Significance
p53 is one of the most highly studied proteins in biomedical research because of its importance in preventing cancer and its direct or indirect role in many biological processes. It is best known as a nuclear protein that is critical for maintaining genomic integrity and regulating gene expression. We have uncovered a molecular mechanism by which p53 translocates into the mitochondria, depending on respiration, and facilitates the repair of oxidative damage to mitochondrial DNA. The dynamic partitioning of p53 between the nuclear and mitochondrial compartments has important implications for cancer and the many other essential functions of p53 in normal physiology.
Keywords: mitochondrial DNA, DNA repair, mutant p53
Abstract
p53, a critical tumor suppressor, regulates mitochondrial respiration, but how a nuclear protein can orchestrate the function of an organelle encoded by two separate genomes, both of which require p53 for their integrity, remains unclear. Here we report that the mammalian homolog of the yeast mitochondrial disulfide relay protein Mia40 (CHCHD4) is necessary for the respiratory-dependent translocation of p53 into the mitochondria. In the setting of oxidative stress, increased CHCHD4 expression partitions p53 into the mitochondria and protects its genomic integrity while decreasing p53 nuclear localization and transcriptional activity. Conversely, decreased CHCHD4 expression prevents the mitochondrial translocation of p53 while augmenting its nuclear localization and activity. Thus, the mitochondrial disulfide relay system allows p53 to regulate two spatially segregated genomes depending on oxidative metabolic activity.
Along with regulating gene transcription in the nucleus, p53 protein also functions in the cytoplasm to regulate such cellular processes as apoptosis and autophagy (1, 2). p53 can bind to the outer membrane of the mitochondria by forming complexes with BclXL and Bcl proteins in response to apoptotic stimulus (3), and also can accumulate in the matrix and interact with cyclophilin D to open the mitochondrial permeability transition pore to trigger necrosis (4). Although best known as the “guardian of the genome,” p53 also has been shown to be important for the repair and maintenance of mitochondrial DNA (mtDNA) (5–10). Cellular stressors, such as reactive oxygen species or even acute exercise, can promote p53 translocation into mitochondria and its interaction with matrix proteins polymerase-γ, mitochondrial transcription factor A, and superoxide dismutase 2 (7, 11, 12). Despite major advances in the field of mitochondrial protein import (13), whether the transfer of p53 through the intermembrane space and the highly charged electrochemical gradient of the inner membrane in normal cellular state occur passively or through an active mechanism with the potential for functional homeostatic regulation, remains largely unexplored.
Recent studies have unveiled a disulfide relay system consisting of the import receptor CHCHD4 and the FAD-dependent sulfhydryl oxidase (GFER) in the intermembrane space that mediates protein translocation into mitochondria (14, 15). Through its N-terminal Cys-Pro-Cys (CPC) motif, CHCHD4 forms a transient intermolecular disulfide bond with cysteine residues in substrate proteins targeted for translocation into the mitochondria (15). Of note, previous studies have shown that the structure and function of p53 is redox-sensitive, and that the oxidation of cysteines in p53 affects its subcellular distribution (16, 17). Under conditions of increased oxidative stress, p53 has two cysteine pairs (C135-C141 and C275-C277) that are capable of forming intramolecular disulfide bonds, a structural feature reportedly present in the typical CHCHD4 substrate (15, 18, 19). Given the sensitivity of p53 to the cellular redox state and its critical role in regulating redox homeostasis (17, 20), we examined whether the importation of p53 into mitochondria could be mediated by the CHCHD4 disulfide relay system.
Results
p53 Interacts and Colocalizes with CHCHD4 in Respiring Mitochondria.
Compared with control lentiviral transduction, the stable overexpression of CHCHD4 in HCT116 cells, a human colon cancer cell line with endogenous WT p53, markedly increased the level of p53 in the mitochondrial fraction (Fig. 1A, lanes 3 and 4). Conversely, the stable knockdown of CHCHD4 using shRNA decreased the relative level of p53 in the mitochondrial fraction (Fig. 1A, lanes 7 and 8). The results of these subcellular fractionation experiments were confirmed using confocal immunofluorescent microscopy to show that overexpression of CHCHD4 increased the mitochondrial colocalization of p53 and CHCHD4, as indicated by the white color on merging with MitoTracker, whereas knockdown of CHCHD4 resulted in loss of the colocalization signal (Fig. 1B, merge all three). Similar subcellular localization of p53 mediated by CHCHD4 was also observed in primary human myoblasts, indicating that this phenomenon is applicable to other cell types as well (Fig. S1A).
Fig. 1.
p53 interacts and colocalizes with CHCHD4 in mitochondria. (A) Western blots of the cytosolic (CYT) and mitochondrial (MITO) fractions of WT HCT116 cells transduced with CHCHD4 cDNA or shRNA lentivirus versus control lentivirus (−) and then transfected with p53. GAPDH and VDAC serve as cyoplasmic and mitochondrial protein loading controls, respectively. (B) Confocal immunofluorescent localization of CHCHD4 (green) and p53 (red) in SCO2+/+ (WT) cells transduced with CHCHD4 cDNA or shRNA lentivirus and then transfected with p53. MitoTracker (blue) indicates mitochondria. (C) Respiring (SCO2+/+) and nonrespiring (SCO2−/−) cells were cotransfected with CHCHD4-His6 and p53, and their lysates were passed over Ni-NTA columns. Total lysate (T) and column-bound (B) fractions were subjected to Western blot analysis. (D) Immunofluorescent imaging of CHCHD4 (green), p53 (red), and mitochondria (MitoTracker, blue) in SCO2−/− cells. (E) Effect of hypoxia (1.5% O2 for 48 h) on the interaction of cotransfected p53 and CHCHD4-His6 by Ni-NTA binding assay in WT cells. (F) p53 interaction with the Cys-Pro-Cys (CPC) motif of CHCHD4 was tested by substituting Cys (C) with Ser (S). (Scale bars: 10 µm.)
The mitochondrial disulfide relay system requires the regeneration of oxidized CHCHD4 via its oxidase GFER, which in turn transfers its electrons to oxidized cytochrome c. Thus, the importation of proteins through this system requires active respiration for the reoxidation of reduced cytochrome c by respiratory complex IV (21). We previously created a nonrespiring HCT116 line by disrupting both copies of Synthesis of Cytochrome c Oxidase 2 (SCO2−/−), a gene essential for complex IV assembly (22, 23). To test whether the p53–CHCHD4 interaction depends on active respiration, we transiently coexpressed p53 with polyhistidine-tagged CHCHD4 (CHCHD4-His6) in isogenic HCT116 cells with (+/+) or without (−/−) SCO2. Western blot analysis of the total (T) cell lysate compared with the nickel-NTA (Ni-NTA) column-bound (B) fraction of CHCHD4-His6–expressing cells revealed a p53–CHCHD4 interaction in the respiring SCO2+/+ cell line that was essentially undetectable in nonrespiring SCO2−/− cells (Fig. 1C, lanes 4 and 8). Confocal immunofluorescent microscopy showed increased accumulation of p53 in the nucleus of SCO2−/− cells, consistent with our previous report of elevated oxidative stress and p53 stabilization in these nonrespiring cells (control in Fig. 1D) (24).
Consistent with the protein–protein interaction data, overexpression of CHCHD4 failed to drive p53 into the mitochondria of the nonrespiring SCO2−/− cell, as indicated by the absence of either yellow (Fig. 1D, merge p53/CHCHD4) or white color (merge all three). Moreover, decreasing respiration by placing SCO2+/+ cells in hypoxia disrupted the p53–CHCHD4 interaction (Fig. 1E, lane 4 vs. lane 8). At the structural level, the specificity of this interaction was further confirmed by substituting the two cysteine residues in the CPC motif of CHCHD4 with serine, which abolished p53 binding (Fig. 1F, lane 10).
CHCHD4 Regulates p53 Nuclear Localization and Activity.
Oxidative stress causes p53 stabilization and accumulation in the nucleus, with subsequent transactivation of its target genes. We hypothesized that modulating the levels of CHCHD4 via cDNA expression or shRNA-mediated knockdown (total cell lysates, Fig. 2A) would shift the distribution of p53 and affect its activity in the nucleus. The treatment of WT HCT116 cells with H2O2 increased the level of p53 in the nucleus, but, as predicted, this increase was attenuated by concomitant expression of CHCHD4 (Fig. 2A, lanes 2 and 4). Conversely, the knockdown of CHCHD4 with shRNA caused a relative increase in the nuclear level of p53 (Fig. 2A, lanes 6 and 8). In these experiments, it is important to note that the total cellular content of p53 was not affected by either overexpression or knockdown of CHCHD4 (Fig. 2A, Right).
Fig. 2.
CHCHD4 expression modulates the subcellular localization and activity of p53 in response to oxidative stress. (A) Nuclear p53 Western blot of WT HCT116 cells transduced with CHCHD4 cDNA or shRNA lentivirus after exposure to oxidative stress (200 µM H2O2 for 1 h). Total cell lysates (Right) show relative CHCHD4 levels compared with control (CTL). (B) Confocal immunofluorescent localization of p53 (red) in CHCHD4-overexpressing cells after oxidative stress versus no treatment (CTL). DAPI stain (blue) shows nuclei. (C) p53 (red) localization in WT cells with knockdown of CHCHD4 versus nonspecific (NS) shRNA. (D) p21 mRNA levels in p53+/+ and p53−/− HCT116 cells transduced with CHCHD4 cDNA (Upper) or shRNA (Lower) lentivirus and treated with H2O2 for 1 h. p21 and p53 protein levels in the corresponding samples of p53+/+ cells are shown as well. (E) p53+/+ (Left) and p53−/− (Right) cells transduced with empty vector (−) or CHCHD4 cDNA lentivirus, treated with 200 µM H2O2 for 1 h, washed, and allowed to recover in normal medium for the indicated times. mtDNA integrity of treated cells relative to nontreated cells (CTL) is shown. (F) mtDNA integrity of WT cells transduced with nonspecific or CHCHD4 shRNA lentivirus, treated with H2O2, and allowed to recover for 24 h. Values are mean ± SE. ns, nonsignificant. (Scale bars: 10 µm.)
We next sought to image this shift in the nuclear level of p53 using confocal immunofluorescent microscopy. The overexpression of CHCHD4 caused exclusion of p53 from the nucleus after H2O2 treatment (Fig. 2B), whereas CHCHD4 depletion using shRNA increased p53 localization in the nucleus (Fig. 2C). Although the total cellular level of p53 protein was not affected by CHCHD4 (Fig. 2D), the subcellular partitioning of p53 affected its nuclear activity, as confirmed by measurement of the mRNA and protein levels of p21, the prototypical p53 target gene. In a p53-dependent manner, CHCHD4 expression decreased the H2O2-stimulated expression of p21, whereas CHCHD4 knockdown increased p21 levels relative to their controls (Fig. 2D). Also consistent with CHCHD4 regulation of p53 nuclear localization and function, overexpression of CHCHD4, which prevents p53 accumulation in the nucleus, decreased nuclear DNA integrity, whereas CHCHD4 depletion increased it (Fig. S2).
CHCHD4 Regulates p53 Function in the Mitochondria.
Because multiple studies have shown that p53 participates in maintaining mtDNA, we investigated whether CHCHD4-mediated p53 importation might affect mtDNA repair as well. To test this idea, we used a PCR-based assay (quantitative long PCR) that has proven to be sensitive in detecting mtDNA breaks (25). In the presence of stable CHCHD4 overexpression or knockdown by lentiviral transduction, the cells were exposed to 200 µM H2O2 for 1 h to introduce mtDNA breaks, and then switched to normal tissue culture medium for the indicated times (Fig. 2 E and F). Compared with empty vector control, the overexpression of CHCHD4 significantly increased the recovery of mtDNA integrity at 6 h after H2O2 treatment in p53+/+ cells, whereas no significant recovery was seen in p53−/− cells even up to 24 h (Fig. 2E). Conversely, CHCHD4 depletion decreased mtDNA integrity at both 0 h and up to 24 h after H2O2 treatment (Fig. 2F). Modulation of CHCHD4 levels caused a similar pattern of mtDNA integrity recovery after oxidative damage in primary human myoblasts, validating the results in the HCT116 cell line (Fig. S1 B and C). These findings demonstrate the importance of the disulfide relay system in maintaining mtDNA, as was also observed in a mitochondrial myopathy syndrome involving mutated CHCHD4 oxidase gene GFER (26). In addition, CHCHD4 promoted mitochondrial respiration and cell growth as reported previously (Fig. S3) (27). Consistent with the interaction between CHCHD4 and p53, the increase in oxidative metabolism associated with CHCHD4 overexpression was dependent on the presence of p53 (Fig. S3A).
Mutated p53 Can Translocate to Mitochondria and Retain mtDNA Repair Activity.
We also wondered whether mutated forms of p53 can translocate into the mitochondria and mediate the repair of damaged mtDNA. We examined two well-characterized mutant forms of p53: the “hotspot” germline mutant p53 R175H and the somatic mutant p53 C135Y. p53 R175H retained its ability to bind CHCHD4 like WT p53, whereas this interaction was abolished in p53 C135Y (Fig. 3A, lanes 4, 8, and 12). It is notable that the C135Y mutation of p53 results in the loss of one of its two intramolecular disulfide bonds typically present in substrates of CHCHD4 (19). Despite similar overexpression levels of both p53 mutants in the nucleus and cytoplasm, confocal immunofluorescent imaging confirmed the lack of p53 C135Y protein colocalization with CHCHD4 in the mitochondria, in marked contrast to p53 R175H, which displayed merged fluorescent signals consistent with CHCHD4 interaction (Fig. 3B, merge). Furthermore, depletion of CHCHD4 prevented p53 R175H localization in the mitochondria and resulted in an immunofluorescent pattern similar to that seen in mutant p53 C135Y protein (Fig. S4, merge images).
Fig. 3.
Effect of p53 mutation on its CHCHD4 interaction, mitochondrial localization, and activity in mtDNA repair. (A) WT, R175H, or C135Y p53 cDNA was transiently cotransfected with CHCHD4-His6 or empty vector plasmid in p53−/− HCT116 cells. CHCHD4-His6 was isolated by passing the cell lysate over a Ni-NTA column. Total lysate (T) and column-bound (B) fractions were evaluated by Western blot analysis. (B) Confocal immunofluorescent imaging of CHCHD4 (green), p53 (red), and mitochondria (MitoTracker, blue). (C) p53−/− cells were transduced with p53 R175H or p53 C135Y lentivirus and then transfected with empty vector or CHCHD4 cDNA. The cells were treated with 200 µM H2O2 for 1 h, washed, and allowed to recover in normal medium for 6 h. mtDNA integrity of treated cells relative to nontreated cells (CTL) is shown. (D) Relative mtDNA integrity in skeletal muscle (SKM), liver, and heart of p53−/− and homozygous p53 R172H (p53H/H) mice compared with p53+/+ mice. Values are mean ± SE. n.s., nonsignificant. *P < 0.05. (Scale bar: 10 µm.)
We next examined the mtDNA repair capacity of the mutant forms of p53. In contrast to p53 C135Y-expressing cells, p53−/− cells transduced with p53 R175H lentivirus gained the capacity to repair mtDNA after H2O2 treatment, which was further augmented by CHCHD4 overexpression (Fig. 3C). To extend the results of these in vitro experiments, we compared the mtDNA integrity of tissues from p53+/+, p53−/−, and homozygous p53 R172H (p53H/H, homologous to the human p53 R175H mutation) mice. The skeletal muscle, liver, and heart tissues of p53−/− mice showed a pattern of lower mtDNA integrity compared with p53+/+ and p53H/H mice, indicating that the p53 R172H mutant retains mtDNA repair activity (Fig. 3D).
In Vivo Effects of CHCHD4-Mediated Translocation of p53 into Mitochondria.
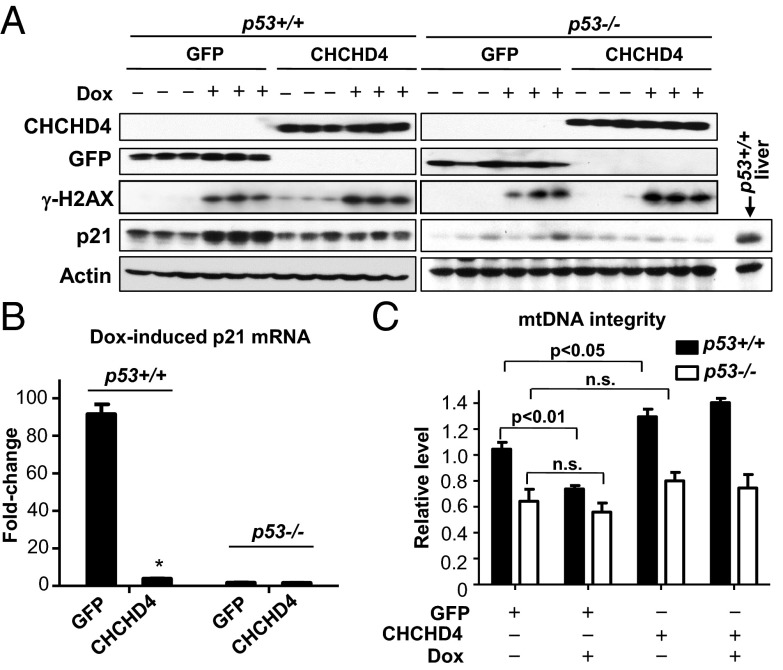
Although our in vitro results show that the subcellular redistribution of p53 by modulation of CHCHD4 expression can affect nuclear p53 activity, we wished to investigate whether such a mechanism may be functional in vivo. To do this, we used adenovirus to deliver control GFP or CHCHD4 cDNA to the liver of mice via tail vein injection. Successful transduction was confirmed by protein expression (Fig. 4A). Subsequent doxorubicin treatment of both groups of mice resulted in similar increases in the levels of DNA damage marker γ-H2AX (Fig. 4A), but the p53-dependent increases in p21 mRNA and protein levels were markedly attenuated by CHCHD4 overexpression (Fig. 4 A and B). In contrast, increased CHCHD4 expression in the liver not only increased the basal level of mtDNA integrity in a p53-dependent manner, but also significantly protected mtDNA from damage by doxorubicin treatment (Fig. 4C). In the p53−/− mice, the low basal level of mtDNA integrity in liver was not further decreased by this doxorubicin treatment regimen (Fig. 4C). The spleen tissue of these mice, which cannot be transduced by i.v. injection of adenovirus, served as a negative control to demonstrate the specific in vivo effect of CHCHD4 on p21 expression (Fig. S5).
Fig. 4.
Effect of CHCHD4 overexpression on p21 expression and mtDNA integrity in liver. (A) At 4 d after tail vein injection of CHCHD4 or control GFP adenovirus to transduce the liver, p53+/+ and p53−/− mice were treated with doxorubicin (Dox) and killed after 18 h. CHCHD4, γ-H2AX, and p21 protein levels in liver were subjected to Western blot analysis to determine CHCHD4 transduction by adenovirus, DNA damage, and p53 activation in the nucleus by doxorubicin treatment, respectively. Three mice in each group are shown. A p53+/+ liver sample was included with the p53−/− samples as a positive control. (B) p21 mRNA level in liver tissue was measured as a marker of nuclear p53 transcriptional activity. (C) Relative mtDNA integrity in the liver tissues was measured as a marker of mitochondrial p53 activity. Values are mean ± SE. *P < 0.001.
Discussion
In the present study, we have delineated a specific molecular mechanism by which p53 can translocate into the mitochondria using a disulfide relay system that is conserved in plants and animals (15). The interaction of p53 with this vectorial import system is dependent on respiration and can be activated by oxidative stress, consistent with the notion that p53 evolved to provide basic adaptive functions against environmental stresses and is involved in redox homeostasis (20, 28). This finding is unique among other proposed p53 translocation mechanisms, in that it is dependent on oxidative metabolism, a function that p53 also has been shown to promote in vivo (29, 30). Furthermore, our identification of a defined molecular mechanism by which p53 can translocate into mitochondria highlights the direct role of p53 in preserving the integrity of the mitochondrial genome, which is more susceptible to oxidative damage owing to its structure and proximity to electron transfer reactions.
The subcellular partitioning of p53 into the mitochondria, with the potential to substantially affect its nuclear activity, has important implications for basic investigations of the function of this widely studied protein. For example, p53 binding to specific genomic DNA sequences has been shown to be disrupted under conditions of severe oxidative stress that involves disulfide bond formation between C135 and C141 (18). Given that CHCHD4 substrates typically contain two nonconsecutive intramolecular disulfide bonds (19), along with our observation that the p53 C135Y mutation abrogrates p53 translocation, it is tempting to speculate that this type of molecular mechanism could seamlessly integrate the redistribution of nuclear DNA-bound p53 to the mitochondria in response to oxidative stress.
Our findings also may provide insight into cancer biology. A recent study elegantly showed that CHCHD4 promotes mitochondrial function and tumor growth, largely attributable to hypoxic HIF1α stabilization, and that its increased expression correlates with poor prognosis in cancer patients (27). We have also found that CHCHD4 promotes cancer cell growth, but that the improvement in mitochondrial metabolism is dependent on p53 in the HCT116 cell line (Fig. S3 A and C). We recently reported that germline mutations of TP53 in humans can increase oxidative metabolism in skeletal muscle, which may be attributed in part to interactions of CHCHD4 with mutated p53, such as the R175H mutation (30). In addition, our study raises the possibility that CHCHD4 also directly impacts p53-regulated pathways important for cancer cell proliferation, such as down-regulation of the cell cycle inhibitor p21. Further exploration of how the partitioning of p53 activity between the nucleus and the mitochondria by CHCHD4 affects cell metabolism, proliferation, or death is likely to provide more insight into p53 regulation in cancer and aging.
Materials and Methods
Cell Culture.
The WT HCT116 cell line was obtained from American Type Culture Collection and cultured in McCoy’s 5A medium with 10% FBS. The p53−/− HCT116 cell line was a generous gift from Bert Vogelstein, Johns Hopkins University, Baltimore, MD. Generation and characterization of the SCO2−/− HCT116 cell line have been described previously (23, 24). Primary skeletal muscle myoblasts expressing WT p53 were obtained from human subjects after approval by the National Institutes of Health internal review board and after participants provided written informed consent (NCT00406445). Myoblasts were isolated and cultured as described previously (30).
Mice.
All mice were maintained and handled in accordance with the National Heart, Lung, and Blood Institute’s Animal Care and Use Committee protocol. The WT, p53−/− (Jackson Laboratories), and p53 R172H [strain 01XL9; National Cancer Institute’s Frederick Mouse Repository (31)] mice were of the C57BL/6 strain or backcrossed at least five generations into C57BL/6 background.
Adenovirus Generation and Injection into Mice.
The adenovirus-expressing mouse CHCHD4 was prepared using the AdEasy Adenoviral Vector system (Agilent Technologies) following the manufacturer’s protocol. The mice were infected with 5 × 107 pfu of adenovirus per gram of body weight. Adenovirus-expressing GFP served as negative control (Ad-GFP; Vector Biolabs).
Gene Knockdown and Overexpression.
In this paper, CHCHD4 refers to the CHCHD4.1 isoform that is homologous to yeast MIA40 and is endogenously expressed by the HCT116 cells (27). The following plasmids were used: nonspecific or human CHCHD4 shRNA for gene knockdown (Open Biosystems), pReceiver-Lv105-human CHCHD4 cDNA (GeneCopoeia) for overexpression, and pReceiver-M77-human CHCHD4-His6 cDNA for His-tag pull-down experiments. Lentiviruses for overexpression and knockdown were prepared using the respective plasmids (Sigma-Aldrich) following the manufacturer’s protocol. Cells were incubated with virus (multiplicity of infection ∼1) for 24 h, followed by selection with 2 µg/mL puromycin. CHCHD4-His6 plasmid was transfected into HCT116 cells using Fugene HD transfection reagent (Roche). The QuikChange II Kit (Stratagene) was used to introduce point mutations into the CPC motif (substitute C53 or/and C55 with serine residues) of pReceiver-M77-human CHCHD4-His6 plasmid. The primer sequences used for site-directed mutagenesis and shRNA are provided below.
Antibodies and Western Blot Analysis.
The following antibodies were used: human p53 in Western blot analysis and immunofluorescent imaging (monoclonal antibody DO-1, sc-126; Santa Cruz Biotechnology), human CHCHD4 in Western blot analysis and immunofluorescent imaging (polyclonal antibody H107, sc-98628; Santa Cruz Biotechnology), actin (Santa Cruz Biotechnology), mouse CHCHD4 (ab87033 Abcam), mouse p21 (OP76; Calbiochem), human p21 (OP64; Calbiochem), GADPH (Ambion), and VDAC (Rockland). Proteins were resolved and subjected to Western blot analysis using standard SDS/PAGE and ECL (GE Healthcare).
Immunofluorescent Imaging.
Cells were allowed to attach to glass-bottomed tissue culture ware (Lab-Tek) for 24 h, after which they were transfected with the indicated constructs for 48 h and then stained with MitoTracker Deep Red FM (Invitrogen). This was followed by fixation in 4% paraformaldehyde PBS (Electron Microscopy Sciences) and permeabilization in 0.1% Triton X-100 PBS. Immunofluorescent labeling was performed by blocking with 5% serum (of secondary antibody species) at room temperature, followed by incubation with p53 or CHCHD4 primary antibody at 4 °C overnight and with Alexa Fluro 488- or Alexa Fluro 555-labeled secondary antibody (Invitrogen) for at 37 °C for 1 h. PBS washes were performed between all steps. Images were captured with a confocal laser scanning microscope (Olympus Fluoview FV10i).
His6-Tag Pull-Down Assay.
HCT116 cells transfected with CHCHD4-His6 or mutated CHCHD4-His6 were lysed in buffer A (1% Triton X-100, 300 mM NaCl, 1 mM PMSF, and 50 mM sodium phosphate; pH 8.0) supplemented with 10 mM imidazole, and then centrifuged at 13,000 × g for 20 min. The supernatant was loaded to a Ni-NTA spin column (Qiagen) for isolating the his6-tagged CHCHD4 and its associated proteins according to the manufacturer’s instructions. The column was sequentially washed with buffer A containing 20 mM, 40 mM, and 80 mM imidazole, then eluted with buffer A containing 150 mM imidazole, which was designated as the binding fraction. Proteins in the total lysate and binding fraction were resolved by SDS/PAGE under reducing conditions (10 mM DTT) and subjected to Western blot analysis with the indicated antibodies.
Real-Time RT-PCR.
Real-time RT-PCR was performed with an ABI HT7900 thermal cycler (Applied Biosystems). The results were normalized by measuring average cycle threshold (Ct) ratios between p21 and the housekeeping gene EIF3F (TIF) (32). PCR primer sequences are provided below.
mtDNA and Nuclear DNA Integrity Assay Using Quantitative Long PCR.
To cause DNA damage, cells were treated with 200 µM H2O2 in serum-free medium at 37 °C for 1 h, or 10 wk-old tumor-free male mice were injected via the tail vein with doxorubicin (20 mg/kg body weight). Total DNA was extracted from cells or mouse liver using the DNeasy Blood and Tissue Kit (Qiagen). For the mtDNA assay, long (8.9 kb in human, 10 kb in mouse) and short mtDNA fragments reflecting mtDNA integrity and copy number, respectively, were amplified using the GeneAmp XL PCR kit as described previously (25). For the nuclear DNA assay, a 13.5-kb human nuclear DNA fragment was amplified using the primer pair listed below. The PCR products were quantified by PicoGreen (Molecular Probes) fluorescence, and relative mtDNA integrity was calculated as a ratio of the amounts of long vs. short PCR fragments. Primers sequences are provided below.
Metabolic Studies.
Whole-cell oxygen consumption and extracellular acidification were measured as markers of oxidative phosphorylation and glycolysis, respectively, using a Seahorse Bioscience XF24 metabolic analyzer (23).
Primer Sequences.
Human CHCHD4 shRNA:
5′-TGGTTACAAATATGATTCG-3′
Human CHCHD4 site-directed mutagenesis:
- CHCHD4 C53S (SPC mutant)
- Forward: 5′-CATTAACTGGAACAGCCCATGCCTTG-3′
- Reverse: 5′-CAAGGCATGGGCTGTTCCAGTTAATG-3′
- CHCHD4 C55S (CPS mutant)
- Forward: 5′-CATTAACTGGAACTGCCCAAGCCTTGGGGGAATGGCCAG-3′
- Reverse: 5′-CTGGCCATTCCCCCAAGGCTTGGGCAGTTCCAGTTAATG-3′
- CHCHD4 C53S and C55S (SPS mutant)
- Forward: 5′-GAAACATTAACTGGAACAGCCCAAGCCTTGGGGGAATGGC-3′
- Reverse: 5′-GCCATTCCCCCAAGGCTTGGGCTGTTCCAGTTAATGTTTC-3′
p21 mRNA quantification:
- Mouse p21
- Forward: 5′-AGGGCAACTTCGTCTGGGAG-3′
- Reverse: 5′-TTGGAGACTGGGAGAGGGCA-3′
- Human p21
- Forward: 5′-CCCGTCTCAGTGTTGAGCCTT-3′
- Reverse: 5′-GTTCCGCTGCTAATCAAAGTGC-3′
mtDNA integrity assay:
- Mouse mtDNA
- 10-kb long fragment
- Forward: 5′-GCCAGCCTGACCCATAGCCATATTAT-3′
- Reverse: 5′-GAGAGATTTTATGGGTGTATTGCGG-3′
- Short fragment
- Forward: 5′-CCCAGCTACTACCATCATTCAAGT-3′
- Reverse: 5′-GATGGTTTGGGAGATTGGTTGATG-3′
- Human mtDNA
- 8.9-kb long fragment
- Forward: 5′-TCTAAGCCTCCTTATTCGAGCCGA-3′
- Reverse: 5′-TTTCATCATGCGGAGATGTTGGATGG-3′
- Short fragment
- Forward: 5′-CCCCACAAACCCCATTACTAAACCCA-3′
- Reverse: 5′-TTTCATCATGCGGAGATGTTGGATGG-3′
Human nuclear DNA integrity assay:
- 13.5-kb long fragment
- Forward: 5′-CGAGTAAGAGACCATTGTGGCAG-3′
- Reverse: 5′-GCACTGGCTTAGGAGTTGGACT-3′
Supplementary Material
Acknowledgments
We thank Alicia M. Evangelista for her kind assistance with adenovirus preparation, members of our laboratory Cory U. Lago and Jie Li for support, and Toren Finkel for helpful advice. This work was supported by the Division of Intramural Research of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310908110/-/DCSupplemental.
References
- 1.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11(3):577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 4.Vaseva AV, et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149(7):1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida Y, et al. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63(13):3729–3734. [PubMed] [Google Scholar]
- 6.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23(39):6559–6568. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- 7.Achanta G, et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24(19):3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787(5):328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JY, et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105(7):705–712. doi: 10.1161/CIRCRESAHA.109.205310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Chen Y, St Clair DK. ROS and p53: A versatile partnership. Free Radic Biol Med. 2008;44(8):1529–1535. doi: 10.1016/j.freeradbiomed.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saleem A, Hood DA. Acute exercise induces p53 translocation to the mitochondria and promotes a p53-Tfam-mtDNA complex in skeletal muscle. J Physiol. 2013;591(Pt 14):3625–3636. doi: 10.1113/jphysiol.2013.252791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesecke N, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121(7):1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Riemer J, Fischer M, Herrmann JM. Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim Biophys Acta. 2011;1808(3):981–989. doi: 10.1016/j.bbamem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Wu HH, Thomas JA, Momand J. p53 protein oxidation in cultured cells in response to pyrrolidine dithiocarbamate: A novel method for relating the amount of p53 oxidation in vivo to the regulation of p53-responsive genes. Biochem J. 2000;351(Pt 1):87–93. doi: 10.1042/0264-6021:3510087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid Redox Signal. 2001;3(4):611–623. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- 18.Augustyn KE, Merino EJ, Barton JK. A role for DNA-mediated charge transport in regulating p53: Oxidation of the DNA-bound protein from a distance. Proc Natl Acad Sci USA. 2007;104(48):18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böttinger L, et al. In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol Biol Cell. 2012;23(20):3957–3969. doi: 10.1091/mbc.E12-05-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: Regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19(1):32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bihlmaier K, et al. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179(3):389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papadopoulou LC, et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat Genet. 1999;23(3):333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto T, et al. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc Natl Acad Sci USA. 2009;106(34):14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung HJ, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1(1):5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 26.Di Fonzo A, et al. The mitochondrial disulfide relay system protein GFER is mutated in autosomal-recessive myopathy with cataract and combined respiratory-chain deficiency. Am J Hum Genet. 2009;84(5):594–604. doi: 10.1016/j.ajhg.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, et al. Human CHCHD4 mitochondrial proteins regulate cellular oxygen consumption rate and metabolism and provide a critical role in hypoxia signaling and tumor progression. J Clin Invest. 2012;122(2):600–611. doi: 10.1172/JCI58780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: Gazing through an evolutionary lens. Nat Rev Cancer. 2009;9(10):758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 29.De S, et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J Cell Sci. 2012;125(Pt 10):2509–2522. doi: 10.1242/jcs.101501. [DOI] [PubMed] [Google Scholar]
- 30.Wang PY, et al. Increased oxidative metabolism in the Li–Fraumeni syndrome. N Engl J Med. 2013;368(11):1027–1032. doi: 10.1056/NEJMoa1214091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell. 2004;119(6):861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Patino WD, et al. Circulating transcriptome reveals markers of atherosclerosis. Proc Natl Acad Sci USA. 2005;102(9):3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.