Abstract
Cations play a large role in stabilizing the native state of RNA in vivo. In addition to Mg2+, putrescine2+ is an abundant divalent cation in bacterial cells, but its effect on the folding of RNA tertiary structure has not been widely explored. In this study, we look at how the stability of four structured RNAs, each with a different degree of dependence on K+ and Mg2+, are affected by putrescine2+ relative to Mg2+. Through the use of thermal melts we observe that (i) at a given concentration, putrescine2+ is less effective than Mg2+ at stabilizing RNA; (ii) the stability imparted to RNA by various diamines is a function of charge density (average separation distance between charges) as well as flexibility of the counterion; and (iii) when Mg2+ is already present in a buffer, further addition of putrescine2+ may either destabilize or stabilize RNA structure, depending on whether the native RNA does or does not chelate Mg2+ ion, respectively. At ion concentrations likely to be found in vivo, the effect of putrescine2+ on the free energy of folding an RNA tertiary structure is probably quite small compared to that of Mg2+, but the ability of mixed Mg2+ - putrescine2+ environments to (in effect) discriminate between different RNA architectures suggests that, in some cells, the evolution of functional RNA structures may have been influenced by the presence of putrescine2+.
Introduction
The ionic composition of the cytoplasm plays a large part in dictating the stability of native RNA structure. In addition to the inorganic ions found in cells (predominantly K+ and Mg2+) organic cations such as putrescine2+ (1,4-diaminobutane) are found in concentrations high enough to warrant attention. (As putrescine is fully protonated near neutral pH, it will be referred to here as putrescine2+ when its cationic properties are being discussed.) Putrescine2+ is the third most abundant cation in bacterial cells with a total concentration that ranges between 10 and 60 mM in E. coli, depending on the growth medium (1). These levels are significant given that the most abundant divalent ion (Mg2+) in the cell has a total concentration of ∼100 mM (2). Despite the importance of understanding putrescine2+-RNA interactions, relatively few studies have focused on this subject (3-5).
In this study, we seek to understand the significance of putrescine2+ ion in regard to RNA tertiary structure stability in vivo. We compare the efficacy of putrescine2+ and Mg2+ at stabilizing the tertiary structure of four different RNAs, chosen to be representative of the diversity of RNA folds found within a bacterial cell. The results are consistent in that Mg2+ stabilizes the native state of all four RNAs to a greater degree than the same concentration of putrescine2+. In the presence of Mg2+, the change in thermal stability upon addition of putrescine2+ is found to either increase or decrease depending on the fold of the RNA. These findings can be attributed to the inability of putrescine2+ to occupy Mg2+ chelation sites as well as a difference in charge density among different divalent cations. The unexpected ability of mixed Mg2+ - putrescine2+ solutions to stabilize or destabilize RNAs of different native architectures suggests that, in some cells, RNA structures have adapted to the presence of putrescine.
Materials and Methods
Chemicals and RNA
All solutions were prepared using water at 18.3 MΩ resistivity. All buffers and salts were ≥ 99.5% purity. MOPS buffer was obtained from Sigma, and brought to pH 6.8 with KOH (K•MOPS). The standard buffer was 40 mM K•MOPS (with 13 mM K+) and 10 μM EDTA (Sigma). KCl (Fluka) was added to give the indicated total concentrations of K+, e.g. 37 mM KCl to give 50 mM K+. MgCl2, and/or diamine were also added as indicated. (At pH 6.8, the diamines used in this study are all greater than 98% of the di-protonated species. . The relatively high concentration of K•MOPS was necessary to maintain pH 6.8 after additions of up to 10 mM of the HCl salts of the diamines used here.) Solutions of MgCl2 (Fluka) were standardized by titration into an EDTA solution (pH 8.0) of known concentration, while monitoring absorbance at 230 nm (6).
The HCl salts of cis-1,4-diaminobutene and trans-1,4-diaminobutene were synthesized as described (7), and the products recrystallized twice from 1:1 methanol-ether. Cis and trans products were obtained in 70% and 58% yield, respectively and dried over phosphorus pentoxide. Proton NMR spectra recorded on a Bruker 400 confirmed the expected products; <1.5% of the total proton signal originated from unassigned peaks. The first acid dissociation constant (pKa) of the 2HCl salts of putrescine, cis-1,4-diaminobutene and trans-1,4-diaminobutene were all determined by titration of the compound (at ∼ 20 mM in 12 ml H2O) with a standardized 100 mM KOH solution (25°C) while monitoring the pH with a calibrated Thermo Scientific, Ross combination 8102BN electrode.
All RNAs were prepared by transcription of linearized plasmid DNA with a histidine-tagged bacteriophage T7 RNA polymerase (8), as described (9). Transcription products were purified by preparative electrophoresis on denaturing, 9-12% polyacrylamide gels. The desired product band was excised from the gel, from which the RNA was electroeluted in an Elutrap Electrophoresis Chamber (Schleicher & Schuell). Centricon filter units (Millipore) were used to equilibrate an RNA in the desired buffer.
Vapor Pressure Osmometry
The osmolalities of putrescine•2HCl solutions were assessed by vapor pressure osmometry (VPO), using a Wescor VAPRO 5520 (Logan, UT) at ambient temperature. Solutions were prepared gravimetrically from dried, crystalline putrescine•2HCl and water. Osmolalities were measured three times on each sample and repeated four times with fresh stocks. Data were fit to a second order polynomial using Kaleidagraph, and compared with literature values for MgCl2 (Figure S4). Putrescine•2HCl clearly deviates more strongly from ideal behavior than does MgCl2 at concentrations greater than ∼100 mmolal. The RNA stability measurements reported here use a mixture of divalent ion(s) and KCl/K•MOPS, the latter monovalent salts usually at a total of 50 mM and in excess over the concentration of Cl− that accompanies the divalent ion. Under these conditions, the mean ionic activity coefficient of the divalent cation is largely determined by the concentration of the anion. A good approximation is that the MgCl2 mean ionic activity coefficient in experiments with 50 mM KCl is approximately the same as that of a MgCl2 solution of the same ionic strength (10). (At I = 50 mM, or ∼17 mM MgCl2, γ ≈ 0.66 (11).) In this region of Figure S4, the MgCl2 and putrescine•2HCl osmolalities are identical within experimental error, and the limiting slopes of the fitted osmolality curves are also approximately the same (2.53 and 2.58).
Although the slope of the osmolality vs. molality curve is related to the activity of the solute, it is necessary to carry out an integration to obtain the activity coefficient. The equation is ln γ2 = (φ − 1) + ∫ [(ϕ − 1) /ϕ]dm2, where m2 is the solute concentration, ϕ is the practical osmotic coefficient (ϕ = Osm/m2), γ2 is the mean ionic activity coefficient, and the integration is carried out from 0 to the m2 concentration of interest (12)(13). The (ϕ−1) term amplifies small errors at low molalities, and in addition the integrand approaches a non-zero value at zero solute concentrations. It is therefore necessary to have accurate measurements of osmolality at very low solute concentrations to obtain activities in the range of interest in these studies. VPO measurements are unable to quantify low solute osmolalities with sufficient accuracy, but we can estimate that about a three-fold lower osmolality for 25 mM putrescine•2HCl over 25 mM MgCl2 would be necessary to generate even a factor of two difference in mean ionic activity coefficient. On this basis, we have compared the effectiveness of Mg2+ and putrescine2+ in stabilizing RNA in terms of their concentration, without introducing any correction for activity differences.
UV melting curves and melt data analysis
Thermal denaturation of RNA samples was carried out with either a Cary 400 spectrophotometer equipped with a six cell thermostatted cuvette holder or an Aviv model 14DS spectrophotometer with a five cell thermostatted cuvette holder. Each instrument was able to reproduce the results of the other within error. UV absorbance was variously monitored at 260, 280 and 295 nm. The temperature schedule for the experiments was as follows: heat from room temperature to 65°C, hold at 65°C for 5 minutes, cool to less than 10°C, then heat to 95°C. Temperature changes in the second and third stages was set between 0.5 and 0.66 °C/min. No significant hysteresis was observed between the cooling and second heating stages; we conclude that renaturation artifacts or RNA degradation has not biased the curves below 65 °C. Absorbance data were plotted as the derivative of absorbance with respect to temperature. The program Global Melt Fit was used to fit all resulting data and estimate errors by a bootstrap method (14). Global fitting of data collected at 260 and 280 nm was used to extract values for both Tm (melting temperature) and ΔH°. The fitted enthalpies of a tertiary unfolding transition did not vary more than 7% over all the conditions tested; the average value was used in calculating ΔG°obs at temperature T0 from the formula
| (3) |
where T0, the reference temperature, is 25 °C.
Plots of ΔΔG°obs, the difference between ΔG°obs in the presence and absence of Mg2+ or putrescine, as a function of divalent ion concentration were fit to a Hill polynomial,
| (4) |
where ΔΔG°obs,max is the maximum ΔΔG°obs at high divalent ion concentration, [2+] is the molar concentration of either Mg2+ or putrescine2+, [2+]0 is the divalent ion concentration at the midpoint (half-maximal -ΔΔG°obs) of the curve, and n is the Hill exponent. Note this y-axis is free energy, instead of the more familiar fraction folded. The number of excess divalent ions taken up in the folding transition is given by the Wyman linkage relation
| (5) |
where R is the gas constant and T the temperature. The value of this derivative at the midpoint of the ΔΔG°obs curve is related to the exponent n by
| (6) |
A similar equation is derived in ref. (15). We emphasize that ΔΓ2+ calculated from eq (6) applies only at the midpoint of the ΔΔG°obs curve; ΔΓ2+ approaches zero at low and high divalent ion concentrations.
Results
RNA Selection
To survey the effects of putrescine on the stability of RNA tertiary structure, four RNAs that differ in their interactions with inorganic ions were selected for study (Figures 1 and S1). In this work, we distinguish “chelated” ions from all other cations interacting with an RNA, which we group together as the “ion atmosphere”. Chelated ions make at least two direct contacts with RNA electronegative groups, and are thus partially dehydrated; they are found in pockets of high negative electrostatic potential (16, 17). The ion atmosphere encompasses both fully hydrated ions at a distance from the RNA surface, and ions whose first-shell waters of hydration may be perturbed by the RNA surface (18).
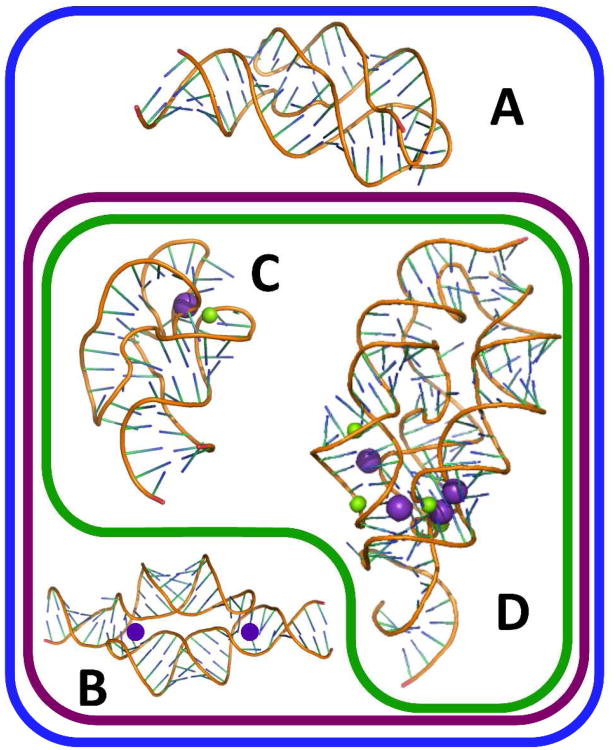
Figure 1.
Venn diagram depicting the RNA structures analyzed in this study and how they interact with cations. Blue outline contains population of RNAs stabilized by cations residing in the ion atmosphere. Purple outline contains RNAs that, in addition, have chelated K+ ions (purple spheres). RNAs within the green outline also have chelated Mg2+ ions (green spheres). A) A-riboswitch, PDB ID: 3G4M. B) TLR RNA, PDB ID:2I7E; (K+ ions have been added as discussed in text). C) 58mer rRNA, PDB ID: 1HC8. D) M-box riboswitch, PDB ID: 2QBZ. See Figure S1 for sequences used in this study.
The aptamer domain of the add adenine riboswitch (A-riboswitch, Figure S1A) was chosen as representative of a structured RNA exclusively stabilized by the ion atmosphere. The A-riboswitch folds into its native structure in the absence of divalent ions upon addition of adenine (or adenine derivatives) to the solution (19, 20). Although the RNA shows increased stability in the presence of Mg2+ (20, 21), crystallographic studies do not resolve any chelated Mg2+ or K+ ions (22, 23). In addition, studies on the monovalent ion dependence of A-riboswitch concluded that this RNA shows no monovalent ion selectivity that might suggest K+ chelation (9).
The homodimeric tetraloop receptor RNA (TLR RNA, Figure S1B) incorporates a motif first resolved in the Tetrahymena group I intron (24-26). This motif is known to chelate a K+ ion (27), and is more stable in the presence of K+ than other group I monovalent ions (9). When the TLR RNA dimerizes, two helical segments come into parallel alignment, a common arrangement in large RNAs. Because the negatively charged helices are brought fairly close together, there is a strong monovalent salt dependence to dimerization; however, Mg2+ is not essential for dimerization (9).
An rRNA fragment (58mer RNA, Figure S1C) and the aptamer domain of the M-box riboswitch (M-box RNA, Figure S1D) were chosen as examples of RNAs that chelate Mg2+. Structural studies of the 58mer RNA have concluded that both K+ and Mg2+ are chelated in close proximity within the native fold of the 58mer (28, 29). This RNA obtains maximum stability in the presence of K+ relative to other group I monovalent ions (30). In addition, electrostatic calculations concluded that a pocket of high negative potential on the RNA is capable of providing enough free energy to partially dehydrate and bind a Mg2+ ion (17). The M-box riboswitch is a sensor of the effective Mg2+ concentration in vivo(31). It is the largest of the RNAs used in this study (160 base pairs), and has a compact core that chelates at least three Mg2+ and three K+ ions.
The two Mg2+-chelating RNAs are distinct in their reliance on Mg2+ for folding to the native state. In the absence of a divalent ion, the 58mer requires 1.6 M NH4Cl (32), and the M-box RNA does not fold even in 2.1 M NaCl (31). In the following sections, the two “Mg2+- chelator” RNAs will be discussed separately from the other two RNAs, which fold to their native structures in modest concentrations of KCl.
Comparison of A-riboswitch and TLR RNA stabilities with putrescine2+ vs. Mg2+
RNA stabilities were assessed using thermal melts monitored by UV absorbance (see Materials & Methods). Conditions are readily found under which both the A-riboswitch and TLR RNA fold to their native structures in the absence of divalent ions. The A-riboswitch folds in the presence of a tight-binding ligand (20 μM 2,6-diaminopurine, DAP) and 50 mM K+; a higher monovalent salt concentration (300 mM K+) is needed to observe the TLR RNA dimer. For the A-riboswitch, a distinctive unfolding transition is only observed in the presence of ligand and corresponds to the loss of tertiary structure (20). The transition shifts to higher Tm upon addition of either putrescine2+ or Mg2+ (Figure S2A). Dimerization of the TLR RNA is accompanied by a small but distinctive transition that is slightly hyperchromic at 260 nm and hypochromic at 280 nm; its Tm is also concentration-dependent as expected for a dimerization reaction (9). This transition also shifts to higher temperature in response to either Mg2+ or putrescine2+ (Figure S2B, S2C). Melting curves obtained under these buffer conditions can thus be analyzed to obtain standard free energies for tertiary structure folding, ΔG°obs, in the presence or absence of divalent ion (see Materials and Methods).
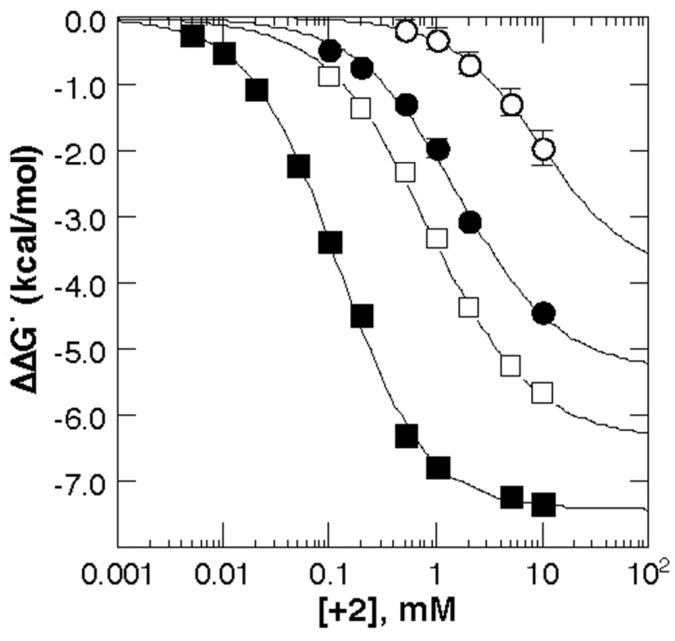
Figure 2 plots the increment in folding free energy due to the divalent ion:
Figure 2.
The increase in stability (ΔΔG°obs, eq 1) upon titration of A-riboswitch (squares) or TLR RNA (circles) with chloride salts of Mg2+ (closed) or putrescine2+ (open). Tetraloop receptor data were collected in K•MOPS - EDTA buffer (see Materials and Methods) with 300 mM K+ while the A-riboswitch data were collected in the same buffer with 50 mM K+ and 20 μM DAP. Either MgCl2 or putrescine•2HCl was added to the concentration indicated by the abscissa, [2+].
| (1) |
where ΔG°obs,ref is the folding free energy in the reference buffer with only monovalent ions, and ΔG°obs,2+ the folding free energy with a given concentration of divalent ion. There is a striking difference in the effectiveness of the two ions; it takes 6 – 8 fold higher putrescine2+ concentration to achieve the same stability increase as with Mg2+.
To contrast the effectiveness of the two different ions in thermodynamic terms, the comparison should be made between RNA stabilities at the same ion activities, rather than ion concentrations. The activity of MgCl2 over a range of concentrations is known (11, 33), and the mean ionic activity coefficient of millilmolar concentrations of Mg2+ in the presence of 50 mM KCl can be estimated as ∼0.66 (10). To our knowledge, there are no similar measurements of putrescine•2HCl activity. We carried out vapor pressure osmometry experiments to address this question (see Materials and Methods, Figure S4). Although it is difficult to measure putrescine osmolalities at low concentrations accurately, we argue from the data that MgCl2 and putrescine•2HCl differ in activity by much less than a factor of two at the concentrations used in our experiments. We conclude that a difference in thermodynamic activity of Mg2+ and putrescine2+ cannot account for the large difference in concentrations needed to stabilize these two RNAs, and that it is reasonable to use a concentration scale when comparing RNA folding free energies measured with these two ions.
Besides the difference in the ability of the two divalent ions to stabilize RNA, another interesting feature of Figure 2 is the difference in uptake of divalent ion that accompanies the folding reaction. We express the number of “excess” divalent ions per RNA that are present in order to neutralize part of the RNA negative charge as ΓMg2+ (for Mg2+) or Γput2+ (for putrescine2+). Upon folding from a partially structured RNA to a more compact native conformation, Γ2+ invariably increases (20, 34). Under the conditions of excess K+ employed here, ΔΓ2+ evaluated at the divalent ion concentration required to fold half of the RNA molecules is identical to the Hill exponent n obtained by fitting the Hill equation to a plot of fraction RNA folded vs. ln[Mg2+] (20). In Figure 2, the slope d(ΔΔG°obs)/d(ln[Mg2+], evaluated at the midpoint of the ΔΔG°obs curve, is related to the same parameter (see Materials and Methods, eq 5). For both RNAs, ΔΓMg2+ is larger than ΔΓput2+, though the large error in ΔΓput2+ for the TLR RNA means the difference is less significant. (The error is from uncertainty in the maximum value of -ΔΔG°obs; data cannot be collected at higher putrescine concentrations without biasing the measurement with a large excess of Cl− ions (20).) ΔΓ2+ is not independent of divalent ion concentration, so a comparison between ΔΓMg2+ and ΔΓput2+ could be misleading because of the large difference in ion concentrations at the midpoints of the two curves. The expectation is that ΔΓ2+ increases with the divalent ion concentration (20), but we find that the ion at higher concentration, putrescine2+, has the smaller value of ΔΓ2+. The difference in ion uptake therefore appears to be a real difference in the behaviors of the ions.
Finally, it appears that putrescine2+ does not stabilize either the A-riboswitch or TLR RNA to the same ΔΔG°obs,max as Mg2+. As noted above, there is a large uncertainty in the maximum stability of the TLR RNA with putrescine2+, but the difference in the A-riboswitch stabilities with the two ions is significant (-1.06 ± 0.20 kcal/mol, see Table 1). Since ΔΓMg2+ and ΔΓput2+ are different for this RNA, it is not surprising that ΔΔG°obs,max is also different. Whether the difference stems from a stronger interaction of Mg2+ with the native state, a stronger interaction of putrescine2+ with partially unfolded RNA conformations, or both those factors, cannot be decided from the present experiments. Ideas about the relation between charge density of ions and their interactions with RNA are taken up in the Discussion.
Table 1. RNA stabilization by divalent ionsa.
| RNA | divalent ion | pKab | [2+]0, mM | ΔΔG°obs, max, kcal/mol | ΔΓ2+, ions/RNA |
|---|---|---|---|---|---|
| A-riboswitch | Mg2+ | 0.104 ± 0.006 | −7.30 ± 0.15 | 3.31 ± 0.20 | |
| A-riboswitch | putrescine | 9.36 (9.35) | 0.87 ± 0.57 | −6.36 ± 0.14 | 2.39 ± 0.11 |
| TLR RNA | Mg2+ | 0.1.62 ± 0.31 | −5.33 ± 0.43 | 2.06 ± 0.30 | |
| TLR RNA | putrescine | 9.80 ± 3.21 | −3.88 ± 0.99 | 1.66 ± 0.47 | |
| A-riboswitch | cadaverine | (10.05) | 1.30 ±0.15 | −5.83 ±0.26 | 2.16 ± 0.17 |
| A-riboswitch | 1,3 diaminopropane | (8.6) | 0.42 ± 0.2 | −6.99 ± 0.12 | 2.58 ± 0.13 |
| A-riboswitch | cis-1,4 diaminobutene | 8.78 | 0.65 ± 0.06 | −7.19 ± 0.26 | 2.41 ± 0.21 |
| A-riboswitch | trans-1,4 diaminobutene | 8.93 | 0.89 ±0.04 | −7.09 ± 0.10 | 2.36 ± 0.07 |
A-riboswitch stability with various diamines
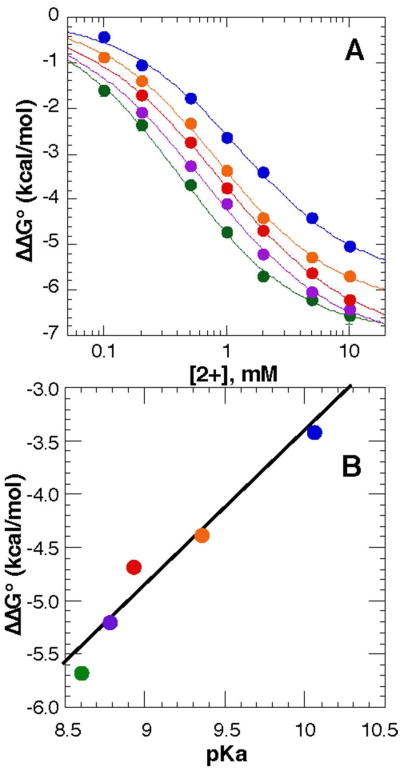
To better understand how putrescine2+ interacts with RNA, several diamines with different carbon linkers were used to ask whether RNA stability depends on either the average distance between amines or the rigidity of the linker. ΔΓput2+ weakly varies between ∼2.2 and 2.4 with different diamines, and is largest with the shortest alkyl linker (Table 1). The midpoint of the titration curve varies more significantly, by about a factor of three (Figure 3A). To ask whether stabilization free energy imparted by a given diamine is correlated with the average intramolecular distance between amine groups in solution, the first acid dissociation constant (pKa) for each diamine was obtained (Table 1). Deprotonation of an amine is energetically less costly if a second positive charge is nearby. Based on Coulomb's law, the first pKa is inversely proportional to the distance between the two charges. A plot of ΔΔG°obs at a specific diamine concentration (2 mM) vs. the first pKa of the cation results in an approximately linear trend (Figure 3B).
Figure 3.
Stability of the A-riboswitch in the presence of various diamines, as derived from thermal stability measurements. A, ΔΔG°obs of the A-riboswitch vs. concentration of cadaverine (blue), putrescine (orange), trans-1,4-diaminobutene (red), cis-1,4-diaminobutene (purple) and 1,3-diaminopropane (green). B, stability of the A-riboswitch at 2 mM concentration of the diamines listed above vs the first acid dissociation constant (pKa, Table 1) of the diamine. The slope of the least squares fit to all the data points (black line) is 1.45 kcal/mol (R=0.975).
A-riboswitch and TLR RNA stability in mixed Mg2+ - putrescine buffers
As shown in Figure 2, Mg2+ stabilizes the A-riboswitch and TLR RNA more effectively than putrescine2+. Under in vivo conditions, though, RNA is stabilized by a mixture of several different counter ions. In order to assess how mixtures of putrescine2+ and Mg2+ influence the stability of RNA, we conducted thermal melts on our set of four RNAs with varying concentrations of the two divalent ions. To help interpret the results, we note that an excess of cations and an exclusion of anions balance the charge of anionic phosphates on the RNA, as required by charge neutrality. Thus, any change in ΔΓput2+ (for instance) must be compensated by a change in the excess of other ionic species in solution (15, 34):
| (2) |
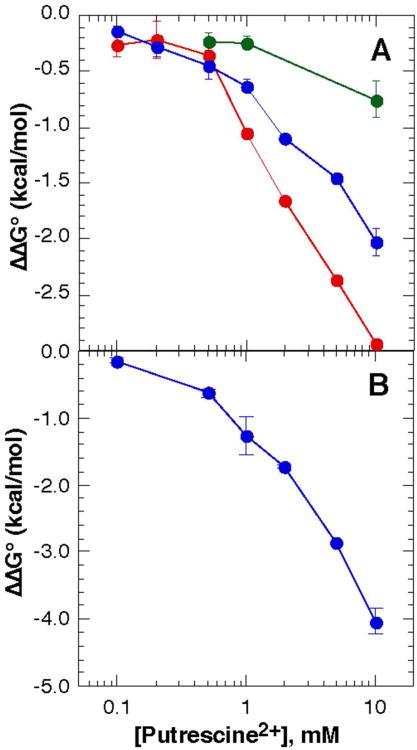
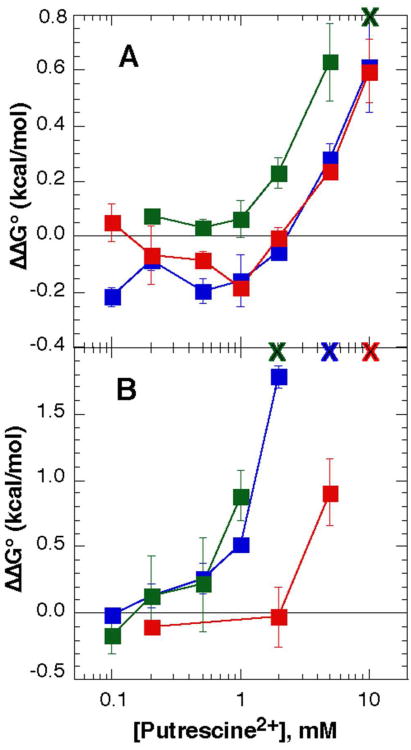
Figure 4A shows the change in A-riboswitch stability vs. concentration of putrescine2+, with the Mg2+ concentration kept constant at the indicated values. The negative slopes of these curves indicate a net uptake of putrescine2+ ions upon folding the RNA to its native state (see Materials and Methods, eq 5). As the Mg2+ concentration increases from 0. 1mM to 0.5 mM the resulting change in free energy upon addition of putrescine2+ becomes less negative, though the maximum uptake (ΔΓput2+ ≈ 1.4 ions/RNA at 2 mM putrescine2+ and 0.1 mM Mg2+, Figure 4A) and -ΔΔG°obs at a given putrescine2+ concentration are never as great as seen with putrescine2+ alone (Figure 2). Thus, the uptake of putrescine2+ is opposed by increased Mg2+ concentrations, as might be expected on the basis of eq (2). The release of Mg2+ can be estimated from the dependence of ΔΔG°obs on Mg2+ concentration at a constant putrescine2+ concentration (eq 1). At 2 mM putrescine and 0.1 mM Mg2+, ΔΓ2+ ≈ -1.4 (Figure 4A). Although it appears that ΔΓMg2+ and ΔΓput2+ are equal and opposite, these estimates are not accurate enough to rule out small accompanying changes in ΔΓK+ and ΔΓCl−.
Figure 4.
Effect of putrescine2+ on the stability of RNAs without Mg2+ chelation sites. ΔΔG°obs for both RNAs was measured in K•MOPS - EDTA buffer with 50 mM K+ and 0.1 mM (red), 0.2 mM (blue) or 0.5 mM (green) MgCl2. 20 μM DAP was included with the A-riboswitch. A, A-riboswitch; B, TLR RNA.
The TLR RNA also shows a net stabilization upon addition of putrescine2+ to a Mg2+-containing buffer (Figure 4B). The trend in ΔΔG°obs is qualitatively similar to the A-riboswitch, although the magnitude of the stabilization is about twice as large for the TLR RNA. (In 0.2 mM MgCl2 and 10 mM putrescine2+, the TLR RNA obtains a maximum stability of -4.03 kcal/mol ± 0.19 kcal/mol, versus −2.03 kcal/mol ± 0.12 kcal/mol for the A-riboswitch.)
Stability of Mg2+ -chelator RNAs in mixed Mg2+ - putrescine buffers
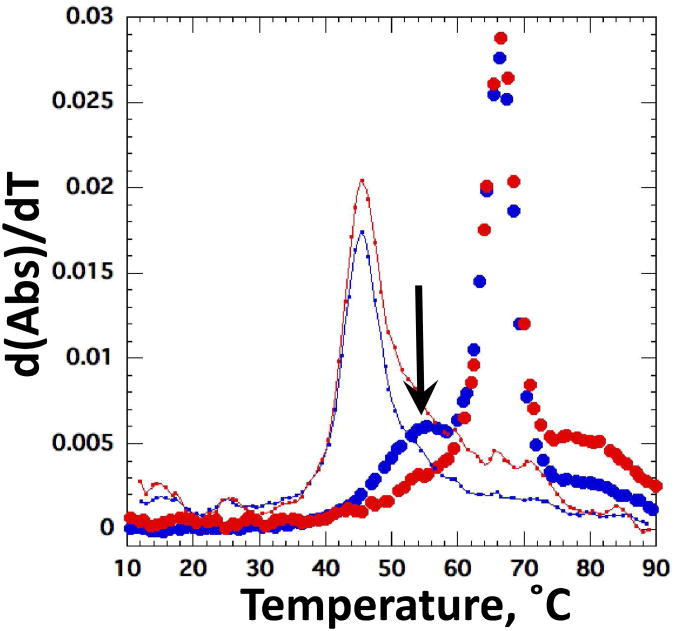
Melting curves of both Mg2+-chelator RNAs show the appearance of a new transition with a distinctive ratio of hyperchromicities at 260 and 280 nm when ∼1 mM MgCl2 is included in the buffer, in addition to 50-60 mM KCl (35) (Figure 5 and S3 D-F). This transition is not observed when putrescine2+ is substituted for Mg2+ at concentrations up to 10 mM; apparently putrescine2+ cannot provide enough free energy of stabilization at the accessible concentrations. When the two Mg2+-chelating RNAs are first induced to fold by Mg2+ and then titrated with putrescine2+, the stability decreases (Figure 6 A, B). The 58mer RNA is destabilized only by putrescine2+ concentrations in excess of ∼2 mM (0.5 to 2.0 mM MgCl2), but the M-box riboswitch is destabilized by >1.8 kcal/mol at 2 mM putrescine2+ and ≤ 1 mM Mg2+. Tertiary structure is undetectable in either RNA at a high enough ratio of putrescine2+ to Mg2+ ions. This behavior is the opposite from our observations with the A-riboswitch and TLR RNA in mixed Mg2+ - putrescine2+ buffers (Figure 4).
Figure 5.
Melting profile of the M-box riboswitch. Data are plotted as the derivative of absorbance with respect to temperature at 260 nm (blue) and 280 nm (red). Closed circles correspond to data collected in buffer with 50 mM K+, 1 mM putrescine2+ and 1 mM Mg2+, in K•MOPS – EDTA buffer. Solid lines correspond to data collected in the same buffer with 50 mM K+ and no divalent ions. Arrow indicates new transition that appears in the presence of Mg2+.
Figure 6.
Stability of the magnesium-chelating RNAs in varying concentrations of putrescine2+. A, stability of the 58mer rRNA fragment relative to stability in buffer with 50 mM K+ and 0.5 mM (green), 1.0 mM (blue), or 2.0 mM (red) MgCl2. The green “X” at the top of the graph represents the concentration of putrescine2+ (with 0.5 mM Mg2+) at which the tertiary structure can no longer be detected in the melting curves. B, stability of the M-box RNA relative to buffer with 50 mM K+ and 0.75 mM (green), 1.0 mM (blue), or 2.0 mM (red) MgCl2. An “X” at the top of graph represents the concentration of putrescine where the tertiary structure is no longer observed with 0.75 mM (green), 1 mM (blue), or 2 mM (red) Mg2+.
Discussion
In vivo relevance of putrescine to RNA folding
In vivo, the stability of an RNA structure may be affected by several different solution components, such as ions, osmolytes (36), and other macromolecules that act as crowding agents (37). Of the three most abundant cations in the bacterial cell, K+, Mg2+ and putrescine2+, relatively little is known about the effects of putrescine2+ on RNA stability. In this work, we try to gain some insight into the in vivo relevance of putrescine2+ to RNA folding by asking what change the ion causes in folding free energy (ΔΔG°obs) and what degree of ion uptake is associated with structure formation (ΔΓput2+) for four different RNA tertiary structures.
The experiments reported here show that putrescine2+ is, in general, far less effective than Mg2+ at stabilizing RNA tertiary structure: two of the RNAs required more than five-fold higher concentrations of putrescine2+ than Mg2+ to reach the same stability (Figure 3), and two RNAs could not be induced to fold to the native structure in buffers with only putrescine2+ and monovalent ions. Putrescine2+ has also been found to be a very weak stabilizer of tRNA tertiary structure, compared to Mg2+ (4). The question of whether these weak effects may influence RNA stability in vivo is difficult to answer definitively. We first consider the range of ion activities that are likely to be found in bacteria in vivo, and then discuss the potential consequences for RNA tertiary folding.
The in vivo activity of Mg2+ is difficult to measure, but several kinds of measurements suggest it is equivalent to that of ∼1.0 mM Mg2+ in the presence of 150 mM KCl, (38-40). (The total cellular concentration of Mg2+ is about 100 fold higher (2), but because the ion interacts so strongly with cellular polynucleotides the effective concentration, or activity, is much lower.) The measurements in the present work were made in a background of 50 to 300 mM K+, with most of the anion as Cl−. At 50 mM KCl, the concentration of Mg2+ needed to reproduce the estimated in vivo activity is ∼0.30 mM. (This calculation is based on MgCl2 and KCl activity coefficients and the principle that the mean ionic activity coefficient of a low concentration of Mg2+ in 50 mM KCl is the same as the mean ionic activity coefficient of Mg2+ in a MgCl2 solution of the same ionic strength as 50 mM KCl, i.e. 17 mM MgCl2(10).) These are rough estimates, but nevertheless, the four RNAs studied here are all significantly stabilized by submillimolar Mg2+ concentrations that are probably comparable to in vivo activities. The M-box RNA is in fact a regulatory switch that turns on or off expression of a Mg2+ transporter gene in response to changes in Mg2+ concentration in vivo, so this RNA might be expected to fold or unfold near physiological Mg2+ activity (31, 41).
We are not aware of any estimates of in vivo putrescine2+ activity. If we take the in vivo total concentration under standard growth conditions to be 60 mM (1), and the same ∼100:1 ratio of total to effective concentration as Mg2+, the activity would be the equivalent of ∼0.2 mM putrescine2+ in 50 mM KCl (or 0.6 mM at 150 mM KCl). However, the apparent affinity of putrescine2+ for helical DNA is 2-3 fold weaker than that of Mg2+ (3), and interactions with RNA may be even weaker, as suggested by the experiments reported here. The ratio of total to effective concentration may therefore be much smaller than 100:1, and the effective concentration in vivo somewhat higher than that of Mg2+. We consider 0.2 –1.0 mM putrescine2+ (50 mM KCl) a reasonable range for reproducing in vivo conditions.
Given the above considerations, it seems likely that, in the context of the bacterial cell, putrescine2+ makes a small but significant contribution to the overall stability of an RNA. For instance, 0.2 to 1.0 mM putrescine2+ stabilizes the A-riboswitch by -1.4 to -3.3 kcal/mol. Though these values drop by a factor of five in the presence of 0.2 mM Mg2+, even a –0.5 kcal/mol free energy gain over Mg2+ alone is enough to shift a folding equilibrium. The M-box RNA is a particularly interesting case, in which putrescine2+ destabilizes the RNA by nearly 2 kcal/mol when present at only twice the Mg2+ concentration (Figure 6B). For a sensor RNA that is tuned to respond to intracellular fluctuations in Mg2+ activity, this free energy change is quite significant. Noting that putrescine2+ concentrations may vary over at least an eight fold range as the osmolality of the growth medium changes (1), there is the intriguing possibility that the responses of the M-box RNA or other riboswitches are modulated to some extent by intracellular putrescine2+ activity.
A caveat about the distribution of putrescine in nature is necessary. Microorganisms such as bacteria and molds contain high concentrations of putrescine2+ (42), and E. coli requires polyamines for growth under anaerobic conditions (43). However, mammalian cells cells typically contain putrescine2+ in approximately 2-3 orders of magnitude lower concentration (42), which are unlikely to affect the folding or stability of any RNAs. The two riboswitches selected for this study are found exclusively in bacteria; consequently their structures may have evolved in response to the high bacterial levels of putrescine. The other two RNAs represent a motif and a structure that are found in all organisms, but it is still possible that their stability is modulated by putrescine in bacteria, and that the specific sequences utilized by the organism or other factors (such as a bound protein, (44)) compensate.
RNA stabilization by divalent ions of the “ion atmosphere”
A question raised by this and other studies of putrescine2+ with RNA (4, 5) is why putrescine2+ is less effective at stabilizing RNA, in comparison to Mg2+; this study additionally poses the question of why putrescine2+ destabilizes some RNAs when Mg2+ is present. For the purposes of the following discussion, it is convenient to use the distinction (see Results) between chelated ions that directly contact two or more RNA groups, and the ion atmosphere, defined here as the remaining cations that interact with an RNA at various distances from the RNA surface. Putrescine2+, or any other cation, is capable of favorable interaction with the RNA as a part of the ion atmosphere, but putrescine2+ is very unlikely to substitute for an inorganic ion at a chelation site. (We know of no RNA that specifically binds putrescine2+ in a way analogous to the chelation of an inorganic ion, though recognition of the amino-butyl side chain of lysine by the lysine riboswitch suggests such a site is feasible (45, 46).) We consider the ion atmosphere first and then discuss the possible effects of putrescine2+ on Mg2+ chelation sites.
Ion atmosphere
The A-riboswitch and TLR RNAs were selected as examples of RNAs for which Mg2+-induced stabilization is entirely by RNA interactions with the ion atmosphere; as detailed in Results, there is no evidence for Mg2+ chelation sites in either RNA. Our conclusions for these two RNAs are that (i) Mg2+ is always more stabilizing than putrescine2+ (Figure 2); (ii) ΔΓ2+ is perhaps slightly less for putrescine2+ than for Mg2+; and (iii) the stabilizing effect of Mg2+ and putrescine2+ together, while not additive, is always as least as strong as that of either of the ions alone (cf. Figures 2 and 4). Thus, with respect to the ion atmosphere, putrescine2+ apparently substitutes to some extent for Mg2+, but is not as effective. An argument has been made that ions with higher charge density (defined as charge divided by Van der Waals volume) are more effective stabilizers of native RNA structures because they may approach the RNA surface (and maximum negative electrostatic potential) more closely, and because of smaller excluded volume effects when ions accumulate in high concentration near the RNA (5). Our results with Mg2+ and putrescine2+ are consistent with this idea.
To further explore the way charge density might affect the interactions of organic ions with RNA, we compared the stability of the A-riboswitch with five different organic diamino compounds. The results (Figure 3 and Table 1) show distinct dependences on the length and saturation of the carbon chain separating the two charges. The first pKa of these ions varies over a range of ∼1.5 pH units; it is a measure of the reciprocal distance between the two charges. To a good approximation, the stabilization imparted by the ion is proportional to a linear charge density, i.e. the charge divided by the average distance of separation (Figure 3B). However, an additional factor to consider is that the linear charge density of an organic ion may be influenced by its environment. In the diamino alkane compounds, the average distance of separation between two amino groups could decrease as the ion approaches the RNA surface, if the decrease in entropy is sufficiently compensated by the simultaneous placement of the two amines in a volume of high negative electrostatic potential. In fact, 1,4 trans-diaminobutene, which is locked in a more extended conformation than either putrescine or 1,4 cis-diaminobutene, imparts 0.45 kcal/mol less negative ΔΔG°obs than expected based on the correlation between pKa and ΔΔG°obs for the diamino-alkanes. Thus, both the flexibility and length of carbon chains may be factors in determining how effectively organic ions (including spermine and spermidine) interact with compact RNAs.
Chelation sites
RNAs that contain Mg2+ chelation sites tend to be strongly dependent on Mg2+ to fold into their native structures, presumably because it is too costly to create the high electrostatic potential of an ion binding pocket if there is no cation capable of occupying the site (17, 47, 48). Steric constraints prevent putrescine2+ from replacing Mg2+ in a chelation site, so it is unsurprising that putrescine2+ (plus monovalent cation) are unable to stabilize the native state of either the 58mer or M-box RNAs. It is less obvious why addition of putrescine2+ should destabilize an RNA that has been driven into its native conformation by Mg2+ (Figure 6). The observation suggests that putrescine2+ weakens the affinity of Mg2+ for chelation sites, but there cannot be direct competition between the ions for occupancy of the site. To rationalize this observation, we turn to a calculation of the repulsive interactions between chelated Mg2+ and the ion atmosphere (17, 48). Occupancy of a chelation site by Mg2+ effectively neutralizes two negative charges of the RNA backbone and results, at high enough Mg2+ concentration, in the displacement of an approximately equivalent positive charge from among the divalent cations of the ion atmosphere. This displacement of favorably-interacting cations is an apparent repulsive (positive free energy) between site-bound Mg2+ and the ion atmosphere. As Mg2+ is titrated, the actual free energy (rather than the standard state free energy) of Mg2+-RNA interactions at the chelated site and in the ion atmosphere change in different ways (cf. Figure 2 of ref. (48)):
The free energy of site-bound Mg2+, ΔGsite, decreases linearly (becomes more negative) with the logarithm of the Mg2+ activity;
The repulsive interaction between the site-bound ion and the ion atmosphere, ΔGIA-S, increases continuously with log[Mg2+].
For 1 mM Mg2+ interacting with the 58mer RNA (60 mM KCl), ΔGsite is estimated from experiments to be −4.5 kcal/mol, which is far larger than the calculated repulsive term (+1.2 kcal/mol under the same conditions (48)). We suggest that the contribution of putrescine2+ to a site-binding term must be nearly zero, as the ion cannot effectively take advantage of the small chelation site. However the second, repulsive term between Mg2+ in the chelation site and putrescine2+ in the ion atmosphere will persist and increase with increasing putrescine2+ concentration. It is this “repulsive” free energy that could reduce the free energy gain from Mg2+ chelation and cause a net destabilization of the RNA at high putrescine2+ concentrations.
Putrescine2+ vs. Mg2+ in vivo
In summary, in comparison to Mg2+, putrescine2+ is most likely a minor contributor to the overall stability of an RNA tertiary structure in vivo. However, putrescine2+ has the curious property of being able to tip the thermodynamic scales towards either folded or unfolded forms of an RNA, depending on the architecture of the specific RNA. This observation leads us to hypothesize that the evolution of functional RNA structures has been influenced by the presence of putrescine2+ in the cellular ionic environment.
Supplementary Material
Acknowledgments
We thank members of the Townsend lab for help in synthesizing both cis and trans-1,4-diaminobutene, and Ryan Hulscher and Andrew Buller for helpful comments on the manuscript.
This work was supported by NIH grants RO1 GM58545 and T32 GM008403 (Program in Molecular Biophysics), and the Francis D. Carlson Fellowship (to RJT).
Abbreviation
- DAP
2,6 diamino-purine
Footnotes
Supporting Information Available: Supporting information contains four figures showing the primary and secondary structures of the four RNAs used in this study, three additional melting curves for each RNA, and vapor pressure osmometry results for putrescine•2HCl. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Capp MW, Cayley DS, Zhang W, Guttman HJ, Melcher SE, Saecker RM, Anderson CF, Record MT., Jr Compensating effects of opposing changes in putrescine (2+) and K+ concentrations on lac repressor-lac operator binding: in vitro thermodynamic analysis and in vivo relevance. J Mol Biol. 1996;258:25–36. doi: 10.1006/jmbi.1996.0231. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn A, Kellenberger E. Productive phage infection in Escherichia coli with reduced internal levels of the major cations. J Bacteriol. 1985;163:906–912. doi: 10.1128/jb.163.3.906-912.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunlin WH, Strick TJ, Record MT., Jr Equilibrium dialysis studies of polyamine binding to DNA. Biopolymers. 1982;21:1301–1314. doi: 10.1002/bip.360210704. [DOI] [PubMed] [Google Scholar]
- 4.Heerschap A, Walters JALI, Hilbers CW. Interactions of some naturally occurring cations with phenylalanine and initiator tRNA from yeast as reflected by their thermal stability. Biophys Chem. 1985;22:205–217. doi: 10.1016/0301-4622(85)80044-2. [DOI] [PubMed] [Google Scholar]
- 5.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge Density of Divalent Metal Cations Determines RNA Stability. J Am Chem Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grilley D, Soto AM, Draper DE. Direct quantitation of Mg2+-RNA interactions by use of a fluorescent dye. Methods Enzymol. 2009;455:71–94. doi: 10.1016/S0076-6879(08)04203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin B, Posseme F, Le Barbier C, Carreaux F, Carboni B, Seiler N, Moulinoux JP, Delcros JG. (Z)-1,4-diamino-2-butene as a vector of boron, fluorine, or iodine for cancer therapy and imaging: synthesis and biological evaluation. Bioorg Med Chem. 2002;10:2863–2871. doi: 10.1016/s0968-0896(02)00147-5. [DOI] [PubMed] [Google Scholar]
- 8.He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 9.Lambert D, Leipply D, Shiman R, Draper DE. The influence of monovalent cation size on the stability of RNA tertiary structures. J Mol Biol. 2009;390:791–804. doi: 10.1016/j.jmb.2009.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harned HS, Robinson RA. Multicomponent Electrolyte Solutions. Vol. 15. Pergamon Press; Oxford: 1968. [Google Scholar]
- 11.Kumbhar RR, Dagade DH, Terdale SS, Patil KJ. Thermodynamic Equilibrium Constant Studies on Aqueous Electrolytic (Alkaline Earth Chlorides) Solutions Containing 18-Crown-6 at 298.15K. J Solution Chem. 2007;36:259–273. [Google Scholar]
- 12.Levine IN. Physical Chemistry. 5th. McGraw Hill; 2002. [Google Scholar]
- 13.Robinson RA, Stokes RH. Electrolyte Solutions. Second Revised. Dover Publications; Minoeola, NY: 2002. [Google Scholar]
- 14.Draper DE, Bukhman YV, Gluick TC. Thermal Methods for the Analysis of RNA Folding Pathways. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. section 11.13. John Wiley & Sons; New York: 2000. p. 13. [DOI] [PubMed] [Google Scholar]
- 15.Leipply D, Lambert D, Draper DE. Ion-RNA interactions thermodynamic analysis of the effects of mono- and divalent ions on RNA conformational equilibria. Methods Enzymol. 2009;469:433–463. doi: 10.1016/S0076-6879(09)69021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 17.Misra VK, Draper DE. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci U S A. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 19.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 23.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 26.Zuo X, Wang J, Foster TR, Schwieters CD, Tiede DM, Butcher SE, Wang YX. Global molecular structure and interfaces: refining an RNA:RNA complex structure using solution X-ray scattering data. J Am Chem Soc. 2008;130:3292–3293. doi: 10.1021/ja7114508. [DOI] [PubMed] [Google Scholar]
- 27.Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferré-D'Amare AR, Strobel SA, Doudna JA. A Specific Monovalent Metal Ion Integral to the A-A Platform of the RNA Tetraloop Receptor. Nature Struc Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- 28.Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 29.Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE. A compact RNA tertiary structure contains a buried backbone-K+ complex. J Mol Biol. 2002;318:963–973. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- 30.Shiman R, Draper DE. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol. 2000;302:79–91. doi: 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- 31.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 32.Bukhman YV, Draper DE. Affinities and Selectivities of Divalent Cation Binding Sites Within an RNA Tertiary Structure. J Mol Biol. 1997;273:1020–1031. doi: 10.1006/jmbi.1997.1383. [DOI] [PubMed] [Google Scholar]
- 33.Robinson RA, Stokes RH. Electrolyte Solutions. 2nd. Butterworth & Co. Ltd; London: 1970. [Google Scholar]
- 34.Grilley D, Soto AM, Draper DE. Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc Natl Acad Sci U S A. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M, Draper DE. Bases defining an ammonium and magnesium ion-dependent tertiary structure within the large subunit ribosomal RNA. J Mol Biol. 1994;244:572–585. doi: 10.1006/jmbi.1994.1753. [DOI] [PubMed] [Google Scholar]
- 36.Lambert D, Draper DE. Effects of Osmolytes on RNA Secondary and Tertiary Structure Stabilities and RNA-Mg2+ Interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 39.London RE. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu Rev Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- 40.Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 42.Tabor CW, Tabor H. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- 43.Chattopadhyay MK, Tabor CW, Tabor H. Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J Bacteriol. 2009;191:5549–5552. doi: 10.1128/JB.00381-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeder C, Conn GL, Draper DE. Optimization of a ribosomal structural domain by natural selection. Biochemistry. 2006;45:6635–6643. doi: 10.1021/bi052544p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J Am Chem Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leipply D, Draper DE. Evidence for a thermodynamically distinct Mg(2+) ion associated with formation of an RNA tertiary structure. J Am Chem Soc. 2011;133:13397–13405. doi: 10.1021/ja2020923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sober HA. CRC Handbook of Biochemistry. CRC Press; Cleveland, OH: 1968. [Google Scholar]
- 50.Dawson RM, Elliot DC, Elliot WH, Jones KM. Data for Biochemical Research. Oxford Sciences Publications; Oxford: 1986. [Google Scholar]
- 51.Perrin DD. Dissociation Constants of Organic Bases in Aqueous Solution (Supplement) Butterworths; London: 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.