Abstract
Transglutaminase (TG) is a family of enzymes that catalyzes cross-linking reactions among proteins. Using fluorescent-labeled highly reactive substrate peptides, we recently developed a system to visualize isozyme-specific in situ enzymatic activity. In the present study, we investigated the in situ activities of TG1 (skin-type) and TG2 (tissue-type) using whole mouse sections of various embryonic developmental stages and neonates. In each case, we also successfully used immunostaining of identical whole mouse sections for protein expression after detection of enzymatic activities. In general, the enzymatic activity was correlated with TG protein expression. However, in some tissues, TG protein expression patterns, which were inconsistent with the enzymatic activities, suggested that inactive TGs were produced possibly by self cross-linking or other modifications. Our method allowed us to simultaneously observe developmental variations in both TG isozyme-specific activities and protein levels in mouse embryonic and neonate tissues.
Keywords: transglutaminase, calcium, embryo, development
Introduction
Transglutaminases (TGs) are a family of enzymes comprising eight isozymes that are widely distributed in tissues and cells (Griffin et al. 2002; Lorand and Graham 2003). These enzymes are involved in multiple biological processes by catalyzing isopeptide bond formation between proteins through glutamine and lysine residues in substrate proteins. In addition, these enzymes catalyze the post-translational modifications such as polyamination or conversion to glutamic acid by incorporating either a primary amine or a water molecule into the glutamine residues, respectively. Among these family members, TG1 (skin-type) and TG2 (tissue-type) are major isozymes that display epidermal and ubiquitous expressions, respectively.
TG1 plays a central role in the formation of the skin barrier, by cross-linking several structural proteins to form a cornified envelope in the most differentiated keratinocytes (Candi et al. 2005; Eckert et al. 2005; Hitomi 2005). This cornified envelope is a 15-nm-thick structure comprising covalently cross-linked products that are deposited beneath the plasma membrane. Mice lacking the gene for TG1 exhibit aberrant skin epidermis formation, which results in water loss due to incomplete cornified envelope formation (Matsuki et al. 1998).
TG2 is expressed in various cells and tissues and has diverse functions. This enzyme reaction was observed to be involved in cell fate decisions, as determined by its post-translational modifications of extracellular matrix proteins, transcription factors, and signaling molecules (Fesus and Piacentini 2002; Beninati and Piacentini 2004; Mehta et al. 2006; Tatsukawa et al. 2009). Although TG2-null mutants exhibit a normal phenotype at birth, aberrant wound healing, mild glucose intolerance, and abnormal phagocytosis have been observed in the tissues of these mice (Sarang et al. 2009). Celiac disease, related to TG2 activity, has been extensively studied; this disease involves a chronic inflammation of the intestinal mucosa triggered by deamidated gluten-derived peptides (Sollid 2002).
Both of these isozymes have been extensively characterized also in terms of their gene expression and substrate specificity (Esposito and Caputo 2005). However, to better understand their physiological significance, the simultaneous detection of their protein expression and activity patterns in various tissues is essential. To date, the tissue distributions of these proteins and their enzymatic activities have not been thoroughly investigated, particularly during embryonic development.
We have recently identified highly reactive glutamine-donor substrate peptides of TGs using a random 12-mer peptide library (Sugimura et al. 2006, 2008; Hitomi et al. 2009). Because these peptides exhibited a highly selective reactivity to their respective isozymes, these appeared to be an effective tool for detecting the enzymatic activities in an isozyme-specific manner. Generally, the use of a fluorescent-labeled substrate has been an efficient tool for the detection of active enzymes (Van Nooden 2010). Using fluorescent-labeled peptides, for both TG1 and TG2, we successfully detected their specific activities in frozen tissue sections (Sugimura et al. 2008; Akiyama et al. 2010; Yamane et al. 2010; Johnson et al. 2012). When a reaction occurred, a lysine-donor substrate in a tissue section covalently incorporated a glutamine-donor peptide and a fluorescent signal represented apparently the presence of active TGs. In particular, experiments using whole mouse sections efficiently provided the results for the expression levels of TG in the active form (Itoh et al. 2011).
In previous studies, the expression patterns of both TG1 and TG2 were investigated through mRNA expression and protein levels in several tissues (Hiiragi et al. 1999; Griffin et al. 2002). There have been some investigations on the developmental variations of TG expression in some tissues (Nagy et al. 1997; Citron et al. 2000; Bailey and Johnson 2004; Lee et al. 2005). However, variations in these enzymatic activities in whole mice have not been thoroughly investigated, even though both enzymes may be essential for body formation by modifying growth factors and/or by cross-linking several structural proteins. Therefore, to acquire more insights into the physiological roles of these TGs during embryonic development, it is necessary to simultaneously analyze both the protein expressions and the enzymatic activity distributions of TGs.
In this study, we used fluorescent-labeled substrate peptides to determine the mouse embryonic expression pattern of the in situ activities of TG1 and TG2. In addition to detecting their enzymatic activity levels, we simultaneously analyzed and compared their protein distributions using antibodies specific for each isozyme. Our results should contribute to understanding the roles of TG activities during mouse body formation.
Materials & Methods
Materials
Fluorescent isothiocyanate (FITC)-labeled peptides were synthesized by Biosynthesis (Lewisville, TX). Rabbit polyclonal anti-TG1 (A018) was purchased from Zedira (Darmstadt, Germany). Rabbit polyclonal anti-TG2 serum was made by Japan Lamb (Hiroshima, Japan) using mouse recombinant TG2 as an antigen that was produced in our laboratory. IgG was affinity purified using NHS-activated Sepharose 4 Fast Flow, which was immobilized with recombinant protein (GE Healthcare Bio-Sciences AB; Uppsala, Sweden). Alexa 555-labeled donkey anti-rabbit IgG (H+L) and Alexa 594-labeled donkey anti-rabbit IgG (H+L) were purchased from Life Technologies (Carlsbad, CA). MAS coated slide glass was purchased from Matsunami Glass Ind., Ltd. (Osaka, Japan). Other chemical reagents were obtained from Sigma-Aldrich (St Louis, MO) or WAKO Chemicals (Osaka, Japan).
Preparation of Tissue Sections
ICR pregnant mice were purchased from Japan SLC, Inc. (Shizuoka, Japan) and were sacrificed to collect embryos at various pregnancy stages (E10.5, E12.5, E14.5, E16.5, and E18.5). Neonatal mice were also used in this analysis. TG1 and TG2 knockout (KO) C57/BL6 mice were maintained by K. Yamanishi and S. Kojima, respectively. TG2 KO mice were originally developed and kindly provided by Dr. R. Graham (Nanda et al. 2001). Embryos were processed for preparation according to the method described by Kawamoto (Kawamoto and Shimizu 2000; Kawamoto 2003) and whole-body sections were produced using a multipurpose cryosection preparation kit (Section Lab Co. Ltd.; Hiroshima, Japan). Briefly, the mouse was frozen in cold hexane (–94C) and then freeze-embedded with Super Cryoembedding Medium (SCEM). Ten-μm-thick sections were prepared with a cryomicrotome (Leica CM3050S; Leica Co. Ltd., Wetzlar, Germany) from the frozen specimen block and collected with cryofilm. All sections were histologically identified by hematoxylin and eosin (H&E) staining. Embryonic sequential sections were used for various reactions for in situ enzymatic activity and immunostaining. All animal experiments were carried out according to the guidelines of each institute (Nagoya University, Hyogo College of Medicine, and RIKEN Institute).
In Situ Detection of the Enzymatic Activity of TG
Unfixed 10-μm cryosections of mice embryos were air-dried and then blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS; 10 mM Na-phosphate, pH 8.0, 150 mM NaCl) at room temperature for 30 min. These sections were subsequently incubated for 60 min at 37C in the substrate reaction solution containing 100 mM Tris-HCl (pH 8.0), 1 mM dithiothreitol, and 5 mM CaCl2 in the presence of FITC-labeled peptide (pepK5, pepT26) at the final concentration of 1 μM. After the enzymatic reaction, PBS containing 25 mM EDTA was added to stop the reaction via chelating calcium ion. Then, after washing with PBS three times, mounting solution SCMM (R2) was added to the sections for observation. As negative controls, the reaction mixture containing EDTA (5 mM) instead of CaCl2 was used. Alternatively, the fluorescent-labeled peptides (FITC-K5QN or FITC-T26QN) in which the reactive glutamine residue was replaced by an asparagine residue were used in the control reaction.
Immunostaining
Ten-μm cryosections of mice embryos were air-dried and then fixed with mixed solution (methanol:acetone = 1:1) for 1 min. After washing with PBS three times, the sections were blocked with 1% BSA in PBS at room temperature for 30 min. The blocked sections were then incubated with isozyme-specific antibodies diluted 1:250 (anti-TG1) and 1:500 (anti-TG2) in 1% BSA in PBS overnight at 4C. The sections were washed three times with PBS containing 0.15% Triton-X100 and 1% BSA and then incubated for 1 hr at 37C with fluorescent-labeled secondary antibodies: Alexa 555-labeled donkey anti-rabbit IgG (TG1) and Alexa 594-labeled donkey anti-rabbit IgG (TG2), diluted 1:1000 in PBS containing 0.15% Triton-X100 and 1% BSA. After washing with PBS, mount medium was dropped onto the sections for observation.
In the case of double staining, immunostaining was performed followed by in situ enzymatic reactions.
Microscopic Observation and Analyses
Samples were observed under a Keyence fluorescence microscope (BZ-9000; Keyence, Osaka, Japan) using a ×4 lens (NA 0.20) for whole mouse sections and a ×20 lens (NA 0.75) for each tissue. H&E-stained images were obtained using the same microscope or scanner (EPSON GT-x750; Epson, Nagano, Japan).
The free software Image J (version 1.43u; image processing and analyzing java; http://rsbweb.nih.gov/ij/) was used for linear adjustment of fluorescent images. This software was also used for image mapping using pseudo-colors from the fluorescent signal as a standard procedure.
Results
Reactivities and Specificities by Comparative Staining Patterns with Fluorescent-Labeled Favorable Substrate Peptides and Antibodies for TG1 and TG2
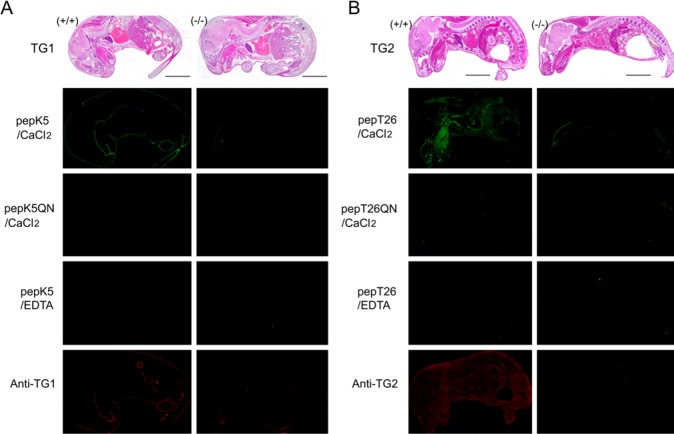
To investigate the relationships between the transamidating activities and protein expression levels of TG1 and TG2, we performed both in situ detection of the activity and immunostaining using favorable substrate peptides and antibodies, respectively. The sequences of pepK5 and pepT26 were used to detect TG1 and TG2 activities, respectively, as shown in Table 1. We initially attempted to detect the activities and protein expressions using sections from TG1 and TG2 KO mice as well as wild type mice (Fig. 1). In this figure, sagittal plane sections of an E19.5 embryo and a neonate were used for TG1 and TG2 analyses, respectively. In the case of TG1 analyses, we used E19.5 embryos instead of neonates throughout the experiments, since KO mice for TG1 died within a couple of hours after birth.
Table 1.
Sequences of Isozyme-specific Substrate Peptides for Transglutaminase Skin-type (TG1) (pepK5) and Tissue-type (TG2) (pepT26).
| Sequence |
|---|
| TG1: YEQHKLPSSWPF (pepK5) |
| YENHKLPSSWPF (pepK5QN) |
| TG2: HQSYVDPWMLDH (pepT26) |
| HNSYVDPWMLDH (pepT26QN) |
pepK5QN and pepT26QN are mutant peptides in which the reactive glutamine residue was substituted with asparagine (indicated in bold).
Figure 1.
In situ detection of enzymatic activities using the preferential substrate peptides and immunostaining analysis using antibodies against transglutaminase skin-type (TG1) and tissue-type (TG2). (A) Sagittal plane sections from the wild-type and TG1-null mice at E19.5 stage were reacted with 1 μM fluorescent isothiocyanate (FITC)-pepK5 in the presence of 5 mM CaCl2. As negative controls, samples were treated with 1 μM of FITC-pepK5QN or in the presence of 5 mM EDTA. Immunofluorescent staining using a polyclonal antibody against TG1 was also performed. (B) Neonate whole-mouse body sagittal plane sections from wild-type and TG2-null mice were reacted with 1 μM FITC-pepT26. As described in the legend to (A), negative controls (1 μM FITC-pepT26QN or EDTA) and immunostaining with an antibody against TG2 were performed. Bars = 0.5 cm.
For an E19.5 stage wild-type mouse embryo (TG1 +/+), significant fluorescent signals resulting from FITC-labeled pepK5 incorporation were observed in epithelial tissues, which was consistent with our previous results (Itoh et al. 2011). No apparent signals could be detected in a reaction with FITC-pepK5QN substituted peptide or in the co-presence of EDTA. In addition, no signals were observed in a section from a TG1-null mouse. These results demonstrated that active TG1 was specifically detected.
When we used a polyclonal antibody against TG1, signals were obtained in the wild-type section corresponding to the in situ enzymatic activity of TG1. In contrast, no signals were detected in a section from a TG1-null mouse. In addition, without the primary antibody, no signals were detected in this section (data not shown). These results indicate that these procedures could be used to detect both the enzymatic activity and protein expression.
As shown in Fig. 1B, TG2 activity analysis in wild-type mouse showed significant fluorescent signals that resulted from cross-linked FITC-pepT26 in various tissues, primarily in connective tissues. No apparent signals were observed for a reaction using FITC-pepT26QN substituted peptide or in the co-presence of EDTA. With the exception of tongue and rectum, no apparent signals were observed in a section from a TG2-null mouse. When using a polyclonal antibody against TG2, signals were detected over a wide area with a pattern similar to that for TG2 enzymatic activity. No signals were detected in a section from a TG2-null mouse. These results demonstrated that both TG2 protein expression and the enzymatic activity could be specifically detected using this procedure simultaneously.
Simultaneous Detection of In Situ TG Activity and Protein Expression Patterns in Embryonic Whole Mouse Sections
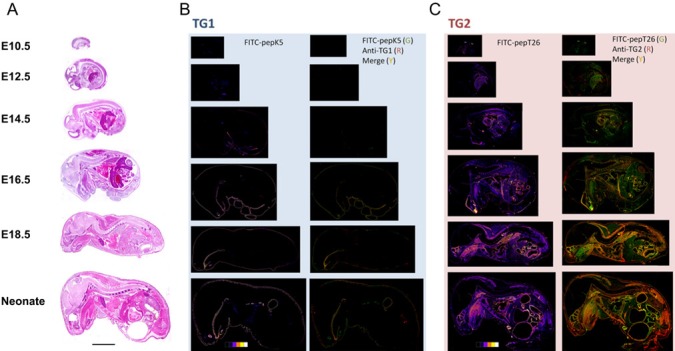
For this experiment, we performed double-staining analyses of frozen sections from embryos at various developmental stages. Sections that reacted with FITC peptides were subsequently fixed and immunostained. For this immunofluorescent analysis, we used secondary fluorescent-conjugated antibodies for staining. These analyses allowed us to compare the enzymatic activities and protein expression patterns of the two major TGs present in the whole body.
First, for TG1 (Fig. 2B), the results for the detection of in situ enzymatic activity showed significant signals primarily in epithelial tissues, including skin, esophagus, forestomach, and tooth. For these epithelial tissues, an E10.5 section showed weak signals, whereas the E12.5 to E14.5 sections exhibited active signals, particularly around the bronchial arch. These signals were dramatically enhanced at the stage of E14.5 to E16.5, as shown by the pseudo-color image data. In a neonate section, signal intensity also began to increase in hair follicles in addition to epithelial areas. Compared with the immunofluorescently stained area, the resulting signals nearly overlapped with those for the signals of active enzyme.
Figure 2.
Simultaneous double staining for in situ enzyme activities and immune detection in whole-mouse body sections. (A) Sections of embryos at various fetal development stages (E10.5, E12.5, E14.5, E16.5, E18.5) and neonatal mice were stained with H&E staining. (B) The sections at midline were subjected to in situ enzymatic activity detection [fluorescent isothiocyanate (FITC)-pepK5] followed by immunostaining for transglutaminase skin-type (TG1), using the same reaction conditions described in the legend to Fig. 1. Results in the left panels show enzymatically active areas indicated by pseudo-color images. Results in the right panels show merged areas (yellow) from the stained areas for the enzymatic activity (green) and immunostaining (red). (C) The sections at midline were subjected to in situ enzymatic activity detection (FITC-pepT26) followed by immunostaining for transglutaminase tissue-type (TG2). Results are in the same order as in (B). Bars = 0.5 cm.
Subsequently, for TG2 (Fig. 2C), the enzymatic activity was particularly higher in liver, heart, and spine compared with other internal organs, on E12.5 to E14.5 sections. An E16.5 section exhibited TG2 activity in other sites, such as muscle. Depending on the developmental stage, the signal pattern gradually changed to reflect that of an adult mouse. With regard to immunofluorescently stained areas, weak enzymatic activity was observed at earlier stages (mostly purple on pseudo-color images). During the embryonic development, the immunofluorescent signals gradually increased.
In Situ Activity and Protein Expression in Specific Tissues
In general, the enzymatic activity level was correlated with the protein expression level. However, in some tissues, the ratios of the levels of enzyme activity and protein levels were varied, as shown by pseudo-color images (Fig. 2). In addition, unique expression patterns were observed in some tissues, as described below (Figs. 3 and 4).
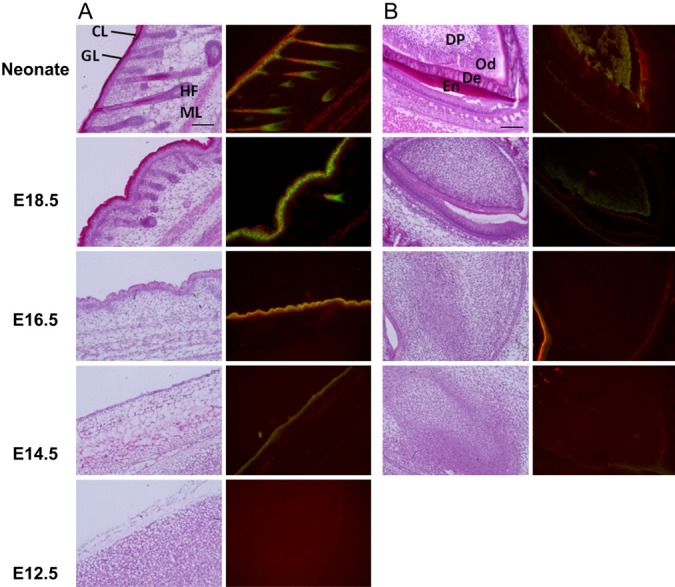
Figure 3.
Double-staining patterns for transglutaminase skin-type (TG1) in various tissues during mouse embryonic development. Enlarged pictures of H&E staining (left) and merged images of in situ activities and immunostaining for TG1, using fluorescent isothiocyanate (FITC)-pepK5 and an anti-TG1 antibody (right), are shown. Results in panels A and B are for the reactions with (A) skin and (B) teeth: CL, cornified layer; GL, granular layer; HF, hair follicle; ML, muscle layer; DP, dental papilla; Od, odontoblast; De, dentin; En, enamel. At E14.5 and E16.5, the surrounding area of teeth was shown.
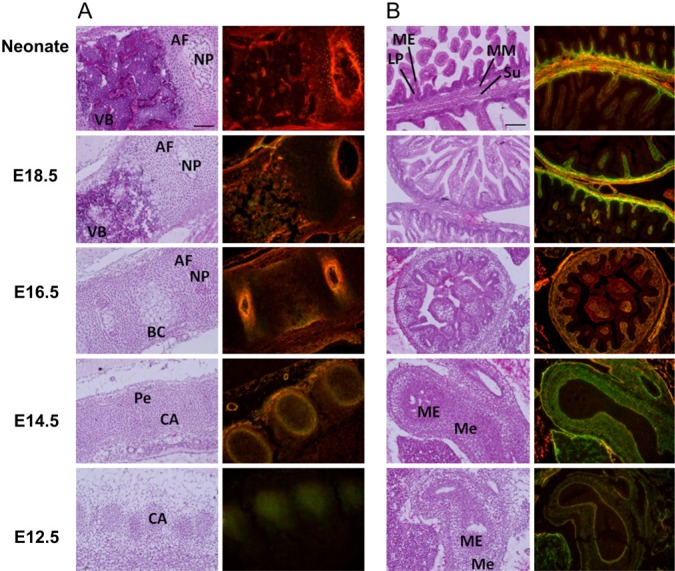
Figure 4.
Double-staining patterns for transglutaminase tissue-type (TG2) in various tissues during mouse embryonic development. H&E staining (left) and merged images of in situ activities and immunostaining (right) with corresponding fluorescent isothiocyanate (FITC)-pepT26 and an anti-transglutaminase tissue-type (TG2) antibody are shown. Results in panels A and B are for reactions. (A) Spine: AF, annulus fibrosus; NP, nucleus pulposus; VB, vertebral body; BC, subperiosteal bone collar; Pe, perichondrium; CA, cartilage anlagen. (B) Intestine: ME, mucosal epithelium; LP, lamina propria mucosae; MM, muscularis mucosae; Su, submucosa; Me, mesenchyme. Bars = 100 μm.
For TG1, which was primarily expressed in skin, the enzyme activity and protein expression levels dramatically increased starting at the E14.5 to E16.5 stage (Fig. 3A). In the outermost cornified layers in the epidermis of the neonate, the immunostaining signal levels were higher than those for enzymatic activity as compared with the patterns in other areas. A similar area (red) was also observed in an E16.5 section, although it was thinner than that in the neonate. This suggested that inactive TG1 itself had possibly accumulated as a substrate in the differentiated keratinocytes by cross-linking. In the neonate, hair follicles also showed apparent signals for TG1 activity and protein expression. Tooth tissues also showed that TG1 was highly expressed in an active form (Fig. 3B). At the E18.5 stage, TG1 activity, which was higher than that of the protein level, was observed around the odontoblast. From the E18.5 stage to the neonatal stage, the protein expression level gradually increased over a wide area containing dentin.
As shown in Fig. 4A, TG2 activity was observed in the cartilage anlagen in the spine at the E12.5 stage. At E14.5, these signals were concentrated in the perichondrium. During further development, from the E16.5 stage to the neonatal stage, and between the annulus fibrosus and nucleus pulposus, TG2 activities were enhanced in accordance with the protein levels. In these areas, the protein level was relatively enhanced compared to the enzyme activity. This pattern was also observed in the hyaline cartilage of neonatal nose chondrocytes, whereas the activity relative to protein expression in the perichondrium was increased at a later stage of development (Suppl. Fig.).
In the intestine, in which higher TG2 activity was detected among the tissues, unique expression variations were observed during development (Fig. 4B). At the E12.5 stage, TG2 was highly expressed with regard to its enzymatic activity in the mesenchymal area of the midgut. Regarding villi development, in the intestinal mucosa, TG2 enzymatic activity and the protein level were enhanced in the layers of the lamina propria mucosae, muscularis mucosae, and submucosa.
Discussion
Post-translational modifications by TGs, such as cross-linking between proteins, deamidation, and the attachment of polyamines, are implicated in various biological events. These enzymes, consisting of the eight isozymes, are involved in these reactions in a calcium-dependent manner. Among them, both TG1 and TG2 are major enzymes that are highly expressed in various tissues and have several physiological roles. However, there is little information regarding the distributions of TG activities during embryonic development among these isozymes. Therefore, we aimed to simultaneously acquire insights into the positional and developmental variations in the activities and protein expressions of both TG1 and TG2.
To detect enzymatic activity in an isozyme-specific manner, we established an in situ detection system using fluorescent-labeled peptides that were isozyme-specific glutamine-donor substrates (Itoh et al. 2011). Mouse whole-body sections were successfully prepared for the detection of in situ activities, and these sections also appeared to be suitable for immunostaining analysis (Fig. 1). Furthermore, as shown in Fig. 2, immunostaining analyses could be subsequently performed after detecting the enzymatic activity. Thus, we could compare the protein expression levels and in situ enzyme activities on the same sections, which established precise tissue distributions for TGs (Table 2). In addition, examining various tissues, including tooth, spine, and nose chondrocytes, provided considerably novel information.
Table 2.
Summary of the Transglutaminase (TG) Protein Expressions and Activity Levels during Mouse Embryo Development.
| TG1 | E12.5 | E14.5 | E16.5 | E18.5 | Neonate |
|---|---|---|---|---|---|
| Tongue (epidermis) | |||||
| A | - | - | ++ | ++ | ++ |
| P | + | + | + | ++ | ++ |
| Skin (granular layer) | |||||
| A | - | + | ++ | +++ | ++ |
| P | - | + | ++ | ++ | ++ |
| Teeth (odontoblast) | |||||
| A | - | - | - | + | + |
| P | - | - | - | - | + |
| TG2 | E12.5 | E14.5 | E16.5 | E18.5 | Neonate |
| Tongue (muscle) | |||||
| A | - | - | - | + | ++ |
| P | - | - | + | ++ | ++ |
| Nose (hyaline cartilage) | |||||
| A | + | + | ++ | ++ | + |
| P | - | + | + | ++ | ++ |
| Spine (vertebra body) | |||||
| A | + | + | + | ++ | ++ |
| P | - | + | + | ++ | +++ |
| Heart (myocardial fiber) | |||||
| A | ++ | ++ | ++ | ++ | ++ |
| P | ++ | ++ | ++ | ++ | ++ |
| Lung (stroma) | |||||
| A | + | + | ++ | ++ | +++ |
| P | - | + | ++ | ++ | +++ |
| Liver (hepatocyte) | |||||
| A | + | + | + | + | + |
| P | - | - | + | + | + |
| Intestine (lamina propria mucosae) | |||||
| A | + | + | ++ | +++ | +++ |
| P | - | - | ++ | ++ | ++ |
| Kidney (nephron) | |||||
| A | - | + | + | + | + |
| P | - | - | + | + | + |
TG protein expressions and activity levels were classified as either undetectable (-), low (+), intermediate (++), or high (+++). This evaluation was carried out by the human operator’s eye. A and P indicate the levels of in situ enzymatic activity and immunologically detected protein, respectively. TG1, transglutaminase skin-type; TG2, transglutaminase tissue-type.
Depending on the tissues and embryonic developmental stages, the ratio of the protein level to activity level is varied. In the liver (Fig. 2, neonate) and the intestine at earlier developmental stages (Fig. 4A), the enzyme activity levels relatively increased but the protein levels did not. In these cases, some activation mechanisms may have been required, such as binding of an activator and limited proteolysis in the case of TG1 (Hitomi 2005). In contrast, as observed by the signals in the developed spine and the outermost cornified layers of the skin (Figs. 3A and 3B), the protein levels were higher at these sites than the enzymatic activity levels. In these cases, TG1 may have been a favorable substrate, and participated in cross-linking activity that would result in polymerization and, also possibly, susceptibility to other modifications such as sumoylation and phosphorylation (Luciani et al. 2009; Wang et al. 2012). This would decrease its enzymatic activity, although the area would still be immunochemically positive. Therefore, our data from this study were significant because active enzymes could be detected in each of these tissues.
In a series of experiments from the present and the previous studies, we could not detect enzyme activity in some tissues such as the brain (TG2) and the lung (TG1), where each enzyme is considered to be actively expressed. This was probably because of the release of the cross-linked products, which would make the signal undetectable. Additionally, during preparation of the tissue sections and the cross-linking reaction, which may have led to a loss in cell membrane integrity, limited activity could be detected, such as in the intracellular areas. Further modifications of our procedures will be required in some tissues.
In this study, the signals detected for TG enzymatic activity were based on lysine-donor (glutamine-acceptor) substrates for each TG. Although several TG substrates have been identified, novel substrates may have been included among these signals, in various tissues. Identifying these proteins using tag-attached peptides would enable the biochemical identification of novel substrates, which would be useful information for understanding their physiological significance in each tissue.
In conclusion, we obtained data for the expression patterns of major TGs at both the protein and transamidation activity levels. In addition to our previously developed detection system for in situ enzyme activity, we established conditions to perform a simultaneous immunostaining analysis. Furthermore, reactions for in situ detection of enzyme activity and subsequent immunofluorescent analysis were successfully performed. These analyses provided information on indexes for both enzyme activity and protein expression levels.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (B) (No. 23380200 to KH; No. 24102533 to SK) and also Grant-in-Aid for Young Scientists Research (No. 24.10612 to MI) from the Ministry of Education, Sports, Science and Technology (JSPS, KAKENHI, Japan) and the Terumo Life Science Foundation (to KH).
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Grant-in-Aid for Scientific Research (B) No. 23380200 (Ministry of Education, Sports, Science and Technology [JSPS], KAKENHI, Japan); Grant-in-Aid for Young Scientists Research No. 24.10612 (JSPS, KAKENHI, Japan); and the Terumo Life Science Foundation, Japan.
References
- Akiyama M, Sakai K, Yanagi T, Fukushima S, Ihn H, Hitomi K, Shimizu H. 2010. Transglutaminase 1 preferred substrate peptide K5 is an efficient tool in diagnosis of lamellar ichthyosis. Am J Pathol. 176:1592–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CD, Johnson GV. 2004. Developmental regulation of tissue transglutaminase in the mouse forebrain. J Neurochem. 91:1369–1379 [DOI] [PubMed] [Google Scholar]
- Beninati S, Piacentini M. 2004. The transglutaminase family: an overview. Amino Acids. 26:367–372 [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. 2005. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Bio. 6:328–340 [DOI] [PubMed] [Google Scholar]
- Citron B, Gregory E, Steigerwalt D, Qin F, Festoff B. 2000. Regulation of the dual function tissue transglutaminase/Gah during murine neuromuscular development: gene and enzyme isoform expression. Neurochem Int. 37:337–349 [DOI] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA. 2005. Transglutaminase function in epidermis. J Invest Dermatol. 124:481–492 [DOI] [PubMed] [Google Scholar]
- Esposito C, Caputo I. 2005. Mammalian transglutaminases. Identification of substrates as a key to physiological function and physiopathological relevance. FEBS J. 272:615–631 [DOI] [PubMed] [Google Scholar]
- Fesus L, Piacentini M. 2002. Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem Sci. 27:534–539 [DOI] [PubMed] [Google Scholar]
- Griffin M, Casadio R, Bergamini CM. 2002. Transglutaminases: nature’s biological glues. Biochem J. 368:377–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiiragi T, Sasaki H, Nagafuchi A, Sabe H, Shen SC, Matsuki M, Yamanishi K, Tsukita S. 1999. Transglutaminase type 1 and its cross-linking activity are concentrated at adherens junctions in simple epithelial cells. J Biol Chem. 274:34148–34154 [DOI] [PubMed] [Google Scholar]
- Hitomi K. 2005. Transglutaminases in skin epidermis. Eur J Dermatol. 15:313–319 [PubMed] [Google Scholar]
- Hitomi K, Kitamura M, Sugimura Y. 2009. Preferred substrate sequences for transglutaminase 2: screening using a phage-displayed peptide library. Amino Acids. 36:619–624 [DOI] [PubMed] [Google Scholar]
- Itoh H, Kawamoto T, Tatsukawa H, Kojima S, Yamanishi K, Hitomi K. 2011. In situ detection of active transglutaminases for keratinocyte type (TGase 1) and tissue type (TGase 2) using fluorescence-labeled highly reactive substrate peptides. J Histochem Cytochem. 59:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KB, Petersen-Jones H, Thompson JM, Hitomi K, Itoh M, Bakker EN, Johnson GV, Colak G, Watts SW. 2012. Vena cava and aortic smooth muscle cells express transglutaminases 1 and 4 in addition to transglutaminase 2. Am J Physiol Heart Circ Physiol. 302:H1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T. 2003. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 66:123–143 [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Shimizu M. 2000. A method for preparing 2- to 50-μm-thick fresh frozen section of large samples and undecalcified hard tissues. Histochem Cell Biol. 113:331–339 [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim YS, Lee YJ, Lee SS, Song IS, Park SC, Chi JG, Chung SI. 2005. Transglutaminase 2 expression in the salivary myoepithelial cell of mouse embryo. Arch Oral Biol. 50:301–308 [DOI] [PubMed] [Google Scholar]
- Lorand L, Graham RM. 2003. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 4:140–156 [DOI] [PubMed] [Google Scholar]
- Luciani A, Villella VR, Vasaturo A, Giardino I, Raia V, Pettoello-Mantovani D’Apolito M, Guido S, Leal T, Quaratino S, et al. 2009. SUMOylation of tissue transglutaminase as link between oxidative stress and inflammation. J Immunol. 183:2775–2784 [DOI] [PubMed] [Google Scholar]
- Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, et al. 1998. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1. Proc Natl Acad Sci USA. 95:1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta K, Fok JY, Mangala LS. 2006. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci. 11:173–185 [DOI] [PubMed] [Google Scholar]
- Nagy L, Thomazy V, Saydak M, Stein J, Davies P. 1997. The promoter of the mouse tissue transglutaminase gene directs tissue-specific, retinoid-regulated and apoptosis-linked expression. Cell Death Differ. 4:534–547 [DOI] [PubMed] [Google Scholar]
- Nanda N, Iismaa SE, Owens WA, Husain A, Mackay F, Graham RM. 2001. Targeted inactivation of Gh/tissue transglutaminase II. J Biol Chem. 276:20673–20678 [DOI] [PubMed] [Google Scholar]
- Sarang Z, Tóth B, Balajthy Z, Köröskényi K, Garabuczi E, Fésüs L, Szondy Z. 2009. Some lessons from the tissue transglutaminase knockout mouse. Amino Acids. 36:625–631 [DOI] [PubMed] [Google Scholar]
- Sollid LM. 2002. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2:647–655 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Hosono M, Kitamura M, Tsuda T, Yamanishi K, Maki M, Hitomi K. 2008. Identification of preferred substrate sequences for transglutaminase 1-development of a novel peptide that can efficiently detect cross-linking enzyme activity in the skin. FEBS J. 275:5667–5677 [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Hosono M, Wada F, Yoshimura T, Maki M, Hitomi K. 2006. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: identification of peptide substrates for TGase 2 and Factor XIIIa. J Biol Chem. 281:17699–17706 [DOI] [PubMed] [Google Scholar]
- Tatsukawa H, Fukaya Y, Frampton G, Martinez-Fuentes A, Suzuki K, Kuo TF, Nagatsuma K, Shimokado K, Okuno M, Wu J, et al. 2009. Role of transglutaminase 2 in liver injury via cross-linking and silencing of transcription factor Sp1. Gastroenterology. 136:1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nooden CJF. 2010. Imaging enzymes at work: metabolic mapping by enzyme histochemistry. J Histochem Cytochem. 58:481–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ande SR, Mishra S. 2012. Phosphorylation of transglutaminase 2 at serine-216 has role in TG2 mediated activation of nuclear factor-kappa B and in the down regulation of PTEN. BMC Cancer. 277:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Fukui M, Itoh M, Alea MP, Thomas V, El Alaoui S, Akiyama M, Hitomi K. 2010. Identification of a preferred substrate peptide for transglutaminase 3 and detection of in situ activity in skin and hair follicles. FEBS J. 277:3564–3574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.