Abstract
The long terminal repeat (LTR) region of leukemia viruses plays a critical role in tissue tropism and pathogenic potential of the viruses. We have previously reported that U3-LTR from Moloney murine and feline leukemia viruses (Mo-MuLV and FeLV) upregulate specific cellular genes in trans in an integration-independent way. The U3-LTR region necessary for this action does not encode a protein but instead makes a specific RNA transcript. Because several cellular genes transactivated by the U3-LTR can also be activated by NFκB, and because the antiapoptotic and growth promoting activities of NFκB have been implicated in leukemogenesis, we investigated whether FeLV U3-LTR can activate NFκB signaling. Here we demonstrate that FeLV U3-LTR indeed upregulates NFκB signaling pathway via activation of Ras-Raf-IκB kinase (IKK) and degradation of IκB. LTR-mediated transcriptional activation of genes did not require new protein synthesis suggesting an active role of the LTR transcript in the process. Using Toll-like receptor (TLR) deficient HEK293 cells and PKR−/− mouse embryo fibroblasts we further demonstrate that although dsRNA activated protein kinase R (PKR) is not necessary, TLR3 is required for the activation of NFκB by the LTR. Our study thus demonstrates involvement of a TLR3 dependent but PKR independent dsRNA mediated signaling pathway for NFκB activation and thus provides a new mechanistic explanation of LTR-mediated cellular gene transactivation.
Keywords: Leukemia virus, LTR, transactivation, NFκB, TLR3
INTRODUCTION
The long terminal repeat (LTR) region of non-transforming retroviruses such as leukemia viruses plays a critical role in their replication and pathogenesis (DesGroseillers and Jolicoeur, 1984; Fan, 1990; Lenz et al., 1984). These repeat regions (U3-R-U5) are generated during reverse transcription of the viral genome and are present at both ends of the proviral sequence integrated in the host genome. The U3-LTR region of leukemia viruses contain multiple transcription factor binding sites that provide tissue-specific expression capacity to the virus (Golemis et al., 1989; Short, Okenquist, and Lenz, 1987). Further, these transcription factor-binding sites can also enhance the transcriptional activity of cellular genes, adjacent to the site of integration of these viruses (Hayward, Neel, and Astrin, 1981). Activation of a proto-oncogene adjacent to the site of integration can result in abnormal expression the gene and may ultimately lead to tumorigenesis. Insertional activation of proto-oncogenes c-myc, pim-1, pvt-1, flvi-2, int-1/wnt-1 and others have been reported in various leukemia virus pathogenesis (Fan, 1997; Morrison, Soni, and Lenz, 1995; Selten, Cuypers, and Berns, 1985).
Long before tumor development, a state of preleukemic hyperplasia is seen in the leukemia virus infected animals. It has been suggested that this preleukemic hyperplasia provides more cells susceptible to virus infection and therefore increases the chance of insertional activation of proto-oncogene (Brightman, Davis, and Fan, 1990; Davis et al., 1987). Interestingly, insertion of polyoma virus transcriptional enhancer element PyF101 into the Moloney murine leukemia virus (Mo-MuLV) U3-LTR greatly reduces the preleukemic hyperplasia and the leukemogenic potential of the virus, but does not interfere with normal replication of the virus (Brightman, Farmer, and Fan, 1993; Brightman et al., 1991; Davis, Linney, and Fan, 1985). This finding suggested that the LTR may have some role in the induction of preleukemic hyperplasia in addition to its activity as an enhancer element. Other studies demonstrated that transient or stable expression of the U3-LTR region of Mo-MuLV or feline leukemia virus (FeLV) in fibroblasts of murine or feline origin, as well as in human lymphoid cell lines, induces expression of specific cellular genes, such as collagenase IV, monocyte chemotactic protein 1 (MCP-1), c-jun and MHC class I (Faller, Weng, and Choi, 1997; Faller et al., 1997; Ghosh and Faller, 1999; Koka, van de Mark, and Faller, 1991; Wilson, Flyer, and Faller, 1987). Upregulation of these genes occur at the level of transcription and is independent of the physical location of the LTR or the responding cellular genes. The LTR from endogenous FeLV however, which is nonpathogenic, does not possess this transactivational activity (Ghosh, Roy-Burman, and Faller, 2000). Whether the cellular gene transactivational activity of the LTR is necessary for preleukemic hyperplasia has not been determined.
The mechanism of LTR-mediated transactivation of cellular genes has not been elucidated. We have previously reported that only the U3 region of the LTR is necessary for this transactivational activity and, in case of FeLV, it can be as small as 210 bp (Choi and Faller, 1995; Ghosh and Faller, 1999; Ghosh, Roy-Burman, and Faller, 2000). The U3-LTR region sufficient for gene transactivational activity does not contain a readable protein-coding frame. A specific RNA transcript for this region has been consistently detected in cells expressing only the U3-LTR as well as cells infected with the full-length virus (Ghosh and Faller, 1999). We have also demonstrated that LTR from nonpathogenic endogenous FeLV, which cannot transactivate cellular genes, does not make LTR specific transcript (Ghosh, Roy-Burman, and Faller, 2000). Since the U3-LTR is unable to make a protein product, it is likely that the RNA transcript generated acts as a mediator of cellular gene transactivation.
Double-stranded RNA (dsRNA) is a strong inducer of several signal transduction pathways including activation of the transcription factor NFκB (Geiss et al., 2001; Ghosh, May, and Kopp, 1998; Siebenlist, Franzoso, and Brown, 1994). NFκB is intimately associated with cellular defense mechanisms as it is activated by variety of inflammatory cytokines, growth factors and stress inducing agents (Karin and Ben-Neriah, 2000; Kopp and Ghosh, 1995). Recent evidences also suggest that NFκB plays an important role in cell survival and anti-apoptotic responses (Baldwin, 2001; Yamamoto and Gaynor, 2001). In non-stimulated cells, NFκB exists as a cytoplasmic heterodimeric complex composed mainly of p50 and RelA proteins bound to inhibitory proteins of IκB family. Upon stimulation, the IκB protein is phosphorylated and is degraded in the proteasome allowing free NFκB heterodimer to migrate to the nucleus where it binds to its cognate binding sites on the promoters of various target genes and enhances their expression. Because of the diverse range of genes that NFκB can modulate, it also has been an important target molecule for many viruses including tumorigenic viruses, such as Epstein Barr virus (EBV), hepatitis C virus (HCV), human T-lymphotropic virus (HTLV), and also human immunodeficiency virus (HIV) (Hiscott, Kwon, and Genin, 2001; Santoro, Rossi, and Amici, 2003). Although different viruses use different strategies to control NFκB, many of them converge to the activation of IκB kinase (IKK). Double stranded RNA-dependent protein kinase R (PKR) and some member of the Toll-like receptor (TLR) family (such as TLR3) have been shown to act as an intermediate in the activation of the NFκB pathway by RNA (Alexopoulou et al., 2001; Williams, 1997; Williams, 2001).
The ability of the LTR to mediate cellular gene transactivation through the production of RNA transcript and the observation that many of the cellular genes transactivated by the LTR are known to be NFκB responsive indicated that LTR might activate NFκB signaling. In this study we investigated such a possibility and report that leukemia virus LTR activates NFκB signaling pathway. We also investigated the role of PKR and RNA activated TLRs in this process. Using cell lines deficient in either PKR or TLRs we found that although PKR is dispensable, TLR3 is necessary for LTR mediated activation of NFκB. Our results thus provide a new insight into the mechanism of leukemia virus LTR-mediated cellular gene activation.
RESULTS
U3-LTR activates NFκB-dependent gene expression
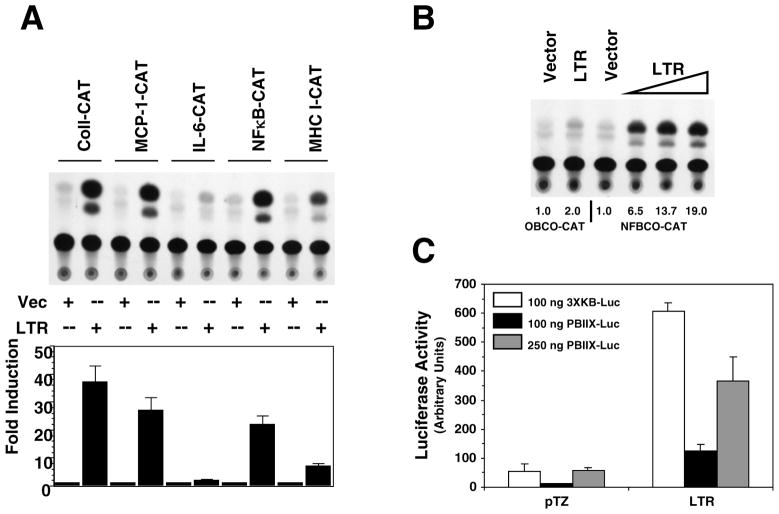
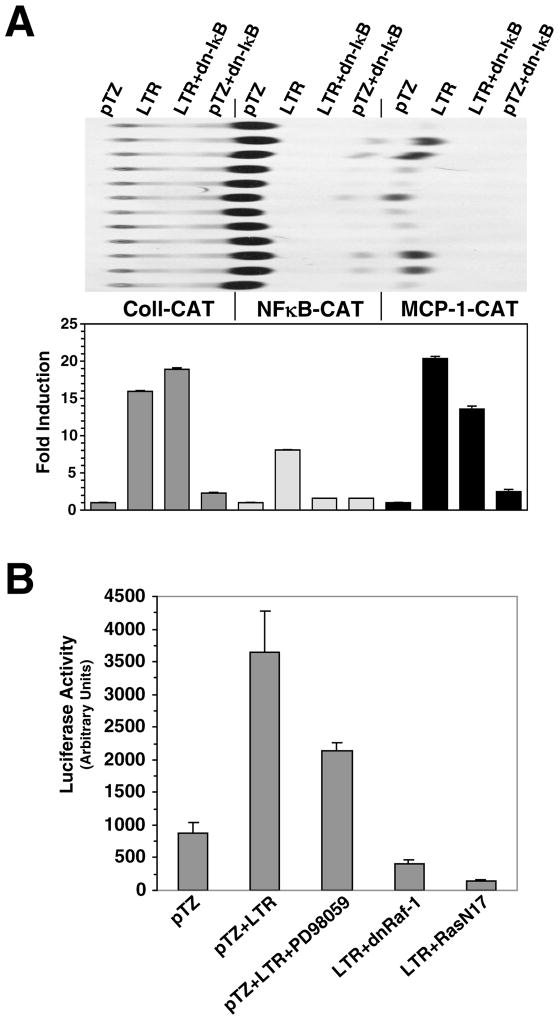
Numerous reports have demonstrated that the leukemia virus LTR directly influences propagation of the virus through the transcription factor binding sites they contain. There are recent indications that the LTR may also play a role in target lymphocyte proliferation during preleukemic hyperplasia. Our previous studies demonstrated that the LTR can upregulate expression of specific cellular genes, some of which can also be activated by NFκB, through the generation of a noncoding RNA transcript (Ghosh and Faller, 1999; Ghosh, Roy-Burman, and Faller, 2000). RNA secondary structure analysis predicted that the putative LTR transcript possessed strong secondary structure (data not shown). Since dsRNA is a strong activator of NFκB, we examined whether NFκB signaling is activated in cells that are expressing LTR. We cotransfected NFκB-dependent reporter plasmid NFBCO-CAT with FeLV U3-LTR construct 61E-LTR into Balb-3T3 cells. The NFBCO-CAT construct contains two NFκB binding sites from β-interferon gene adjacent to a minimal β-globin promoter (Richardson and Gilmore, 1991). We compared upregulation of this NFBCO-CAT reporter by the 61E-LTR along with other promoter-reporters we have previously shown to be transactivated by the LTR. In Figure 1A, we show that the 61E-LTR can activate expression of NFBCO-CAT reporter up to 22-fold. In parallel, we show that the LTR construct can induce expression of reporters containing promoters for genes such as collagenase IV, MCP-1, MHC-I, but not IL-6, thus corroborating our previously published report (Ghosh and Faller, 1999). We next tested whether activation of NFκB-dependent gene expression by the LTR was directly related to increased DNA-binding activity of NFκB protein. To address this issue, in transient transfection assays we used another reporter construct OBCO-CAT, which is same as the NFκB dependent reporter NFBCO-CAT except that the two NFκB binding sites are absent (Richardson and Gilmore, 1991). In Figure 1B we demonstrate that 61E-LTR can activate NFBCO-CAT but not the NFκB binding site-deleted reporter OBCO-CAT suggesting that these sites are necessary for NFκB activation by the LTR. This activation is dose-dependent as the fold induction of NFBCO-CAT increases with increasing amount of 61E-LTR.
Figure 1. Activation of NFκB dependent gene expression by the LTR.
A. Analysis of transactivational activity of the 61E-LTR towards CAT-reporters with promoters from different genes. Balb-3T3 cells were cotransfected in 100mm plates with 7.5 μg of 61E-LTR or the vector plasmid pTZ19 and 7.5 μg of various CAT-reporter constructs by DEAE-dextran method and were harvested 48 hrs post-transfection for reporter assays. This experiment was performed three times and a representative chromatogram is presented. For quantitation purpose, individual spots on the chromatogram were collected by scraping and radioactivity was measured by liquid scintillation counting. Fold-induction for any particular CAT construct was expressed as the ratio of acetylated 14C-chloramphenicol generated for that construct by the LTR to that generated by the vector. B. Requirement of NFκB binding-site in the reporter for activation by the LTR. Two different CAT-reporter plasmids, with or without NFκB binding sites in the promoter (NFBCO-CAT or OBCO-CAT, respectively) were cotransfected separately with 61E-LTR or pTZ19 plasmid in Balb-3T3 cells and CAT assays were performed. To determine dose dependence, 2.5, 5.0 or 7.5 μg of 61E-LTR were transfected. One representative chromatogram and fold-induction values are presented. C. Analysis of transactivational activity of 61E-LTR towards two different NFκB-dependent luciferase reporter constructs. Six-well plates of Balb-3T3 cells were cotransfected with 61E-LTR or pTZ19 (300 ng) and 3XKB-luc (100 ng) or PBIIX-Luc reporter (100 or 250 ng) by lipofectamine plus method and harvested 48 hrs post-transfection for luciferase assays. Total amount of plasmid transfected in each well in both transfection methods was maintained equal using pTZ19 vector plasmid.
We next tested whether LTR can act on other NFκB-dependent reporter constructs. We used two luciferase reporter constructs, 3XKB-Luc (containing three copies of MHC class I κB element upstream of a minimal c-fos promoter) and PBIIX-luc (containing two copies of immunoglobulin light chain κB element upstream of a minimal c-fos promoter) (Kopp and Ghosh, 1994; Mitchell and Sugden, 1995). As shown in Figure 1C, both these reporters were also strongly induced by 61E-LTR (7-10 fold). It may be noted however; the basal activity with PBIIX-luc reporter was significantly lower and higher amounts of reporter plasmid than that of the 3XKB-luc reporter was necessary to achieve comparable basal activity.
LTR enhances nuclear translocation of p65
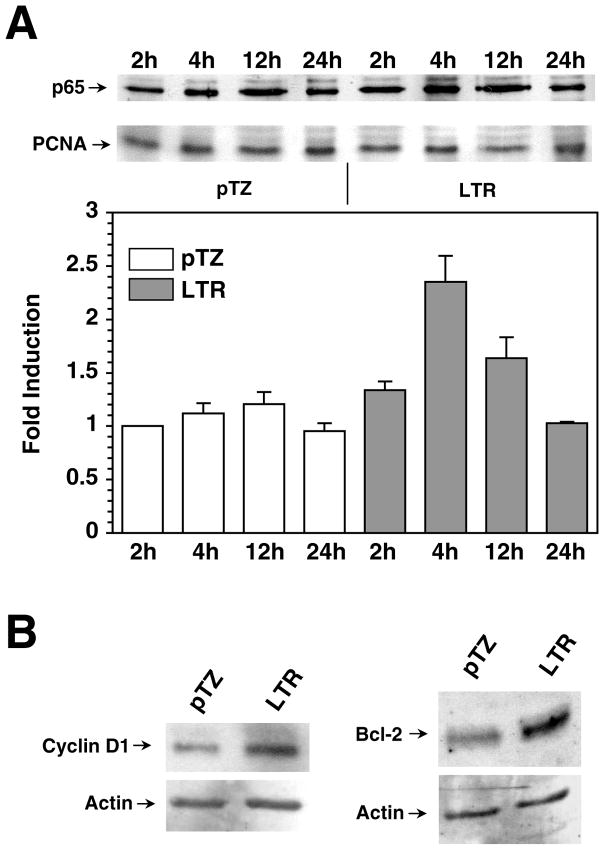
We determined whether upregulation of NFκB dependent gene expression by the LTR was due to the increase of NFκB protein in the nucleus. We measured the RelA protein (p65, most common member of NFκB family of proteins) level in the nucleus following expression of LTR. Nuclear extracts were prepared from Balb-3T3 cells at multiple time points following transfection with 61E-LTR or vector plasmid pTZ and were analyzed for the p65/RelA protein by western immunoblotting. The level of common nuclear protein PCNA in these samples was tested for sample loading control. Figure 2A demonstrates that p65/RelA protein level in LTR transfected cells increased up to 2.2 times compared to pTZ vector transfected cells. Transfection efficiency in these experiments was approximately 25%. The increase in p65 level could be seen within 2 hr post-transfection, reached a peak by 4 hrs, and decreased gradually over time. Transient increase of NFκB level of this nature following virus infection has been previously reported with vesicular stomatitis virus (VSV) and measles virus (MeV) (Boulares, Ferran, and Lucas-Lenard, 1996; Donze et al., 2004; Helin et al., 2001). The p65 level in pTZ vector transfected cells increased insignificantly during the course of the study. These results show that expression of LTR increased the level of p65/RelA protein in the nucleus.
Figure 2. Translocation of p65 and expression of endogenous NFκB dependent genes by the LTR.
A. Analysis of nuclear p65 following expression of LTR. Nuclear extracts were prepared from Balb-3T3 cells transfected with 61E-LTR or pTZ19 by lipofectamine method at indicated time post-transfection. The time denotes period after the initial 3 hr incubation with DNA and lipofectamine. Twenty micrograms of protein for each sample were separated in 10% SDS-PAGE and analyzed for p65 or PCNA by western immunoblotting. Band intensities were determined directly on the film using LabWorks Image Analysis Program (UVP Inc, Upland, CA). Fold inductions were calculated after the band intensities were normalized for equal PCNA loading. B. Activation of endogenous cyclin D1 and Bcl-2 protein. Twenty microgram of whole cell lysates from Balb-3T3 cells transfected with either 61E-LTR or pTZ19 vector were analyzed for cyclin D1 or Bcl-2 level by western immunoblotting. Lysates were prepared 40 hrs after transfection. Same blots were further analyzed for β-actin protein following stripping of the antibodies used in first immunoblotting.
To test whether NFκB activation by the LTR indeed affects expression of endogenous NFκB dependent genes we tested the level of two genes, cyclin D1 and Bcl-2, in 61E-LTR transfected cells. Cyclin D1 helps progression of cell cycle in G1 phase and Bcl-2 acts as an anti-apoptotic protein, both of which are regulated by NFκB (Guttridge et al., 1999; Wang et al., 1998). As shown in Figure 2B, both cyclin D1 and Bcl-2 levels were increased significantly in LTR expressing cells over backbone vector expressing cells. This result therefore, clearly demonstrates that NFκB activation by the LTR leads to upregulation of endogenous NFκB dependent genes.
Full-length FeLV also activates NFκB
We tested whether LTR would activate NFκB in the context of full-length virus. To address this issue we first tested whether full-length FeLV or MuLV could activate NFκB. We cotransfected Balb-3T3 cells with NFBCO-CAT and either 61E-LTR or the full-length FeLV clone 61E and determined NFκB activation by CAT assay. As shown in Figure 3A, NFBCO-CAT was activated up to 5.7-fold by the 61E. We also tested NFκB activation by the U3-LTR construct and full-length clone of Mo-MuLV (GMNX and Mov9, respectively). The NFBCO-CAT reporter expression was activated up to a 29.1− and 9.4-fold by the U3-LTR and full-length Mo-MuLV constructs, respectively. Our finding thus corroborated previous observation that full-length Mo-MuLV activates NFκB dependent gene expression (Pak and Faller, 1996). It is noteworthy that the fold-induction of NFκB activation by the LTR in the context of full-length virus was several-fold lower than that with the isolated LTR. This effect is due to lower net LTR copy number per absolute amount of DNA transfected and is consistent with our previous publication (Ghosh and Faller, 1999). Next, to determine if the U3-LTR is the responsible factor in full-length virus for NFκB activation we used a mutant FeLV U3-LTR construct EDD2, which has 8 base substitution mutation. We also used a mutant full-length molecular clone of FeLV, 61E-Mut, which has EDD2-specific mutations in both of its U3-LTR. We have previously demonstrated that this mutant virus replicates with wild-type kinetics yet cannot transactivate collagenase or MCP-1 genes like the wild-type virus (Abujamra, Faller, and Ghosh, 2003). As shown in Figure 3B, both the EDD2 and the 61E-Mut were unable to activate NFκB dependent luciferase reporter gene expression in transient transfection experiment. This result demonstrated that U3-LTR portion of the full-length FeLV is responsible for activation of NFκB dependent gene expression by the virus.
Figure 3. Full length FeLV activates NFκB in an LTR-dependent manner.
A. Analysis of NFκB activation by full-length and U3-LTR constructs from FeLV and Mo-MuLV. Balb-3T3 cells were cotransfected with 7.5 μg of NFBCO-CAT reporter and 7.5 μg of 61E-LTR (FeLV U3-LTR) or GMNX (Mo-MuLV U3-LTR) or 61E (full-length FeLV) or Mov9 (full-length Mo-MuLV) or pTZ19 vector plasmid by DEAE-dextran method. Cells were harvested 48 hrs post-transfection for CAT assays as described in Figure 1. Similar results were obtained in three independent experiments and the fold-induction values for the presented chromatogram are indicated. B. Analysis of NFκB activation by the full-length FeLV containing mutation in the U3-LTR. Balb-3T3 cells were cotransfected with 100 ng of 3XKB-luc reporter and 300 ng of either wild type or mutant FeLV U3-LTR constructs (61E-LTR or EDD2, respectively), or wild type or mutant full-length FeLV constructs (61E or 61E-Mut, respectively). Cells were harvested 48 hrs post-transfection for luciferase assays as in Figure 1.
Proteasomal degradation of the inhibitory protein IkB is necessary for NFκB activation by the LTR
Activation of NFκB dependent gene expression primarily depends on phosphorylation of inhibitory protein IκB followed by its proteasomal degradation in the cytoplasm and subsequent translocation of free NFκB heterodimer to the nucleus. To determine whether proteasomal activity is necessary for the activation of NFκB dependent gene expression by the LTR we used the drug lactacystin that inhibits chymotrypsin- and trypsin-like activities of proteasome (Dick et al., 1997). In transient transfection experiments with 61E-LTR and NFκB dependent luciferase reporter 3XKB-luc we tested the effect of 5 μM Lactacystin on reporter activity. As shown in Figure 4A, lactacystin, but not the DMSO solvent, inhibited induction of luciferase reporter by the LTR significantly by 24 hr post-transfection. As a control we tested upregulation of the same reporter by p65/RelA expression plasmid and effect of lactacystin on it. Because proteasomal activity is not necessary for p65/RelA-mediated upregulation of 3XKB-Luc, as expected, similar concentrations of lactacystin did not affect reporter activation. This result demonstrated that active proteasome function is necessary for NFκB activation by the LTR.
Figure 4. Activation of NFκB by the LTR require proteasomal activity, phosphorylation and degradation of IκB.
A. Effect of proteasomal inhibitor lactacystin on LTR-mediated activation of NFκB. Balb-3T3 cells were cotransfected with 100 ng 3XKB-luc reporter and 300 ng 61E-LTR or p65/relA expression plasmid by lipofectamine plus method. Lactacystin (5μM) or DMSO solvent control was added on to the cells along with DNA-lipofectamine mixture. Cells were harvested 24 hr post-transfection for luciferase assay. B. Induction of IκB-α degradation by the LTR. Balb-3T3 cells transfected with 61E-LTR or pTZ19 similary as in Figure 2A but instead cytoplasmic extracts were prepared at indicated time period and analyzed for IkB-α by western immunoblotting. Same blots were later analyzed for β-actin. C. Effect of expression of super repressor dn-IκB. Balb-3T3 cells were cotransfected with 100 ng 3XKB-luc reporter and 300 ng 61E-LTR and various amounts of dn-IκB plasmid (50 ng, 100 ng or 200 ng) by lipofectamine plus method as indicated in Figure 1. Cells were harvested 24 hrs post-transfection for luciferase assay. D. Effect of inhibition of IκB-kinase (IKK) activity. Balb-3T3 cells were cotransfected with 3XKB-luc reporter and 61E-LTR along with expression plasmids for dominant negative form of IKK subunits (dn-IKK1 and dn-IKK2, 300 ng) as appropriate and processed for luciferase assay.
To determine whether NFκB activation by the LTR actually leads to degradation of the inhibitory protein IκB, we analyzed cytoplasmic IκB-α level over time in 61E-LTR transfected cells. As shown in Figure 4B, IκB-α level decreased within 1 hr after transfection, reaching a minimum level within 2-3 hrs and went back to steady state level thereafter. This trend was very much reciprocal to the increase of nuclear p65 level following LTR expression as reported in Figure 2A. This result thus demonstrated that LTR expression leads to IκB-α degradation, which facilitates subsequent translocation of p65 to the nucleus.
To determine whether NFκB activation by the LTR requires phosphorylation of the inhibitory protein IκB, we conducted two different experiments. First, we used a dominant negative form of IκB (dn-IκB) that has serine to alanine mutation at positions 32 and 36, which prevent its phosphorylation, polyubiquitinylation and subsequent proteasomal degradation. Overexpression of this dn-IκB thus sequesters endogenous NFκB in the cytoplasm, blocking its dissociation and translocation into the nucleus. As shown in Figure 4C, dn-IκB expression strongly inhibits LTR-mediated activation of NFκB dependent reporter 3xKB-Luc in transient transfection experiments. In the second approach, we inhibited the upstream kinase that phosphorylates IκB and determined how it affects LTR-mediated transactivation. In response to stimulation, IκB is phosphorylated by IκB-kinase (IKK) complex. IKK complex is composed of two kinases, IKKα, IKKβ and another modulatory component IKKγ. Different external stimuli for activation of NFκB pathway preferentially activate either IKKα or IKKβ although they both can phosphorylate IκB. We used dominant negative forms of IKKα (serine to alanine mutation at positions 176 and 180, dn-IKK1) and IKKβ (serine to alanine mutation at positions 177 and 181, dn-IKK2) which can interact with upstream regulators but are unable to phosphorylate IκB (Mercurio et al., 1997). As shown in Figure 4D, dn-IKK2 but not dn-IKK1, inhibits the ability of the LTR to activate NFκB-dependent reporter. This result collectively showed that phosphorylation of IκB is necessary for NFκB activation by the LTR.
Gene transactivation function of the LTR does not require new protein synthesis
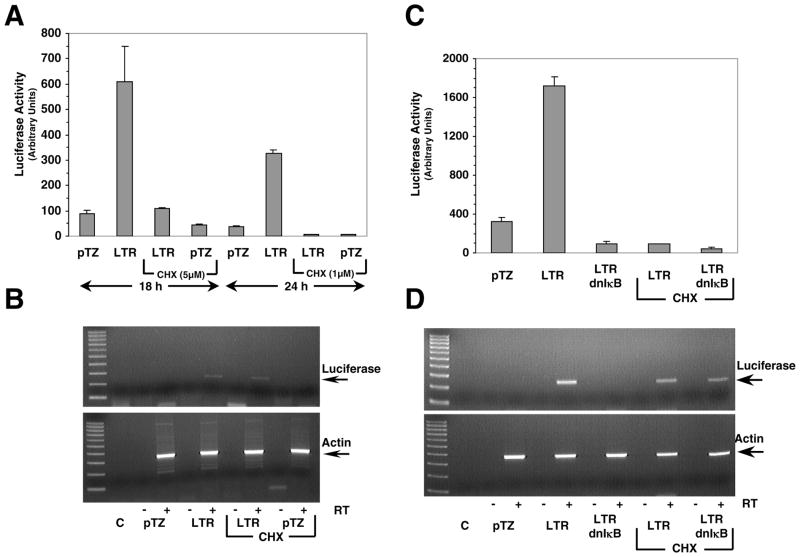
To determine if synthesis of any new protein is necessary for NFκB activation by the LTR we cotransfected 61E-LTR and 3XKB-luc reporter in Balb-3T3 cells as before but in presence or absence of the protein synthesis inhibitor cycloheximide, and measured transactivation by luciferase assay. In parallel, we analyzed transcription of the luciferase gene by measuring luciferase mRNA production by RT-PCR analysis. We chose two different concentrations of cycloheximide, 5 μM for 18 hr or 1 μM for 24 hr. Cycloheximide was added to the cells at the same time as the lipofectamine-DNA complexes. Both treatments of cycloheximide inhibited >95% new protein synthesis in Balb-3T3 cells as determined by 35S-methionine incorporation (data not shown). As shown in Figure 5A, cycloheximide at both 5 μM and 1 μM concentrations almost completely inhibited LTR-mediated activation of NFκB-dependent luciferase activity as expected, demonstrating effective suppression of protein synthesis. In untreated cells, however, 7-fold activation of luciferase reporter by the LTR was observed, as in previous experiments. RT-PCR analysis performed on the total RNA extracted from cycloheximide untreated cells showed that there was strong induction of luciferase RNA expression in cells transfected with 61E-LTR whereas no such induction was seen in cells transfected with vector pTZ19 (Figure 5B). Interestingly, presence of 1 μM cycloheximide did not inhibit induction of luciferase RNA expression by the LTR. Similar result was also obtained with cells treated with 5 μM cycloheximide (data not shown). PCR performed on the same RNA samples without any reverse transcription confirmed the absence of any contaminating luciferase reporter DNA. The production of β-actin RNA was similar in all samples irrespective of cycloheximide treatment. This result demonstrated that new protein synthesis was not required for LTR-mediated activation of NFκB.
Figure 5. New protein synthesis is not necessary for LTR-mediated gene transactivation.
A. Effect of cycloheximide on the activation of NFκB-dependent luciferase reporters by the LTR. Cotransfection experiments were carried out with 3XKB-luc reporter and 61E-LTR or vector pTZ19 plasmid in Balb-3T3 cells by lipofectamine plus method. Cycloheximide (5 μM or 1μM CHX) was added to cells in appropriate wells 30 min after the DNA-lipofectamine complex was added onto the cells. Cells were harvested for luciferase assay at 18 hr (for 5 μM CHX treatment) or 24 hr (for 1 μM CHX treatment) post-transfection. B. Effect of cycloheximide on transcription of luciferase reporter gene by the LTR. Total cellular RNA (2 μg) from a second set of Balb-3T3 cells cotransfected and cycloheximide treated essentially similarly as above, were subjected to RT-PCR analysis for luciferase mRNA. RT-PCR figure shows data for 24 hr treatment group only. Essentially similar results were obtained with 18 hr treatment group. In order to demonstrate absence of DNA contamination in RNA preparations, one set of PCR amplification for each sample was carried out without any RT and loaded onto the lanes marked as ‘-’. All RNA samples were also tested for β-actin mRNA by RT-PCR performed in similar manner as for luciferase mRNA. Lane C represents PCR control with no RNA sample. Products were separated on 2% agarose gel. The 100 bp DNA ladder was included in the left lane. The amplified product for luciferase mRNA (250 bp) and β-actin mRNA (353 bp) are indicated by an arrow. C. Effect of cycloheximide on the inhibitory effect of dn-IκB. Balb-3T3 cells were cotransfected with 3XKB-luc reporter and 61E-LTR or vector pTZ19 plasmid as above. In addition, some wells were also cotransfected with 200 ng of dn-IκB as indicated. One set of LTR and LTR+dn-IκB transfected cells were treated with 1 μM cycloheximide. Cells from all transfected wells were harvested at 24 hr post transfection for luciferase assay. D. Effect of cycloheximide and dn-IκB on transcriptional activation of luciferase reporter gene by the LTR. Total cellular RNA from a similar set of transfected cells as in section C were subjected to RT-PCR analysis for luciferase and β-actin mRNA essentially as described in section B.
We designed another experiment to determine the role of new protein synthesis in transactivation by using dn-IκB, which strongly inhibits LTR-mediated activation of NFκB. We reasoned that since protein synthesis is necessary for dn-IκB mediated effect, dn-IκB should not be able to inhibit LTR-mediated luciferase RNA production in presence of cycloheximide. Result presented in Figure 5C shows that activation of the luciferase reporter activity by 61E-LTR, in presence of dn-IκB, was abolished both in presence and in absence of cycloheximide, as expected. RT-PCR experiment showed expression of dn-IκB completely inhibited LTR-mediated luciferase RNA production in the absence of cycloheximide. However, luciferase RNA production was detected in cycloheximide treated cells cotransfected with LTR and dn-IκB, as predicted (Figure 5D). This data demonstrated further that new protein synthesis is not necessary for transactivation. Further, complete inhibition of luciferase RNA production by the LTR in presence of dn-IκB also suggested against any mechanism by which LTR may act directly as a co-activator of gene transcription.
PKR−/− MEFs support activation of NFκB by the LTR
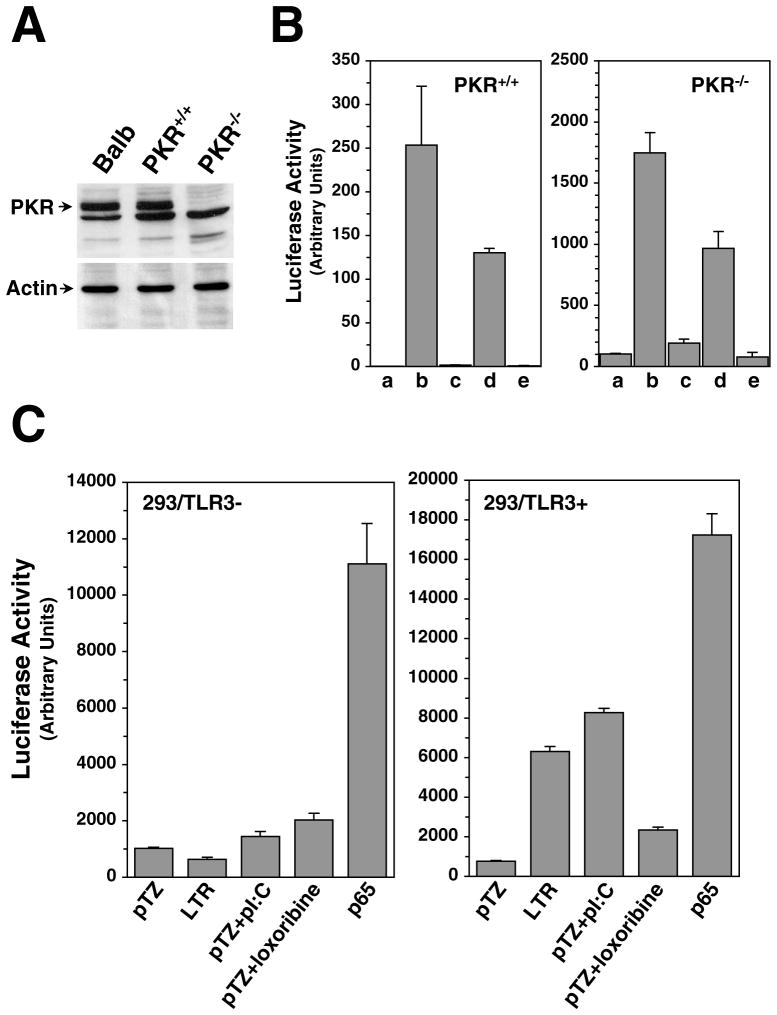
RNA folding analysis indicated that LTR transcript can assume stable stem-loop structure, making dsRNA dependent kinase PKR, a known activator of NFκB, a potential target in the LTR-mediated transactivation process. To investigate potential involvement of the PKR in this signaling cascade, we analyzed the activation of NFκB dependent reporter by the LTR in PKR−/− MEFs and compared to normal PKR+/+ MEFs and Balb-3T3 cells. We verified the status of PKR protein in these cells by western blot analysis of whole cell lysates. As expected, PKR−/− MEFs did not express PKR, whereas the other two cell lines did express the protein (Figure 6A). Next, we co-transfected these three cell lines with wild type or mutant U3-LTR (61E-LTR or EDD2), full-length virus (61E or 61E-Mut) or pTZ19 vector and 3XKB-Luc and measured luciferase activity in the cell lysates 48 hrs after transfection. Cells were also transfected with 100 ng of green florescence protein in separate wells to compare transfection efficiency. As shown in Figure 6B, even in the absence of PKR expression 61E-LTR was able to activate the NFκB reporter, and the level of induction (6–8 fold) was comparable to that seen in either PKR+/+ MEFs and Balb-3T3 cells (as shown in Figure 3). Comparison of GFP expression in these three cells showed that transfection efficiency in PKR−/− cells were 40–50%, compared to 25–30% in Balb-3T3 cells and 15–20% in PKR+/+ MEFs. Lower transfection efficiency in PKR+/+ MEFs compared to PKR−/− lines has also been reported in other studies (Elia et al., 2004). These results demonstrated that PKR is not necessary for NFκB activation by the LTR.
Figure 6. Role of PKR and TLR3 in the activation of NFκB by the LTR.
A. Analysis of PKR expression in the cell lines used in this study. Whole cell lysates (15 μg total protein) from actively growing Balb-3T3, PKR+/+ or PKR−/− MEFs were separated on 10% SDS-PAGE, and western immunoblotting analysis was performed using polyclonal anti-PKR serum. Lysates were also analyzed in separate SDS-PAGE for β-Actin using specific monoclonal antibody. B. Analysis of NFκB-dependent reporter gene activation in PKR−/− cells. PKR−/− and PKR+/+ cells were cotransfected by lipofectamine plus method with 100 ng 3XKB-luc and 300 ng of either wild type or mutant FeLV U3-LTR constructs (61E-LTR or EDD2, respectively), wild type or mutant full-length FeLV constructs (61E or 61E-Mut, respectively) or pTZ19 vector plasmid. Cells were harvested for luciferase assay 48 hrs after transfection. As a control, same set of transfection was also carried out on Balb-3T3 cells as in Figure 3B. C. Role of TLR3 in the activation of NFκB pathway by the LTR. TLR deficient regular HEK293 cells and HEK293 cells stably expressing mouse TLR3 (from Invivogen) were cotransfected by lipofectamine plus method with 100 ng of 3XKB-Luc reporter and 300 ng of pTZ19 vector, 61E-LTR or p65 expression plasmid as indicated. Synthetic dsRNA polyI:C (100 μg/ml) or loxoribine (1μM) were added to the media 8 hrs before harvesting the cells for luciferase assay at 24 hrs post-transfection.
TLR3 acts as an intermediate in LTR-mediated signaling
As an alternate mechanism of activation of RNA mediated signaling event, we looked into the possibility that LTR may activate TLR signaling. TLRs are integral part of cellular innate immune response, which are activated by various extracellular ligands of microbial origin including dsRNA, single stranded RNA (ssRNA), lipopolysaccharides, and CpG DNA (Kawai and Akira, 2005; Takeda, Kaisho, and Akira, 2003). Activation of these receptors initiates number of complex signaling cascade leading to activation of both NFκB and MAPK pathways. Of the eleven members of the TLR family identified to date, TLR3, TLR7 and TLR8 are activated by RNA molecule (Heil et al., 2004). However, dsRNA is the primary activator for TLR3 (Alexopoulou et al., 2001). Although, TLRs are primarily cell surface molecules, TLR3, −7, −8 and −9 are also retained in endocytic compartment (Heil et al., 2003). To test whether TLR3 signaling is activated by the LTR we tested HEK293 cells (which are negative for all TLR expression) and HEK293 cells stably expressing mouse-TLR3 for their ability to transactivation gene expression. As shown in Figure 6C, parental HEK293 cells did not support activation of NFκB dependent reporter expression by 61E-LTR but HEK293 that expresses m-TLR3 (HEK293/m-TLR3) strongly activated expression of the reporter. Transfection of p65 expression plasmid in both cells resulted in huge activation of the reporter in both cell types suggesting that the failure of reporter activation in HEK293 cells by the LTR is due to the absence of TLR3 and not because of transfection efficiency. That HEK293/m-TLR3 cells were really expressing m-TLR3 was verified by the fact that synthetic dsRNA polyI:C strongly activated NFκB reporter in these cells but not in HEK293 cells. Synthetic agonist for ssRNA-activated TLR7 and TLR8, loxoribine, however, failed to activate reporter similarly in either cell lines. These results strongly suggested that TLR3 mediated signaling is involved in LTR-mediated activation of NFκB dependent genes.
Ras plays a role in NFκB activation by the LTR
We have previously demonstrated that inhibitor of MEK1/2 abrogates collagenase IV gene transactivation by the LTR which suggested a necessary role of MAPK pathway in the process. To determine whether NFκB activation is required for upregulation of MAPK pathway by the LTR we tested the effect of expression of dn-IκB (which inhibits NFκB activation) on LTR mediated activation of collagenase IV and MCP-1 genes. In transient transfection assays dn-IκB was cotransfected with −517 coll-CAT or MCP-1-CAT and 61E-LTR in Balb-3T3 cells. As shown in Figure 7A, dn-IκB strongly inhibited activation of NFκB by the LTR as expected, but not of collagenase IV or MCP-1 gene expression. This data demonstrated that activation of NFκB is not necessary for collagenase IV or MCP-1 gene upregulation by the LTR.
Figure 7. Analysis of upstream signaling pathways leading to the activation of NFκB by the LTR.
A. Inhibition of NFκB activation does not affect collagenase IV and MCP-1 activation by the LTR. Balb-3T3 cells were cotransfected with −517coll-CAT, NFBCO-CAT or MCP-1-CAT together with pTZ19 vector or 61E-LTR and dn-IκB as described in Figure 1A. B. Analysis of role of Ras-Raf-MAPK pathway in NFκB activation. Balb-3T3 cells were cotransfected with 3XKB-luc and 61E-LTR or pTZ19 vector along with inhibitors as indicated. The dominant negative Ras (RasN17) and dnRaf-1 (Raf-BXB-K375W) constructs were used at 300 ng per well. The MEK1/2 inhibitor PD98059 (50 μM) was added to the cells in appropriate wells along with DNA-lipofectamine mixture.
Since upstream regulators of MAPK pathway such as Ras and Raf have been shown to activate NFκB signaling, we wanted to test if they have any role in the NFκB activation by the LTR. We tested functional inhibitors of various upstream regulators of Ras-Raf-MAPK pathway for their ability to abrogate LTR mediated NFκB activation. We cotransfected 61E-LTR and 3XKb-Luc reporter along with dominant negative Ras (N17-Ras) or dominant negative Raf-1 (dnRaf-1/Raf-BXB-K375W), or treated the cells with MEK1/2 inhibitor PD98059. As shown in Figure 7B, both N17-Ras and dnRaf-1 strongly inhibited LTR mediated activation of 3XKB-Luc reporter. On the other hand, PD98059 only had a modest effect on inhibiting 3XKB-Luc reporter. These data demonstrated that functional Ras and Raf-1 are necessary for NFκB activation by the LTR. However, inability of PD98059 to block such activity effectively suggested that LTR does not require active MAPK for activating NFκB dependent gene expression.
DISCUSSION
In this study we demonstrated that the U3-LTR region of FeLV activates NFκB-dependent gene expression. Previous studies have shown that insertion of unrelated sequences into the leukemia virus U3-LTR region leads to defect in preleukemic events in pathogenesis such as, hematopoietic hyperplasia and production of recombinant mink cell focus-inducing viruses but without affecting virus replication and spread (Brightman, Farmer, and Fan, 1993). This suggested that U3-LTR might influence target cell growth by some mechanism intrinsic to LTR. We have previously demonstrated that the U3-LTR region of Mo-MuLV and FeLV can transactivate expression of specific cellular genes and this activity correlated with the ability of the LTR to make specific RNA transcript (Ghosh and Faller, 1999; Ghosh, Roy-Burman, and Faller, 2000). In this study, we investigated whether this transactivating property of the LTR might function through activation of NFκB. Further, we focused on NFκB signaling pathway because it can be activated by dsRNA and could potentially mediate the induction of many of the host genes regulated by the LTR. Although NFκB is an important regulator of host immune response against variety of external stimuli, its role in various cancers has also been described. Reticuloendotheliosis virus encoded Rel protein, whose cellular counterpart c-Rel is a member of NFκB complex, causes lymphoid tumors in chickens (Chen, Wilhelmsen, and Temin, 1983; Wilhelmsen, Eggleton, and Temin, 1984). Further, the genes for c-Rel, NFκB2 (p100/52), and Bcl-3 proteins, all members of NFκB complex, are located within regions of chromosomes that are often involved in rearrangement or deletion (Fracchiolla et al., 1993; McKeithan et al., 1997; Rayet and Gelinas, 1999). NFκB is also known to activate genes that are involved in the control of cell growth and apoptosis. Antiapoptotic genes such as Bcl-2 family members (Bcl-XL and A1/Bfl-1), cellular inhibitors of apoptosis (c-IAP and IXAP) and TRAFs or genes such as cyclin D1 which promote cell cycle progression, can be directly activated by NFκB and thus facilitate tumorigenesis (Guttridge et al., 1999; Wang et al., 1999; Wang et al., 1998; Wu et al., 1998). In fact, many tumor cell lines express high level of NFκB in the nucleus and conversely, inhibition of NFκB expression in many transformed cells induces apoptosis (Sovak et al., 1997; Wang, Mayo, and Baldwin, 1996). Thus, our finding that the LTR alone can activate the NFκB pathway is significant and suggests that this activation could be a mechanism by which LTR may exert cell proliferative activity.
NFκB is activated by various external stimuli, including growth factor and mitogens, stress-inducing agents, cytokines, bacterial lipopolysaccharides and viral infection. The signaling pathways activated by these stimuli are diverse and include a variety of secondary signal transducer molecules such as TNF-α receptor associated factors (TRAFs), NFκB-inducing kinase (NIK), TLRs, TGF-β activated kinase (TAK1), MAPK kinase kinase 1 (MEKK1), protein kinase C, and PKR (Karin and Lin, 2002; Yamamoto and Gaynor, 2004). Most of these signaling molecules ultimately activate IKK. Although phosphorylation of IκB by IKK and its subsequent proteasomal degradation followed by nuclear translocation of dimeric NFκB is the mechanism of NFκB activation by most stimuli, two other atypical pathways have also been described. NFκB activation by hypoxia treatment or UV radiation does not involve IκBα phosphorylation at serine 32 and 36 (Imbert et al., 1996; Li and Karin, 1998). In our experiments the super-repressor dn-IκB completely abrogated LTR-mediated NFκB activation, indicating that IκBα phosphorylation at position 32 and 36 was required and no atypical method of activation was involved. Similarly, the ability of lactacystin to inhibit LTR-mediated transactivation, as shown in Figure 4, indicated that proteasomal activity is also an integral part of this transactivation process. Transactivation experiments with dominant-negative IKKs demonstrated that IKKβ is specifically required for NFκB activation by the LTR, linking it to the same pathway as that activated by proinflammatory cytokines (Li et al., 1999; Sizemore et al., 2002; Tanaka et al., 1999).
The central importance of NFκB activation in cellular physiology has made it an attractive target for viruses and many viruses have developed various strategies to alter the NFκB signaling pathway. In most of these strategies, virus-encoded proteins interact with different signaling molecules that ultimately activate IKK. For example, EBV latent membrane protein (LMP1), HCV core protein, rotavirus VP4 protein, HBV HBx protein, HTLV-1 tax protein, HIV gp120 and tat protein and influenza virus proteins can all activate IKK (Cahir-McFarland et al., 2000; Demarchi, Gutierrez, and Giacca, 1999; Flory et al., 2000; Flory et al., 1998; Harhaj and Sun, 1999; Kim et al., 2001; LaMonica et al., 2001; Sylla et al., 1998; You, Chen, and Lee, 1999). On the other hand, measles virus and vesicular stomatitis virus mediated activation of NFκB has been attributed to cellular PKR activation possibly by viral RNA molecules (Donze et al., 2004). A recent comprehensive review of various pathways to modulate NFκB activity employed by viruses is available (Santoro, Rossi, and Amici, 2003). We demonstrated in this report that the full-length wild type FeLV, but not the virus with a mutation in U3-LTR, can activate NFκB. We also demonstrated that activation of NFκB dependent gene expression by the LTR takes place in the absence of any new protein synthesis. We have previously demonstrated a causal relationship between transcript production and transactivational ability of the LTR (Ghosh, Roy-Burman, and Faller, 2000). Taken together, our data shows that NFκB activation by FeLV is a RNA-mediated event.
Several studies have implicated an essential role of PKR in the activation of NFκB by dsRNA. For example, dsRNA has been shown to activate NFκB binding to the β-interferon promoter in many different cell lines and the nucleoside analog 2-aminopurine (2-AP), a PKR inhibitor, blocked such activity (Visvanathan and Goodbourn, 1989). Further, selective degradation of PKR RNA in HeLa cells by antisense oligonucleotide also inhibited dsRNA mediated activation of NFκB (Maran et al., 1994). Synthetic dsRNA (pI:C) mediated activation of NFκB was also perturbed in PKR−/− cells, suggesting further the role of PKR in the process (Yang et al., 1995). In other experiments, IKKβ has been shown to be essential for PKR mediated signaling in response to dsRNA or vesicular stomatitis virus (Chu et al., 1999). Possible involvement of LTR transcript and requirement of IKKβ for NFκB activation in our study thus suggested that PKR could be involved in the process. However, by using PKR−/− MEFs we demonstrate here that LTR mediated activation of NFκB is independent of PKR activity. Two previous studies demonstrated that such PKR independent alternate dsRNA dependent NFκB activation pathways indeed exist. One such study demonstrated that dsRNA-mediated activation of β-IFN and inflammatory cytokine does not require PKR or RNase-L (Iordanov et al., 2001). In another study, both PKR-dependent and -independent pathways have been demonstrated for dsRNA-mediated activation of macrophages (Maggi et al., 2000). Furthermore, it has been shown that PKR is not required for dsRNA-induced NFκB activation in rat islets (Blair et al., 2001).
Several recent studies demonstrated that association of TLRs with their specific ligands activate both NFκB and MAPK pathways (Kawai and Akira, 2005; Wang et al., 2001). Various virus-associated molecular patterns such as, dsRNA, ssRNA, CpG DNA, and envelope glycoproteins have already been reported to be the targets of various TLRs (Boehme and Compton, 2004). Our demonstration that TLR3 is required for LTR mediated activation suggests that LTR transcript is another member of these targets. Most importantly, the necessity of TLR3 in NFκB activation by the LTR suggests that this could be the missing link between LTR transcript and cellular gene transactivation.
Since LTR also activates other cellular genes such as collagenase IV and MCP-1, and there are evidences that NFκB can modulate their expression (Takeshita et al., 1999; Ueda et al., 1997), it was possible that NFκB acts a mediator for their upregulation in LTR expressing cells. But we could not demonstrate any effect of dn-IκB on the LTR mediated expression of collagenase IV or MCP-1. However, our studies with dominant negative forms of MAPK pathway members showed that in fact, Ras and Raf-1 acts as upstream regulator of NFκB signaling by the LTR. Ras and Raf-1 have been previously shown to activate NFκB (Arsura et al., 2000; Baumann et al., 2000; Finco and Baldwin, 1993). Rat liver epithelial cells transformed with oncogenic Ras or Raf shows upregulation of NFκB and this activity could be inhibited by the expression of dn-IKK1 or dn-IKK2. Specifically, dn-IKK2 was more potent in inhibiting Raf-induced NFκB activation in Raf-transformed cells, whereas both Raf and PI3-kinase mediated pathways were involved in Ras transformed cells and both IKK1 and IKK2 were necessary for the latter. In our system RasN17, Raf-BXB-K375W and dn-IKK2 (but not dn-IKK1) inhibited LTR-mediated activation of NFκB. This suggested a Ras-Raf-IKK-IκB pathway of NFκB activation. Our finding of Ras as an upstream regulator of LTR mediated cellular gene transactivation is consistent with our previous finding which demonstrated that MEK1/2 inhibitor PD98059 inhibits activation of collagenase IV gene expression by the LTR (Ghosh and Faller, 1999). It is therefore conceivable that both NFκB and MAPK activation by the LTR acts in synergy in disease pathogenesis.
Data presented in this report also suggests a mechanism by which both NFκB as well as MAPK pathway can be activated following interaction of LTR transcript with TLR3. Such association could lead to activation of Ras as has been previously demonstrated for CpG DNA and TLR9 interaction (Yeo, Yoon, and Yi, 2003). Activated Ras subsequently initiates Ras-Raf-IKK-IκB signaling cascade that liberates NFκB, facilitating its translocation to the nucleus and ultimately enhances NFκB dependent gene expression. Activated Ras and Raf can also independently upregulate MAPK signaling pathway.
Materials and methods
Cells
Balb-3T3 and HEK293 cells were obtained from American Type Culture Collection and maintained in DMEM containing penicillin (100 U/ml) and streptomycin (100 μg/ml) with 10% donor calf serum or fetal calf serum, respectively, at 37°C in a humidified incubator under 5% CO2. Wild type and PKR-negative mouse embryo fibroblasts (MEF) (PKR+/+ and PKR−/−, respectively) were provided by B. Williams from the Cleveland Clinic Foundation and were maintained in DMEM containing 10% fetal calf serum and antibiotics. HEK293 cells stably expressing mouse TLR3 were purchased from Invivogen (San Diego, CA) and maintained in DMEM containing 10% fetal calf serum, penicillin, streptomycin and blasticidin (10 μg/ml).
Plasmids
Replication competent FeLV full-length clone 61E and U3-LTR construct 61E-LTR (which contain sequences from −307 to +34) have been described previously (Ghosh and Faller, 1999). Replication competent Mo-MuLV full-length clone Mov9 and U3-LTR construct GMNX (which contain sequences from −419 to −147) also have been described (Choi and Faller, 1995). EDD2 is a derivative of 61E-LTR with 8 base substitutions that abrogates it transactivational activity towards collagenase IV promoter. 61E-Mut is a full-length FeLV as 61E but has EDD2 specific mutations in both 5′- and 3′-LTR. Both EDD2 and 61E-Mut have been described before (Abujamra, Faller, and Ghosh, 2003). The eukaryotic expression plasmid for a “dominant negative” form of IκB (dn-IκB), also known as super-repressor of NFκB, was provided by M. Karin and contains serine to alanine mutation at positions 32 and 36. Dominant-negative forms of IκB kinases, IKKα (dn-IKK1) and IKKβ (dn-IKK2) were gifts from G. Sonenshein. Dominant-negative form of Ras (RasN17) and Raf-1 (Raf-BXB-K375W) were obtained from G. Denis and C. Chen, respectively. Reporter plasmids NFBCO-CAT, OBCO-CAT and 3XKB-Luc were from T. Gilmore and PBIIx-Luc from S. Ghosh. All other reporter constructs (−517/+62 coll-CAT, -543 JE-CAT, KbHN-MHC-CAT, and IL-6-CAT) have been described in our previous work (Ghosh and Faller, 1999). Plasmid pTZ19 is a 2.86 Kb cloning vector from United States Biochemicals.
Transfection and reporter assays
Actively growing cells were transfected with CsCl purified plasmids by either the DEAE-dextran method as described previously (Ghosh and Faller, 1999) or by Lipofectamine Plus reagent (Invitrogen) according to manufacturer’s protocol. For chloramphenicol acetyl transferase (CAT) assays, cells were harvested at indicated times and lysed by quick freezing and thawing three times. Equal amounts of protein from clarified supernatant fractions were then assayed for CAT activity using 14C-Chloramphenicol and acetyl-CoA, followed by chromatographic separation of acetylated 14C-chloramphenicol. For luciferase assays, cells were washed once in PBS and lysed on the plate using cell culture lysis reagent (Promega). To ensure complete lysis, harvested lysate was subjected to rapid freezing and thawing once. Clarified supernatant was then used for luciferase assay using assay reagents from Promega in a TD-20/20 luminometer (Turner Designs). Transfection efficiency was monitored by cotransfection of either 1 μg (for DEAE-dextran transfection) or 0.1 μg (for lipofectamine plus transfection) of an expression plasmid for green fluorescence protein (EGFP) from Stratagene. In some transfection experiments, Lactacystin (Clasto-Lactacystin β-lactone, Calbiochem), cycloheximide (Sigma Chemicals), PD98059 (Calbiochem), polyI:C (Sigma Chemicals) or Loxiribine (Invivogen) were used as described in the results section and figure legends. All transfection experiments were repeated at least three times and standard deviations were determined from these replicates.
RT-PCR
Analysis of requirement for new protein synthesis during gene transactivation was performed by RT-PCR. RNA was extracted from various transfected cells, untreated or treated with cycloheximide. Cells were washed once with PBS, harvested and total cellular RNA was extracted by Trizol extraction method according to manufacturer’s protocol (Invitrogen). All RNA preparations were digested with RQ1-RNase free DNase (Promega) at a concentration of 0.1 unit/μl, followed by phenol-chloroform extraction, ethanol precipitation and suspension in DEPC-treated water. DNase digestions were repeated to ensure complete removal of any contaminating DNA. First strand cDNA synthesis for luciferase and β-actin mRNA was performed on 2 μg of total RNA at 50°C using Thermoscript cDNA synthesis kit (Invitrogen). Antisense primers used for reverse transcription had following sequences: 5′-ATAAATGTCGTTCGCGGGCG-3′(luciferase) and 5′-CAAACATGATCTGGGTCATCTTCTC-3′(β-actin). One-twentieth fraction of the RT product was used in PCR reactions (30 sec denaturation at 94°C, 1 min annealing at 55°C, and 1 min extension at 72°C) using antisense primers as above and following sense primers: 5′-GCATAAGGCTATGAAGAGAT-3′(luciferase) and 5′-GCTCGTCGTCGACAACGGCTC-3′ (β-actin). RNA samples without any reverse transcription were used as a negative control in all PCR reactions. In order to maintain a linear range during amplification, PCR was carried out for only 25 cycles.
Nuclear Extracts
Cells were harvested by scraping, washed once with PBS and twice with hypotonic buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl) at 4°C. Cells were then allowed to swell in presence of 1 ml hypotonic buffer (per 107 cells) for 20 min on ice. Cells were harvested and resuspended in 0.4 ml hypotonic buffer containing 0.5 mM DTT, 0.5 mM PMSF, 1 μg/ml aprotinin and 0.2% NP-40 and left on ice for another 5 min. Cytoplasmic membrane was disrupted by vortexing vigorously for 30 sec and the nuclei were pelleted by high speed centrifuge at 4°C for 20 sec. Nuclei were suspended in high salt extraction buffer (20 mM HEPES, pH 7.9; 1.5 mM MgCl2; 420 mM NaCl; 0.5 mM DTT; 0.5 mM PMSF; 1 μg/ml aprotinin; 25% glycerol) and incubated for 30 min on ice for nuclear proteins to leach out. The nuclear extract was clarified by centrifugation in microfuge for 10 min at 4°C.
Immunoblotting
Whole cell extracts (WCE) were prepared in RIPA buffer (25 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.1% SDS; 1.0% sodium deoxycholate; 1.0% Triton X-100) containing proteinase inhibitor cocktail from Roche. Twenty micrograms of nuclear extract or WCE were separated on a 10% SDS-polyacrylamide gel and transferred electrophoretically to a nitrocellulose membrane. Membranes were blocked with 5% non-fat dry milk in PBS containing 0.05% Tween-20 and were incubated overnight with following antibodies as appropriate: rabbit polyclonal anti-p65/RelA (Santa Cruz sc-372), anti IκB-α (Santa Cruz sc-371), anti-PKR (Santa Cruz sc-708), mouse monoclonal anti-PCNA (Santa Cruz sc-56), and anti-β-actin (Oncogene CP01). Goat anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase (Oncogene) were used as secondary antibody. Bound antibodies were detected by Western Lightning Chemoluminescence kit (PerkinElmer Life Sciences). Prestained molecular weight protein markers (Benchmark marker, Invitrogen) were used to determine molecular weight of the detected bands.
Acknowledgments
We thank Patricia Kessler, Bryan Williams and Brunella Taddeo for providing PKR+/+ and PKR−/− MEFs. We also thank Michael Karin, Tom Gilmore, Gerald Denis, Changmin Chen, and Sankar Ghosh for plasmid constructs used in this study. This work was supported by a New Investigator Research Grant from Massachusetts Division of American Cancer Society (S.K.G.), an Institutional Research Grant IRG7200124 from the American Cancer Society (S.K.G.), and a National Institutes of Health grant CA079397 (D.V.F.). Partial support also came from Department of Defense grant W81XWH-04-1-0734 (S.K.G.).
References
- Abujamra AL, Faller DV, Ghosh SK. Mutations that abrogate transactivational activity of the feline leukemia virus long terminal repeat do not affect virus replication. Virology. 2003;309(2):294–305. doi: 10.1016/s0042-6822(03)00069-2. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Arsura M, Mercurio F, Oliver AL, Thorgeirsson SS, Sonenshein GE. Role of the IkappaB kinase complex in oncogenic Ras- and Raf-mediated transformation of rat liver epithelial cells. Mol Cell Biol. 2000;20(15):5381–91. doi: 10.1128/mcb.20.15.5381-5391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107(3):241–6. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B, Weber CK, Troppmair J, Whiteside S, Israel A, Rapp UR, Wirth T. Raf induces NF-kappaB by membrane shuttle kinase MEKK1, a signaling pathway critical for transformation. Proc Natl Acad Sci U S A. 2000;97(9):4615–20. doi: 10.1073/pnas.080583397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LA, Heitmeier MR, Scarim AL, Maggi LB, Jr, Corbett JA. Double-stranded RNA-dependent protein kinase is not required for double-stranded RNA-induced nitric oxide synthase expression or nuclear factor-kappaB activation by islets. Diabetes. 2001;50(2):283–90. doi: 10.2337/diabetes.50.2.283. [DOI] [PubMed] [Google Scholar]
- Boehme KW, Compton T. Innate sensing of viruses by toll-like receptors. J Virol. 2004;78(15):7867–73. doi: 10.1128/JVI.78.15.7867-7873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulares AH, Ferran MC, Lucas-Lenard J. NF-kappaB activation Is delayed in mouse L929 cells infected with interferon suppressing, but not inducing, vesicular stomatitis virus strains. Virology. 1996;218(1):71–80. doi: 10.1006/viro.1996.0167. [DOI] [PubMed] [Google Scholar]
- Brightman BK, Davis BR, Fan H. Preleukemic hematopoietic hyperplasia induced by Moloney murine leukemia virus is an indirect consequence of viral infection. Journal of Virology. 1990;64 (9):4582–4. doi: 10.1128/jvi.64.9.4582-4584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman BK, Farmer C, Fan H. Escape from in vivo restriction of Moloney mink cell focus-inducing viruses driven by the Mo+PyF101 long terminal repeat (LTR) by LTR alterations. Journal of Virology. 1993;67(12):7140–8. doi: 10.1128/jvi.67.12.7140-7148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman BK, Rein A, Trepp DJ, Fan H. An enhancer variant of Moloney murine leukemia virus defective in leukemogenesis does not generate detectable mink cell focus-inducing virus in vivo. Proc Natl Acad Sci U S A. 1991;88(6):2264–8. doi: 10.1073/pnas.88.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A. 2000;97(11):6055–60. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IS, Wilhelmsen KC, Temin HM. Structure and expression of c-rel, the cellular homolog to the oncogene of reticuloendotheliosis virus strain T. J Virol. 1983;45(1):104–13. doi: 10.1128/jvi.45.1.104-113.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Faller DV. A transcript from the long terminal repeats of a murine retrovirus associated with trans activation of cellular genes. Journal of Virology. 1995;69(11):7054–60. doi: 10.1128/jvi.69.11.7054-7060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11(6):721–31. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- Davis B, Linney E, Fan H. Suppression of leukaemia virus pathogenicity by polyoma virus enhancers. Nature. 1985;314(6011):550–3. doi: 10.1038/314550a0. [DOI] [PubMed] [Google Scholar]
- Davis BR, Brightman BK, Chandy KG, Fan H. Characterization of a preleukemic state induced by Moloney murine leukemia virus: evidence for two infection events during leukemogenesis. Proc Natl Acad Sci USA. 1987;84(14):4875–4879. doi: 10.1073/pnas.84.14.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi F, Gutierrez MI, Giacca M. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-kappaB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J Virol. 1999;73(8):7080–6. doi: 10.1128/jvi.73.8.7080-7086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L, Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. Journal of Virology. 1984;52:945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick LR, Cruikshank AA, Destree AT, Grenier L, McCormack TA, Melandri FD, Nunes SL, Palombella VJ, Parent LA, Plamondon L, Stein RL. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272(1):182–8. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- Donze O, Deng J, Curran J, Sladek R, Picard D, Sonenberg N. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. Embo J. 2004;23(3):564–71. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Vyas J, Laing KG, Clemens MJ. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein-Barr virus small RNA EBER-1. Eur J Biochem. 2004;271(10):1895–905. doi: 10.1111/j.1432-1033.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Faller DV, Weng H, Choi SY. Activation of collagenase IV gene expression and enzymatic activity by the Moloney murine leukemia virus long terminal repeat. Virology. 1997;227(2):331–42. doi: 10.1006/viro.1996.8345. [DOI] [PubMed] [Google Scholar]
- Faller DV, Weng H, Graves DT, Choi SY. Moloney murine leukemia virus long terminal repeat activates monocyte chemotactic protein-1 protein expression and chemotactic activity. Journal of Cellular Physiology. 1997;172(2):240–52. doi: 10.1002/(SICI)1097-4652(199708)172:2<240::AID-JCP11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Fan H. Influences of the long terminal repeats on retroviral pathogenicity. Seminars in Virology. 1990;1:164–174. [Google Scholar]
- Fan H. Leukemogenesis by Moloney murine leukemia virus: a multi-step process. Trends in Microbiology. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- Finco TS, Baldwin AS., Jr Kappa B site-dependent induction of gene expression by diverse inducers of nuclear factor kappa B requires Raf-1. J Biol Chem. 1993;268(24):17676–9. [PubMed] [Google Scholar]
- Flory E, Kunz M, Scheller C, Jassoy C, Stauber R, Rapp UR, Ludwig S. Influenza virus-induced NF-kappaB-dependent gene expression is mediated by overexpression of viral proteins and involves oxidative radicals and activation of IkappaB kinase. J Biol Chem. 2000;275(12):8307–14. doi: 10.1074/jbc.275.12.8307. [DOI] [PubMed] [Google Scholar]
- Flory E, Weber CK, Chen P, Hoffmeyer A, Jassoy C, Rapp UR. Plasma membrane-targeted Raf kinase activates NF-kappaB and human immunodeficiency virus type 1 replication in T lymphocytes. J Virol. 1998;72(4):2788–94. doi: 10.1128/jvi.72.4.2788-2794.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracchiolla NS, Lombardi L, Salina M, Migliazza A, Baldini L, Berti E, Cro L, Polli E, Maiolo AT, Neri A. Structural alterations of the NF-kappa B transcription factor lyt-10 in lymphoid malignancies. Oncogene. 1993;8(10):2839–45. [PubMed] [Google Scholar]
- Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276(32):30178–82. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Ghosh SK, Faller DV. Feline leukemia virus long terminal repeat activates collagenase IV expression through AP-1. Journal of Virology. 1999;73(6):4931–4940. doi: 10.1128/jvi.73.6.4931-4940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Roy-Burman P, Faller DV. Long Terminal Repeat Regions from Exogenous but not Endogenous Feline Leukemia Viruses Transactivate Cellular Gene Expression. Journal of Virology. 2000;74(20):9742–9748. doi: 10.1128/jvi.74.20.9742-9748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E, Li Y, Fredrickson TN, Hartley JW, Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. Journal of Virology. 1989;63(1):328–37. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19(8):5785–99. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Sun SC. IKKgamma serves as a docking subunit of the IkappaB kinase (IKK) and mediates interaction of IKK with the human T-cell leukemia virus Tax protein. J Biol Chem. 1999;274(33):22911–4. doi: 10.1074/jbc.274.33.22911. [DOI] [PubMed] [Google Scholar]
- Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290(5806):475–80. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Heil F, Ahmad-Nejad P, Hemmi H, Hochrein H, Ampenberger F, Gellert T, Dietrich H, Lipford G, Takeda K, Akira S, Wagner H, Bauer S. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol. 2003;33(11):2987–97. doi: 10.1002/eji.200324238. [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Helin E, Vainionpaa R, Hyypia T, Julkunen I, Matikainen S. Measles virus activates NF-kappa B and STAT transcription factors and production of IFN-alpha/beta and IL-6 in the human lung epithelial cell line A549. Virology. 2001;290(1):1–10. doi: 10.1006/viro.2001.1174. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Kwon H, Genin P. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J Clin Invest. 2001;107(2):143–51. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert V, Rupec RA, Livolsi A, Pahl HL, Traenckner EB, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle PA, Peyron JF. Tyrosine phosphorylation of I kappa B-alpha activates NF-kappa B without proteolytic degradation of I kappa B-alpha. Cell. 1996;86(5):787–98. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Wong J, Bell JC, Magun BE. Activation of NF-kappaB by double-stranded RNA (dsRNA) in the absence of protein kinase R and RNase L demonstrates the existence of two separate dsRNA-triggered antiviral programs. Mol Cell Biol. 2001;21(1):61–72. doi: 10.1128/MCB.21.1.61-72.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–63. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–7. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor downstream signaling. Arthritis Res Ther. 2005;7(1):12–9. doi: 10.1186/ar1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee YH, Won J, Yun Y. Through induction of juxtaposition and tyrosine kinase activity of Jak1, X-gene product of hepatitis B virus stimulates Ras and the transcriptional activation through AP-1, NF-kappaB, and SRE enhancers. Biochem Biophys Res Commun. 2001;286(5):886–94. doi: 10.1006/bbrc.2001.5496. [DOI] [PubMed] [Google Scholar]
- Koka P, van de Mark K, Faller DV. Trans-activation of genes encoding activation-associated human T lymphocyte surface proteins by murine retroviral sequences. Journal of Immunology. 1991;146(7):2417–25. [PubMed] [Google Scholar]
- Kopp E, Ghosh S. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 1994;265(5174):956–9. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- Kopp EB, Ghosh S. NF-kappa B and rel proteins in innate immunity. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- LaMonica R, Kocer SS, Nazarova J, Dowling W, Geimonen E, Shaw RD, Mackow ER. VP4 differentially regulates TRAF2 signaling, disengaging JNK activation while directing NF-kappa B to effect rotavirus-specific cellular responses. J Biol Chem. 2001;276(23):19889–96. doi: 10.1074/jbc.M100499200. [DOI] [PubMed] [Google Scholar]
- Lenz J, Celander D, Crowther RL, Patarca R, Perkins DW, Haseltine WA. Determination of the leukemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature. 1984;308:467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci U S A. 1998;95(22):13012–7. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J Exp Med. 1999;189(11):1839–45. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB, Jr, Heitmeier MR, Scheuner D, Kaufman RJ, Buller RM, Corbett JA. Potential role of PKR in double-stranded RNA-induced macrophage activation. Embo J. 2000;19 (14):3630–8. doi: 10.1093/emboj/19.14.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maran A, Maitra RK, Kumar A, Dong B, Xiao W, Li G, Williams BR, Torrence PF, Silverman RH. Blockage of NF-kappa B signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265(5173):789–92. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- McKeithan TW, Takimoto GS, Ohno H, Bjorling VS, Morgan R, Hecht BK, Dube I, Sandberg AA, Rowley JD. BCL3 rearrangements and t(14;19) in chronic lymphocytic leukemia and other B-cell malignancies: a molecular and cytogenetic study. Genes Chromosomes Cancer. 1997;20(1):64–72. [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278(5339):860–6. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Mitchell T, Sugden B. Stimulation of NF-kappa B-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69(5):2968–76. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HL, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. Journal of Virology. 1995;69(1):446–55. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Faller DV. Moloney murine leukemia virus activates NF-kappa B. Journal of Virology. 1996;70(6):4167–72. doi: 10.1128/jvi.70.6.4167-4172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18 (49):6938–47. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Gilmore TD. vRel is an inactive member of the Rel family of transcriptional activating proteins. J Virol. 1991;65(6):3122–30. doi: 10.1128/jvi.65.6.3122-3130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MG, Rossi A, Amici C. NF-kappaB and virus infection: who controls whom. Embo J. 2003;22(11):2552–60. doi: 10.1093/emboj/cdg267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G, Cuypers HT, Berns A. Proviral activation of the putative oncogene pim-1 in MuLV-induced T cell lymphomas. EMBO Journal. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short MK, Okenquist SA, Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. Journal of Virology. 1987;61(4):1067–72. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu Rev Cell Biol. 1994;10:405–55. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- Sizemore N, Lerner N, Dombrowski N, Sakurai H, Stark GR. Distinct roles of the Ikappa B kinase alpha and beta subunits in liberating nuclear factor kappa B (NF-kappa B) from Ikappa B and in phosphorylating the p65 subunit of NF-kappa B. J Biol Chem. 2002;277(6):3863–9. doi: 10.1074/jbc.M110572200. [DOI] [PubMed] [Google Scholar]
- Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100(12):2952–60. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla BS, Hung SC, Davidson DM, Hatzivassiliou E, Malinin NL, Wallach D, Gilmore TD, Kieff E, Mosialos G. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc Natl Acad Sci U S A. 1998;95(17):10106–11. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Takeshita H, Yoshizaki T, Miller WE, Sato H, Furukawa M, Pagano JS, Raab-Traub N. Matrix metalloproteinase 9 expression is induced by Epstein-Barr virus latent membrane protein 1 C-terminal activation regions 1 and 2. J Virol. 1999;73(7):5548–55. doi: 10.1128/jvi.73.7.5548-5555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, Goeddel DV. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity. 1999;10(4):421–9. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- Ueda A, Ishigatsubo Y, Okubo T, Yoshimura T. Transcriptional regulation of the human monocyte chemoattractant protein-1 gene. Cooperation of two NF-kappaB sites and NF-kappaB/Rel subunit specificity. J Biol Chem. 1997;272(49):31092–9. doi: 10.1074/jbc.272.49.31092. [DOI] [PubMed] [Google Scholar]
- Visvanathan KV, Goodbourn S. Double-stranded RNA activates binding of NF-kappa B to an inducible element in the human beta-interferon promoter. Embo J. 1989;8(4):1129–38. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Wang CY, Guttridge DC, Mayo MW, Baldwin AS., Jr NF-kappaB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19(9):5923–9. doi: 10.1128/mcb.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274(5288):784–7. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–3. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Eggleton K, Temin HM. Nucleic acid sequences of the oncogene v-rel in reticuloendotheliosis virus strain T and its cellular homolog, the proto-oncogene c-rel. J Virol. 1984;52(1):172–82. doi: 10.1128/jvi.52.1.172-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem Soc Trans. 1997;25(2):509–13. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- Williams BR. Signal integration via PKR. Sci STKE. 2001;(89):RE2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- Wilson LD, Flyer DC, Faller DV. Murine retroviruses control class I major histocompatibility antigen gene expression via a trans effect at the transcriptional level. Molecular and Cellular Biology. 1987;7(7):2406–15. doi: 10.1128/mcb.7.7.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MX, Ao Z, Prasad KV, Wu R, Schlossman SF. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science. 1998;281(5379):998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107(2):135–42. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Gaynor RB. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem Sci. 2004;29(2):72–9. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. Embo J. 1995;14(24):6095–106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SJ, Yoon JG, Yi AK. Myeloid differentiation factor 88-dependent post-transcriptional regulation of cyclooxygenase-2 expression by CpG DNA: tumor necrosis factor-alpha receptor-associated factor 6, a diverging point in the Toll-like receptor 9-signaling. J Biol Chem. 2003;278(42):40590–600. doi: 10.1074/jbc.M306280200. [DOI] [PubMed] [Google Scholar]
- You LR, Chen CM, Lee YH. Hepatitis C virus core protein enhances NF-kappaB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J Virol. 1999;73(2):1672–81. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]