Significance
Insight into the mechanisms that underlie the process of faithful chromosome segregation is of importance for understanding the place of humans in the living world. Bacteria are the most abundant free-living organisms and have important roles in human health and disease. Nevertheless, molecular understanding of bacterial chromosome segregation is in its infancy. In this paper, the action of one conserved molecular machine that acts to coordinate the late steps of chromosome segregation with cell division is analyzed molecule by molecule in real time, using three complementary single-molecule techniques simultaneously. By using normal and mutated proteins, the complete reaction pathway is revealed from assembly of the initial nucleoprotein complex, to its activation by a DNA motor, and to the subsequent steps that lead to chromosome unlinking.
Keywords: tethered fluorophore motion, site-specific DNA recombination, protein–DNA interaction, chromosome segregation, single-molecule FRET
Abstract
Three single-molecule techniques have been used simultaneously and in tandem to track the formation in vitro of single XerCD–dif recombination complexes. We observed the arrival of the FtsK translocase at individual preformed synaptic complexes and demonstrated the conformational change that occurs during their activation. We then followed the reaction intermediate transitions as Holliday junctions formed through catalysis by XerD, isomerized, and were converted by XerC to reaction products, which then dissociated. These observations, along with the calculated intermediate lifetimes, inform the reaction mechanism, which plays a key role in chromosome unlinking in most bacteria with circular chromosomes.
Multiprotein molecular machines mediate most biochemical processes, whose mechanistic understanding requires the combined use of structural and biochemical analyses of wild-type and mutant complexes. Ensemble biochemical and structural analyses lose spatiotemporal resolution through population averaging, thereby limiting mechanistic insight. In contrast, single-molecule techniques provide real-time observation of the dynamics of individual complexes and allow detection of intermediates undetectable with other methods. However, a limitation of many single-molecule techniques is that they only address a single observable and may need considerable information about a system mechanism for the observable to be mechanistically interpreted. Site-specific recombination reactions are ideally suited to single-molecule analysis because they involve discrete complexes that undergo predictably orchestrated catalytic steps. A range of important DNA rearrangements in prokaryotes and simple eukaryotes are mediated by site-specific recombination, which is also widely exploited as a tool in genome engineering (1–3). Tyrosine family recombinases facilitate the propagation of plasmids and viruses, and have key roles in plasmid and chromosome stability and in the horizontal transfer of pathogenicity and antibiotic resistance determinants. The widely distributed XerCD tyrosine recombination system and its orthologs act in chromosome and plasmid segregation and stability in most bacteria, by converting chromosome and plasmid dimers to monomers and by acting in the decatenation of linked chromosomes (1, 2, 4, 5).
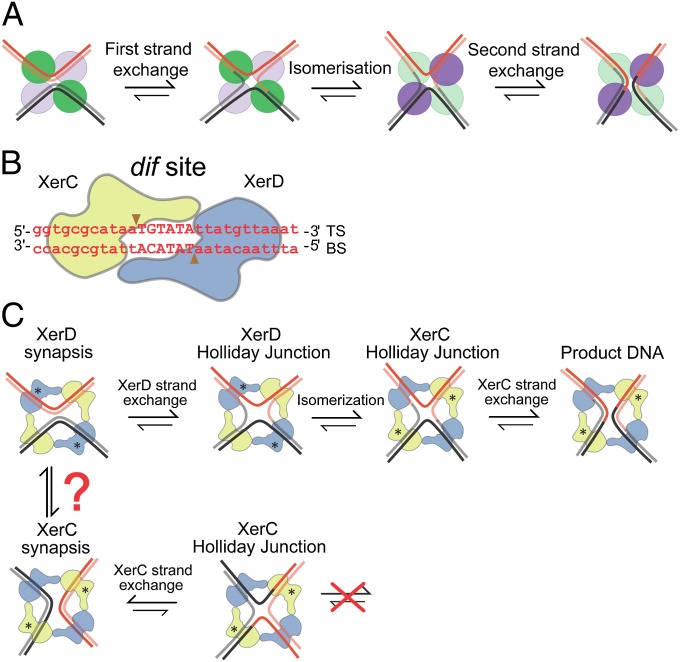
Extensive ensemble biochemical and structural studies have shown that tyrosine recombinases share a common reaction mechanism in which a tyrosine nucleophile catalyzes recombination in two steps, with Holliday junctions (HJs) as essential reaction intermediates. Reciprocal activation–inactivation of recombinase pairs occurs during HJ isomerization as the reaction proceeds (2, 6, 7). The paradigm for this mechanism comes from biochemical and structural studies of Cre–loxP recombination (2, 6, 8–10) (Fig. 1A).
Fig. 1.
Tyrosine family site-specific recombination. (A) Schematic showing tyrosine recombinase pathway. The green and violet circles indicate recombinase protomers, with the color being highlighted when a given recombinase pair is active. Isomerization of the HJ intermediate plays an essential role in the reciprocal catalytic switches in recombinase activity. (B) XerCD–dif core site. XerC (yellow) and XerD (blue) bind to their respective DNA dif half-sites. TS and BS are the “top” and “bottom” strands, respectively. The scissile phosphates, indicated by the arrowheads, flank the central region, TGTATA. (C) XerCD–dif recombination at dif as envisaged using the Cre–loxP paradigm (6) (active monomers are labeled with asterisks). FtsK activated pathway is shown at the Top; an XerD-active synapsis (XerD*) leads to a XerD–HJ, which is subsequently isomerized to a XerC–HJ, which is resolved by XerC to recombinant products. In the absence of FtsK, XerCD–dif synaptic complexes (XerC*) can form and undergo cycles of XerC-mediated strand exchanges (Bottom). XerC and XerD synaptic complexes can only be interconverted by changing the path of the DNA helix in a complex, or by breaking the recombinase–recombinase interactions involved in synapsis and reforming the complementary synaptic complex de novo.
XerCD recombination systems typically use two different yet related recombinases, XerC and XerD, each of which mediates exchange of a specific pair of strands: “top” and “bottom” strands, respectively (Fig. 1B). In some bacteria, XerCD has been replaced by a single polypeptide, XerS (11). XerCD can act on a variety of recombination sites naturally present in plasmids and chromosomes with different requirements and outcomes. Recombination at dif, a site located in the replication terminus region of the chromosome, requires activation by the DNA translocase, FtsK.
Although the XerCD–dif–FtsK reaction has been studied extensively using ensemble biochemical experiments, the lack of structures of the recombinases bound to DNA has limited progress in understanding the detailed reaction mechanism. In the absence of FtsK, XerC can catalyze HJ formation and resolution in vivo and in vitro with no detectable formation of product by XerD (12, 13). In contrast, in the presence of FtsK, recombination is initiated by XerD, giving HJs that can isomerize to form a substrate that allows completion of recombination by XerC (12, 14) (Fig. 1C). Until now, it has been unclear how FtsK action gives rise to synaptic structures that can be acted on by XerD, although it has been proposed previously that FtsK actively remodels a synaptic complex that was initially poised to undergo catalysis by XerC (14). Such a remodeling would require dissociation of XerCD–dif complexes and reassembly in a different conformation, if the reaction follows the Cre–loxP paradigm.
Here, we report a detailed analysis of XerCD–dif–FtsK recombination within individual synaptic complexes in real time. By using three simultaneous and independent observables, Förster resonance energy transfer (FRET), tethered fluorophore motion (TFM) and protein-induced fluorescence enhancement (PIFE), we were able to follow the long-range and nanoscale dynamics of the complete recombination reaction. We observed the arrival of the FtsK translocase at an initial synaptic complex, and the conformational change that FtsK induced during recombination activation, forming a transient activated complex in which XerD exchanged a pair of strands to form an HJ intermediate. Isomerization of the HJ formed a substrate for catalysis by XerC, thereby leading to complete recombinant product, which then dissociated into its duplex components.
Results
XerCD–dif Synaptic Complexes.
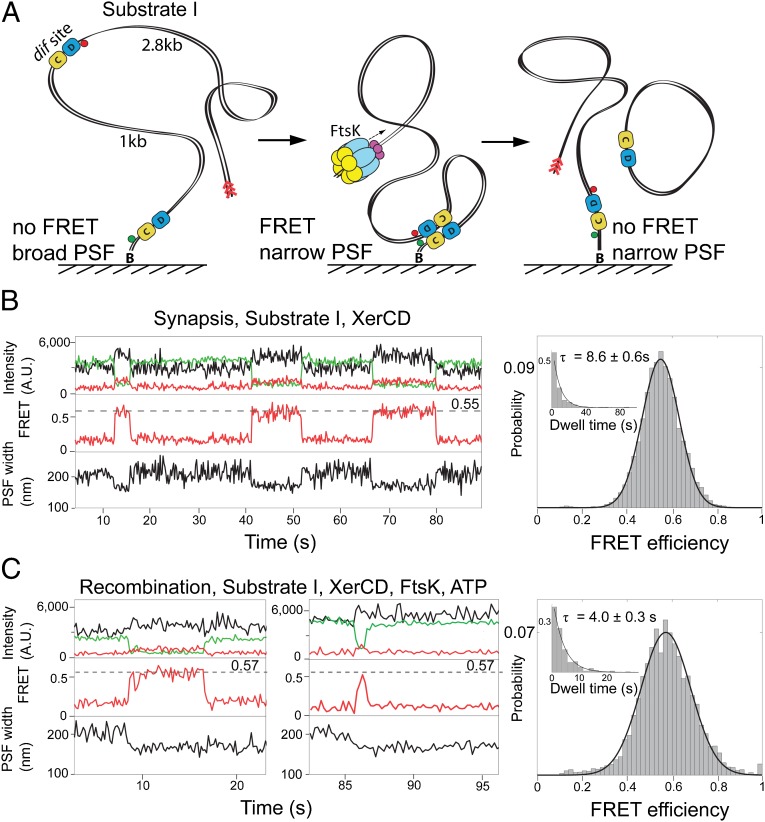
To characterize the assembly and architecture of synaptic complexes, we used TFM in combination with single-molecule FRET by implementing alternating laser excitation on a total internal reflection microscope (15, 16). This allowed simultaneous monitoring of large-scale conformational changes in DNA and nanoscale changes within the nucleoprotein complexes. The substrates were 4-kbp–long DNA molecules, which contained two repeated dif sites separated by 1 kbp; they were tethered to a PEG-passivated glass coverslip using a biotin–NeutrAvidin interaction. A 2.8-kbp DNA extension containing an FtsK loading site beyond the distal dif site facilitated efficient loading of FtsK onto DNA. The substrates were labeled adjacent to the dif sites with a FRET donor (Cy3B) and acceptor (Cy5) (Figs. 2A, 3A, and 4A, and Fig. S1). In all substrates, the acceptor fluorophore was separated from the surface by >1 kbp of linear B-DNA (with a contour length of ∼300 nm). The persistence length of DNA is around 50 nm, so the surface distal fluorophore is allowed considerable diffusional freedom around the tether point. Because this motion occurs on a millisecond timescale, the in-plane motion of the fluorophore about the surface is evidenced by a blurring of the associated point spread function (PSF) in the 100-ms camera exposures (15). XerCD-mediated formation of a synaptic complex between two dif sites was expected to reduce this diffusional freedom of the distal fluorophore, which would reduce the PSF width. This reduction in PSF width is the main TFM observable.
Fig. 2.
XerCD–dif synapsis and recombination. (A) Schematic of the recombination reaction with substrate I. Substrate I has the fluorophores attached to DNA adjacent to each dif site; the positions of the acceptor and the donor fluorophores are indicated with red and green circles, respectively (Fig. S1A). Progression through the recombination was monitored using two observables: PSF width of the acceptor and FRET. Recombination between two dif sites leads to a formation of product DNA molecules, one of them remains attached to the slide and has fluorophores flanking dif on both sides. The preferential binding site for FtsK, KOPS (FtsK orientation polarizing sequence), is indicated with red arrowheads. (B) Nonproductive synaptic complexes (Top). Representative time trace of the intensities of donor (green), and acceptor (red) under donor excitation; and acceptor under acceptor excitation (black). FRET efficiency (Middle) and PSF width (Bottom) are shown. The nonproductive events are defined by the ultimate broadening of the PSF coincident with the disappearance of the FRET signal. All data were acquired at a frame rate of 10 Hz, unless otherwise stated. Histogram (Right) of FRET efficiency (E*) and dwell time (Inset) of XerCD–dif synaptic complexes (n = 380). Dwell times were fit to single exponentials. (C) Recombining complexes. The complexes were identified by the persistence of a low PSF after the disappearance of FRET. Two representative time traces are shown, along with histograms of FRET efficiency and dwell times of recombining complexes (n = 249; Right).
Fig. 3.
XerCD–dif–FtsK recombination intermediates. (A) A cartoon of the recombination reaction within substrate II (Fig. S1B). Recombination of this substrate generates a circular DNA product, carrying both fluorophores; its dissociation was assayed by the simultaneous disappearance of both red and green fluorescence emission. (B) Initial synaptic complexes in the absence of FtsK. A representative time and a histogram of FRET efficiency are shown. The mean lifetime of the synapses, τ, is indicated (n = 382; Fig. S3A). (C) Productive recombination events in the presence of FtsK–ATP. Intermediates of recombination were distinguished by their different apparent FRET efficiencies. The following intermediates could be identified: initial synaptic complex (highlighted yellow), XerD–active synaptic complexes, XerD*, and/or XerD HJ (green), and XerC HJ/XerC product (XerC–P) (cyan). Dissociation of the complex upon completion of recombination is indicated in the time trace with an arrow. Color-coded histograms of FRET efficiencies for each step and the mean lifetimes, τ, are shown on the Right (n = 134; Fig. S3A). We note good agreement between the lifetime of the high FRET state between substrates I and II, shown in this figure and in Fig. 2C. The dwell times indicated are for individual transitions, not the entire reaction. To complete the kinetics of the pathway, it is necessary to combine rates from all three substrates. (D) Complexes in the presence of XerCKQD and FtsK. Data were taken with a frame rate of 20 Hz. The complexes were transiently trapped at the HJ intermediate stage, and exhibited a rapid interconversion between E* ≈ 0.22 and ≈ 0.37, suggesting rapid isomerization between XerC–HJ and XerD–HJ. These complexes assembled in the initial conformation (highlighted yellow) and adopted the same conformation before dissociation (purple). Histograms of FRET efficiencies are shown on the Right (n = 84). (E) Complexes in the presence of XerCDKQ and FtsK. The representative time traces display multiple reversible reductions in FRET signal to E* ≈ 0.44, suggesting the transient formation of XerD*. A histogram of FRET efficiencies is shown on the Right (n = 66).
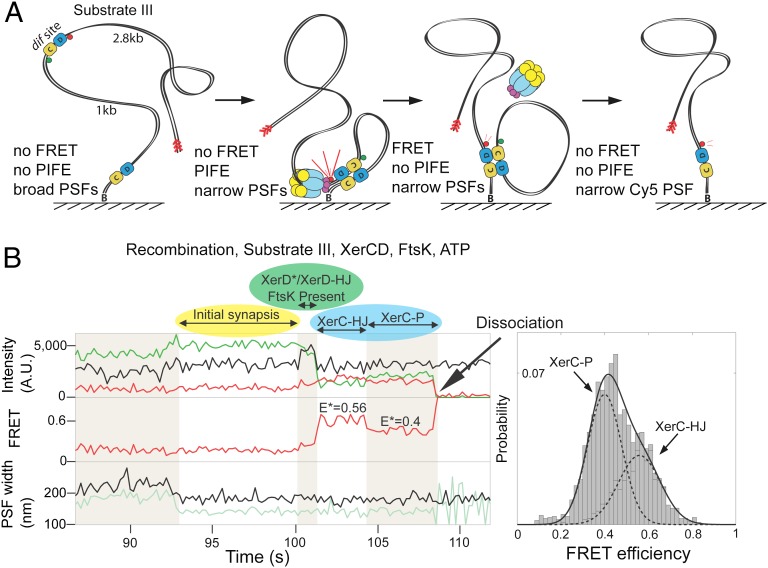
Fig. 4.
Activation by FtsK. (A) Schematic of recombination within substrate III (Fig. S1C). The fluorophores are flanking a distal dif site. On completion of recombination and complex dissociation, the donor fluorophore is lost. Multiple steps during recombination can be distinguished using three observables: PSF, FRET, and PIFE. (B) Representative time trace showing the progression of the recombination with this substrate. The color coding of various states is the same as in Fig. 2C: initial complex, yellow; XerD*/XerD–HJ, green; XerC–HJ/XerC–P, cyan. The increase in acceptor intensity (Top, black line, at ≈100 s), as a consequence of PIFE, is indicative of the presence of FtsK at XerCD–dif and is preceded by simultaneous narrowing of PSFs for both donor (pale green) and acceptor (black), indicative of initial synapse formation. A histogram showing FRET efficiency was fit with two Gaussians (n = 111), having separated the two states using HMM (Materials and Methods). The dwell times for XerC–HJs and XerC–Ps are shown in Fig. S4B.
Substrate I design was informed by the Cre–loxP crystal structure (2HOI) (17), and was similar to that used by Pinkney et al. (15) (Fig. 2A and Fig. S1A). It was expected to give recombination intermediates having interfluorophore distances of less than 90 Å during early stages of the recombination reaction, thereby giving rise to apparent FRET efficiencies of E* > 0.2 (Table S1). When this substrate was reacted with XerCD in the absence of FtsK, we observed reversible cycles of transient narrowing of PSF, consistent with the assembly and disassembly of synaptic complexes that did not undergo recombination, as expected, because FtsK is required for initiation of catalysis by XerD (Fig. 2B, Bottom). The narrowing of the PSF was accompanied by the appearance of FRET, with E* ≈ 0.55 (Fig. 2B, Middle), corresponding to an interfluorophore distance of ≈59 Å (Tables S2 and S3 and Materials and Methods). The mean lifetime τ = 8.6 s of these initial synaptic complexes was obtained from a single exponential fit to the dwell time distribution (Fig. 2B, Inset). A minority (<9%) of narrowing events were not accompanied by any apparent FRET, which we attribute to nonspecific sticking of protein–DNA complexes to the surface.
FtsK Activates Recombination on Initial Synaptic Complexes.
To test whether the above initial synaptic complexes were substrates for the activation of recombination by FtsK, we repeated the same experiments using substrate I in the presence of FtsK and ATP. We observed two predominant complexes: ones similar to those in Fig. 2B, exhibiting simultaneous disappearance of FRET, and PSF broadening, indicative of synaptic complex formation and dissociation (Fig. S2A); and complexes indicative of successful recombination (Fig. 2C), in which FRET was lost but a narrow PSF remained. The lifetime of these latter complexes was significantly shorter (τ = 4.0 s; Fig. 2C, Inset) than that of initial complexes that formed in the absence of FtsK (τ = 8.6 s), indicating efficient activation of synaptic complexes by FtsK.
To gain further insight into the recombination mechanism, we used substrate II (Fig. 3A and Fig. S1B), which using the Cre–loxP crystal structure (2HOI) as a template was labeled at sites that should allow detection of intermediates preceding HJ isomerization (Table S1). Additionally, upon completion of recombination and dissociation of the product, both fluorophores should disappear from view simultaneously (Fig. 3A). Due to the different labeling position, the initial complexes in the absence of FtsK had an E* ≈ 0.72 (Fig. 3B), higher than in substrate I; nevertheless, the lifetime of these complexes without FtsK (τ = 8.1 s; Fig. S3A) was comparable to those observed with substrate I (τ = 8.6 s).
In the presence of FtsK and ATP, we observed discrete transitions from the initial synaptic complex with E* ≈ 0.72 and a narrow PSF (Fig. 3C, yellow highlight) to a short-lived intermediate that retained the narrow PSF, but had E* ≈ 0.37 (Fig. 3C, green highlight); followed by a low FRET state with E* ≈ 0.18 (Fig. 3C, cyan highlight); followed by simultaneous loss of both fluorophores, indicative of the completion of recombination and dissociation of the reaction products (Fig. 3C, black arrow). We interpret these three FRET states as follows: E* ≈ 0.72 defines the initial synaptic complexes; E* ≈ 0.37 describes synaptic complexes in which XerD is active (XerD*) and/or the XerD–HJ resulting from strand exchange by XerD on XerD*; finally E* ≈ 0.18 complexes consist of HJ intermediates following isomerization, where XerC is active (XerC–HJs), and the product DNA complexes (XerC–Ps), resulting from strand exchange by XerC on XerC–HJs. Note that the XerC–HJ and the XerC–P states (Fig. 3C) are sufficiently close to the background E* ≈ 0.17 (Materials and Methods) that they are essentially indistinguishable.
To validate this interpretation, we analyzed reactions using catalytically inactive mutants of XerC and XerD [XerCKQ (K172Q) and XerDKQ (K172Q); Fig. 3 D and E] in the presence of their wild-type protein partners using substrate II. The reactions containing XerCKQ and XerD gave synaptic complexes with E* ≈ 0.72, indistinguishable from the wild-type XerCD complexes (Fig. S3B). In the presence of FtsK and ATP, these complexes yielded two rapidly interconverting FRET states (Fig. 3D). Analysis of the interconverting FRET states with hidden Markov modeling (HMM) (Materials and Methods) (18) revealed that the interconversion occurred between E* ≈ 0.37 and E* ≈ 0.22 (Fig. S3D). We interpret these interconversions as isomerization between XerD–HJ and XerC–HJ states, because XerCKQ cannot catalyze the final HJ resolution to product and accumulates HJs in bulk assays (13). Furthermore, resolution of the interconverting FRET states always progressed through a high FRET state, indicative of the initial synaptic complex (E* ≈ 0.72; Fig. 3D, purple). This shows that the initial complex (E* ≈ 0.72) is an obligatory intermediate in XerCD–dif recombination and that XerD–HJs can be resolved by XerD to initial synaptic complexes, at least in the presence of XerCKQ.
Reaction of XerC with XerDKQ in the absence of FtsK and ATP produced synaptic complexes with E* ≈ 0.72 (Fig. S3C), similar to the initial synaptic complexes formed by wild-type XerCD. We expected that these would be susceptible to FtsK-mediated remodeling to XerD*, but that they would not be able to progress to XerD–HJ. Indeed, an FtsK-dependent interconversion between states with E* ≈ 0.72 and E* ≈ 0.44 was observed (Fig. 3E and Fig. S3E). These results established that remodeling of the initial complex precedes catalysis by XerD. We note that the E* ≈ 0.44, assigned here for XerD*, is significantly different from the E* ≈ 0.37 for XerD–HJ determined in complexes with XerCKQD (Fig. 3D) and again different from XerD*/XerD–HJ observed in recombining complexes (Fig. 3C, E* ≈ 0.37). We propose that the E* ≈ 0.37 state, observed in recombining complexes, represents mostly XerD–HJ with XerD* being transient. Because XerD* and XerD–HJ differ in E*, we suggest that they differ in overall architecture. We converted E* ≈ 0.37 and E* ≈ 0.44 into distances (Materials and Methods and Table S2), and found that the interfluorophore distance differed by 5 ± 15 Å, with the majority of the uncertainty in all FRET-derived distances in our work attributed to uncertainty in the orientation factor of the fluorophores, κ2.
PIFE Monitors the Arrival of FtsK at the Initial Synaptic Complex.
To study the final steps of recombination we designed a third substrate within which the fluorophores flanked the distal dif site and again, using the Cre–loxP structure as a template, were positioned to show FRET after HJ isomerization, with dissociation of product DNA leading to a loss of FRET and loss of the donor fluorophore (Fig. 4A, Fig. S1C, and Table S1). In the absence of FtsK and ATP, we detected the formation of synaptic complexes, inferred from simultaneous narrowing of both donor and acceptor PSFs (Fig. S4A). In the presence of FtsK and ATP, equivalent synaptic complexes were observed, as well as complexes indicating productive recombination. The latter showed narrowing of PSF subsequently followed by a transition to an E* ≈ 0.56 state (Fig. 4B); followed by a transition to an E* ≈ 0.40 state; and then recombinant product dissociation monitored by the loss of donor fluorescence. Transition back to the E* ≈ 0.56 state from the E* ≈ 0.40 state were also occasionally observed (Fig. S4B). Because XerC–HJs precede XerC–Ps and the E* ≈ 0.56 state preceded the lower E* ≈ 0.40 state, we conclude that E* ≈ 0.56 corresponds to XerC–HJ and E* ≈ 0.40 corresponds to XerC–P. This is supported by experiments using XerCKQ and wild-type XerD with this substrate (Fig. S5). We infer from the above reversible transitions that XerC–P can be converted to XerC–HJ by XerC-mediated catalysis. Because XerC–HJ and XerC–P differ in E*, we infer that they differ in architecture. By converting E* to distance (Materials and Methods and Table S2), we found that the interfluorophore distance changed by 8 ± 13 Å between XerC–HJ and XerC–P. Taken together with the difference in E* between XerD* and XerD–HJ discussed in the previous section, these changes show that structural rearrangements occur after strand exchange by both XerC and XerD.
The proximity of a protein to an organic fluorophore can enhance or quench its fluorescence. Tryptophan residues can participate in a photoinduced electron transfer process, quenching fluorescence (19). However, for dyes that can undergo a cis–trans isomerization, the proximity of a protein can reduce the rate of isomerization to the photoinactive cis state, increasing the quantum yield of the fluorophore. This enhancement is termed PIFE (20). Substrate III allowed us to monitor the arrival of FtsK at synaptic complexes using PIFE of the acceptor fluorophore (Fig. 4A). We observed that in 77% of the recombination events monitored, the transition between the initial synaptic complex and the XerC–HJ was accompanied by PIFE of the acceptor fluorophore (Fig. 4B, Top, highlighted green, and Fig. S4C). This confirms that FtsK activates preformed initial complexes. Disappearance of PIFE was concomitant with the emergence of the FRET signal hallmarking isomerization of the HJ (Fig. 4B, Top and Middle, and Fig. S4D). The loss of PIFE upon HJ isomerization to form XerC–HJ, could result either from FtsK moving away from the vicinity of the recombining complex (for example, by dissociation, or by reversing translocation), or from a change in conformation of the FtsK-recombining complex. Reversals of translocation have been reported previously (21–23). Observation of PIFE with substrate I suggested the same sequence of events (Fig. S4E).
Discussion
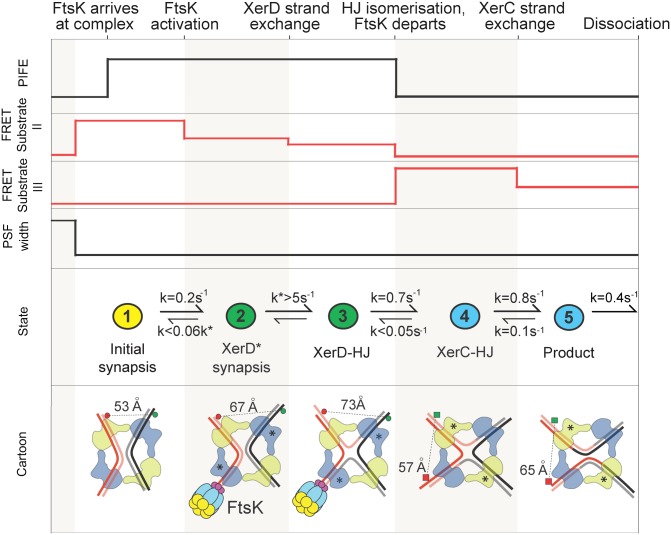
The use of three independent observables, FRET, PSF width, and PIFE, has allowed us to monitor long-range substrate DNA movements that occur during the formation of single XerCD–dif synaptic complexes, along with the nanoscale conformational transitions that occur within the nucleoprotein complexes as the recombination reaction proceeds. This analysis has provided direct information on the recombination pathway. Taken together, the results lead us to propose the reaction pathway shown in Fig. 5. Spontaneously formed initial synaptic complexes can be acted on directly by FtsK, leading to a conformational change that results in the formation of a transient synaptic complex (XerD*) in which XerD is poised to undergo catalysis, thereby generating the XerD–HJ intermediate. Isomerization of this HJ intermediate leads to a XerC–HJ, which can then be reacted on by XerC, giving a recombinant product synaptic complex that dissociates quickly. Following the initial activation step, the proposed recombination pathway is broadly similar to the established Cre–loxP paradigm for tyrosine recombinases (6, 8–10). Nevertheless, we note that in the Cre–loxP structures, the architecture of a synaptic complex is almost identical to the HJ intermediate that is generated from it (8–10). In contrast, our FRET measurements indicate discrete conformational changes in architecture as the HJ forms and resolves (Fig. 5), perhaps committing the reaction to a more directional outcome.
Fig. 5.
FtsK-activated XerCD–dif recombination. (Top and Middle) Summary of the changes in PSF, FRET, and PIFE states of the reactions on substrates II and III, with rate constants deduced from intermediate lifetimes (Materials and Methods). The colors in the Middle correspond to those used in Figs. 2–4. (Bottom) The proposed architectures of the initial, intermediate, and final complexes based on the interfluorophore distances derived from analyses with substrate II (fluorophore pairs indicated with circles) and substrate III (fluorophore pairs indicated with squares). Limits on reverse rates have been estimated given the rarity of backward transitions (2%).
Our observation that PIFE resulting from FtsK arrival at a synaptic complex is transient and disappears as XerC–HJs form, indicates that a functional change occurs in the complex at this point of the reaction. This could be a local change in FtsK position or conformation, or one that leads to FtsK dissociation. Nevertheless, we note that in reactions of XerDKQ and XerC on substrate II, FtsK is able to make repeated cycles of apparent activation, suggesting that in situations where recombination is nonproductive, FtsK remains in the proximity of the synaptic complex. Similarly, our observation that XerD can catalyze the resolution of HJs in the presence of XerCKQ and FtsK lends support to the view that FtsK can remain in the vicinity of the synaptic complex as recombination proceeds, consistent with the multiple rounds of recombination that are expected to occur during decatenation (5).
Our experiments have specifically addressed the pathway of recombination activated by FtsK, in which XerD initiates catalysis. The requirement for the FtsK activation step distinguishes this pathway from that undertaken in Cre–loxP recombination. The observation that FtsK appears to act on preformed synaptic complexes is consistent with an earlier report showing that a single translocating FtsK hexamer is sufficient to activate recombination on a XerCD–dif synaptic complex (24). The observation argues against models in which activation occurs on duplexes before synapsis (for example, by promoting XerD* synaptic complex formation), or in which FtsK actively remodels a complex that was initially poised to undertake catalysis by XerC (14). Because FtsK acts on preexisting synaptic complexes that undergo directly a conformational change to form XerD* complexes, the substrate for catalysis by XerD, suggests that XerC* synaptic complexes form independently. The presence and action of XerC* complexes has been inferred from ensemble studies in vivo and in vitro by the demonstration of XerC-catalyzed HJ formation and resolution in the absence of FtsK (12, 14, 25). Our assays have not revealed insight into XerC* formation, because dif–XerCD synaptic complexes of narrow PSF and the expected FRET values (Fig. 3B and Fig. S4A) were not detected, suggesting they are too infrequent to be investigated with our assay. Nevertheless, we inferred XerC-catalyzed formation of XerC–HJs from XerC–P complexes (k = 0.1 s−1) in reactions initiated by XerD-mediated catalysis (Fig. 5); note that XerC* complexes are expected to be identical to XerC–Ps.
Studies of the “XerC-first” pathway in which XerC* complexes generate XerC–HJs have not previously yielded evidence that FtsK can promote the isomerization of XerC–HJs to XerD–HJs, which then undergo catalysis by XerD (25). Nevertheless, the observation that HJs made by XerD, in the presence of XerCKQ and FtsK, can be converted back to substrate by XerD-mediated catalysis, shows that this pathway is possible, although it may not be favorable with wild-type XerCD. In our view, the XerC-first pathway is unlikely to be significant in vivo.
Given the intracellular concentrations of XerCD and their avid binding to dif sites, we believe it likely that dif sites in vivo are largely recombinase bound. Therefore, sister dif sites may synapse with high efficiency immediately after their duplication at the termination of replication. Those in a conformation suitable for activation by FtsK will still synapse, whereas those in the XerC* conformation will be stabilized by cycles of XerC-mediated HJ formation and resolution. We would not expect XerC* and XerD* complexes to be interconvertible without dissociation into their duplex components (Fig. 1). Together, these processes may help sister dif sites remain in proximity.
Once FtsK loads onto a chromosome, its translocation toward XerCD-bound dif will lead to the formation of activated XerD* complexes that undergo a complete recombination reaction. The temporal and spatial organization of FtsK activity in the region of dif will ensure that complete recombination reactions on the initial complexes, revealed in this work, will be limited to the septal region of the cell and to late stages of the cell cycle, thereby safeguarding against inappropriate recombination. Future work will address further mechanistic details of the FtsK activation mechanism and how this relates to the simplification of topology during FtsK-activated XerCD–dif recombination on circular DNA substrates.
Materials and Methods
Standard techniques for DNA labeling and protein purification were used and are described in detail in SI Text. Single-molecule experiments are described in SI Text. Details and procedures for data analysis and accurate FRET and distance calculation and simulations are also presented in SI Text.
Supplementary Material
Acknowledgments
Work in the D.J.S. laboratory was supported by Wellcome Trust Program Grant WT083469MA. J.N.M.P. was supported by the UK Engineering and Physical Sciences Research Council, and P.F.J.M. was supported by MathWorks. A.N.K. was supported by European Commission Seventh Framework Programme Grant FP7/2007-2013 HEALTH-F4-2008-201418, Biotechnology and Biological Research Council Grant BB/H01795X/1, and European Research Council Starter Grant 261227.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311065110/-/DCSupplemental.
References
- 1.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Hallet B, Sherratt DJ. Transposition and site-specific recombination: Adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21(2):157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 3.Akopian A, Marshall Stark W. Site-specific DNA recombinases as instruments for genomic surgery. Adv Genet. 2005;55:1–23. doi: 10.1016/S0065-2660(05)55001-6. [DOI] [PubMed] [Google Scholar]
- 4.Sherratt DJ, et al. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 1995;347(1319):37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 5.Grainge I, et al. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26(19):4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Duyne GD. A structural view of Cre-loxP site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 7.Nunes-Düby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26(2):391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389(6646):40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 9.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17(14):4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc Natl Acad Sci USA. 1999;96(13):7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolivos S, Pages C, Rousseau P, Le Bourgeois P, Cornet F. Are two better than one? Analysis of an FtsK/Xer recombination system that uses a single recombinase. Nucleic Acids Res. 2010;38(19):6477–6489. doi: 10.1093/nar/gkq507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barre F-X, et al. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14(23):2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39(12):5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aussel L, et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108(2):195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 15.Pinkney JNM, et al. Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc Natl Acad Sci USA. 2012;109(51):20871–20876. doi: 10.1073/pnas.1211922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101(24):8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J Biol Chem. 2007;282(33):24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 18.Uphoff S, Gryte K, Evans G, Kapanidis AN. Improved temporal resolution and linked hidden Markov modeling for switchable single-molecule FRET. ChemPhysChem. 2011;12(3):571–579. doi: 10.1002/cphc.201000834. [DOI] [PubMed] [Google Scholar]
- 19.Doose S, Neuweiler H, Sauer M. A close look at fluorescence quenching of organic dyes by tryptophan. ChemPhysChem. 2005;6(11):2277–2285. doi: 10.1002/cphc.200500191. [DOI] [PubMed] [Google Scholar]
- 20.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108(18):7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pease PJ, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307(5709):586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 22.Saleh OA, Pérals C, Barre F-X, Allemand J-F. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23(12):2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc Natl Acad Sci USA. 2012;109(17):6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massey TH, Aussel L, Barre FX, Sherratt DJ. Asymmetric activation of Xer site-specific recombination by FtsK. EMBO Rep. 2004;5(4):399–404. doi: 10.1038/sj.embor.7400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J Mol Biol. 2000;299(2):391–403. doi: 10.1006/jmbi.2000.3762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.