Significance
The AMP-activated protein kinase (AMPK) of eukaryotes has been called “the cellular fuel gauge” because it is a central regulator of carbon metabolism that senses cellular energy charge. We show that Snf1, the catalytic subunit of AMPK of the yeast Saccharomyces cerevisiae, is modified by attachment of the small ubiquitin-like modifier SUMO, which inhibits Snf1 function. This process provides yet another way cells regulate function of this highly conserved protein kinase.
Keywords: protein kinase regulation, protein modification, signal transduction
Abstract
The AMP-activated protein kinase (AMPK) is a major stress sensor of mammalian cells. AMPK’s homolog in the yeast Saccharomyces cerevisiae, the SNF1 protein kinase, is a central regulator of carbon metabolism that inhibits the Snf3/Rgt2-Rgt1 glucose sensing pathway and activates genes involved in respiration. We present evidence that glucose induces modification of the Snf1 catalytic subunt of SNF1 with the small ubiquitin-like modifier protein SUMO, catalyzed by the SUMO (E3) ligase Mms21. Our results suggest that SUMOylation of Snf1 inhibits its function in two ways: by interaction of SUMO attached to lysine 549 with a SUMO-interacting sequence motif located near the active site of Snf1, and by targeting Snf1 for destruction via the Slx5-Slx8 (SUMO-directed) ubiquitin ligase. These findings reveal another way SNF1 function is regulated in response to carbon source.
Glucose is the preferred carbon source of most cells, including Saccharomyces cerevisiae, which ferments it to ethanol and CO2, producing only two ATPs, even when oxygen is available to drive production of much more ATP. This preference for fermentation (which cancer cells share), is known as the Crabtree or Warburg effect (1, 2). Because of the energetic inefficiency of fermentation, yeast cells must be adroit in sensing glucose. S. cerevisiae has three well-known glucose sensing pathways: (i) the Gpa1/2-Ras2-PKA pathway that regulates stress response (through Msn2/4) and other things; (ii) the SNF1 pathway, which regulates respiratory metabolism and other processes; and (iii) the Snf3/Rgt2-Rgt1 (SRR) pathway that regulates expression of genes encoding hexose transporters (3).
The SRR (sensor/receptor-repressor) pathway begins at the cell surface with high-affinity (Snf3) (4) and low-affinity (Rgt2) glucose sensors (5) that are coupled to the casein kinases Yck1 and Yck2, which catalyze phosphorylation of the corepressor proteins Mth1 and Std1 (6), leading to their ubiquitinylation by SCFGrr1 (7, 8). The subsequent destruction of Mth1 and Std1 inactivates the Rgt1 transcriptional repressor, resulting in derepression of HXT genes encoding hexose transporters (7, 9). In response to glucose, Yck1/2 also mediates inactivation and degradation of transporters of alternative carbon sources, such as maltose (Mal61) (10) and lactate/pyruvate/acetate (Jen1) (11). Achieving this glucose-induced switching of transporters seems to be the main purpose of the SRR pathway (12).
The SNF1 protein kinase—the ortholog of the AMP-activated protein kinase (AMPK), a major stress-activated protein kinase in mammalian cells (13, 14)—is a central regulator of carbon metabolism (15, 16). This kinase is an activator of Adr1 and Cat8, which activate expression of genes involved in the diauxic shift, ethanol, and lactate uptake and catabolism, gluconeogenesis, and respiration (17–21), and is an inhibitor of the Mig1 repressor of glucose-repressed genes (22). SNF1 is a heterotrimer of the Snf1 catalytic subunit, the Snf4 activation subunit (14), and one of three subunits (Sip1, Sip2, and Gal83) that localize SNF1 to different cellular compartments (14, 23).
In yeast cells grown in the absence of glucose, Snf1 is active, phosphorylated on its activation loop threonine 210 primarily by Sak1, but also by Tos1 and Elm1 (16, 24, 25). Addition of glucose to cells results in a reduction in ADP levels that causes the Glc7-Reg1 protein phosphatase to dephosphorylate T210 and thereby inactivate SNF1 (26–28). Reg1 is required for glucose-induced destruction of Mth1 and HXT gene expression (10, 29, 30), a requirement that is relieved by deletion of SNF1 (29), suggesting that SNF1 inhibits glucose sensing through the SRR pathway. T210 can also be dephosphorylated by the Sit4 protein phosphatase (although insufficiently to compensate for reg1Δ when glucose sensing is perturbed by diversion of glucose to glycogen) (31). Deleting both SIT4 and REG1 is lethal because it results in overactive SNF1, which is toxic to cells (31, 32).
Many proteins that become modified by the small ubiquitin-like modifier SUMO, encoded in yeast by SMT3, regulate diverse processes. One role for SUMOylation is to promote interaction with other proteins via a SUMO-interacting motif (SIM) (33–35). Another role is to direct ubiquitinylation of SUMOylated proteins by the SUMO-targeted Ubiquitin E3 ligases (StUbL) Slx5-Slx8 and Ris1 (36), resulting in substrate degradation.
SUMO is conjugated to its target proteins by a mechanism analogous to ubiquitin conjugation. SUMO is activated by ATP-dependent thioester bond formation with the E1 activator Aos1-Uba2 (37), transferred to the E2 conjugator Ubc9 (38), then conjugated to a lysine on a substrate protein, usually in the sequence ΦKxD/E (where Φ is a hydrophobic amino acid, and x is any amino acid), with the help of an E3 ligase.
S. cerevisiae has four SUMO-E3 ligases. Siz1 is responsible for the majority of SUMOylation during vegetative growth, with Siz2 conducting most of the remainder (39); Mms21 directs SUMOylation of proteins involved in chromosome maintenance and recombination (40, 42–46). MMS21 is an essential gene because Mms21 plays a critical role in the structural maintenance of chromosomes (SMC) protein complex. However, mutations affecting the Mms21 RING finger domain that abolish its SUMO-ligase activity are not lethal (40, 46, 47), suggesting that the essential function Mms21 executes in the SMC complex is not related to its SUMO ligase activity. To date, all known Mms21 substrates are involved in DNA metabolism and repair. Here we present evidence that Snf1 is negatively regulated by its SUMOylation, catalyzed by Mms21.
Results
SUMOylation Affects the SRR Pathway and ADH2 Expression.
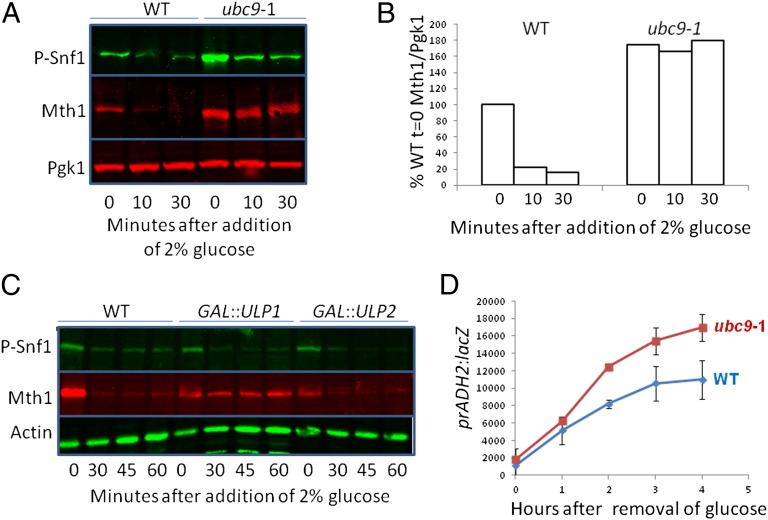
A screen of yeast mutants missing each of the 12 ubiquitin and ubiquitin-like (E2) ligases revealed that the SUMO (E2) ligase Ubc9 (38) is required for glucose-induction of HXT1 and HXT3 expression, suggesting that SUMO plays a role in the SRR pathway (48). We confirmed this finding by showing that Ubc9 is required for glucose-induced Mth1 destruction (Fig. 1 A and B). (Note that the basal levels of Mth1 and phospho-Snf1 are elevated in ubc9-1 cells.) Conversely, overexpression of the Ulp1 (but not the Ulp2) deSUMOylase prevents glucose-induced Mth1 destruction (Fig. 1C). The rate of induction of ADH2 expression, which is dependent upon SNF1 activation of the Adr1 transcription factor (49), is increased in a ubc9 mutant (Fig. 1D). These results suggest that SUMO influences function of a protein that regulates the SRR pathway and ADH2 expression.
Fig. 1.
SUMO is required for glucose sensing. (A) Cells were grown in 2% galactose at 24 °C overnight, then for 1 h at 37 °C. Preheated glucose was added to a final concentration of 2% and cells were processed for immunoblots (Materials and Methods) at the indicated times. “P-Snf1” is visualized with antibody that detects Snf1 phosphorylated on its activation loop T210 (Materials and Methods). P-Snf1 is displayed above Mth1, although they occupy the same space on the membrane; each was detected with a different channel of the imager. (B) Quantification of Mth1 from A. The Mth1/Pgk1 ratio is relative to wild-type cells grown in galactose (t = 0). (C) Cells were grown overnight at 30 °C in 3% glycerol. Galactose was added to 2% for 2 h to induce Ulp1 expression followed by addition of glucose to 2%; samples were processed for immunoblots at the indicated times. (D) Cells were grown at 24 °C overnight in 4% glucose, then for 1 h at 37 °C to allow for inactivation of the temperature-sensitive Ubc9 (ubc9-1). Cells were washed three times with 37 °C water, and resuspended in 3% glycerol medium at 37 °C. Samples were taken for β-galactosidase assays at the indicated times. n = 3.
Identification of the SUMO (E3) Ligase Required for Glucose Sensing.
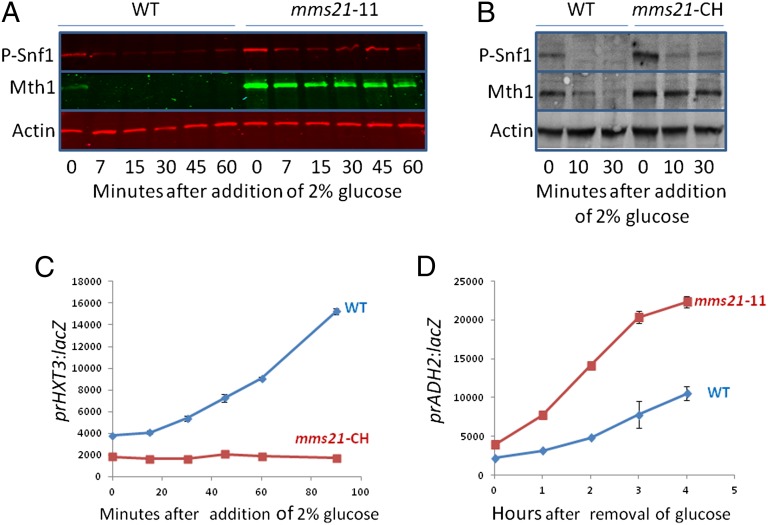
Of the three SUMO (E3) ligases, only Mms21 is necessary for glucose-induced destruction of Mth1: the mms21-11 and mms21-CH mutations that inactivate the SUMO E3 ligase function of Mms21 [but not its essential interaction with the SMC complex (40, 45)], increases basal levels of Mth1 and abolishes (or nearly so) its destruction (Fig. 2 A and B), and glucose-induction of HXT3 expression (Fig. 2C). In further experiments we used the mms21-CH mutant or the mms21-11 mutant interchangeably. [Note that the amount of phospho-Snf1 is elevated in the mms21-11 mutant, because of increased Snf1 levels in this mutant (Fig. S1 A and B). Glucose-induced dephosphorylation of Snf1 is unaffected by mutation of mms21.] Induction of ADH2 expression is enhanced by the mms21-11 mutation (Fig. 2D). The Siz1 and Siz2 SUMO (E3) ligases are not required for Mth1 degradation (Fig. S1 C and D).
Fig. 2.
Mms21 regulates Snf1 activity. (A) Cells were grown in 2% galactose at 30 °C overnight. Glucose was added to 2% and samples were processed for immunoblots at the indicated times. (B and C) Cells were grown in 2% galactose at 30 °C overnight, then at 34 °C for 1 h before addition of preheated glucose to 2%. Samples were processed for immunoblots (B) and β-galactosidase assays (C) at the indicated times. All further experiments with the mms21-CH mutant were conducted at 34 °C to ensure Mms21 was maximally inactivated. (D) The mms21-11 mutant was grown overnight at 30 °C in 4% glucose, cells were washed three times with water and resuspended in 3% glycerol medium at 30 °C. Samples were taken for β-galactosidase assays at the indicated times. n = 3.
Placement of Mms21 in the SRR Glucose-Sensing Pathway.
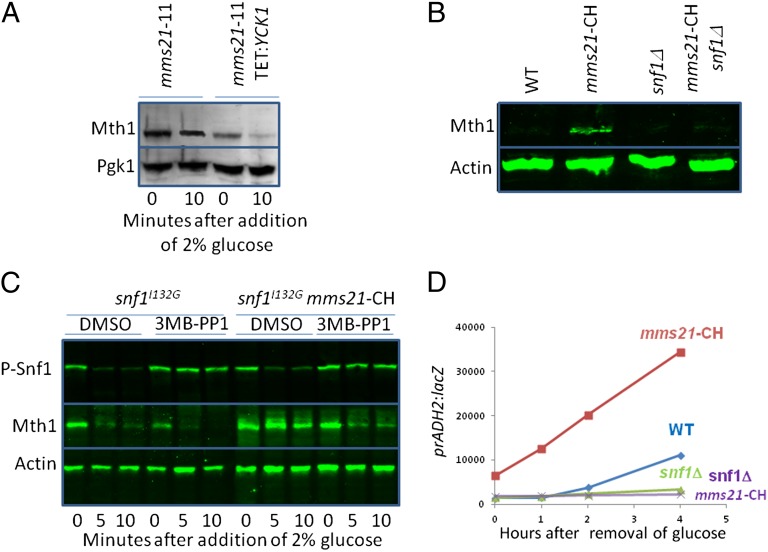
To determine where Mms21 acts in the SRR pathway, we tested genetic interactions of mms21 mutations with alterations of SRR pathway components. Overexpression of Yck1 fused to the tail of Rgt2 (6) restores Mth1 degradation in mms21-11 cells (Fig. 3A), suggesting that Mms21 acts upstream of Yck1 in the SRR pathway. Mutations that reduce SNF1 function suppress the glucose sensing defects caused by an mms21 mutation: deletion of SNF1 restores Mth1 degradation in mms21-CH cells grown on glucose (Fig. 3B), as does inhibiting an ATP analog-sensitive SNF1 [Snf1I132G (50)] with the ATP analog 3MB-PP1 (Fig. 3C). Deletion of SNF1 reverses the increase in ADH2 expression caused by the mms21-CH mutation (Fig. 3D). These results point to Snf1 as a (direct or indirect) target of Mms21, and suggest that SUMOylation inhibits Snf1 function.
Fig. 3.
Mms21 acts upstream of Snf1 in glucose sensing. (A) Cells were grown at 30 °C overnight in 2% galactose; 2 μg/mL doxycycline was added 30 min before adding glucose to 2%. Samples were processed for immunoblots at the indicated times. (B) Cells were grown overnight in 4% glucose at 30 °C, the temperature was raised to 34 °C for 1 h, and samples were processed for immunoblots. (C) Cells were grown at 30 °C overnight in 2% galactose, then for 1 h at 34 °C then pretreated with 25 mM 3MB-PP1 or DMSO for 2 h before addition of preheated glucose to 2%. Samples were processed for immunoblots at the indicated times. Because Mth1 used in this experiment carries the S-tag (72), it does not comigrate in the gel with Snf1 as it does in the other figures. (D) Cells were grown at 30 °C overnight in 4% glucose, then for 1 h at 34 °C, then washed three times with water at 34 °C, resuspended in 3% glycerol at 34 °C. Samples were taken for β-galactosidase assays at the indicated times.
Snf1 Is the Target of Mms21.
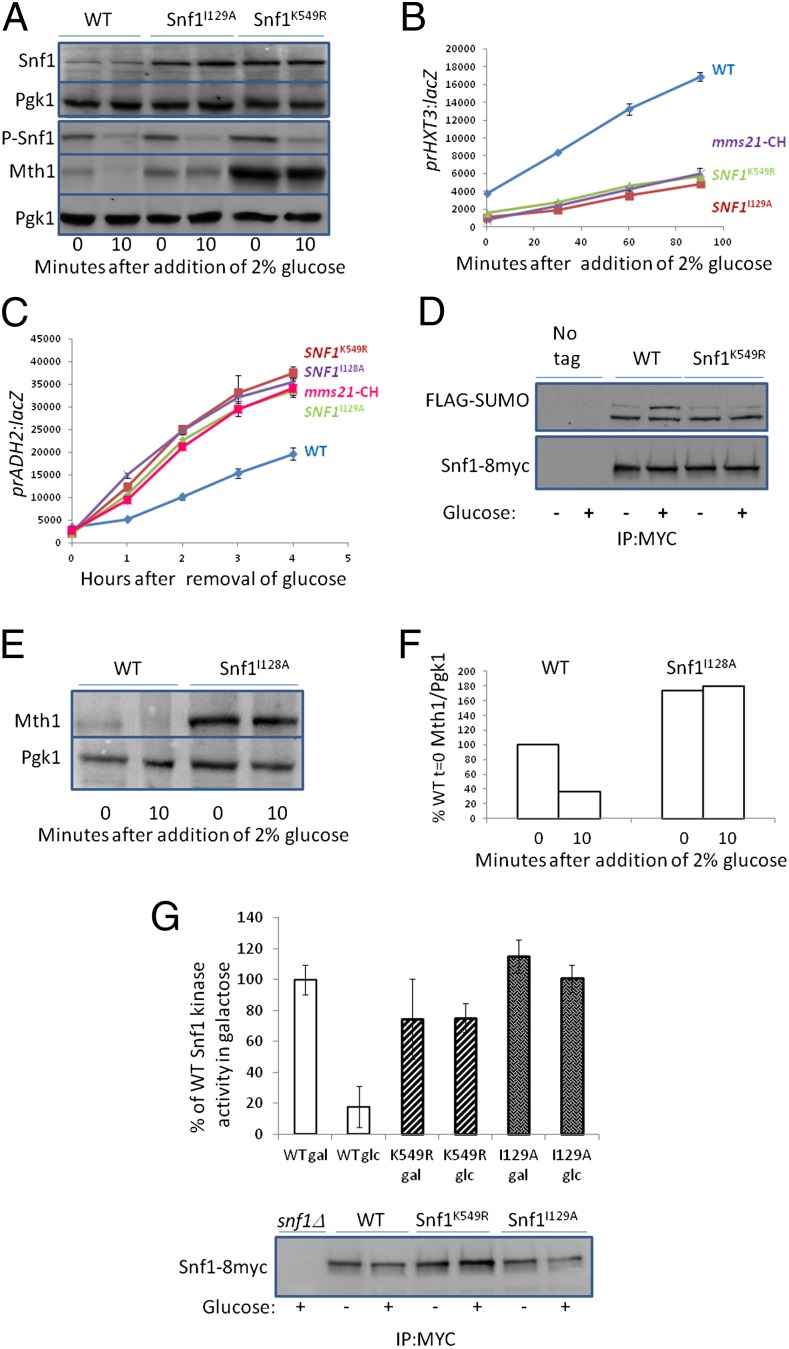
Snf1 has a robust consensus SUMOylation site [K/R-x-x-Φ-K*-x-D/E (51), where x is any amino acid and Φ is hydrophobic, and K* is linked to SUMO] at K549 in its C-terminal regulatory domain (Fig. S3). Changing lysine 549 to arginine prevents Mth1 destruction in response to glucose (Fig. 4A) (note that it also increases the basal levels of Mth1), and reduces HXT3 expression (Fig. 4B), and enhances induction of ADH2 expression (Fig. 4C). Mutations altering each of four lysines in weak consensus SUMOylation sites [Φ-K*-x-D/E (52)] in Snf1 do not cause these phenotypes (Fig. S2 A and B).
Fig. 4.
Snf1 is SUMOylated. (A) snf1Δ cells with plasmids bearing wild-type SNF1 or SNF1 with the indicated mutations were grown overnight at 30 °C in 2% galactose. Glucose was added to 2% and samples processed for immunoblots at the indicated times. A separate blot shows Snf1 levels. (B and C) snf1Δ cells with plasmids bearing wild-type SNF1 or SNF1 with the indicated mutations, or snf1Δmms21-CH cells with a plasmid bearing wild-type SNF1 were grown overnight at 30 °C in 2% galactose (B), or in 4% glucose (C), then for 1 h at 34 °C. Preheated glucose was added to 2% (B), or (C) cells were washed three times with water at 34 °C and resuspended in 3% glycerol at 34 °C, and samples were taken for β-galactosidase assays at the indicated times. n = 3. (D) ulp1-ts cells [strain 1274 (59)] expressing Snf1-8myc (71), Snf1K549R-8myc or Snf1 (with no tag), together with a plasmid containing GAL:His6-FLAG-Smt3 were grown overnight at 24 °C in 2% galactose then shifted to 37 °C for 3 h before addition of glucose to 4% for 1 h. Samples were processed for immunoprecipitation with anti-Myc and immunodetection with anti-FLAG. [We believe the lower band is cross-reaction with the antibody, as has been observed for Smc5, another substrate of Mms21 (40)]. (E) snf1Δ cells with plasmids bearing wild-type SNF1 or SNF1I128A were grown overnight at 30 °C in 2% galactose. Glucose was added to 2% and samples processed for immunoblots at the indicated times. (F) Quantification of E using ImageJ. The Mth1/Pgk1 ratio is relative to wild-type cells grown in galactose (t = 0). (G) Cells were grown in media containing 2% galactose (gal) or 2% glucose (glc). Snf1-8myc was immnoprecipitated and protein kinase assays performed on Snf1 immobilized on beads using SAMS peptide as the phosphate acceptor (see Materials and Methods). Activities were normalized to Snf1 abundance and are shown to the activity of wild-type Snf1 from cells grown in galactose.
The fact that the SNF1K549R mutation causes the same phenotypes as the mms21-11 and mms21-CH mutations suggests that K549 of Snf1 is modified by SUMO. Indeed, immunoprecipitated Snf1 (from cells with a temperature-sensitive Ulp1 to prevent removal of SUMO) can be detected with antibody that recognizes SUMO (Fig. 4D). The amount of SUMOylated Snf1 (Fig. 4D, Upper) is increased by the addition of glucose to cells, and is decreased by the K549R mutation.
A Potential SUMO-Interacting Motif in Snf1 Is Necessary for SNF1 Inhibition.
The N-terminal protein kinase domain of Snf1 contains two overlapping potential SIMs [defined as V/I-x-V/I-V/I or V/I-V/I-x-V/I/L, with acidic residues in close proximity (51)], at D126-V131 (SIM1) and I129-E133 (SIM2) (Fig. S3). Changing isoleucine 129, which is shared by these two SIMs, to alanine (I129A), reduces glucose-induced degradation of Mth1 (Fig. 4A), severely reduces the rate of glucose induction of HXT3 expression (Fig. 4B), and enhances induction of ADH2 expression (Fig. 4C), the same phenotypes caused by the mms21-11/CH (Fig. 2 and Fig. S1 A and B) and SNF1K549R (Fig. 4 A–C) mutations. Mutation of I128, which only affects SIM1, protects Mth1 from glucose-induced degradation (Fig. 4E) (note that it also increases the basal level of Mth1), and increases ADH2 expression, similar to the effect of the mms21-CH mutation (Fig. 4C), but changing isoleucine 132 in SIM2 to glycine [the snf1as1 mutation (50)] neither reduces Mth1 degradation (Fig. 3C) nor enhances ADH2 expression (Fig. S2C) (it actually reduces ADH2 expression, probably because the mutation reduces SNF1 activity). Thus, SIM1, but not SIM2, seems to be required for inhibition of Snf1 function by SUMOylation. The reduction in protein kinase activity of SNF1 (53, 54) caused by glucose depends on both the SUMO (K549R) and the SIM1 (I129A) sites of Snf1 (Fig. 4G), suggesting that SUMOylation of Snf1 inhibits its enzymatic activity.
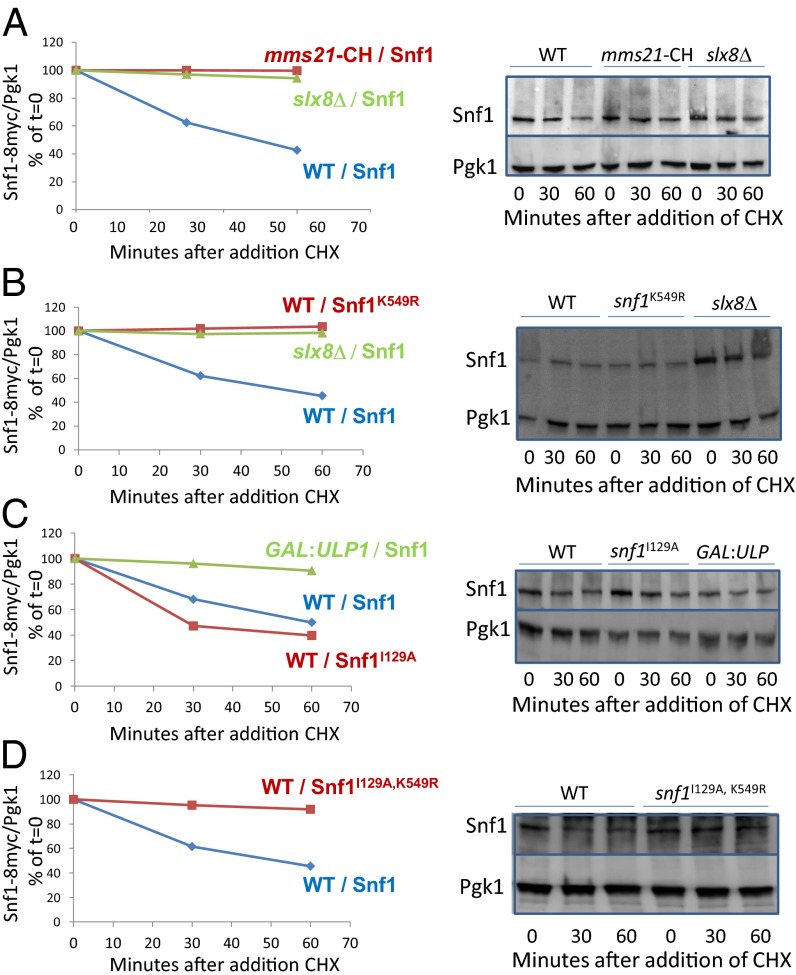
SUMOylation Destabilizes Snf1.
Snf1 is ubiquitinylated, and its consequent degradation is best detected in a mutant missing the Ubp8 deubiquitinylase (55). We measured Snf1 levels in ubp8Δ cells to explore the effect of SUMOylation of Snf1 on its stability. The level of Snf1 diminishes when protein synthesis is inhibited with cycloheximide (Fig. 5), but Snf1 is stable in mms21-CH (Fig. 5A) and SNF1K549R mutants (Fig. 5B), revealing that Snf1 degradation depends on its SUMOylation. Indeed, deletion of SLX8, which encodes a SUMO-targeted ubiquitin ligase (36), stabilizes Snf1 (Fig. 5 A and B), as does overexpression of the Ulp1 deSUMOylase (Fig. 5C). These results suggest that SUMO directs Slx8 to ubiquitinylate Snf1. Snf1I129A may be degraded more rapidly than wild-type Snf1 (Fig. 5C), perhaps because this mutation prevents the SUMO attached to Snf1 at K549 from interacting with the N-terminal SIM1 motif of Snf1, thereby exposing the SUMO to Slx8. If so, we would expect the increased destruction of Snf1I129A to be dependent upon SUMOylation of K549, and indeed, Snf1I129A, K549R is stable (Fig. 5D).
Fig. 5.
Effects of SUMOylation and ubiquitinylation on Snf1 stability. Snf1 levels were measured in ubp8Δ cells carrying no other mutation (WT), or also carrying slx8∆ or mms21-CH, or a plasmid carrying GAL1::ULP1, as indicated before the slash (/) for each set of data; the nature of the Snf1 expressed in each mutant (Snf1-8myc or Snf1K549R or Snf1I129A, or Snf1I129A,K549R) is indicated after the slash. Cells were grown in 2% galactose at 34 °C (A) or 30 °C (B–D). Samples were processed for immunoblots at the indicated times after addition of cycloheximide (CHX) to 200 μg/mL and glucose to 2%. Pgk1 and Snf1 were quantified using ImageJ.
Discussion
Our results suggest that conjugation of SUMO to K549 of Snf1 is likely catalyzed by the Mms21 SUMO (E3) ligase. Snf1 was found to be SUMOylated in glucose-grown cells in one comprehensive screen for SUMOylated proteins (56), but not in others (57–59). Those incongruent results may be a result of the prolonged inactivation of Ulp1 required to detect SUMOylated Snf1, which may reflect rapid removal of SUMO under normal conditions.
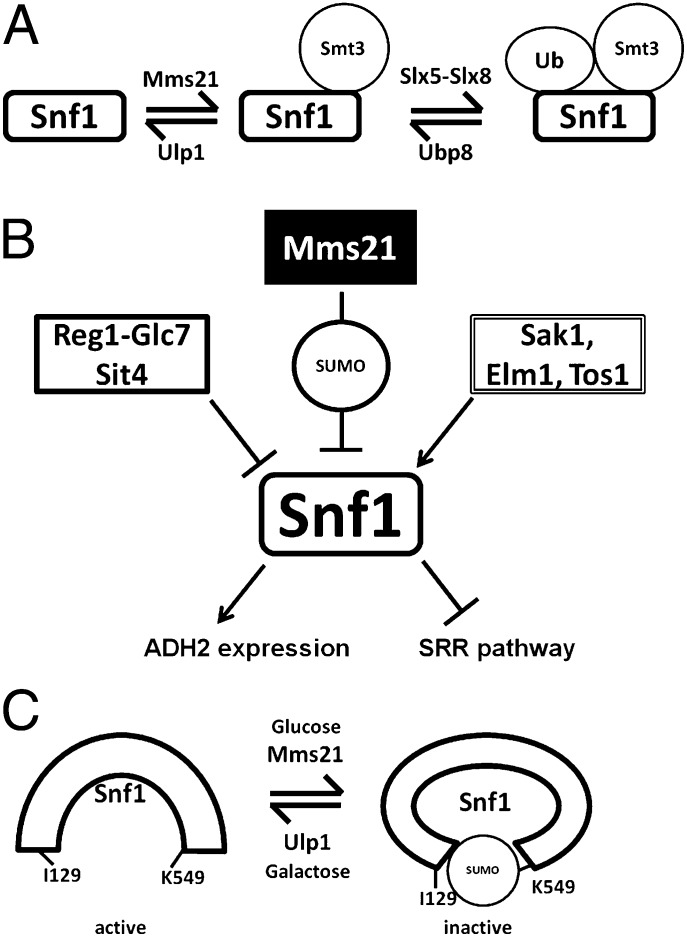
SUMOylation of Snf1 reduces its activity, thereby preventing activation of Adr1 and ADH2 expression and relieving SNF1 inhibition of glucose sensing by the SRR pathway. SNF1 also regulates the basal level (i.e., in the absence of glucose) of Mth1. One way SUMOylation of Snf1 reduces its function is by causing it to be degraded (Fig. 5). Inhibition of Snf1 SUMOylation results in an increase in Snf1 levels. These results expand the identified players in the SUMO/Ubiquitin cycle of Snf1 from the previously reported deubiquitinylase Ubp8 (55) to include a SUMO-E3 ligase (Mms21), a SUMO-targeted ubiquitin E3 ligase (Slx5-Slx8), and a deSUMOylase (Ulp1) (Fig. 6A), and explain the observation that Snf1Δ381–608 is more abundant than full-length Snf1 (60). Our results reveal another input into regulation of Snf1 (summarized in Fig. 6B).
Fig. 6.
Model for regulation of Snf1 by SUMO and ubiquitin. (A) The SUMO/Ubiquitin cycle of Snf1. Ubp8 was previously identified as a deubiquitinylase for Snf1 (55). (B) Summary of Snf1 regulatory inputs and outputs described here. Arrows indicate activation; perpendicular lines indicate inhibition. (C) Our interpretation of the effects of SUMOylation on the conformation of Snf1.
SUMOylation of Snf1 may also inhibit its activity through interaction of SUMO attached to K549 with the SIM (SIM1) in Snf1, which could contribute to forming or stabilizing the inactive conformation of Snf1 (14, 61) (Fig. 6C). In this view, the active conformation of Snf1 caused by removal of glucose from cells or by the I128A and I129A mutations exposes the SUMO moiety to the Slx5-Slx8 SUMO-directed ubiquitin ligase, resulting in an increased rate of Snf1 degradation. Indeed, the acceleration of Snf1 destruction caused by the SNF1I129A mutation depends upon K549 (Fig. 5C). However, we cannot exclude the possibility that another SUMOylated protein interacts with the SIM in Snf1, or that the SUMO linked to Snf1 K549 interacts with other SIMs, as has been suggested for the homologous recombination pathway (34). It is interesting that preventing SUMOylation of Snf1 or interaction of SUMO with the SIM of Snf1 results in increased SNF1 activity in glucose-grown cells (Fig. 4G), even though the level of T210-phosphorylated Snf1 diminishes. Perhaps interfering with SUMO’s effects on Snf1 has unmasked complexity in regulation of Snf1 function.
We suggest that a possible role for Snf1 degradation is to attenuate its levels in the cell, especially under conditions in which Snf1 activity may be deleterious to cell growth. Because destruction of Snf1 occurs on a timescale of hours, whereas glucose sensing is rapid, we imagine that SUMO inhibits Snf1 function in the short term through a SUMO–SIM interaction, and in the long-term results in increased turnover of Snf1. AMPKα2 levels in mouse myoblast cells are similarly down-regulated in response to glucose by the Ubiquitin E3-ligase Wwp1 (62).
Although a connection between SNF1 and the response to MMS or hydroxyurea has been reported (63), we believe our finding that Snf1 is SUMOylated via Mms21 is a unique example of an Mms21 substrate not directly involved in DNA repair or chromosome maintenance. These findings suggest a link between carbon and DNA metabolism. We wonder if the down-regulation of Snf1 by its SUMOylation may be connected to the switch from respiration to fermentation that occurs during S-phase (64) and when DNA is damaged (65).
Materials and Methods
Yeast Strains and Media.
Most experiments were conducted with strains of the W303 genetic background (66). The Snf1 stability experiments and immunoprecipitations used BY4741/2. Yeasts were transformed using the frozen lithium acetate method (67). Strains used are listed in Table S1.
SNF1 yeasts were grown for HXT3 expression and Mth1 degradation assays in synthetic media containing 2% (wt/vol) galactose with the necessary nutrient supplements; snf1Δ cells were grown in media containing 2% glucose. For ADH2 expression assays, cells were grown in 4% glucose, washed three times with water, and resuspended in media containing 3% glycerol. For measuring Snf1 degradation, cycloheximide (AG Scientific) was added to 200 μg/mL from a 10-mg/mL stock solution in 50% ethanol. For experiments with the snf1as1 (I132G) mutant (50), 25 μM 3MB-PP1 in DMSO (gift of K. Shokat, Department of Cellular and Molecular Pharmacology, University of California, San Francisco) was added 2 h before addition of glucose.
Plasmids.
Selective markers in plasmids were exchanged by gap repair (66). Plasmids were mutagenized using Quickchange (Alligent) and confirmed by Sanger sequencing. Plasmids used are listed in Tables S2 and S3.
β-Galactosidase Assays.
The β-galactosidase assay kit (Pierce, cat. no. 75768) was used in a 96-well plate format. Cell concentration was read at 600 nm. Reaction time was typically 5 min at room temperature. The plate reader was a BIO-TEK instruments incorporated synergy HT multidetection microplate reader. β-Galactosidase activity is given in Miller units.
Immunopreciptations.
40–50 mL of cells were centrifuged for 2 min at 4,400 × g in an Eppendorf 5702 (which provides very rapid acceleration and deceleration; 2 min was found to be the minimal amount of time for maximal cell pelleting). Cells were resuspended in 600 μL ice-cold B60 (68) containing 480 mM KAc, 0.1% Triton X-100, 1 mM NEM, HALT protease, and phosphatase inhibitors (Pierce #1861280), and pepstatin A (Sigma #P4265), with 500-μL glass beads (Biospec Products, Cat No. 11079105). Cells were vortexed at 4 °C for 30 min with 1-min rests every other minute and spun at 13,000 rpm for 30 min at 4 °C. The supernatant was transferred to a new tube and spun for an additional 5 min. Protein concentration was determined by BCA (Pierce #23228/1859078). Extracts containing 17 mg of total protein were incubated with 40 μL Preconjugated EZ-View anti-Myc beads (Sigma #E6654) for 2 h and washed three times with lysis buffer and once with B60 containing 60 mM KAc without inhibitors before eluting by boiling into nonfluorescent sample buffer (Pierce #39001).
Immunoblots.
To prevent activation of Snf1 by processing of the cells (69), cells were killed before centrifugation by adding 5 mL of cells to 1 mL 100% TCA. Cells were vortexed with glass beads, pelleted, and resuspended in nonfluorescent sample buffer (Licor 928–40004). Protein concentration was determined by the Coomassie elution with SDS method (70), except that destaining was done with water. Protein extracts were run on 10% TGS gels (Bio-Rad) and transferred to nitrocellulose or PVDF membranes. Membranes were probed with mouse anti-Myc (9E10; Santa Cruz), rabbit anti–S-tag (Abcam), rabbit anti-phospho T172 AMPKα (Cell Signaling), which also detects phosphorylation of the orthologous T210 of Snf1, mouse anti-HA (Roche), mouse anti-FLAG (M2, Sigma), mouse anti-GFP (Roche), or rabbit anti-GFP (Sigma) antibodies at 1,000× dilution in blocking buffer (Rockland MB-070). Loading controls were detected with rabbit antiactin (Epitomics; 500×) or mouse anti-Pgk1 (Invitrogen; 10,000×) antibodies. Blots were visualized with a LiCOR odyssey or a Bio-Rad imager. Proteins of similar sizes were visualized simultaneously using anti-mouse or anti-rabbit secondary antibodies labeled with fluorescent dyes that emit at different wavelengths [anti-mouse 680LT and anti-rabbit 800CW (LiCOR) or anti-mouse Dylight 488 and anti-rabbit Dylight 549 (Epitomics)]. Phospho-Snf1T210 is presented above Mth1, although they occupied the same space in the gel. Slices of the same gel are enclosed in boxes and joined together; slices of different gels (always with their respective loading controls) are separated by a space. Quantification of all blots (using ImageJ) are presented in Fig. S4.
Protein Kinase Assays.
Snf1-8myc was immunoprecipitated from extracts of snf1Δ cells carrying SNF1-8myc on a plasmid [pYL199 (71) and derivatives]. Pelleted cells were resuspended in 600 μL of ice-cold lysis buffer containing 60 mM KAc (68) with 0.1% Nonidet P-40 and protease/phosphatase inhibitors, as described above. Extracts containing 27 mg of total protein were incubated with 40 μL preconjugated EZ-View anti-Myc beads (Sigma #E6654) (prepared as described above but with 60 mM KAc in the lysis buffer) for 2 h, washed three times with B60 containing 60 mM KAc with protease/phosphatase inhibitors, once with kinase buffer from ADP-Glo assay kit (Promega #V9101), and resuspended in 400 μL kinase buffer and protein kinase activity was determined using the ADP-Glo kit in a 96-well plate format (reaction volume scaled up five times) using SAMS peptide (5 nanomoles) (Signalchem) as the substrate. Protein from 100 μL of the beads was extracted into nonfluorescent sample buffer (Pierce #39001) and run on a 10% polyacrylamide gel, which was transferred to PVDF membranes. Snf1-8myc was detected with anti-myc (9E10; Santa Cruz), and the relative Snf1 abundance was quantified using ImageJ. Protein kinase activity was normalized to Snf1 protein levels in wild-type cells grown in galactose (n = 3).
Supplementary Material
Acknowledgments
We thank M. Carlson for critical review of the manuscript and for yeast strains and plasmids; X. Zhao, K. Arndt, E. Young, S. Prakash, S. Berger, K. Tatchell, E. O’Shea, M. Hochstrasser, J. Rutter, T. Ravid, and B. Johnson for strains and plasmids; K. Shokat for 3MB-PP1; and Y. Lavy, J. Dover, M. Matecic, and S. Creat for helpful discussions. This work was supported by National Institutes of Health Grant GM32540.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304839110/-/DCSupplemental.
References
- 1.De Deken RH. The Crabtree effect: A regulatory system in yeast. J Gen Microbiol. 1966;44(2):149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 3.Busti S, Coccetti P, Alberghina L, Vanoni M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors (Basel) 2010;10(6):6195–6240. doi: 10.3390/s100606195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisson LF, Neigeborn L, Carlson M, Fraenkel DG. The SNF3 gene is required for high-affinity glucose transport in Saccharomyces cerevisiae. J Bacteriol. 1987;169(4):1656–1662. doi: 10.1128/jb.169.4.1656-1662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozcan S, Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996;16(10):5536–5545. doi: 10.1128/mcb.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101(6):1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flick KM, et al. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14(8):3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol Cell Biol. 2004;24(20):8994–9005. doi: 10.1128/MCB.24.20.8994-9005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5(1):167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadura N, Robinson LC, Michels CA. Glc7-Reg1 phosphatase signals to Yck1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006;172(3):1427–1439. doi: 10.1534/genetics.105.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paiva S, et al. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: Role of lysine 63-linked ubiquitin chains. J Biol Chem. 2009;284(29):19228–19236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3(1):221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardie DG, Ross FA, Hawley SA. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17(4):2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghillebert R, et al. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J. 2011;278(21):3978–3990. doi: 10.1111/j.1742-4658.2011.08315.x. [DOI] [PubMed] [Google Scholar]
- 16.Hedbacker K, Hong SP, Carlson M. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol Cell Biol. 2004;24(18):8255–8263. doi: 10.1128/MCB.24.18.8255-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bojunga N, Entian KD. Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p) Mol Gen Genet. 1999;262(4-5):869–875. doi: 10.1007/s004380051152. [DOI] [PubMed] [Google Scholar]
- 18.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 19.Tachibana C, et al. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25(6):2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther K, Schüller HJ. Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology. 2001;147(Pt 8):2037–2044. doi: 10.1099/00221287-147-8-2037. [DOI] [PubMed] [Google Scholar]
- 21.Young ET, Dombek KM, Tachibana C, Ideker T. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J Biol Chem. 2003;278(28):26146–26158. doi: 10.1074/jbc.M301981200. [DOI] [PubMed] [Google Scholar]
- 22.Vallier LG, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137(1):49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 1994;13(24):5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elbing K, McCartney RR, Schmidt MC. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem J. 2006;393(Pt 3):797–805. doi: 10.1042/BJ20051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA. 2003;100(15):8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95(11):6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrashekarappa DG, McCartney RR, Schmidt MC. Subunit and domain requirements for adenylate-mediated protection of Snf1 kinase activation loop from dephosphorylation. J Biol Chem. 2011;286(52):44532–44541. doi: 10.1074/jbc.M111.315895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer FV, et al. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 2011;14(5):707–714. doi: 10.1016/j.cmet.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasula S, Jouandot D, 2nd, Kim JH. Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett. 2007;581(17):3230–3234. doi: 10.1016/j.febslet.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomás-Cobos L, Sanz P. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem J. 2002;368(Pt 2):657–663. doi: 10.1042/BJ20020984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz A, Xu X, Carlson M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci USA. 2011;108(16):6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz A, Liu Y, Xu X, Carlson M. Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc Natl Acad Sci USA. 2012;109(22):8652–8657. doi: 10.1073/pnas.1206280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JL, Ulrich HD. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res. 2012;40(22):11380–11388. doi: 10.1093/nar/gks892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151(4):807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 35.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101(40):14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uzunova K, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282(47):34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 37.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16(18):5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272(43):26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 39.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106(6):735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102(13):4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20(15):2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chavez A, George V, Agrawal V, Johnson FB. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J Biol Chem. 2010;285(16):11922–11930. doi: 10.1074/jbc.M109.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan X, et al. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell. 2009;35(5):657–668. doi: 10.1016/j.molcel.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai R, et al. Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J Biol Chem. 2011;286(16):14516–14530. doi: 10.1074/jbc.M110.157149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santa Maria SR, Gangavarapu V, Johnson RE, Prakash L, Prakash S. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(23):8409–8418. doi: 10.1128/MCB.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi Y, et al. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 2008;4(10):e1000215. doi: 10.1371/journal.pgen.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reindle A, et al. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci. 2006;119(Pt 22):4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- 48. Li F (1998) Analysis of yeast Grr1 protein. PhD dissertation (Washington Univ, St. Louis, MO)
- 49.Ratnakumar S, Kacherovsky N, Arms E, Young ET. Snf1 controls the activity of adr1 through dephosphorylation of Ser230. Genetics. 2009;182(3):735–745. doi: 10.1534/genetics.109.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirra MK, et al. A chemical genomics study identifies Snf1 as a repressor of GCN4 translation. J Biol Chem. 2008;283(51):35889–35898. doi: 10.1074/jbc.M805325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minty A, Dumont X, Kaghad M, Caput D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem. 2000;275(46):36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 52.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147(5):981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies SP, Carling D, Hardie DG. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem. 1989;186(1-2):123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- 54.Woods A, et al. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269(30):19509–19515. [PubMed] [Google Scholar]
- 55.Wilson MA, et al. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol Cell Biol. 2011;31(15):3126–3135. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 57.Denison C, et al. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4(3):246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279(40):41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 59.Wykoff DD, O’Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4(1):73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Leech A, Nath N, McCartney RR, Schmidt MC. Isolation of mutations in the catalytic domain of the snf1 kinase that render its activity independent of the snf4 subunit. Eukaryot Cell. 2003;2(2):265–273. doi: 10.1128/EC.2.2.265-273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459(7250):1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 62.Lee JO, et al. E3 ubiquitin ligase, WWP1, interacts with AMPKα2 and down-regulates its expression in skeletal muscle C2C12 cells. J Biol Chem. 2013;288(7):4673–4680. doi: 10.1074/jbc.M112.406009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubacq C, Chevalier A, Mann C. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol Cell Biol. 2004;24(6):2560–2572. doi: 10.1128/MCB.24.6.2560-2572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316(5833):1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 65.Kitanovic A, et al. Metabolic response to MMS-mediated DNA damage in Saccharomyces cerevisiae is dependent on the glucose concentration in the medium. FEMS Yeast Res. 2009;9(4):535–551. doi: 10.1111/j.1567-1364.2009.00505.x. [DOI] [PubMed] [Google Scholar]
- 66.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 67.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15(10B):963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 68.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol. 2001;21(9):3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6(11):1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 70.Marbach I, Licht R, Frohnmeyer H, Engelberg D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J Biol Chem. 2001;276(20):16944–16951. doi: 10.1074/jbc.M100383200. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Xu X, Carlson M. Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot Cell. 2011;10(3):313–319. doi: 10.1128/EC.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raines RT, McCormick M, Van Oosbree TR, Mierendorf RC. The S.Tag fusion system for protein purification. Methods Enzymol. 2000;326:362–376. doi: 10.1016/s0076-6879(00)26065-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.