Abstract
Since its discovery as an oncogene carried by the avian acute leukemia virus MC29 in myelocytomatosis (Roussel et al. 1979) and its cloning (Vennstrom et al. 1982), c-MYC (MYC), as well as its paralogs MYCN and MYCL1, has been shown to play essential roles in cycling progenitor cells born from proliferating zones during embryonic development, and in all proliferating cells after birth. MYC deletion induces cell-cycle exit or cell death, depending on the cell type and milieu, whereas MYC and MYCN amplification or overexpression promotes cell proliferation and occurs in many cancers. Here, we review the relationship of MYC family proteins to the four molecularly distinct medulloblastoma subgroups, discuss the possible roles MYC plays in each of these subgroups and in the developing cells of the posterior fossa, and speculate on possible therapeutic strategies targeting MYC.
MYC proteins appear to play different roles in each of the four medulloblastoma subgroups. These differences appear to reflect the distinct roles that MYC genes play in hindbrain development.
Medulloblastoma (MB), the most common malignant brain tumor of childhood, is a diverse and heterogeneous disease. Most frequent in children between the ages 0 and 9, this aggressive tumor can occur at any age (Louis et al. 2007). Histopathological variants also exist and are associated with different outcomes (Rutkowski et al. 2005; Ellison et al. 2011b). Most recently, transcriptional profiling identified four major MB subgroups: two associated with pathogenic abnormalities in the Wingless (WNT) and Sonic Hedgehog (SHH) developmental signaling pathways (the WNT and SHH subgroups), and two that are less well molecularly characterized and referred to as group 3 and group 4 tumors. These transcriptional subgroups relate to differences in age at diagnosis, sex, histopathology, incidence of metastatic disease, somatic variations, and provide an improved prediction of clinical outcome (Fig. 1A) (Thompson et al. 2006; Kool et al. 2008; Cho et al. 2011; Northcott et al. 2011; Taylor et al. 2012). Despite this heterogeneity, medulloblastomas are still treated uniformly. Patients receive surgery, radiation, and adjuvant chemotherapy, and although curative at about 70%, this regimen leaves survivors with debilitating side effects and fails to cure all comers (Mulhern et al. 2005; Gajjar et al. 2006; Packer et al. 2006). Before this suboptimal but broadly effective treatment can be changed, an improved understanding of the pathogenesis of the subtypes is needed. If therapy is to become truly tailored to each molecular subgroup, bona fide tumorigenic targets for each subgroup must be identified and drugs developed to hit those targets.
Figure 1.
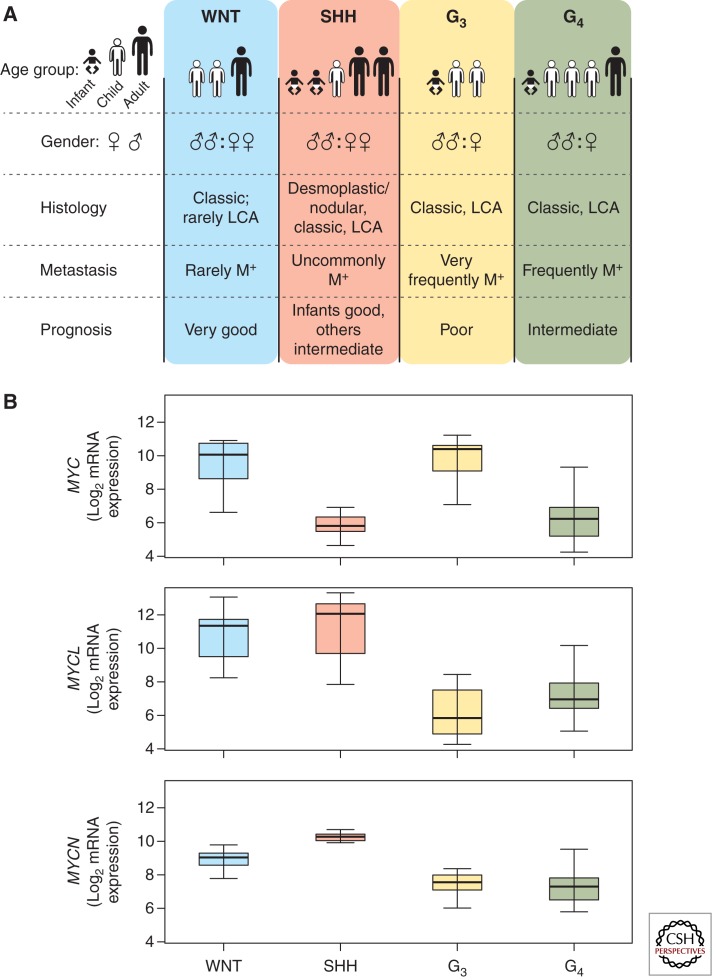
Features of the human medulloblastoma subgroups and variable expression of MYC genes. (A) Clinical and demographic features of medulloblastoma subgroups (LCA, large cell anaplastic; M+, positive for metastatic disease). Number of human figures in age group row and symbols in gender row represent the relative distribution in the four subgroups (for example, Group 4 [G4] medulloblastoma is most common in children and in males) (based on data from Taylor et al. 2012). (B) MYC, MYCL1, and MYCN Log2 mRNA expression across the four subgroups of medulloblastoma (expression data from Robinson et al. 2012 [n = 74]). Group 3, G3.
One such target is the MYC family of proteins (MYC, MYCN, and to a lesser degree MYCL1). These proteins are of interest because they appear to play different roles in each of the medulloblastoma subgroups. This relationship to the different subtypes underscores the diversity of these proteins and promises to reveal clues about medulloblastoma tumorigenesis, especially if we can gain a better understanding of the relationship of MYC protein function to normal and transformed cells of the posterior fossa, the intracranial cavity that houses the cerebellum and the brain stem within which medulloblastomas arise.
MYC AND THE MEDULLOBLASTOMA SUBGROUPS
MYC proteins are associated with many cancers and medulloblastoma is no exception. MYC, MYCN, and MYCL1 amplifications have all been described in medulloblastomas (Northcott et al. 2012b). MYC and MYCN amplification and expression have been intensely scrutinized in medulloblastoma because highly aggressive tumors frequently harbor MYC or MYCN amplification and/or overexpression (McManamy et al. 2007; Pfister et al. 2009; Cho et al. 2011). Classification schemes correlating expression and amplification of these proteins to poor outcome have been proposed (de Haas et al. 2008; Park et al. 2012). However inconsistencies, such as high expression in a subset of good responders, have made these criteria difficult to apply. These discrepancies arise because MYC proteins relate differently to each subgroup. When viewed in this context, the relationship of MYC to prognosis becomes clearer.
MYC in the WNT Subgroup
The WNT subgroup of medulloblastoma is the most curable with >90% of patients surviving on current therapy (Ellison et al. 2005; Clifford et al. 2006; Gajjar et al. 2006; Ellison et al. 2011a). It is also the least common medulloblastoma subtype, accounting for only 10%. Patients are generally older, with an average age of about 10 years. There is an even distribution of females to males. The histology is overwhelmingly classic, and the tumors are very infrequently metastatic (Fig. 1A) (Kool et al. 2012; Taylor et al. 2012). Transcriptional profiling reveals a high expression of WNT pathway genes in these tumors compared with the other subgroups (Thompson et al. 2006; Kool et al. 2008; Northcott et al. 2011). Next-generation sequencing studies show that ∼90% of these tumors harbor activating mutations in β-CATENIN (CTNNB1), the central orchestrator of the canonical WNT pathway (Jones et al. 2012; Northcott et al. 2012a; Pugh et al. 2012; Robinson et al. 2012). The only mouse model of this disease requires an activating mutation in Ctnnb1 and loss of Trp53 (Gibson et al. 2010). Partners of CTNNB1 and members of the canonical WNT pathway are also frequently mutated in this subgroup (Robinson et al. 2012). These data strongly support the constitutive activation of the WNT pathway as causal to this medulloblastoma subgroup.
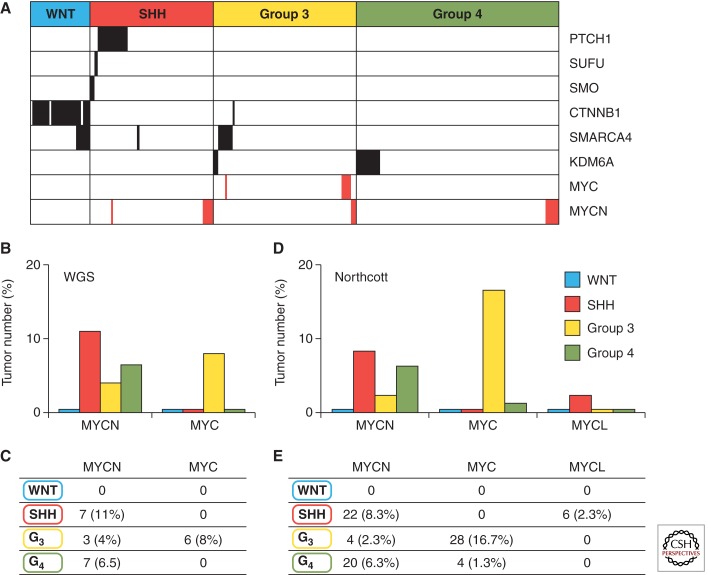
In the WNT subgroup, although MYCN and MYCL1 have a moderately high overall expression level in comparison to group 3 and group 4, MYC expression is very high and comparable only to that of group 3 (Fig. 1B). This is interesting because, as previously noted, MYC overexpression has been associated with a worse prognosis (de Haas et al. 2008; Park et al. 2012). High expression in the subgroup with the best overall survival refutes this idea and is the primary reason that MYC expression alone is a poor prognosticator. Moreover, unlike the other subgroups, MYC, MYCN, and MYCL1 amplifications almost never occur in the WNT subgroup (Fig. 2A–E) (Jones et al. 2012; Northcott et al. 2012b; Pugh et al. 2012; Robinson et al. 2012).
Figure 2.
Amplifications of MYC genes in human medulloblastoma. (A) MYC and MYCN amplifications, shown in red, and the most common recurrent subgroup mutations (missense, indels, frameshift mutations), shown in black, relative to the four medulloblastoma subgroups. Results are reported across three genomic studies (Jones et al. 2012; Pugh et al. 2012; Robinson et al. 2012). (B,C) MYCN and MYC amplifications relative to four medulloblastoma subgroups across three whole genome sequencing (WGS) studies (Jones et al. 2012; Pugh et al. 2012; Robinson et al. 2012). (D,E) MYCN and MYC amplifications relative to four medulloblastoma subgroups in 827 medulloblastomas (Northcott et al. 2012b).
MYC is a downstream target of WNT signaling and its expression is increased on the formation of a transcription factor complex through the interaction of CTNNB1 with TCF/LEF family proteins (He et al. 1998). It is not, therefore, surprising that MYC expression is elevated in tumors driven by constitutive WNT signaling. Furthermore, many mouse models of medulloblastoma have been developed through enforced MYC and MYCN expression in various cell types throughout the posterior fossa and none of these results in the formation of WNT subgroup (Zindy et al. 2007; Swartling et al. 2010; Kawauchi et al. 2012; Swartling et al. 2012). These data suggest that MYC's relationship to the WNT subgroup is not tumorigenic or progressive, but rather a marker of enhanced WNT pathway activity.
MYC in the SHH Subgroup
The SHH subgroup of medulloblastoma has an intermediate prognosis and makes up ∼25%–30% of all medulloblastomas. SHH subgroup patients tend to be very young (<5 years old) or older (>16 years old). Both males and females are equally likely to be diagnosed with SHH medulloblastoma. All three major histologic variants (nodular desmoplastic, classic, and large cell/anaplastic [LCA]) are described within this group (Northcott et al. 2011; Kool et al. 2012; Taylor et al. 2012). Outcome correlates with histology, and nodular desmoplastic SHH tumors have a good overall survival, whereas LCA tumors show a worse outcome (McManamy et al. 2007; Ellison et al. 2011a). Metastatic disease is uncommon but associates with other poor prognostic features such as LCA histology and older age at diagnosis (Fig. 1A) (Ellison et al. 2011a). Transcriptional profiling shows significant overexpression of SHH signaling genes in this group relative to the other subgroups (Thompson et al. 2006; Kool et al. 2008; Cho et al. 2011; Northcott et al. 2011). WGS and other studies have identified numerous mutations in genes that regulate the SHH pathway, all of which result in its constitutive activation (Jones et al. 2012; Northcott et al. 2012a; Pugh et al. 2012; Robinson et al. 2012). Numerous mouse models of this disease have been developed by engineering mutations that activate the SHH pathway (Goodrich et al. 1997; Hallahan et al. 2004; Uziel et al. 2005; Zindy et al. 2007; Hatton et al. 2008; Ayrault et al. 2010).
MYCN and MYCL1 are highly expressed in the SHH subgroup relative to the other subgroups (Fig. 1B). Moreover MYCN and MYCL1 amplification both occur in SHH medulloblastoma (Fig. 2A–E) (Northcott et al. 2012b). MYCN amplification in this subgroup is associated with poor prognosis (Ellison et al. 2011a). Additionally, combined data from WGS studies show a strong association between MYCN amplification, LCA histology, and TP53 mutational status; each are markers of poor prognosis (Jones et al. 2012; Pugh et al. 2012; Rausch et al. 2012; Robinson et al. 2012). These data suggest that although MYCN expression is uniformly high in this subgroup, MYCN amplification occurs in more aggressive, drug-resistant tumors.
Parallels are seen in mouse models of SHH medulloblastoma. Activation of the SHH pathway promotes the expression and stabilization of MYCN (Oliver et al. 2003; Thomas et al. 2009). Enforced expression of MYCN in granule neural precursors (GNPs) collaborates with the loss of one copy of the Ptch1 mutant and Cdkn2c (p18Ink4c), or with the loss of Trp53 and Cdkn2c to accelerate SHH MB development in mice from 9 to 3 months after birth and to increase penetrance from 14% to 60% (Uziel et al. 2005; Zindy et al. 2007; Ayrault et al. 2009; Kawauchi et al. 2012). Moreover, enforced MYCN expression produces more invasive, drug-resistant tumors (Kessler et al. 2009).
To the SHH subgroup, therefore, MYCN is both a marker of enhanced SHH activity and a harbinger of aggressivity. These activities can be distinguished by copy number but not expression. Analogous to MYC in the WNT subgroup, MYCN expression increases on SHH pathway activation, meaning that all SHH tumors show high MYCN expression including highly curable nodular desmoplastic tumors. However, more predictive of prognosis, is the copy number because, when amplified, these tumors associate with higher risk features and respond poorly to therapy. These data suggest that MYCN, especially if amplified and intrinsically deregulated, will induce and maintain SHH tumor progression.
MYC in the Group 3 Subgroup
Group 3 medulloblastomas have the worst overall survival at <50% survival on many protocols (Cho et al. 2011; Northcott et al. 2011; Taylor et al. 2012). These tumors account for ∼25%–30% of medulloblastoma. They occur in infants (<3 years old) and children but not adults. Patients are twice as likely to be male as female. Metastases are more prevalent in this subgroup than any other, exceeding 40% in some studies, and LCA histology—another high-risk feature—is also prevalent (Fig. 1A) (Kool et al. 2012; Northcott et al. 2012a; Taylor et al. 2012). Transcriptional profiling analysis did not reveal appreciable signaling pathways but enrichment for genes involved in ribosomal biosynthesis, nucleotide metabolism, photoreceptor differentiation, and GABAergic function (Kool et al. 2008; Cho et al. 2011). Next-generation sequencing of 76 group 3 primary tumors in three independent studies yielded few recurrent mutations in this group (Jones et al. 2012; Northcott et al. 2012a; Pugh et al. 2012; Robinson et al. 2012). However, numerous mutations have been identified in genes involved in the reading, writing, and erasing of epigenetic marks. These mutations overlapped with those found in group 4 suggesting a common transforming pathway (Fig. 3A,B) (Robinson et al. 2012).
Figure 3.
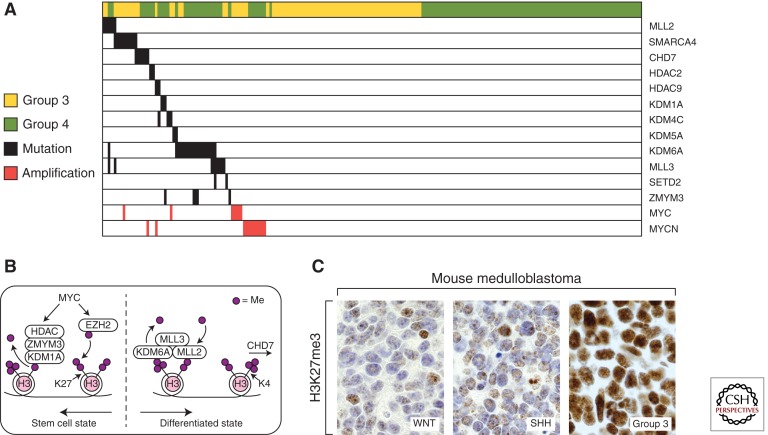
MYC genes and the epigenetic aberrations seen in human group 3 and group 4 medulloblastoma. (A) MYC and MYCN amplifications, shown in red, relative to the most common recurrent epigenetic mutations (missense, indels, frameshift mutations) in group 3 and group 4 reported across three genomic studies (Jones et al. 2012; Pugh et al. 2012; Robinson et al. 2012). (B) A diagram of epigenetic regulators altered in group 3 and group 4 medulloblastoma that influence the methylation of H3K27 and H3K4 histone marks. Somatic aberrations found in group 3 and group 4 medulloblastoma drive cell toward a stem-cell-like epigenetic state by favoring H3K27 trimethylation. (C) H3K27me3 expression in WNT, SHH, and group 3 mouse models (From Robinson et al. 2012; adapted, with permission, from the author.)
MYC is more intricately linked to this subgroup than to any other. In group 3 tumors, MYC is significantly more highly expressed compared with SHH and group 4, but not the WNT subgroup (Fig. 1B). MYC amplifications occur at a much higher frequency (10%–17%) than in any other subgroup (Fig. 2A–E) (Northcott et al. 2012b). MYCN amplifications also occur in this group albeit much more infrequently (2%–4%) (Fig. 2A–E) (Northcott et al. 2012b). These MYC and MYCN amplifications occur in a mutually exclusive fashion suggesting that they may have overlapping functions in this setting (Fig. 2A) (Northcott et al. 2012a).
Two MYC-driven mouse models of group 3 MBs have been generated. Our model was generated by enforced expression of the wild-type MYC gene in GNPs purified from Trp53-null mice (Kawauchi et al. 2012); the other relied on enforced expression of a partially stabilized mutant of MYC (MYCT58A) with a dominant-negative form of Trp53 in CD133/prominin 1-positive neural stem cells (NSCs) (Pei et al. 2012). The resultant tumors showed that the LCA phenotypes were highly aggressive, and transcriptionally resembled group 3 MB. MYC withdrawal caused complete tumor regression (Pei et al. 2012). These data suggest that MYC is a significant contributor to the initiation, maintenance, and progression of this disease (see Gabay et al. 2013).
Of note, many mouse models of MB, including these MYC-driven group 3 models, require the loss of Trp53 function; yet somatic mutations of TP53 occur mostly in WNT and SHH medulloblastoma and are associated with MYCN rather than MYC amplification (Pfaff et al. 2010; Rausch et al. 2012). The absence of these mutations in group 3 tumors means that TP53 loss is not required for human tumor initiation, that another cooperative genetic event assists tumor initiation, or that the TP53 pathway is in some other way compromised. To clarify this question, an analysis of TP53 protein expression and function and its regulation in human primary medulloblastoma samples grown as neurospheres or xenografts is warranted (Milde et al. 2012; Zhao et al. 2012).
MYC in the Group 4 Subgroup
Group 4 medulloblastomas have a similar prognosis to SHH tumors. They account for ∼40%–50% of medulloblastoma, occur mainly in children but can occur in adults, and, similar to group 3, predominantly occur in males. Most group 4 MBs have a classic histology (Fig. 1A) (Kool et al. 2012; Northcott et al. 2012a; Taylor et al. 2012). Transcriptional profiling revealed enrichment for genes of neuronal differentiation and glutamatergic receptors (Kool et al. 2008; Cho et al. 2011). Next-generation sequencing identified recurrent mutations in the molecular machinery that reads, writes, and erases epigenetic marks (described below), some overlapping with those found in group 3 tumors (Fig. 3A,B) (Jones et al. 2012; Northcott et al. 2012a; Pugh et al. 2012; Robinson et al. 2012).
Group 4 tumors generally have low MYC and MYCN expression compared with the other subgroups (Fig. 1B). However, mean MYCN expression levels are still comparatively high when compared with mature cerebellum and exist at comparable levels to fetal cerebella (Swartling et al. 2010). This implies that a certain level of MYCN may be required for tumor maintenance. In addition, MYCN amplifications occur in some group 4 tumors and these, like group 3, appear to be mutually exclusive with epigenetic mutations (Fig. 3A) and to other amplifications (Northcott et al. 2012a; Robinson et al. 2012). This suggests that, although rare, MYCN amplification may initiate group 4 medulloblastoma.
Mouse cerebellar NSCs from postnatal day 0, but not from embryonic day 16, transduced with a partially stabilized form of MYCN (MYCNT58A) produce SHH-independent medulloblastoma (Swartling et al. 2012). Similarly, when conditional MYCNT58A expression is targeted to neural cells from postnatal day 1 through adulthood, mice develop group 4-like MBs (Swartling et al. 2010, 2012). These tumors enter senescence on removal of stabilized MYCNT58A (Swartling et al. 2010). These experiments show the influence of the timing of the oncogenic stimulus and its requirement for tumorigenesis (see Gabay et al. 2013).
This temporal association draws parallels to the peak age of this disease occurring in older children rather than infants and implies that there may be a finite period during which certain cell pools are particularly vulnerable to tumorigenesis. These data suggest that MYCN plays a role in initiation and maintenance of group 4 medulloblastoma.
MYC IN THE DEVELOPING POSTERIOR FOSSA
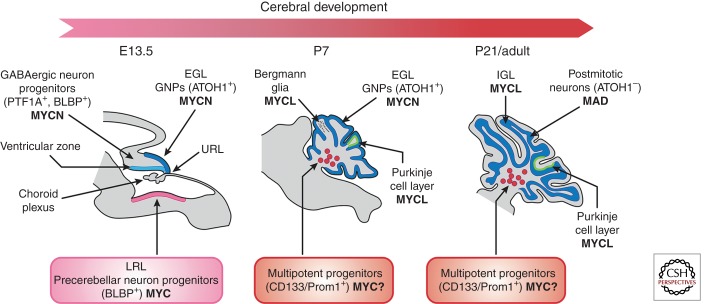
The cerebellum is a unique brain structure because, unlike the rest of the brain, granule neural progenitors actively proliferate after birth (Roussel and Hatten 2011). Cerebellar progenitor neurons arise during embryogenesis from the ventricular zone and the upper rhombic lip (URL) to form the cerebellar anlage (Fig. 4). During this time, neural progenitors are also born in the lower rhombic lip (LRL) on the floor of the fourth ventricle and express the Blbp gene (Gibson et al. 2010). The neural progenitors within the ventricular zone of the cerebellar anlage migrate radially outward and give rise to Bergman glia and Purkinje cells, which are postmitotic by birth. Conversely neural progenitors from the upper rhombic lip give rise to GNPs that rapidly proliferate after birth in the external granule layer (EGL) of the developing cerebellum, exit cycle, and migrate inward. Once the GNPs cross the Purkinje cell layer they come to rest and form the internal granule layer (IGL) as postmitotic neurons (Roussel and Hatten 2011). The white matter within the core of the mature cerebellum contains neuronal stem cells expressing CD133/prominin that are capable of generating new neurons during the life of the organism.
Figure 4.
Expression of MYC genes during mouse cerebellar development. At embryonic day E13.5, GNPs that migrate from the upper rhombic lip (URL) and form the external granule layer (EGL) and GABAergic neural progenitors in the cerebellar ventricular zone (VZ) express MYCN. MYC is found expressed in precerebellar neuron progenitors in the lower rhombic lip. At P7, whereas the GNPs in the EGL continue to express MYCN, the Purkinje cell layer, composed of Bergman glia and Purkinje cells, expresses MYCL and CD133/prominin-positive multipotent progenitors. At P21, all GNPs have exited cycle, no longer express MYCN but do express MXD (see Eisenman 2013), and have migrated through the Purkinje cell layer to reside as postmitotic neurons in the internal granule layer (IGL) that, like Purkinje cells, express MYCL (www.gensat.org; www.cdtdb.neuroinf.jp/CDT/Top.jsp; Allen Brain Atlas). CD133/prominin-positive multipotent progenitors are marked with a “MYC?” because this is suspected but not known. Prom1, prominin 1.
MYC, MYCN, and MYCL expression in the developing posterior fossa appears to be cell context dependent (Fig. 4). GNPs express MYCN, but not MYC, in their proliferative phase in response to SHH. Neuronal progenitors in the Purkinje cell layer and in the ventricular zone express MYCL but not MYCN or MYC (www.cdtdb.neuroinf.jp/CDT/Top.jsp). MYC is expressed in brain lipid-binding protein (BLBP)-positive glial and LRL neural progenitors and possibly in CD133/prominin NSCs that reside in the white matter (Wey et al. 2010). This differential expression of MYC and MYCN by distinct cerebellar progenitors lends credence to the argument that the different medulloblastoma subgroups arise from distinct cerebellar progenitors. Indeed SHH subgroup medulloblastomas originate from GNPs and overexpress MYCN, whereas WNT subgroup medulloblastomas emanate from LRL precursors and overexpress MYC. However, attempts to identify precursors of group 3 and group 4 medulloblastomas through enforced expression of MYC or MYCN into identical cerebellar precursor cells yielded some intriguing differential effects depending on the gene used, which suggests that the two paralogs influence the cellular program in different ways (Kawauchi et al. 2012).
Rapid proliferation of GNPs is mediated by SHH, which is produced by Purkinje cells. By binding to its receptor PATCHED (PTCH1), SHH directly activates MYCN transcription via GLI transcription factors (Kenney et al. 2003). The MYCN requirement for cerebellum development was shown in a Nestin-Cre transgenic mouse. Conditional deletion of Mycn in early neural stem and progenitor cells of this mouse led to profound cerebellum defects, in part owing to the failure to suppress two cyclin-dependent kinase inhibitory proteins (CKIs), p18Ink4c and p27Kip1 (Knoepfler et al. 2002). Remarkably, these cerebellar defects could be partially rescued by the loss of the two CKIs suggesting that MYCN inhibition of p18Ink4c and p27Kip1 expression is required for proper cerebellar development (Knoepfler et al. 2002; Zindy et al. 2006; Hurlin 2013).
Although MYC is not expressed in GNPs, the conditional loss of Mycn in neural progenitors induces Myc in GNPs, which partially compensates for the loss of Mycn, whereas loss of both Myc and Mycn leads to the complete absence of the EGL and GNPs (Zindy et al. 2006). This functional redundancy implies that MYC and MYCN may have similar roles. These homologs contain conserved MYC boxes, and similar basic helix-loop-helix (bHLH)-zipper structures that recognize the same DNA motif when dimerized with MAX (see Eisenman 2013; Rahl and Young 2013). Knock-in of MYCN into the MYC locus rescues murine development, strongly suggesting major functional redundancy (Malynn et al. 2000).
However, MYC and MYCN also possess long stretches of nonrelated amino acid sequences, which raises the possibility that they are functionally diverse. Indeed, enforced expression of MYC or MYCN in Trp53-null GNPs induced two distinct groups of MBs: group 3 and SHH, respectively (Kawauchi et al. 2012). Why these two very similar proteins induce two completely different MB groups is not known. Experiments swapping different regions of the MYC and MYCN proteins to assess which regions are responsible for MB outcome and identifying differences in MYC- versus MYCN-associated cofactors will be required to establish the basis for distinct functions of MYCN and MYC. In addition, chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) could be used to identify different MYC and MYCN genomic-binding sites, which will be highly informative toward understanding the emergence of distinct MBs.
MYC PROTEINS AND THE EPIGENETIC LANDSCAPE IN MEDULLOBLASTOMA
Only recently has a role for epigenetic modifiers in regulating medulloblastoma genesis been suggested. First, high-resolution single-nucleotide polymorphism (SNP) genotyping identified novel genetic events in genes that target H3 lysine 9 (H3K9), a repressive chromatin mark found in postmitotic cells consistent with transcriptional silencing (Northcott et al. 2009b). Second, MLL2 and MLL3 mutations were found to be the next most common mutations in a cohort of medulloblastoma after PTCH1 (Parsons et al. 2011). When WGS was combined with transcriptional profiling, subgroup-specific epigenetic events became apparent (Jones et al. 2012; Northcott et al. 2012a; Pugh et al. 2012; Robinson et al. 2012). Most epigenetic mutations identified in the WNT subgroup can be functionally linked to CTNNB1, substantiating that WNT pathway genetic anomalies remain the central driver (Robinson et al. 2012). Similarly, the epigenetic events identified in the SHH subgroup might be expected to cooperate with activating mutations in the SHH signaling pathway. However, it is the overlapping epigenetic events in group 3 and group 4 that will shed light on the mechanism by which these tumors develop; a mechanism that appears to involve the MYC genes.
The highest percentage of mutations in histone modifiers was found in KDM6A, the histone H3 lysine 27 (H3K27me) demethylase (Fig. 3A) (Northcott et al. 2012a). KDM6A associates with the MLL2 and MLL3 H3-K4 methyltransferases, which are also recurrently mutated along with CHD7, and ZMYM3 whose function further influences H3K4 methylation (Robinson et al. 2012). This clustering of mutations around the machinery that initiates, maintains, and erases H3K27 and H3K4 methyl marks appears to drive a stem cell-like phenotype of preserved H3K27 trimethylation coupled with impaired H3K4 methylation (Fig. 3B). An investigation of other genes that influence these histone signals revealed that EZH2, the H3K27 methyltransferase, was overexpressed in a large subset of groups 3 and 4 medulloblastomas. As expected of enzymes with opposing functions, gain of EZH2 expression was seen in tumors that lacked inactivating KDM6A mutations (Robinson et al. 2012). Furthermore, both group 3 and group 4 human tumors and MYC-driven group 3 murine MBs had higher levels of H3K27 methylation than SHH or WNT human or murine tumors (Fig. 3C) (Robinson et al. 2012; Campbell and White 2013; Dubuc et al. 2013).
Thus, group 3 and group 4 MBs retain the H3K27 trimethyl marks by loss of the demethylator KDM6A, gain of the methylator EZH2, and interference with H3K4 methylation by mutating MLL2, MLL3, ZMYM3, and CDH7 (Fig. 3B). The predicted effect is the preservation or promotion of a stem-cell-like state preventing tumor cells from differentiating. The amplification of MYC and MYCN is mutually exclusive with these recurrent mutations in epigenetic regulators raising the possibility that MYC and MYCN amplification could affect epigenetic marks that promote maintenance of the undifferentiated state (Fig. 3A) (see Chappell and Dalton 2013).
Indeed MYC genes have been implicated in chromatin and epigenetic regulation. They regulate transcription by recruiting histone acetyltransferases (HAT) (McMahon et al. 2000) and other chromatin-modifying factors (Eilers and Eisenman 2008). Changes in MYC levels have been shown to have widespread effects on chromatin marks (Knoepfler et al. 2006). Importantly, MYC has also been shown to induce EZH2 expression in other cancers (Koh et al. 2011; Salvatori et al. 2011). Given that MYC and EZH2 maintain self-renewal, and that binding of MYC induces robust regulation of Sox2 (Lin et al. 2009), it was not surprising to find “stemness” markers such as Oct4, Sox2, and Nanog, all associated with aggressive cancers (Chang and Hung 2012), in mouse group 3 MB cells (Kawauchi et al. 2012).
MYC AND MicroRNAS IN MEDULLOBLASTOMA
MYC genes and microRNAs regulate each other. MicroRNAs (miRs) are 20- to 22-nucleotide-long RNA molecules that repress translation by interacting with the 3′-untranslated region of target mRNAs (He and Hannon 2004). Although the principal consequence of MYC activation is microRNA repression, MYC also induces microRNAs that, in part, mediate its function (see Thomas-Tikhonenko and Psathas 2013).
MYC Regulation of MicroRNAs
MicroRNAs encoded by the miR-17 ∼ 92 and miR-106b ∼ 25, but not the miR-106a ∼ 363, clusters are overexpressed specifically in human and mouse SHH MBs (Ferretti et al. 2009; Northcott et al. 2009a; Uziel et al. 2009). The miR-17 ∼ 92 cluster, also called OncomiR1, was first identified in Eµ-MYC-induced B-cell lymphomas (He and Hannon 2004) and found to be regulated by Mycn in SHH-treated cerebellar neural progenitors (Northcott et al. 2009a) and by MYC proteins in other cancers (Mendell 2008). The miR-17 ∼ 92 cluster is a member of a family of three clusters including two paralogs, miR-106b ∼ 25 and miR-106a ∼ 363, which are encoded on different chromosomes and express microRNAs that share seed sequences (Ventura and Jacks 2009). Consistent with its high expression in SHH MB, enforced expression of the miR-17 ∼ 92 cluster, together with mutation of Ptch1+/−, accelerates the onset and penetrance of SHH MB (Uziel et al. 2009). Although the true direct targets of miRs from the miR-17 ∼ 92 cluster remain unknown in medulloblastoma, we found that the miR-17 ∼ 92 cluster, but not the other two paralogs, was dispensable for embryonic cerebellar development but essential for SHH MB development in the mouse because its loss in Ptch1+/− mice completely suppressed tumor development (F Zindy and MF Roussel, unpubl.). Tumor progression can be inhibited by using anti-miR 8-mer tiny locked nucleic acids (LNAs) that target the seed sequence of miR-17/20 and miR-19a/b in SHH MB cells in vitro and in animals bearing flank or cortical SHH MB allografts (Murphy et al. 2013). SHH MBs with the highest expression of these miRs are the most sensitive to LNA treatment, indicating that tumor cells are addicted to miR expression. These data suggest that LNAs targeting the miR-17 ∼ 92 cluster family could be used therapeutically in SHH MB and other cancers that overexpress members of this cluster family. Besides the miR-17 ∼ 92 cluster family, the miR-183 ∼ 96 ∼ 182 cluster was also found in MYC-amplified MBs to regulate cell survival, proliferation, and migration (Weeraratne et al. 2012). Interestingly, miR-182 promotes metastasis, a hallmark of aggressive MBs (Bai et al. 2012).
Three other microRNAs are differentially regulated in MYC-overexpressing MBs compared to MBs in which MYC is not overexpressed (Ferretti et al. 2009). However, whether these microRNAs are directly regulated by MYC is as yet unknown.
MicroRNA Regulation of MYC
Assessment of somatic copy number aberrations across 1087 unique primary MBs also identified translocations fusing MYC to microRNAs, including PVT1 (encoding microRNA miR-1204) in group 3 MB, clearly linking MYC and microRNA expression. In this translocation, the first exon/intron of PVT1 that includes miR-1204 is linked to exons 2 and 3 of MYC leading to high levels of expression of both miR-1204 and MYC in these MBs (Northcott et al. 2012b). MiR-1204 has been implicated as a candidate oncogene in combination with MYC (Shtivelman and Bishop 1989; Carramusa et al. 2007; Northcott et al. 2012b).
MYC can also be regulated by microRNAs in MBs. The miR-33b gene located at 17p11.2, a genomic locus frequently deleted in group 3 MB, was recently found to be up-regulated by lovastatin and, in turn, to repress MYC expression in two human cell lines (DAOY and D283) derived from MYC-driven primary MB patient samples (Takwi et al. 2012). In DAOY cells, loss of miR-512.2 correlated with increased MYC expression (Lv et al. 2012). Primary human medulloblastomas should be similarly investigated.
THERAPEUTIC STRATEGIES FOR MEDULLOBLASTOMA
Since its first identification, MYC has resisted all attempts to be “drugged.” However, because deprivation of MYC and MYCN in group 3 and group 4 models either induce senescence or apoptotic cell death (Swartling et al. 2010; Pei et al. 2012), several strategies are being considered to suppress MYC function either by regulating its expression or preventing its binding to partner proteins.
Because MYC requires MAX for efficient transcription (Blackwood and Eisenman 1991) preventing the interaction between MYC and MAX may suppress MYC function as was shown in MYC-transformed chicken fibroblasts (Berg et al. 2002). Unfortunately, inhibitors of the MYC/MAX interaction have not been suitable for therapy and further efforts are warranted to identify therapeutically viable small molecule antagonists of MYC/MAX dimerization (see Eisenman 2013).
Epigenetic modifiers are being considered as targets for the treatment of different cancers, including MBs. As described above, many epigenetic modifiers that regulate the function of histones, including histone H3, are mutated or overexpressed in human MBs. EZH2 inhibitors have already been shown to be effective in treating lymphomas (McCabe et al. 2012). Given the high expression of EZH2 in many group 3 and group 4 MBs, it will be important to establish the effectiveness of EZH2 inhibitors in preclinical testing in existing mouse models and in primary human xenografts. Histone deacetylase (HDAC) inhibitors, including vorinostat, are currently in clinical trials in pediatric patients with malignant brain tumors, and a recent study found that a novel human group 3 MB cell line was highly sensitive to HDAC inhibitors alone and in combination with radiation (Milde et al. 2012).
Another intensely studied potential therapeutic target is BRD4, a BET-family bromodomain protein, which was identified as the top hit from a shRNA screen of epigenetic modifiers in MYC-driven lymphomas and in acute myeloid leukemia (AML) (Blobel et al. 2011; Zuber et al. 2011a,b). BRD4 is recruited to acetylated histones to stimulate transcriptional elongation and modify chromatin-regulating complexes (Rahman et al. 2011). BRD4 positively regulates MYC transcription by binding to the MYC promoter. BRD4 inhibition by JQ1, which competitively binds to the acetyl-lysine recognition motif, effectively induced regression of MYC-driven multiple myeloma and B-cell lymphomas (Filippakopoulos et al. 2010; Nicodeme et al. 2010; Delmore et al. 2011). Other BRD4 inhibitors are also being evaluated as potential therapeutics for the treatment of MYC-driven cancers (see Bradner 2013).
Because MYC up-regulates cell metabolism by, for example, increasing folate pathway activity and nucleotide synthesis, small molecule inhibitors of these pathways may also be suitable. This approach will require the screening of libraries of small molecules using MYC-driven mouse models of MB, which should be forthcoming (see Dang 2013).
Finally, microRNAs that are overexpressed or underexpressed in MYC-driven medulloblastoma might be used therapeutically in these tumors. MicroRNAs are not predicted to cross an uncompromised blood–brain barrier and alternative modes of delivery, such as nanoparticles or intraventricular administration, should be investigated.
CONCLUDING REMARKS
The relationship of the MYC family of genes to medulloblastoma is widespread but subgroup specific. Although promoting proliferation in all the subgroups, the MYC genes play different roles in each. Fittingly, these differences appear to reflect the distinct roles MYC genes play in hindbrain development. The role in the initiation, maintenance, and progression of the aggressive group 3 medulloblastomas as well as toward the progression of SHH subgroup make the MYC genes and their regulators very inviting therapeutic targets for MBs for which current therapies are inadequate.
ACKNOWLEDGMENTS
We thank Susan Watson for scientific editing; Betsy Williford from the Department of Biomedical Communication for illustrations; and David Solecki, Mary-Elizabeth Hatten, and Paul Knoepfler for helpful discussions. We apologize to colleagues whose work was not included here owing to space limitations. This work is funded by a National Institutes of Health grant CA-096832 (M.F.R.), a Cancer Core Grant CA-21765 (M.F.R. and G.W.R.), and the American Lebanese-Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital (SJCRH).
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF 2009. Two tumor suppressors, pp27Kip1 and patched-1, collaborate to prevent medulloblastoma. Mol Cancer Res 7: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrault O, Zhao H, Zindy F, Qu C, Sherr CJ, Roussel MF 2010. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res 70: 5618–5627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai AH, Milde T, Remke M, Rolli CG, Hielscher T, Cho YJ, Kool M, Northcott PA, Jugold M, Bazhin AV, et al. 2012. MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol 123: 529–538 [DOI] [PubMed] [Google Scholar]
- Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK 2002. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci 99: 3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN 1991. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kalota A, Sanchez PV, Carroll M 2011. Short hairpin RNA screen reveals bromodomain proteins as novel targets in acute myeloid leukemia. Cancer Cell 20: 287–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bradner J 2013. Inhibiting Myc. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014266 [DOI] [Google Scholar]
- *.Campbell KJ, White RJ 2013. MYC regulation of cell growth through control of RNA polymerae I and III activities. Cold Spring Harb Perspect Med 10.1101/cshperspect.a018408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, Feo S 2007. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol 213: 511–518 [DOI] [PubMed] [Google Scholar]
- Chang CJ, Hung MC 2012. The role of EZH2 in tumour progression. Br J Cancer 106: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chappell J, Dalton S 2013. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L, Eberhart CG, et al. 2011. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 29: 1424–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW 2006. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle 5: 2666–2670 [DOI] [PubMed] [Google Scholar]
- *.Dang CV 2013. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas T, Hasselt N, Troost D, Caron H, Popovic M, Zadravec-Zaletel L, Grajkowska W, Perek M, Osterheld MC, Ellison D, et al. 2008. Molecular risk stratification of medulloblastoma patients based on immunohistochemical analysis of MYC, LDHB, and CCNB1 expression. Clin Cancer Res 14: 4154–4160 [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, Kool M, Jones DT, Unterberger A, Morrissy AS, et al. 2013. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 125: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, Pearson AD, Clifford SC 2005. β-Catenin status predicts a favorable outcome in childhood medulloblastoma: The United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol 23: 7951–7957 [DOI] [PubMed] [Google Scholar]
- Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, et al. 2011a. Medulloblastoma: Clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol 121: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Kocak M, Dalton J, Megahed H, Lusher ME, Ryan SL, Zhao W, Nicholson SL, Taylor RE, Bailey S, et al. 2011b. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol 29: 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, et al. 2009. MicroRNA profiling in human medulloblastoma. Int J Cancer 124: 568–577 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gabay M, Li Y, Felsher DW 2013. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin MJ, et al. 2006. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol 7: 813–820 [DOI] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, et al. 2010. Subtypes of medulloblastoma have distinct developmental origins. Nature 468: 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, et al. 2004. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res 64: 7794–7800 [DOI] [PubMed] [Google Scholar]
- Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, Hansen S, Knoblaugh SE, Lee D, Eberhart CG, et al. 2008. The Smo/Smo model: Hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res 68: 1768–1776 [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- *.Hurlin PJ 2013. Control of vertebrate development by MYC. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. 2012. Dissecting the genomic complexity underlying medulloblastoma. Nature 488: 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW, et al. 2012. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 21: 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130: 15–28 [DOI] [PubMed] [Google Scholar]
- Kessler JD, Hasegawa H, Brun SN, Emmenegger BA, Yang ZJ, Dutton JW, Wang F, Wechsler-Reya RJ 2009. N-myc alters the fate of preneoplastic cells in a mouse model of medulloblastoma. Genes Dev 23: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 16: 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM 2011. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget 2: 669–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, et al. 2008. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PloS ONE 3: e3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, et al. 2012. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN 2009. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS ONE 4: e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv SQ, Kim YH, Giulio F, Shalaby T, Nobusawa S, Yang H, Zhou Z, Grotzer M, Ohgaki H 2012. Genetic alterations in microRNAs in medulloblastomas. Brain Pathol 22: 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev 14: 1390–1399 [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, et al. 2012. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492: 108–112 [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol 20: 556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManamy CS, Pears J, Weston CL, Hanzely Z, Ironside JW, Taylor RE, Grundy RG, Clifford SC, Ellison DW 2007. Nodule formation and desmoplasia in medulloblastomas-defining the nodular/desmoplastic variant and its biological behavior. Brain Pathol 17: 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT 2008. miRiad roles for the miR-17–92 cluster in development and disease. Cell 133: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde T, Lodrini M, Savelyeva L, Korshunov A, Kool M, Brueckner LM, Antunes AS, Oehme I, Pekrun A, Pfister SM, et al. 2012. HD-MB03 is a novel Group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. J Neurooncol 110: 335–348 [DOI] [PubMed] [Google Scholar]
- Mulhern RK, Palmer SL, Merchant TE, Wallace D, Kocak M, Brouwers P, Krull K, Chintagumpala M, Stargatt R, Ashley DM, et al. 2005. Neurocognitive consequences of risk-adapted therapy for childhood medulloblastoma. J Clin Oncol 23: 5511–5519 [DOI] [PubMed] [Google Scholar]
- Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kaupinnen S, Roussel MF 2013. Silencing of the miR-17∼92 cluster family inhibits medulloblastoma progression. Cancer Res (to be published) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, et al. 2009a. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res 69: 3249–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, et al. 2009b. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet 41: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. 2011. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 29: 1408–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, et al. 2012a. Medulloblastomics: The end of the beginning. Nat Rev Cancer 12: 818–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. 2012b. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ 2003. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci 100: 7331–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer RJ, Gajjar A, Vezina G, Rorke-Adams L, Burger PC, Robertson PL, Bayer L, LaFond D, Donahue BR, Marymont MH, et al. 2006. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol 24: 4202–4208 [DOI] [PubMed] [Google Scholar]
- Park AK, Lee SJ, Phi JH, Wang KC, Kim DG, Cho BK, Haberler C, Fattet S, Dufour C, Puget S, et al. 2012. Prognostic classification of pediatric medulloblastoma based on chromosome 17p loss, expression of MYCC and MYCN, and Wnt pathway activation. Neurooncol 14: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. 2011. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, Witt H, Korshunov A, Read TA, Sun JL, et al. 2012. An animal model of MYC-driven medulloblastoma. Cancer Cell 21: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff E, Remke M, Sturm D, Benner A, Witt H, Milde T, von Bueren AO, Wittmann A, Schottler A, Jorch N, et al. 2010. TP53 mutation is frequently associated with CTNNB1 mutation or MYCN amplification and is compatible with long-term survival in medulloblastoma. J Clin Oncol 28: 5188–5196 [DOI] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, Wittmann A, Devens F, Gerber NU, Joos S, et al. 2009. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 27: 1627–1636 [DOI] [PubMed] [Google Scholar]
- *.Psathas JN, Thomas-Tikhonenko A 2013. MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. 2012. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488: 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2013. Myc and transcription elongation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. 2012. Novel mutations target distinct subgroups of medulloblastoma. Nature 488: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Hatten ME 2011. Cerebellum development and medulloblastoma. Curr Topics Dev Biol 94: 235–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel M, Saule S, Lagrou C, Rommens C, Beug H, Graf T, Stehelin D 1979. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature 281: 452–455 [DOI] [PubMed] [Google Scholar]
- Rutkowski S, Bode U, Deinlein F, Ottensmeier H, Warmuth-Metz M, Soerensen N, Graf N, Emser A, Pietsch T, Wolff JE, et al. 2005. Treatment of early childhood medulloblastoma by postoperative chemotherapy alone. N Engl J Med 352: 978–986 [DOI] [PubMed] [Google Scholar]
- Salvatori B, Iosue I, Djodji Damas N, Mangiavacchi A, Chiaretti S, Messina M, Padula F, Guarini A, Bozzoni I, Fazi F, et al. 2011. Critical role of c-Myc in acute myeloid leukemia involving direct regulation of miR-26a and histone methyltransferase EZH2. Genes Cancer 2: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Bishop JM 1989. The PVT gene frequently amplifies with MYC in tumor cells. Mol Cell Biol 9: 1148–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sorrell MC, McFerrin L, Eisenman RN 2013. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, et al. 2010. Pleiotropic role for MYCN in medulloblastoma. Genes Dev 24: 1059–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, et al. 2012. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 21: 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takwi AA, Li Y, Becker Buscaglia LE, Zhang J, Choudhury S, Park AK, Liu M, Young KH, Park WY, Martin RC, et al. 2012. A statin-regulated microRNA represses human c-Myc expression and function. EMBO Mol Med 4: 896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, et al. 2012. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol 123: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WD, Chen J, Gao YR, Cheung B, Koach J, Sekyere E, Norris MD, Haber M, Ellis T, Wainwright B, et al. 2009. Patched1 deletion increases N-Myc protein stability as a mechanism of medulloblastoma initiation and progression. Oncogene 28: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, et al. 2006. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol 24: 1924–1931 [DOI] [PubMed] [Google Scholar]
- Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S, Rehg JE, Calabrese C, Solecki D, Eberhart CG, et al. 2005. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev 19: 2656–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie SQ, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, et al. 2009. The miR-17 similar to 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci 106: 2812–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennstrom B, Sheiness D, Zabielski J, Bishop JM 1982. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol 42: 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Jacks T 2009. MicroRNAs and cancer: Short RNAs go a long way. Cell 136: 586–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, et al. 2012. Pleiotropic effects of miR-183∼96∼182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol 123: 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS 2010. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum 9: 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Liu Z, Yu L, Zhang Y, Baxter P, Voicu H, Gurusiddappa S, Luan J, Su JM, Leung HC, et al. 2012. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro-oncol 14: 574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF 2006. N-Myc and the cyclin-dependent kinase inhibitors pp18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci 103: 11579–11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Uziel T, Ayrault O, Calabrese C, Valentine M, Rehg JE, Gilbertson RJ, Sherr CJ, Roussel MF 2007. Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res 67: 2676–2684 [DOI] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. 2011a. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25: 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011b. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]