deduced that 4% glucose is a suitable concentration to accurately evaluate the degree of glucose sensitivity of hyl1-2 and ago1-25 (Supplementary Fig. S1). Under this condition and after 2 d of light exposure, only 50% of Ler and 20% of Col-0 seedlings germinated (Fig. 1B), while 80% of ago1-25 and nearly all hyl1-2 seeds germinated (Fig. 1B). This result indicates a significant lower sensitivity to 4% glucose of the two miRNA-deficient genotypes as compared to the wild type. Moreover, their level of glucose hyposensitivity was comparable to the one exhibited by gin2-1 (Fig. 1B). The same differences in glucose sensitivity were essentially maintained among the different genotypes for both the post-germination and seedling establishment stages until after 5 d of light exposure (Fig. 1B). The glucose hyposensitivity of the mutants was not due to differential capacity to germinate since in the absence of glucose, early seedling development was indistinguishable between genotypes (Supplementary Fig. S2). In addition, 4% mannitol (i.e. the osmotic control) was unable to reproduce the effects of 4% glucose (Fig. 1C, Supplementary Fig. S2A), indicating that the glucose-mediated delay of seedling development is not due to osmotic stress. As a means to further support this set of results, glucose sensitivity of the relatively weak mutant allele, dcl1-11, was also evaluated. As expected, it was found to be hyposensitive to 4% glucose (Supplementary Figs S2B and S3). Together, these observations imply that a miRNA-related machinery is involved in glucose-induced development delay from germination to seedling establishment.
Since the effect of glucose can be mediated by the ABA signalling pathway during early seedling development (León and Sheen, 2003; Dekkers et al., 2008), it was important to verify whether seedling development in the miRNA-deficient mutants would also be hyposensitive to ABA (Supplementary Fig. S4). As a control, the abi4-1 mutant was shown to be ABA hyposensitive (Supplementary Figs S2C and S4; Finkelstein, 1994). While ago1-25 and hyl1-2 were found to be more sensitive to ABA than the wild type from germination to establishment, hypersensitivity of dcl1-11 was evident in the post-germination and establishment stages (Supplementary Figs S2C and S4), as previously described (Lu and Fedoroff, 2000; Kim et al., 2008; Zhang et al., 2008; Earley et al., 2010). The data suggest that the glucose hyposensitivity phenotype of ago1-25, dcl1-11 and hyl1-2 is at least partly independent on ABA-related signalling pathway.
Expression of miRNA genes in response to glucose
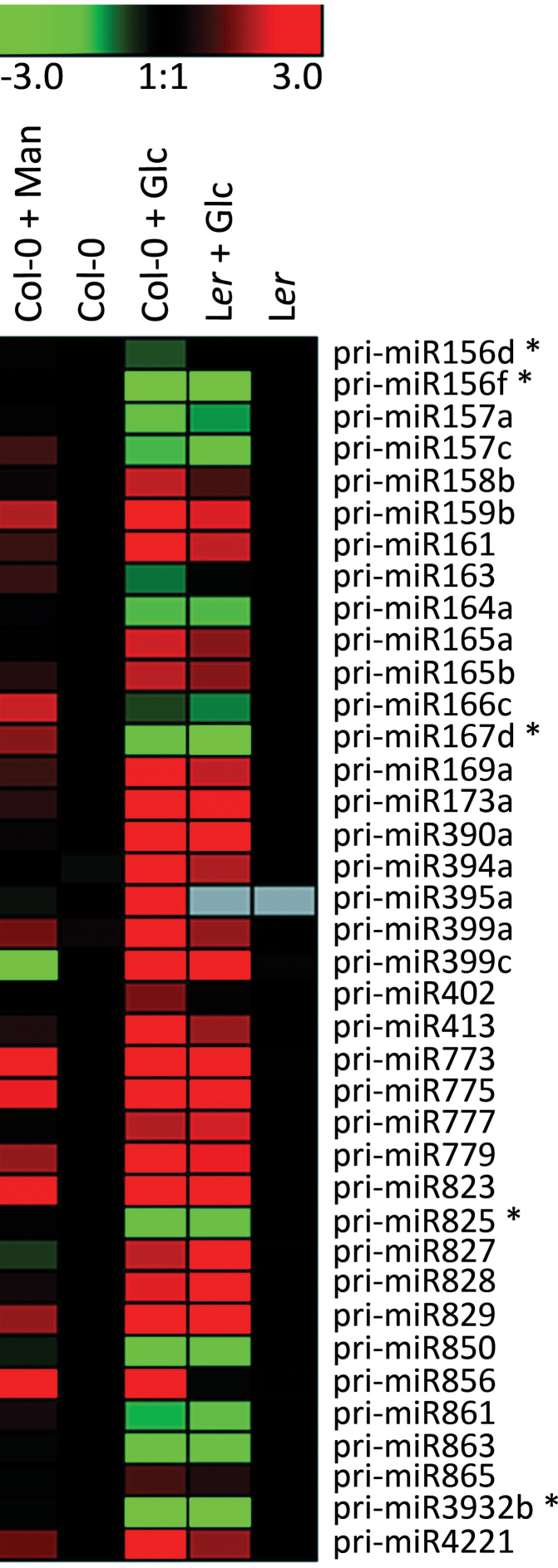
To identify which miRNAs may be involved in the glucose-induced early seedling development delay, a platform established for quantification of 200 pri-miR transcripts by qRT-PCR was used (Pant et al., 2009). Although pri-miRs are not the biologically active molecules, their abundances often reflect the levels of the mature miRNA (Pant et al., 2009). While this approach allows for the verification of expression changes for a specific MIRNA locus, special assays are required to quantify the mature miRNAs. These assays, however, cannot always distinguish mature miRNA species of the same miRNA family due to their extensive sequence homology. The expression analysis was carried out with RNAs from Col-0, Ler, and gin2-1 seedlings grown for 3 d in light in media with or without 4% glucose or mannitol (osmotic control). A total of 38 pri-miRs belonging to 32 families were found to be differentially expressed in the presence of 4% glucose as compared to the control in Col-0. Of these, 33 showed the same response in Ler accession (Fig. 2, Supplementary Table S2). Since the pri-miR platform was designed based on Col-0 background, primer mismatches may explain the differences observed between Col-0 and Ler profiles. In addition, five of the 38 glucose-responsive miRNA precursors—pri-miR156d, pri-miR156f, pri-miR167d, pri-miR3932b, and pri-miR825—were found to require the glucose-sensing activity of HXK1, as revealed by the comparison between pri-miR accumulation in gin2-1 and Ler seedlings. pri-miR172, which is a downstream component of a regulatory cascade that involves miR156 (Martin et al., 2010; Nonogaki, 2010) was found to be upregulated, yet it did not pass the threshold to be considered significant. Quite noticeably, the mature miRNAs derived from several of the identified glucose-responsive pri-miRs are known to target mRNAs that participate directly or indirectly in different aspects of plant development (Table 1, Supplementary Table S2).
Fig. 2.
pri-miR levels in Col-0 and Ler seedlings after 3 d of light exposure grown in control media, 4% glucose (Glc) media, and 4% mannitol (Man) media. Potential HXK1-sensing dependent pri-miRs are marked with an asterisk. Seed dormancy was previously broken by imbibition for 2 d at 4° C in the dark. This set of glucose-responsive pri-miRs (column 3) presented at least 1.5-fold expression difference to non-treated control samples (column 2), at least 2-fold difference to mannitol-treated samples (column 1), and were considered to be significantly different according to Student’s t-test (P < 0.05). The colours represent the log2 values of the relative quantification between (Col-0 vs. Col-0 + treatment) and (Ler vs. Ler + treatment); green indicates repression, red indicates induction. Mannitol-treated Ler is not shown.
Table 1.
Long-term 4% glucose-regulated pri-miRs related to development control
| Relative quantification by qRT-PCR | ||||||
|---|---|---|---|---|---|---|
| pri-miRNA | 4% Glucose | 4% Mannitol | Development | Target gene family | Developmental process | Reference |
| pri-miR156d† | 0.25±0.05a | 0.76±0.10b | 0.18±0.05 | SPL | Embryo development/ vegetative leaves emergence | Wu et al. (2009); Nodine and Bartel (2010) |
| pri-miR156f† | 0.11±0.11a | 0.80±0.26b | nd | |||
| pri-miR159b† | 5.26±0.24a | 2.14±0.09b | 2.10±0.47c | MYB | Seedling growth and development/ABA response | Reyes and Chua (2007) |
| pri-miR166c† | 0.27±0.02a | 2.12±0.29b | 0.08±0.03c | HD-ZIP III | Leaf flattening | Husbands et al. (2009) |
| pri-miR169a† | 2.05±0.28a | 1.18±0.26b | 1.33±0.19c | NF-Y | Embryo development/ ABA response | Li et al. (2008); Yamamoto et al. (2009) |
| pri-miR390a† | 4.00±0.92a | 1.59±0.65b | 2.52±0.24c | TAS3 | Lateral root development/l eaf polarity | Marin et al. (2010) |
| pri-miR773 | 19.81±5.74a | 3.87±1.33b | 3.93±0.52c | MET2 | Biotic stress response | Fahlgren et al. (2006); Li et al. (2010) |
| pri-miR775 | 21.76±3.54a | 3.98±0.86b | 9.80±1.84c | GT | Arabinogalactan- protein biosynthesis | TarBase: Vergoulis et al. (2012) |
| pri-miR823 | 18.98±1.71a | 4.24±2.17b | 5.24±1.14c | CMT3 | Embryo development | TarBase: Vergoulis et al. (2012) |
| pri-miR828 | 8.44±1.19a | 5.07±0.85b | 2.04±0.51c | MYB | Nutrient availability control | Luo et al. (2012) |
Relative quantification values for glucose and mannitol are from samples grown in media supplied with 4% of the respective sugar in comparison with the untreated control, all grown for 3 d in light; relative quantification values for development are from samples grown for 1 d in light compared with samples grown for 3 d in light, both under control conditions. All samples are Col-0; values are mean ± SD of at least three biological replicates. Target gene families are based on the indicated published results or according to TarBase database (Vergoulis et al., 2012). †Pri-miRs that had the correspondent miRNA/miRNA family sequence identified as responsive to short-term (i.e. 4h) 2% glucose, based on deep sequencing analysis (Supplementary Table S3); a–cChanges in transcript accumulation were considered significant for differences with fold-change ≥|1.5| and according to Student’s t-test (P < 0.05): aglucose-treated vs. untreated; bmannitol-treated vs. glucose-treated; cdevelopment vs. glucose-treated samples; nd, not detected.
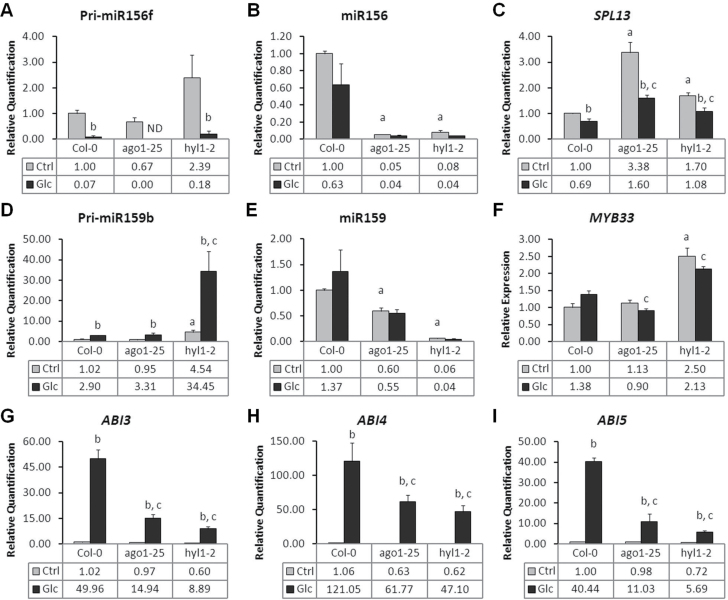
The differential expression induced by glucose was confirmed by qRT-PCR for 10 of the 12 development-related pri-miRs that were evaluated (83%; Table 1), essentially validating the high-throughput pri-miR quantification data. Since 4% glucose delays seedling development, it was also necessary to distinguish true glucose effect from developmental cues that may influence pri-miR expression. To this end, the expression of the 10 validated pri-miRs was compared between wild-type seedlings grown in control conditions for 1 d in light, which corresponds to the developmental stage reached by seedlings grown for 3 d in light in presence of 4% glucose (~30% germination; Fig. 1), and seedlings grown for 3 d in light in media not supplied with glucose, which stands for the control conditions used for the pri-miR profiling (100% germination; Fig. 2, Supplementary Table S2). All pri-miRs tested except pri-miR156d showed significant differences between glucose- and development-promoted changes (Table 1). Thus, for these nine pri-miR, developmental signals may contribute to no more than half of the expression changes observed in 4% glucose-grown seedlings (Table 1). pri-miR156f could not be detected in seedlings grown for 1 d in light, indicating a very low expression level, while 4% glucose repressed its expression by 9-fold (Table 1).
To further support genuine glucose-mediated responses, changes of pri-miR expression promoted by short-term 4% glucose treatments were evaluated in Col-0. pri-miR expression alterations resulting from treatment with 4% glucose applied for 4h on seedlings grown for 3 d in light (Supplementary Fig. S5) followed the same expression trends as those observed for seedlings grown for 3 d in light in 4% glucose media (Table 1). The only exception was pri-miR169a (Supplementary Fig. S5), which presented an opposite response that may reflect an alternative regulation pattern between short- and long-term responses. These data, therefore, essentially corroborate the notion of true glucose-promoted responses. The same tendency of glucose-promoted responses between short- and long-term treatments was also observed for the target genes (data not shown).
The subset of nine pri-miRs whose expression was specifically affected by 4% glucose and could not be attributed to developmental cues (Table 1) was used to evaluate the assumption that pri-miR accumulation may reflect the levels of the corresponding biologically active mature miRNA (Pant et al., 2009). The mature miRNA of the families corresponding to the validated pri-miRs were quantified in Col-0, ago1-25 and hyl1-2 seedlings grown for 3 d in light in the presence and absence of 4% glucose. This analysis revealed that the pattern of mature miRNA accumulation follows the trend of pri-miR expression changes in response to glucose (Fig. 3, Supplementary Fig. S6). However, an opposite pattern was observed for pri-miR169a/miR169, which may explain the increase of the NF-YA5 target transcript in response to glucose, indicating that glucose may also have effects on miRNA maturation (Supplementary Fig. S6B). As expected, a clear dependence on HYL1 activity was seen for the production of all miRNAs evaluated. AGO1 also appeared to be required for proper accumulation of miR156, miR159, miR166, miR169, miR773, miR775, and miR823 (Fig. 3, Supplementary Fig. S6), which is in agreement with previous reports (Vaucheret et al., 2004; Tang et al., 2012). These data suggest that pri-miR level indeed reflect the abundance of the corresponding miRNA and that glucose can influence the pool of miRNA species.
Fig. 3.
Expression of (A) pri-miR156f, (B) miR156, (C) SPL13, (D) pri-miR159b, (E) miR159, (F) MYB33, (G) ABI3, (H) ABI4, and (I) ABI5 in Col-0, ago1-25 and hyl1-2 seedlings grown in 4% glucose or control media after 3 d of light exposure. All expression values are in comparison to untreated Col-0. Values are mean±SD of three biological replicates. Below each graph, the relative transcript abundance is given. Changes in transcript accumulation were considered significant for differences with fold-change ≥|1.5| and according to Student’s t-test (P < 0.05) for the following comparisons: a, untreated ago1-25 or hyl1-2 vs. untreated Col-0; b, glucose-treated vs. untreated samples (same genotype); c, glucose-treated mutant vs. glucose-treated Col-0. nd, not detected.
Finally, glucose-promoted changes in miRNA accumulation were evaluated during a short-term (i.e. 4h) treatment with a reduced sugar concentration (i.e. 2%), which is known not to induce ABA accumulation (Matiolli et al., 2011). Based on deep sequencing of Arabidopsis Col-0 sRNAs, 29 known unique miRNA sequences out of 155 detected in seedlings grown in liquid MS/2 media for 6 d in light were found to be altered by the short-term treatment with 2% glucose (Supplementary Table 3; Table 1). This result further supports the notion that miRNA accumulation can be regulated by glucose. A comparison of these short-term, glucose-mediated miRNA profile alterations with the pri-miR/miRNA changes detected in seedlings grown for 3 d in light in the presence of 4% glucose (i.e., long-term treatment; Fig. 2, Supplementary Table S2) revealed a common set of 12 miRNAs, six of them showing similar responses to glucose, while the other six exhibited opposite responses (Supplementary Table 3; Table 1). Such differences may be due to differences in developmental stages or glucose concentration between the experiments, but also suggest that short- and long-term glucose-mediated regulation on miRNA expression can be different.
mRNA misregulation resulting from miRNA-pathway deficiency may be associated with glucose hyposensitivity of ago1-25, dcl1-11, and hyl1-2 from germination to seedling establishment
In an attempt to get insights into how miRNA-related regulatory pathways may mediate sensitivity to glucose during early seedling development, the set of nine validated glucose-responsive pri-miRs (Table 1), whose mature forms are known to be involved in developmental cues (miR156, miR159, miR166, miR169, miR390, miR823), biotic stress response (miR773), adaptation to mineral nutrient availability (miR828), and arabinogalactan-protein biosynthesis (miR775), were analysed in more detail. The analysis consisted of evaluating the long-term glucose-mediated expression changes of these nine selected MIRNA genes together with their respective target genes in the Col-0, ago1-25, and hyl1-2 mutant backgrounds.
Deficiencies in HYL1 or AGO1 were shown to differently affect the accumulation of the nine selected pri-miRs (Fig. 3, Supplementary Fig. S6). Since HYL1 is required for pri-miR processing (Szarzynska et al., 2009; Voinnet, 2009), higher accumulation of pri-miRs in hyl1-2 would be expected. All but one (pri-miR166c) of the nine pri-miRs tested were found to be more abundant (pri-miR156f, pri-miR159b, pri-miR169a, pri-miR390a, pri-miR773) or present at equal levels (pri-miR775, pri-miR823, pri-miR828) in hyl1-2 as compared to the wild type, in the absence and presence of glucose (Fig. 3, Supplementary Fig. S6). pri-miR166c was found to be significantly reduced in hyl1-2, indicating the existence of an alternative role for HYL1 or a feedback regulation affecting pri-miR166c expression (Supplementary Fig. S6).
In untreated ago1-25 plants, the accumulation of most (seven out of nine) of the pri-miRs was not affected when compared to the wild type, a result that was expected since AGO1 is assumed not to be directly involved in miRNA biogenesis (Fig. 3, Supplementary Fig. S6). However, pri-miR169a and pri-miR773 levels were reduced in ago1-25, possibly reflecting an indirect effect, such as feedback regulation (Supplementary Fig. S6). Most glucose-induced pri-miR accumulation in ago1-25 appears to be attenuated in comparison with Col-0. In contrast, both the pri-miRs that are repressed by glucose, pri-miR156f and pri-miR166c, responded more strongly in ago1-25 in comparison to Col-0 (Fig. 3, Supplementary Fig. S6). The reasons for these opposite responses are unclear, but could possibly derive from altered gene expression programmes due to AGO1 deficiency (Tang et al., 2012).
An inverse correlation between changes in pri-miR levels and the accumulation of the respective target mRNA in response to glucose in the wild type was observed for pri-miR166c/PHV, pri-miR390/TAS3, pri-miR775/At1g53290, and pri-miR773/MET2 (Supplementary Fig. S6). For the remaining five pri-miRs and their targets (pri-miR828/TAS4, pri-miR823/CMT3, pri-miR169a/NF-YA5, pri-miR156f/SPL13, and pri-miR159/MYB33), the glucose-induced modulation of pri-miR levels was correlated with parallel changes of their targets (Fig. 3, Supplementary Fig. S6). Overall, the wild-type pattern of target mRNA accumulation in control conditions and/or in response to glucose was found to be qualitatively similar in the ago1-25 and hyl1-2 mutants (Fig. 3, Supplementary Fig. S6). For PAP1 mRNA accumulation, which is target of TAS4-derived trans-acting small interfering RNA (ta-siRNA) as part of a feedback regulatory loop (Luo et al., 2012), quantitative differences were observed in both mutants. Accumulation of mRNA was also deregulated in comparison to the wild type for TAS4 and MYB33 in hyl1-2 and for NF-YA5 and SPL13 in ago1-25 (Fig. 3, Supplementary Fig. S6). Additionally, the mRNA level of the transcriptional factor SPL13, whose accumulation was reported to lead to a delay in the emergence of vegetative leaves (Martin et al., 2010; Nonogaki, 2010), was found to be significantly higher in both ago1-25 and hyl1-2 as compared to Col-0 (Fig. 3C).
Short-term responses promoted by 4% glucose on pri-miR expression in Col-0 were found to be basically the same as long-term responses (Fig. 3, Supplementary Figs S5 and S6). The same trend was also found for ago1-25 and hyl1-2 (Fig. 3, Supplementary Figs S5 and S6). In addition, most responses to glucose in dcl1-11 follow the responses observed in hyl1-2 (Supplementary Fig. S5). However, in control conditions, six pri-miRs (pri-miR159b, pri-miR166c, pri-miR169a, pri-miR775, pri-miR823 and pri-miR828) were present at lower levels in dcl1-11 than in Col-0 (Supplementary Fig. S5). It is unclear however what causes the reduction of these pri-miR levels in dcl1-11, but could be related to this specific dcl1 allele.
How these observed quantitative differences in mRNA levels of these miRNA target genes in the miRNA-deficient mutants are involved in their glucose hyposensitive phenotype remains elusive. A potential candidate to explain the glucose hyposensitive phenotype of ago1-25 is the transcription factor gene MYB33 which is regulated by miR159b and mediates ABA-induced inhibition of seedling development (Reyes and Chua, 2007). The lower level of MYB33 mRNA in ago1-25 after long-term treatment with 4% glucose compared with the wild type may partly explain the attenuated development delay of the mutant in such conditions (Fig. 3F). However, the higher amount of MYB33 mRNA in response to glucose in hyl1-2 in comparison with the wild type would be expected to result in glucose-promoted development delay, which is the opposite of the observed glucose hyposensitive phenotype of hyl1-2 (Fig. 3F). MYB33 is part of an ABA-related network which involves ABI3 and ABI5 and retards seedling development under abiotic stress conditions (Reyes and Chua, 2007). Mutants for ABI3, ABI4, and ABI5 have been described as gin mutants (reviewed in Gibson, 2005), so it was important to evaluate the glucose-induced modulation of the expression of these three genes in the miRNA-pathway mutants. ABI3, ABI4, and ABI5 expression were found to be strongly induced by 4% glucose during long-term treatment in Col-0 seedlings grown for 3 d in light (Fig. 3G–I) but not by mannitol, discounting the possibility of an osmotic effect (Supplementary Fig. S7A). Moreover, the fold-change observed for ABI3, ABI4, and ABI5 in 4% glucose-grown seedlings could only partially be attributed to developmental signals, as significantly different responses were observed between seedlings 1 and 3 d after light exposure (Supplementary Fig. S7B). This glucose-specific effect was significantly attenuated in the ago1-25 and hyl1-2 mutants (Fig. 3G–I). This result suggests that while ABI3, ABI4, and ABI5 are not known to be direct miRNA targets, it is possible that the lower levels of the mRNAs of these three ABA signalling elements are, at least partly, responsible for the glucose hyposensitive phenotype of miRNA-pathway mutants. The observed lower accumulation of ABI3, ABI4, and ABI5 mRNAs in ago1-25 and hyl1-2 could be explained by an unknown upstream transcriptional activator that is target of a glucose-dependent miRNA.
Discussion
Developmental processes and responses to environmental changes may rely on fast and fine adjustments of mRNA or protein profiles, which can be partially achieved through miRNA-mediated control of mRNA decay and/or translation (Voinnet, 2009; Floris et al., 2009). Additionally, miRNAs in plants have been proposed to control developmental transitions by preventing premature mRNA expression (Nodine and Bartel, 2010). Thus, the regulation of MIRNA gene expression is crucial for proper growth and development. The modulation of the expression of up to 38 pri-miR genes by long-term treatment with glucose shown in this study, mainly via an HXK1-sensing independent pathway, reveals a new aspect of how sugar signalling could impact on seedling growth, development, and physiology through miRNA-related regulatory schemes (Fig. 2, Supplementary Table S2). This notion is in agreement with previous reports (Yang et al., 2013) and was further supported by the observation that the pattern of glucose-mediated regulation of seven among eight tested pri-miRs reflects the profile of the corresponding mature miRNAs (Fig. 3, Supplementary Fig. S6). Only one case of inverse correlation was observed for pri-miR169a/miR169 (Supplementary Fig. S6), which might reflect antagonist transcriptional and post-transcriptional control (Ramachandran and Chen, 2008; Viswanathan et al., 2008; Ren et al., 2012; Manavella et al., 2012).
In wild-type seedlings, the consequences of pri-miR regulation by glucose on the accumulation of their target mRNA was not found to be fully predictable based solely on the activity of miRNA-mediated mRNA degradation. Among nine target mRNAs that were analysed in more detail, four showed an inverse correlation between the levels of glucose-promoted changes of pri-miR levels and of the mRNA in the wild type (Fig. 3, Supplementary Fig. S6). The trend observed for the remaining five target mRNAs was to follow the expression pattern of the pri-miR (Fig. 3, Supplementary Fig. S6). Such a finding could reflect control of miRNA activity, which seems to occur despite its accumulation and may be related to the developmental stage (Alonso-Peral et al., 2012). Feedforward (Vidal et al., 2010), dampening regulatory mechanisms or differential tissue-specific expression patterns of the miRNA and its target (Voinnet, 2009) are other possible explanations. For example, miR390 and TAS3 are part of a ta-siRNA regulatory loop and are known to be specifically expressed at sites of lateral root initiation (Marin et al., 2010), implying that changes in ARF3 levels promoted by TAS3-derived ta-siRNA may only be detectable locally (Supplementary Fig. S6C).
In comparison to the wild type, ago1-25 and hyl1-2 mutants had variable consequences on target mRNA accumulation in both control and 4% glucose conditions (Fig. 3, Supplementary Fig. S6). Although target mRNA would be expected to accumulate to higher levels in miRNA mutants, it was found to be true only for a limited set of targets in ago1-25 and/or hyl1-2 either in control conditions and/or in response to glucose, (NF-YA5, TAS3, At1g53290, MYB33, and SPL13; Fig. 3, Supplementary Fig. S6). The absence of systematic increase of target mRNA levels in these mutants may be due to different reasons, including the strength of the mutation, which is leaky in the case of ago1-25, and redundancy, which has been reported for AGO proteins (Mi et al., 2008) and HYL1 (Vazquez et al., 2004; Szarzynska et al., 2009).
The few differences in target gene mRNA levels that were observed in ago1-25 and hyl1-2 as compared to the wild type did not provide a clear-cut reason for the glucose hyposensitive phenotype of these mutants. For instance, MYB33 may partly be involved in the ago1-25 glucose hyposensitive phenotype but cannot be responsible for the reduced sensitivity to glucose in hyl1-2 (Fig. 3F). It is, however, possible that rather than a single causal change it is the combination of small or subtle alterations that underlies the glucose hyposensitive phenotype of the miRNA-deficient mutants. For instance, dcl1 embryo patterning defects cannot be phenocopied solely by miR156-resistant SPL10 and SPL11 transgenes, despite their great derepression in dcl1 (Nodine and Bartel, 2010). In addition, besides their activity in target mRNA cleavage, translational inhibition mediated by miRNA may also have an important functional role (Brodersen et al., 2008; Yang et al., 2012). Alternatively, since ABI3, ABI4, and ABI5 are known to modulate sensitivity of seedling development to glucose (Arenas-Huertero et al., 2000; Cheng et al., 2002; Arroyo et al., 2003; Dekkers et al., 2008), the weaker accumulation of ABI3, ABI4, and ABI5 mRNA in response to long-term treatment with glucose in ago1-25 and hyl1-2 mutants could also be directly involved in the glucose hyposensitive phenotype of these mutants during this developmental window (Fig. 3G–I). How the glucose-mediated regulation of these ABA signalling elements is altered in the miRNA-deficient mutants is unknown but is likely to be indirectly related to inappropriate expression of developmental programmes.
To conclude, the regulation of miRNA expression by glucose supports the notion that control of mRNA decay and/or translation represents another mechanistic aspect involved in tuning the glucose regulatory network. A key issue now is to identify the gene expression programmes related to glucose-promoted developmental responses.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences.
Supplementary Table S2. Glucose-regulated pri-miRs and corresponding miRNA targets.
Supplementary Table S3. Known miRNAs present in control and short-term 2% glucose-treated seedlings.
Supplementary Fig. S1. Effects of different concentrations of glucose on early seedling development of wild-type Col-0, miRNA-deficient mutants ago1-25 and hyl1-2, and glucose-insensitive mutant abi4-1/gin6.
Supplementary Fig. S2. Statistical analyses of germination and development efficiencies.
Supplementary Fig. S3. Effects of glucose on early seedling development of Col-0, Ler, hyl1-2, gin2-1, and dcl1-11.
Supplementary Fig. S4. Effects of ABA on early seedling development of Col-0, aba2-1, abi4-1, ago1-25, dcl1-11, and hyl1-2.
Supplementary Fig. S5. Short-term effects of 4% glucose on accumulation of MIRNA genes pri-miRs in Col-0, ago1-25, dcl1-11, and hyl1-2 seedlings after 3 d of light exposure.
Supplementary Fig. S6. Long-term effects of glucose on accumulation of MIRNA genes pri-miR, their corresponding miRNA family and respective targets in Col-0, ago1-25 and hyl1-2 seedlings grown in 4% glucose or control media after 3 d of light exposure.
Supplementary Fig. S7. Validation of glucose-promoted changes on ABI3, ABI4 and ABI5 accumulation.
Acknowledgements
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (grant 2008/54529-4) and the BIOEN programme (2008/52071-0). B.D.P. and W.R.S. received support from the German Federal Ministry for Education and Research (BMBF-Fkz 0315462). The authors thank S. A. Martins for technical support, D. Newman for reviewing the manuscript, H. Vaucheret for kindly providing the Arabidopsis mutants, the M. Stitt and W. R. Scheible groups for experimental support, and B. A. Buzatto for support with the statistical analyses.
References
- Alonso-Peral MM, Sun C, Millar AA. 2012. MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis . PLoS ONE 7, e34751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. 2000. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Development 14, 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein RR, León P. 2003. Three genes that affect sugar sensing (Absicisc Acid Insensitive 4, Abscisic Acid Insensitive 5, and Constitutive Triple Response 1) are differentially regulated by glucose in Arabidopsis . Plant Physiology 133, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P. 2009. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. The Plant Journal 59, 359–374 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. 2008. Widespread translational inhibition by plant miRNAs and siRNAs. Science 30, 1185–1190 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell 14,–2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD. 2011. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana . PLOS Genetics 7, e1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiology 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers BJW, Schuurmans JAMJ, Smeekens SCM. 2008. Interaction between sugar and abscisic acid signaling during early seedling development in Arabidopsis . Plant Molecular Biology 67, 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H. 2011. Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiology 157, 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. 2009. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Smith MR, Weber R, Gregory BD, Poethig RS. 2010. An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana . Silence 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. 2012. Sugars, signaling, and plant development. Journal of Experimental Botany 63, 3367–3377 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. 2006. Regulation of AUXIN RESPONSE FACTOR 3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis . Current Biology 16, 939–944 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. 1994. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. The Plant Journal 5, 765–771 [Google Scholar]
- Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B. 2009. Post-transcriptional regulation of gene expression in plants during abiotic stress. International Journal of Molecular Sciences 10, 3168–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. 2005. Control of plant development and gene expression by sugar signaling. Current Opinion in Plant Biology 8, 93–102 [DOI] [PubMed] [Google Scholar]
- Husbands AY, Chitwood DH, Plavskin Y, Timmermans MCP. 2009. Signals and prepatterns: new insights into organ polarity in plants. Genes and Development 23, 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. 2009. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signaling in Arabidopsis thaliana . The Plant Journal 59, 316–328 [DOI] [PubMed] [Google Scholar]
- Karve A, Xia X, Moore Bd. 2012. Arabidopsis Hexokinase-Like1 and Hexokinase1 form a critical node in mediating plant glucose and ethylene responses. Plant Physiology 158, 1965–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. 2008. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant and Cell Physiology 49, 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research 39, D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León P, Sheen J. 2003. Sugar and hormone connections. TRENDS in Plant Science 8, 110–116 [DOI] [PubMed] [Google Scholar]
- Li P, Wind JJ, Shi X, Zhang H, Hanson J, Smeekens SC, Teng S. 2011. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane-bound domain. Proceedings of the National Academy of Sciences, USA 108, 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. 2008. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plan Cell 20, 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. 2006. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Research 16, 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. 2010. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiology 152, 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. 2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin and cytokinin. The Plant Cell 12, 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QJ, Mittal A, Jia F, Rock CD. 2012. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis . Plant Molecular Biology 80, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D. 2012. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151, 859–870 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant Cell 22, 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Asahina M, Liu PP, et al. 2010. The microRNA156 and microRNA172 gene regulation cascades at post-germinative stages in Arabidopsis . Seed Science Research 20, 79–87 [Google Scholar]
- Matiolli CC, Tomaz JP, Duarte GT, et al. 2011. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiology 157, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. 1989. Generalized linear models. London: Chapman and Hall; [Google Scholar]
- Mi S, Cai T, Hu Y, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. 2003. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light and hormonal signaling. Science 300, 332–336 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. The Plant Cell 14, 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2010. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes and Development 24, 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. 2010. MicroRNA gene regulation cascades during early stages of plant development. Plant and Cell Physiology 51, 1840–1846 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR. 2009. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiology 150, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li TC, Kang SG, Na JK, Jang JC. 2003. Mechanisms of glucose signaling during germination of Arabidopsis . Plant Physiology 132, 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. 2009. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiology 150, 1356–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. 2008. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis . Science 321, 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B. 2012. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis . Proceedings of the National Academy of Sciences, USA 109, 12817–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. 2007. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal 49, 592–606 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annual Review of Plant Biology 57, 675–709 [DOI] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K. 2012. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis . The Plant Cell 24, 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Léon-Kloosterziel KM, Koornneef M, Zeevaart JAD. 1997. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana . Plant Physiology 114, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. 2010. Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology 13, 274–279 [DOI] [PubMed] [Google Scholar]
- Stewart JL, Maloof JN, Nemhauser JL. 2011. PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS ONE 6, e19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Zeeman SC. 2012. Starch turnover: pathways, regulation and role in growth. Current Opinion in Plant Biology 15, 282–292 [DOI] [PubMed] [Google Scholar]
- Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WR, Mueller-Roeber B, Jarmolowski A, Szweykowska-Kulinska Z. 2009. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Research 37, 3083–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Bian S, Tang M, et al. 2012. MicroRNA-mediated repression of the seed maturation program during vegetative development in Arabidopsis . PLoS Genetics 8, e1003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. 2008. Eleven golden rules of quantitative RT-PCR. The Plant Cell 20, 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes and Development 18, 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR. 2002. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proceedings of the National Academy of Sciences, USA 99, 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not post-transcriptional transgene silencing. Current Biology 14, 346–351 [DOI] [PubMed] [Google Scholar]
- Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. 2012. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Research 40, D222–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 107, 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. 2008. Selective blockade of microRNA processing by Lin28. Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis and activity of plant microRNAs. Cell 136, 669–687 [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana . Science 339, 704–707 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis . Cell 138, 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T. 2009. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. The Plant Journal 58, 843–856 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. 2003. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525 [DOI] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS. 2012. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis . Proceedings of the National Academy of Sciences, USA 109, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS. 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C . eLife 2, e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Yuan LJ, Shao Y, Du W, Yan DW, Lu YT. 2008. The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis . Plant, Cell and Environment 31, 562–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.