Significance
Understanding the evolution of malaria parasites and their phylogenetic context is key to understanding this important human disease. We report an unexpected high diversity of malaria parasite genera in bats from West African forest ecosystems. Two lineages are closely related to Plasmodium parasites from rodents, which are common laboratory model systems, and the results are consistent with switches between these hosts over their evolutionary history. Bats are considered important reservoir hosts for many pathogens, particularly viruses, and have unusually high immunological tolerances. The abundant malaria parasite infections are consistent with this exceptional immunology and suggest that in bats the parasites repeatedly evolved life cycles away from disease-causing replication in red blood cells to less pathogenic propagation in liver tissue.
Keywords: Haemosporida, Chiroptera, vector-borne disease, molecular phylogeny, host–pathogen coevolution
Abstract
As the only volant mammals, bats are captivating for their high taxonomic diversity, for their vital roles in ecosystems—particularly as pollinators and insectivores—and, more recently, for their important roles in the maintenance and transmission of zoonotic viral diseases. Genome sequences have identified evidence for a striking expansion of and positive selection in gene families associated with immunity. Bats have also been known to be hosts of malaria parasites for over a century, and as hosts, they possess perhaps the most phylogenetically diverse set of hemosporidian genera and species. To provide a molecular framework for the study of these parasites, we surveyed bats in three remote areas of the Upper Guinean forest ecosystem. We detected four distinct genera of hemosporidian parasites: Plasmodium, Polychromophilus, Nycteria, and Hepatocystis. Intriguingly, the two species of Plasmodium in bats fall within the clade of rodent malaria parasites, indicative of multiple host switches across mammalian orders. We show that Nycteria species form a very distinct phylogenetic group and that Hepatocystis parasites display an unusually high diversity and prevalence in epauletted fruit bats. The diversity and high prevalence of novel lineages of chiropteran hemosporidians underscore the exceptional position of bats among all other mammalian hosts of hemosporidian parasites and support hypotheses of pathogen tolerance consistent with the exceptional immunology of bats.
Malaria is a mosquito-borne epidemic human disease caused by protozoan parasites of the genus Plasmodium. Four different species known to cause human malaria have been studied intensively over several decades, and in the recent past two additional species have also been verified as human malaria parasites (1, 2). However, human-infecting Plasmodium species represent only a small fraction of over 550 species in the order Haemosporida that are classified into 17 extant genera (3). One hallmark of all hemosporidian parasites is the obligate host switch between a vertebrate intermediate host and an arthropod vector as a definitive host. However, across this family, a diverse array of intermediate hosts are used, including several orders of mammals, birds, squamate reptiles, turtles, and crocodilians (4). Based on solitary reports over the last century, it is thought that parasites belonging to at least seven hemosporidian genera can infect bats (Chiroptera), most of which are likely exclusive to this order (5).
Bats have an almost worldwide distribution, feature diverse life history traits, and play important ecological roles (6). Chiroptera is the second largest order of mammals after the Rodentia, with an estimated 1,232 living species and 18 families, which represent ∼20% of all living mammalian species (7). Bats are also important reservoir hosts for numerous emerging and highly pathogenic viruses (8, 9). In marked contrast, the hemosporidian parasites of bats remain largely unstudied, despite the first records dating back to the late 19th century (10). The corresponding vectors for most bat parasites remain unknown. Similarly, the phylogenetic relationships for the majority of these parasites remain enigmatic.
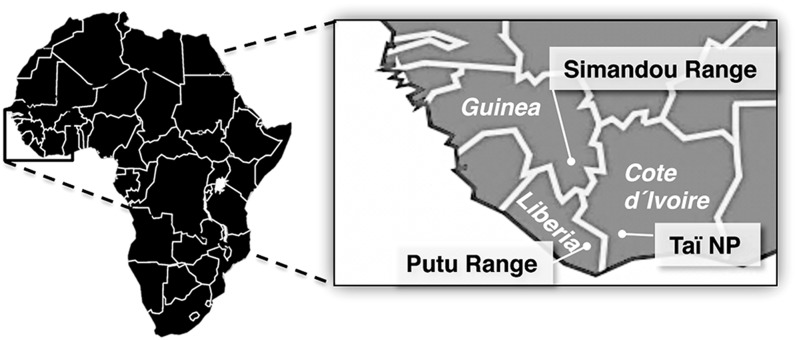
Here, we present a unique systematic analysis of Haemosporida in a diverse species assemblage of bats. For this study, we performed surveys in three remnants of the Upper Guinean forest ecosystem, considered one of the world’s biologically most diverse, but also one of the most endangered terrestrial ecosystems (Fig. 1). Our analysis highlights the overall diversity of chiropteran hemosporidian parasites and reveals distinct host–parasite associations.
Fig. 1.
Bat sampling areas in West Africa. Bats were captured during the dry season between November and December 2006 in Taï National Park, Côte d’Ivoire, in December 2008 in the Forêt Classée de Pic de Fon in the Simandou range of Guinea, and between November and December 2010 in the Putu range in southeastern Liberia.
Results
We captured a total of 274 bats belonging to 7 families and 31 species. Based on thorough and multiple microscopic examinations of thin blood smears, we observed hemosporidian parasites in 111 individuals, corresponding to an overall prevalence of 40% (Fig. S1). All individuals identified as infected by microscopy were then confirmed by diagnostic PCR and sequence analysis (111 of 111), establishing that there were no false-positive samples. Fifty samples scored as negative by microscopy were also tested by PCR, and all proved negative, indicating the false-negative rate is low. Collectively, infected bats belonged to 13 different host species from five families: Pteropodidae, Vespertilionidae, Miniopteridae, Hipposideridae, and Rhinolophidae (Table 1).
Table 1.
Investigated bat species
| Bat suborder | Bat family | Species* | Parasite genus |
| Yinpterochiroptera | Pteropodidae | Myonycteris angolensis† (3/3) | Plasmodium |
| Epomophorus gambianus (1/1), Epomops buettikoferi (7/25), Hypsignathus monstrosus (5/10), Micropteropus pusillus (68/72), Myonycteris leptodon‡ (11/63), Nanonycteris veldkampii (6/8) | Hepatocystis | ||
| Eidolon helvum (0/2), Megaloglossus azagnyi§ (0/12), Rousettus aegyptiacus (0/6), Scotonycteris ophiodon (0/5), Scotonycteris zenkeri (0/5) | |||
| Rhinolophidae | Rhinolophus alcyone (1/1), Rhinolophus landeri (1/2) | Nycteria | |
| Rhinolophus guineensis (0/5) | |||
| Hipposideridae | Hipposideros cyclops (3/4) | Plasmodium | |
| Hipposideros jonesi (0/2), Hipposideros cf ruber (0/10) | |||
| Yangochiroptera | Miniopteridae | Miniopterus villiersi (3/12) | Polychromophilus |
| Vespertilionidae | Neoromicia capensis (1/3), Pipistrellus aff. grandidieri¶ (1/3) | Polychromophilus | |
| Neoromicia guineensis (0/1), Pipistrellus bellieri (0/3), Pipistrellus nanulus (0/2) | |||
| Molossidae | Chaerephon nigeriae (0/1), Mops condylurus (0/1), Mops leonis (0/2), Mops thersites (0/7), Otomops martiensseni (0/1) | ||
| Nycteridae | Nycteris arge (0/1), Nycteris hispida (0/1) |
Infected species are highlighted in bold and numbers of infected per captured individuals are shown in brackets.
Bat species synonyms: †Rousettus smithi/ Lissonycteris angolensis, ‡Myonycteris torquata, §Megaloglossus woermanni.
The West African Pipistrellus aff. grandidieri might represent a yet undescribed species.
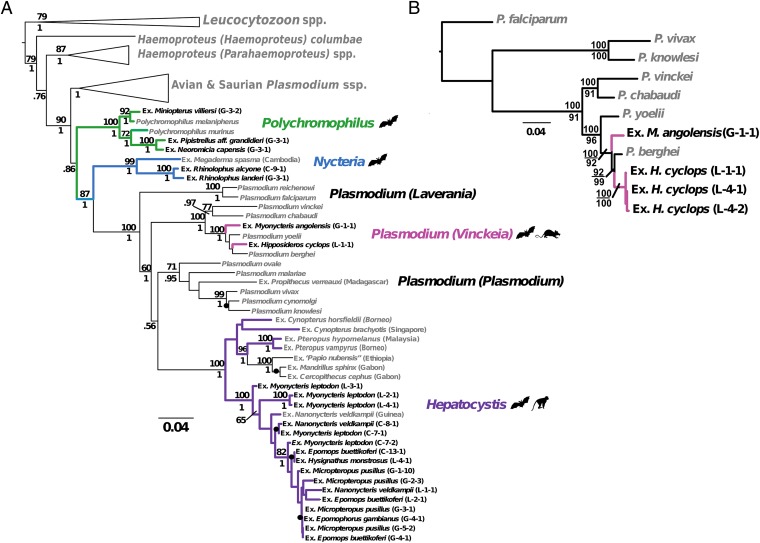
To classify the parasites, we used morphological characteristics (SI Text), a multigene phylogenetic approach from all three parasite genomes, consulted previous records, and considered the corresponding hosts and ecological localities. Robust phylogenetic analyses using both maximum-likelihood and Bayesian algorithms, including multiple previously published sequences from diverse hemosporid genera from other bat hosts, as well as rodents, primates, birds, and lizards, showed clear separation of the West African parasites into four well-supported, distinct genera: Plasmodium, Polychromophilus, Nycteria, and Hepatocystis (Fig. 2A and Fig. S2). We did not observe any individual bat host to be infected with more than one parasite genus or morphologically identified species, nor did the molecular data suggest that there were mixed-species or mixed-genus infections present in these bats.
Fig. 2.
Haemosporida from West African bats separate into four distinct genera. (A) Three-genome phylogeny of the hemosporidian parasites of the bats of this study, including published sequences for context, obtained by ML. Phylogeny was constructed with partitioned analysis of mitochondrial (cytb, cox1), apicoplast (clpc), and nuclear (ef2a) genes, rooted with Leucocytozoon taxa. ML bootstrap values (100 replicates) and Bayesian posterior probabilities are indicated above and below nodes, respectively. Recent divergences with high nodal support are indicated by black dots. Genera of bat malaria parasites are labeled to the right. Hemosporidian parasites from this study are highlighted in bold type. (B) ML phylogeny of bat and rodent Plasmodium species obtained via analysis of four genes as in A, plus nine additional nuclear genes (actin-1, actin-2, adenylosuccinate lyase, cysteine proteinase, dihydrofolate reductase/thymidylate synthase, histone H2A, inosine monophosphate-dehydrogenase, ookinete surface protein P25, and polyubiquitin), with three primate taxa included as outgroups.
Plasmodium species were restricted to the fruit bat species Myonycteris angolensis (Pteropodidae; formerly Lissonycteris angolensis) and the insectivorous Hipposideros cyclops (Hipposideridae) and showed high prevalences in these species (Fig. 2 and Table 1). M. angolensis harbored parasites classified as Plasmodium voltaicum based on morphological features (Fig. 3A and Fig. S3A), a finding that is consistent with previous records of this parasite species in this host (11). Individuals of H. cyclops were infected with parasites identified as Plasmodium cyclopsi (Fig. 3A and Fig. S3B), a species that has been reported from the same host species in Gabon (12). Unexpectedly, in the phylogenetic analysis, both of these Plasmodium species fell within the clade of rodent malaria parasites and showed patterns of multiple switches between chiropteran and rodent hosts (Fig. 2 and Fig. S4). Sequencing of nine additional nuclear-encoded genes from these chiropteran Plasmodium samples yielded slightly different within-clade relationships, but consistently showed a close relationship with Plasmodium yoelii and Plasmodium berghei (Fig. 2B and Fig. S4). This tight link was entirely maintained when we included sequences from all known rodent Plasmodium strains in our analysis (Fig. S4). These rodent malaria species are widely used model parasites to study Plasmodium biology, disease progression, and novel malaria intervention strategies (13). We conclude that the surprising and tight relationship of bat Plasmodium species with those infecting rodents suggests multiple host switches between the two host orders.
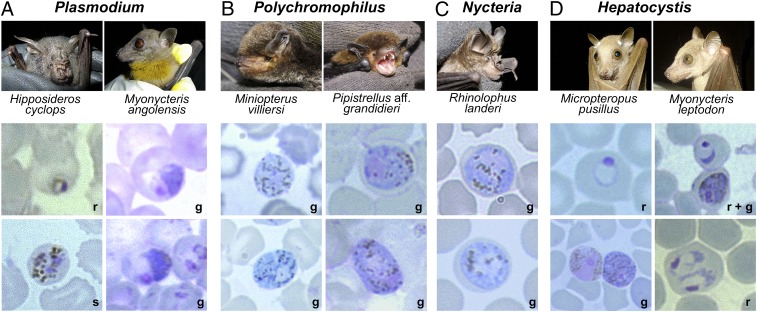
Fig. 3.
Hemosporidian parasites and their host species. Shown are captured bats and representative micrographs of Giemsa-stained thin blood films of their respective hemosporidian parasite blood stages (r, ring stage; s, schizont; g, gametocyte). (A) P. cyclopsi and P. voltaicum blood stages isolated from H. cyclops and M. angolensis, respectively. (B) Polychromophilus gametocytes isolated from miniopterid and vespertilionid bats. (C) Nycteria gametocytes isolated from two rhinolophid bats. (D) Hepatocystis blood stages isolated from six pteropodid bats. Shown are two (Micropteropus pusillus, Myonycteris leptodon) of six host species and different blood stages of their parasites. Micrographs were taken at 1,000× magnification.
Several infections in insectivorous bats were morphologically identified as belonging to the genus Polychromophilus (Fig. 3B and Fig. S3C). The samples from Miniopterus villiersi possessed slightly coarser-grained pigment, a characteristic attributed to Polychromophilus melanipherus (10) and are part of the same clade as P. melanipherus samples from Miniopterus schreibersii (Fig. 2A and Fig. S2). Similarly, the parasites of Pipistrellus aff. grandidieri and Neoromicia capensis are very similar but do not group closely with P. murinus samples from European vespertilionid hosts (14), and thus may represent a distinct species. All samples showed very low parasitemias (Table S1), consisting only of mature gametocytes, most likely because Polychromophilus infections were chronic in the captured hosts. Taken together, our findings support the notion that Polychromophilus is restricted to insectivorous bats, but has a wide cross-equatorial geographic range.
Nycteria parasites were found in two horseshoe bats, Rhinolophus alcyone and Rhinolophus landeri (Fig. 3C and Fig. S3 D and E). Only sexual stages were found in the blood, consistent with the characteristics of this genus (5). Our analyses place Nycteria as a very distinct sister clade to the mammalian Plasmodium/Hepatocystis clade (Fig. 2A), suggesting that the origin of the mammalian hemosporidians may have been in African bats. A parasite found in Megaderma spasma from Cambodia (15), included in our phylogenetic dataset, grouped with the Nycteria sequences. Collectively, this first molecular analysis of Nycteria parasites underscores the evolutionary distance of these Haemosporida and implies still unrecognized biological peculiarities in this parasite genus.
The vast majority of parasites we observed in our sampling were Hepatocystis species. All were found in fruit bats (Pteropodidae) with a high overall prevalence of 58% (Fig. 3D and Table 1). Apparently, Hepatocystis infections are almost universal in several West African pteropodid bats, indicative of continuous and highly efficient transmission cycles. The Hepatocystis group of the West African samples might comprise different species and phylogenetically clearly places as a sister group to another Hepatocystis clade, which contains parasites in both African primate (baboon and mandrill) and Asian pteropodid fruit bat hosts (Fig. 2A). As with previous reports (16, 17), Hepatocystis appears to be a derived clade from the mammalian Plasmodium parasites. The remarkable phylogenetic diversity of Hepatocystis correlates with the diverse array of vertebrate hosts. Based on our data, we hypothesize that bats were also likely the first hosts of Hepatocystis, although our phylogeny also supports one invasion into African primates. However, this clade was more closely related to Asian bat Hepatocystis parasites than the West African bat Hepatocystis (Fig. 2A).
Discussion
The close phylogenetic relationship of the chiropteran Plasmodium species with those of rodents offers a fresh look at the evolution of rodent malaria, which is used as an experimental model for human disease (13) and evolutionary biology (18). All four known murine Plasmodium species, Plasmodium chabaudi, P. yoelii, P. berghei and Plasmodium vinckei naturally infect African thicket rats (Muridae: Grammomys), which are arboreal. The Plasmodium-infected bat species, H. cyclops, forms small colonies in cavities in large hollow trees, and M. angolensis usually roosts in caves, but is also occasionally found in hollow trees (19, 20). These distinct roosting behaviors might attract the same dipteran vectors as those seeking tree-dwelling rodents, and provides a plausible explanation for the phylogenetic relationship of their blood parasites. An additional link might be scaly-tailed flying squirrels (Anomaluridae) (19), which sometimes share their roosts with H. cyclops and have been found infected with Plasmodium anomaluri (12), a species that morphologically appears to be the closest to P. cyclopsi (12). Based on our data, we propose that P. cyclopsi and P. voltaicum should be readily transferrable from naturally infected bats to laboratory mice and mosquito colonies. Adaptation of these Plasmodium species to experimental murine models could offer unprecedented insights into phenotypic and genetic evolution of malarial parasites. The first identification of rodent Plasmodium species that can eventually be transmitted to laboratory rodents was by Vincke and Lips, who found sporozoites of what is now known as P. berghei in the salivary glands of infected Anopheles dureni millecampsi (21). This landmark discovery marked the beginning of the widespread use of murine malaria parasites as in vivo models for biomedical malaria research. These model systems became invaluable tools for large-scale drug testing, experimental vaccine development, and molecular and cellular studies of malaria-associated pathology and host immune responses (22, 23). The importance of these murine hosts is such that the complete genome of one of them, P. yoelii, was sequenced in parallel with that of P. falciparum (13). However, many currently used strains of rodent malaria used in experimental studies derive from isolates that have been adapted to inbred laboratory hosts for over three decades, thus representing a somewhat unnatural parasite–host system.
Recent studies (24) addressed the exceptional ability of bats to endure numerous viruses, some of which are highly pathogenic for humans. Tolerating high prevalences of hemosporidian parasites along with high parasitemias, as shown for Hepatocystis infections in this study, might be another example of the well-adapted immune responses of bats. It is possible that even the change in the parasites’ life cycle away from erythrocytic schizogony in three of the bat-infecting genera (Hepatocystis, Nycteria, and Polychromophilus) could have been an evolutionary “compromise” of using these metabolically active hosts and driven by the immune responses of the bat hosts. Similarly, the Plasmodium species infecting bats could offer a unique comparative system with the rodent malaria parasites to investigate the response of these parasites to the very different immunological environment of chiropterans.
Our phylogenetic analysis also challenges several traditional and recently proposed ideas in hemosporidian relationships. This analysis, rooted with Leucocytozoon species, places the avian and squamate Plasmodium species as polyphyletic to the mammalian Plasmodium species, and further suggests that bats were likely the first mammalian hosts of Plasmodium before the parasites began using rodents and primates, including humans, as vertebrate hosts. The topology of the phylogeny (Fig. 2 and Fig. S2) also suggests that once Plasmodium parasites invaded mammals, they never switched back into nonmammalian hosts. We note that this interpretation contrasts with suggestions that across-clade switching might have occurred multiple times (15), or that Polychromophilus parasites are more closely related to the parasites of birds and squamate reptiles (25). To eventually resolve which of these two scenarios is more likely, additional taxa, perhaps including lineages external to Haemosporida but more closely related than other sequenced Apicomplexa, will likely be required (26).
The sampling of our study presents only a snapshot of the diversity of bats and thus of potential host species. Although the pace of discovery of novel mammalian parasite lineages slowed over the past few decades (3), more recently there has been a resurgence of investigating wild hosts, particularly primates in both the New and Old Worlds, using molecular screening techniques (e.g., refs. 27–30). These studies have broadened our conceptions of the diversity of hemosporidian lineages in these hosts and, in some cases, will challenge our concepts of the origin of the human-infecting species and their natural epidemiology (31, 32). These surveys, like our study, illustrate that minimally invasive sampling in biodiversity hotspots is likely to reveal numerous previously unrecognized parasitic infections in most mammalian host taxa. Renewed investigation of hemosporidian parasites in natural hosts is critical to eventually gain a better understanding of host–pathogen coevolution.
The exceptional phylogenetic and ecological diversity of bats have certainly contributed to the diversity of malaria parasites in these hosts. Bats have a deep evolutionary history, with an inferred origin of 66 Mya, and it has been proposed that the biogeographic center of origin of the order was in Africa, but with numerous dispersal events (33). The diverse array of hemosporidian genera currently present in African bats may thus be a reflection of the long and complex history of their hosts. The taxa sampled in this study include members of both chiropteran suborders (Table 1). The arthropod vectors, where sexual recombination occurs, should also be included in future considerations, because they likely play a major role in parasite diversification (34). In conclusion, our findings reveal an intriguing correlation between exceptional diversity and phylogenetic relationships of arthropod-borne, obligate intracellular blood parasites and the unique ecological niche and species range that distinguish bats from all other mammals as potential hosts.
Materials and Methods
Bats were captured in Côte d’Ivoire, Guinea, and Liberia (Fig. 1, Fig. S1, and Table S2). For molecular analysis, blood and wing punches were sampled. For morphological parasite identification thin blood smears were prepared for Romanowsky–Giemsa staining and microscopic analysis. Confirmatory diagnosis at the genus and, where possible, species level was defined as combination of microscopy, PCR, and sequencing. Parasitemias were recorded for each infected bat (Table S1) by direct counting of parasite cells in 40 microscopic fields (see SI Materials and Methods for full details), except in eight cases, where the quality of the thin blood smear was too poor to obtain an accurate count. Four signature genes representing the three parasite genomes (mitochondrial: cytochrome b, cytochrome oxidase I; plastid: caseinolytic protease C; nuclear: elongation factor 2A) were selected for the phylogenetic analyses. For just the Plasmodium isolates, nine additional nuclear genes were amplified and sequenced (actin-1, actin-2, adenylosuccinate lyase, cysteine proteinase, dihydrofolate reductase/thymidylate synthase, histone H2A, inosine monophosphate-dehydrogenase, ookinete surface protein P25, and polyubiquitin). Primers are listed in Table S3; GenBank accession numbers are listed in Tables S4 and S5. After Sanger sequencing and assembly, alignments were done using MUSCLE (35). We evaluated the phylogenetic relationships using both maximum-likelihood (ML) and Bayesian inference methods. Data were divided into four partitions according to genes. For ML analysis, we used raxmlGUI (36). Nodal support was evaluated using 100 rapid bootstrap pseudoreplicates (37). Bayesian inference was conducted in MrBayes v3.2.0 (38), with two runs of four chains (three heated, one cold, temperature = 0.20) each for 10 million generations. The GTR + I + Γ type model was used for each independent partition. Reversible rate matrices, partition-specific rate multipliers and stationary state frequencies had a Dirichlet prior. The α and proportion of invariant sites had uniform priors. A prior of all topologies equally likely was used for τ and the prior on branch lengths was set as unconstrained exponential (parameter 10). Convergence was tested in the program AWTY (39).
Supplementary Material
Acknowledgments
We thank Stefan Pettersson and Siv Aina Leendertz, who captured the bats in Côte d’Ivoire; Rainer Hutterer, for identifications and preservation of the bat vouchers from Guinea at the Zoological Museum Koenig in Bonn; Kevin Olival for early contributions to the study; and Frieder Mayer for discussions. We also thank SNC-Lavalin Group Inc.; RioTinto; the Putu Iron Ore Project, which permitted sampling during the consulting jobs of J.S. and J.D. in Guinea, and J.S. in Liberia; the Guinean Ministère en charge de l’Agriculture de l’Elevage de L'Environnement et des Eaux et Fôrets; the Liberian Forest Development Authority; the authorities in Côte d’Ivoire for long-term support, especially the Ministry of Environment and Forests and the Ministry of Research; the directorship of the Taï National Park; the Office Ivoirien des Parcs et Réserves; and the Swiss Research Center in Abidjan. This research was supported by a Research Coordination grant from the US National Science Foundation (see http://malariarcn.org), the Annette Kade Fellowship from the American Museum of Natural History, and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. are reported in Tables S4 and S5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311016110/-/DCSupplemental.
References
- 1.Sutherland CJ, et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis. 2010;201(10):1544–1550. doi: 10.1086/652240. [DOI] [PubMed] [Google Scholar]
- 2.Singh BK, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 3.Martinsen ES, Perkins SL. In: Malaria Parasites: Comparative Genomics, Evolution and Molecular Biology. Carlton JM, Perkins SL, Deitsch KW, editors. Norfolk, United Kingdom: Caister Academic; 2013. pp. 1–15. [Google Scholar]
- 4.Levine N. The Protozoan Phylum Apicomplexa. Boca Raton, FL: CRC; 1988. [Google Scholar]
- 5.Garnham PCC. Malaria Parasites and Other Haemosporidia. Oxford: Blackwell Scientific; 1966. [Google Scholar]
- 6.Kalka MB, Smith AR, Kalko EK. Bats limit arthropods and herbivory in a tropical forest. Science. 2008;320(5872):71. doi: 10.1126/science.1153352. [DOI] [PubMed] [Google Scholar]
- 7.Wilson D, Reeder DM. In: Mammal Species of the World. A Taxonomic and Geographic Reference. 3rd Ed. Wilson D, Reeder DM, editors. Baltimore: Johns Hopkins Univ Press; 2005. [Google Scholar]
- 8.Leroy EME, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 9.Calisher CHC, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dionisi A. Un parassita del globulo rosso in una specie di pipistrello (Miniopterus schreibersii Kuhl.) Atti Reale Accad Lincei. 1898;7:214–215. [Google Scholar]
- 11.Van Der Kaay HJ. Description of a new Plasmodium, Plasmodium voltaicum sp. nov., found in a fruit-bat, Roussettus smithi, in Ghana. Ann Trop Med Parasitol. 1964;58:261–264. doi: 10.1080/00034983.1964.11686241. [DOI] [PubMed] [Google Scholar]
- 12.Landau II, Chabaud AG. [Description of P. cyclopsi n. sp. a parasite of the microchiropteran bat Hipposideros cyclops in Gabon (author’s transl)] Ann Parasitol Hum Comp. 1978;53(3):247–253. doi: 10.1051/parasite/1978533247. [DOI] [PubMed] [Google Scholar]
- 13.Carlton JM, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419(6906):512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 14.Witsenburg F, Salamin N, Christe P. The evolutionary host switches of Polychromophilus: A multi-gene phylogeny of the bat malaria genus suggests a second invasion of mammals by a haemosporidian parasite. Malar J. 2012;11:53. doi: 10.1186/1475-2875-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duval L, et al. Multiple host-switching of Haemosporidia parasites in bats. Malar J. 2007;6:157. doi: 10.1186/1475-2875-6-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88(5):972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Escalante AAA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95(14):8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reece SE, Drew DR, Gardner A. Sex ratio adjustment and kin discrimination in malaria parasites. Nature. 2008;453(7195):609–614. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decher J, Fahr J. Hipposideros cyclops. Mamm Species. 2005;763:1–7. [Google Scholar]
- 20. Rosevear DR (1965) The Bats of West Africa [Trustees of the British Museum (Natural History), London]
- 21.Vincke JH, Lips M. Un noveau Plasmodium d’un rongeur suvage du Congo Plasmodium berghei n.sp. Ann Soc Belg Med Trop. 1948;28:97–104. [PubMed] [Google Scholar]
- 22.Janse CJ, et al. A genotype and phenotype database of genetically modified malaria-parasites. Trends Parasitol. 2011;27(1):31–39. doi: 10.1016/j.pt.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Matuschewski K. Murine infection models for vaccine development: The malaria example. Hum Vaccin Immunother. 2012;9(3):450–456. doi: 10.4161/hv.23218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Outlaw DC, Ricklefs RE. Rerooting the evolutionary tree of malaria parasites. Proc Natl Acad Sci USA. 2011;108(32):13183–13187. doi: 10.1073/pnas.1109153108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagner SC, Misof B, Maier WA, Kampen H. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 2007;101(3):493–503. doi: 10.1007/s00436-007-0499-6. [DOI] [PubMed] [Google Scholar]
- 27.Singh BK, Simon Divis PC. Orangutans not infected with Plasmodium vivax or P. cynomolgi, Indonesia. Emerg Infect Dis. 2009;15(10):1657–1658. doi: 10.3201/eid1510.090364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser MM, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg Infect Dis. 2010;16(12):1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bueno MG, et al. Survey of Plasmodium spp. in free-ranging neotropical primates from the Brazilian Amazon region impacted by anthropogenic actions. EcoHealth. 2013;10(1):48–53. doi: 10.1007/s10393-012-0809-z. [DOI] [PubMed] [Google Scholar]
- 30.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107(4):1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prugnolle F, et al. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7(2):e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eick GN, Jacobs DS, Matthee CA. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (chiroptera) Mol Biol Evol. 2005;22(9):1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- 34.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47(1):261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvestro D, Michalak I. RaxmlGUI: A graphical front-end for RAxML. Org Divers Evol. 2012;12(4):335–337. [Google Scholar]
- 37.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 38.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24(4):581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.