Most loss-of-function single mutants show no visible phenotype differences compared with the wild type. This work shows that mutations in duplicate transcription factor genes of the root epidermis elicit dramatic changes in transcript profiles despite the lack of a morphological defect, illustrating the importance of in-depth phenotypic analyses and the complexity of duplicate gene diversification.
Abstract
Traditional genetic analysis relies on mutants with observable phenotypes. Mutants lacking visible abnormalities may nevertheless exhibit molecular differences useful for defining gene function. To examine this, we analyzed tissue-specific transcript profiles from Arabidopsis thaliana transcription factor gene mutants with known roles in root epidermis development, but lacking a single-gene mutant phenotype due to genetic redundancy. We discovered substantial transcriptional changes in each mutant, preferentially affecting root epidermal genes in a manner consistent with the known double mutant effects. Furthermore, comparing transcript profiles of single and double mutants, we observed remarkable variation in the sensitivity of target genes to the loss of one or both paralogous genes, including preferential effects on specific branches of the epidermal gene network, likely reflecting the pathways of paralog subfunctionalization during evolution. In addition, we analyzed the root epidermal transcriptome of the transparent testa glabra2 mutant to clarify its role in the network. These findings provide insight into the molecular basis of genetic redundancy and duplicate gene diversification at the level of a specific gene regulatory network, and they demonstrate the usefulness of tissue-specific transcript profiling to define gene function in mutants lacking informative visible changes in phenotype.
INTRODUCTION
The analysis of gene function using mutant individuals is a powerful approach for understanding biological phenomena. However, this approach relies on the availability of mutants that exhibit observable differences in phenotype. Unfortunately, the majority of single-gene mutants lack detectable alterations, due to genetic redundancy, other compensatory mechanisms, or difficulties in phenotypic assessment, which limits the usefulness of a genetic approach for defining gene function (Pickett and Meeks-Wagner, 1995; Bouché and Bouchez, 2001; Hanada et al., 2009a; Pérez-Pérez et al., 2009; Lloyd and Meinke, 2012).
The development of large-scale assays for transcript and metabolite accumulation in specific organs, tissues, and environments provides the opportunity for detailed characterization of plant phenotypes. Indeed, genome-wide transcriptome analyses have uncovered locus-associated gene expression variation between different strains with normal phenotypes, indicating an unappreciated degree of molecular variation (Kliebenstein et al., 2006; Keurentjes et al., 2007; Fu et al., 2009; Hanada et al., 2009b; Terpstra et al., 2010; Brady et al., 2011). The observation of molecular differences in plants with similar outward appearance implies that single-gene mutants lacking visible phenotypes may nevertheless exhibit molecular alterations useful for defining gene function.
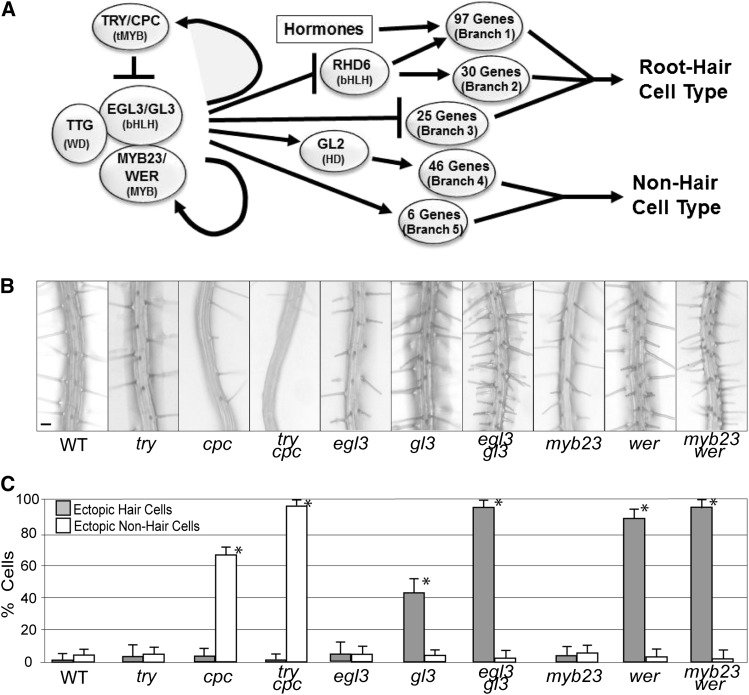
The gene regulatory network controlling the differentiation of root epidermal cells in Arabidopsis thaliana provides a model for studying the roles of duplicate genes in a specific gene network. The specification of the two cell types in the root epidermis, root hair cells, and non-hair cells is controlled by a suite of positive and negative transcriptional regulators (Schiefelbein et al., 2009; Tominaga-Wada et al., 2011; Bruex et al., 2012; Grebe, 2012). Among these are three sets of transcription factors encoded by presumed paralogous genes, including one-repeat Myb type proteins TRIPTYCHON (TRY) and CAPRICE (CPC) that promote the hair cell fate, basic helix-loop-helix proteins ENHANCER OF GLABRA3 (EGL3) and GLABRA3 (GL3) that promote the non-hair fate, and R2R3-Myb-type proteins MYB23 and WEREWOLF (WER) that promote the non-hair fate (Figure 1). The WER/MYB23 and EGL3/GL3 proteins appear to participate in a multimeric transcriptional regulatory complex with the TTG WD-repeat protein to induce non-hair cell differentiation, as well as expression of the TRY/CPC one-repeat Mybs (Lee and Schiefelbein, 1999; Ryu et al., 2005; Wang et al., 2008). The family of one-repeat Mybs engages in a lateral inhibition mechanism by moving from cell to cell and hindering formation of the MYB23/WER-EGL3/GL3-TTG complex, which permits expression of hair cell differentiation genes (Wada et al., 1997; Schellmann et al., 2002; Kurata et al., 2005; Tominaga et al., 2007; Wang et al., 2007, 2008; Song et al., 2011). The relative abundance of the MYB23/WER-EGL3/GL3-TTG transcriptional complex in the presumptive hair cells and non-hair cells of the developing root epidermis appears to be controlled by cell position–dependent SCRAMBLED receptor kinase signaling (Kwak et al., 2005; Kwak and Schiefelbein, 2008; Savage et al., 2008).
Figure 1.
Genes Controlling Epidermal Cell Fate in the Arabidopsis Root.
(A) Simplified depiction of the gene regulatory network for Arabidopsis root epidermal cell differentiation. The TRY/CPC, EGL3/GL3, and MYB23/WER transcriptional regulators influence the expression of downstream genes in five distinct branches (Branches 1 to 5), based on their GL2, RHD6, and/or hormone dependence (for details, see Bruex et al., 2012).
(B) Root phenotype of single and double mutants affecting the try/cpc, egl3/gl3, or myb23/wer loci. WT, the wild type. Bar = 50 µm.
(C) Quantitative analysis of cell type formation in single and double mutants, showing mean proportion of cells in the N position that differentiate as hair cells (ectopic hair cells) and cells in the H position that differentiate as non-hair cells (ectopic non-hair cells). Bars indicate ± sd for three independent trials. Asterisks indicate significant deviation from the wild type (P < 0.05).
The availability of three pairs of paralogous genes with defined roles in the root epidermal system provided an opportunity to analyze the molecular basis of redundancy and duplicate gene function at the level of a specific gene regulatory network. In particular, we reasoned that tissue-specific transcript profiling of single and double mutants in the root epidermal system might enable a comparative quantitative analysis of gene expression changes across the network. Our results show that even mutants lacking a visible phenotype can possess significant alterations in tissue-specific transcript profiles useful for defining gene function and that high-resolution transcriptome analysis of single and double mutants provides insight into the relative functional roles of paralogous genes in a specific gene network.
RESULTS
The try, egl3, and myb23 Mutants Alter Root Epidermis Gene Expression
As a foundation for this study, we quantified the cell-type specification in the root epidermis of each of the single and double mutants affecting the TRY-CPC, EGL3-GL3, and MYB23-WER gene sets. For each of these paralogous gene pairs, homozygous recessive presumed loss-of-function mutations in one member (try, egl3, or myb23) cause no visible alteration in root epidermis development using standard analysis methods, mutations in the other members (cpc, gl3, or wer) cause significant cell specification defects, and the double mutant combinations (try cpc, egl3 gl3, or myb23 wer) have extreme phenotypes, essentially lacking one of the two cell types (Figures 1B and 1C; see Supplemental Table 1 online), consistent with previously published data (Lee and Schiefelbein, 1999; Schellmann et al., 2002; Bernhardt et al., 2003, 2005; Simon et al., 2007; Kang et al., 2009; Tominaga-Wada et al., 2012).
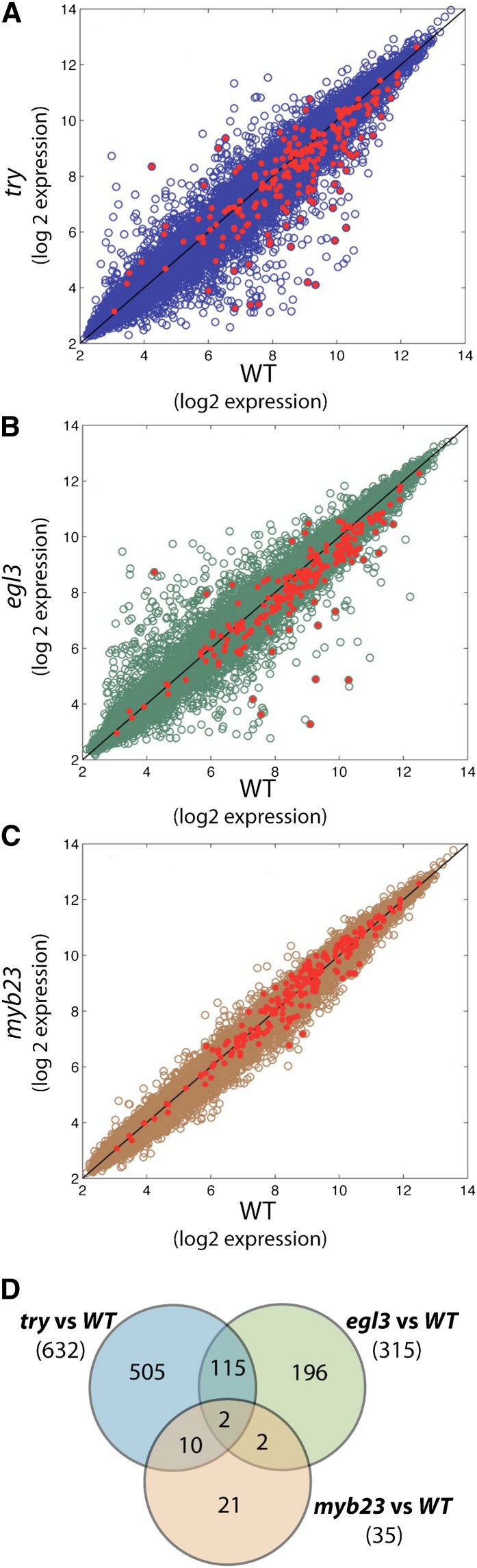
To determine whether the single mutants lacking a visible phenotype (try, egl3, and myb23) exhibit transcriptional changes associated with root epidermis cell differentiation, an epidermal green fluorescent protein (GFP) marker, WER:GFP (Lee and Schiefelbein, 1999), was introduced into each mutant by crossing to enable fluorescence-activated cell sorting (FACS)–based enrichment for cells from the developing root epidermis tissue followed by gene expression assays using ATH1 microarray chips (Birnbaum et al., 2005; Bruex et al., 2012). Comparing each single mutant to the wild type (three biological replicates/line), we discovered a substantial degree of differential gene expression (Figures 2A to 2C; see Supplemental Data Set 1 and Supplemental Table 2 online). The differentially expressed gene sets (try versus the wild type, 632 genes; egl3 versus the wild type, 315 genes; myb23 versus the wild type, 35 genes; using fold change [FC] > 2.0, significance analysis of microarrays [SAM; Tusher et al., 2001]; t-statistic > 2.5, q-value < 0.05) exhibited significant overrepresentation for genes known to be preferentially expressed in differentiating root hair cells (a 487 COBL9:GFP-sorted gene set [Brady et al., 2007]; P values: 7.1E-16 [try], 9.4E-7 [egl3], and 0.04 [myb23]) and genes known to be differentially expressed in the root epidermis of hairy versus hairless mutants (a 208-member gene set, designated the “core root epidermal gene set” [Bruex et al., 2012]; P values: 2.0E-23 [try], 4.7E-7 [egl3], and 0.29 [myb23]). A significant number of these differentially expressed genes was affected in more than one of the mutants (Figure 2D; P value < 0.002 for each two-way and three-way comparison), and the overlapping set of genes are overrepresented for members of the core root epidermal gene set (P value < 0.001; see Supplemental Data Set 1 online). Moreover, as a group, the core root epidermal gene set exhibited significantly greater deviation in transcript accumulation (relative to the wild type) than the set of all other genes (Figures 2A to 2C; P values: 1.2E-17 [try], 3.9E-8 [egl3], and 1.1E-5 [myb23]). These data show that mutations of root epidermis regulatory genes lacking a morphological effect nevertheless elicit transcript changes linked to root epidermis development.
Figure 2.
Genome-Wide Root Epidermal Gene Expression in the try, egl3, and myb23 Mutants Relative to the Wild Type.
(A) to (C) Gene expression is plotted on a log2 scale for each gene transcript represented on the Affy ATH1 chip. In each plot, the red-colored dots indicate the 208 genes of the core root epidermal gene set (Bruex et al., 2012). WT, the wild type.
(D) Overlap in differentially expressed genes between the try versus the wild type, egl3 versus the wild type, and myb23 versus wild-type sets.
Differential Effects of Paralogous Regulatory Gene Mutants on the Root Epidermal Network
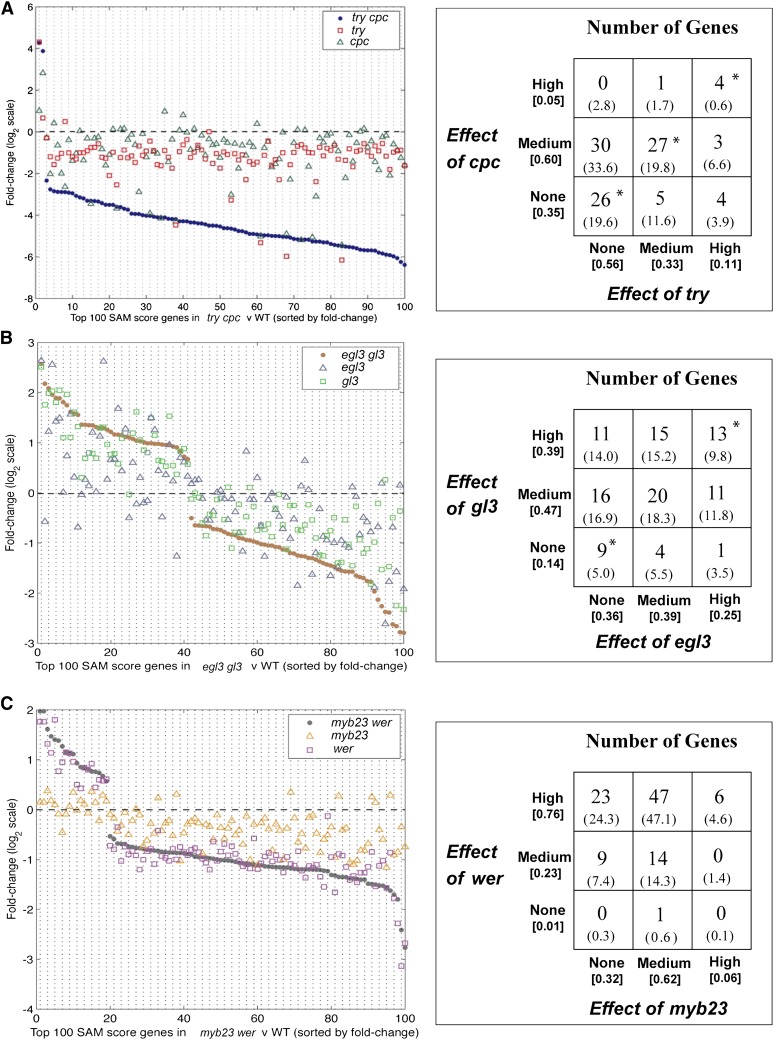
To evaluate the molecular effect of the try, egl3, and myb23 mutations in relation to their corresponding double mutants (try cpc, egl3 gl3, and myb23 wer), we compared our single mutant transcriptome data to the WER:GFP cell-sorted transcript profiles from double mutants (Bruex et al., 2012), focusing on the 100 genes in each double mutant that exhibit the greatest differential expression relative to the wild type (SAM-derived; see Supplemental Data Set 2 online). In general, each single mutant tended to alter transcript accumulation of these genes in the same direction (i.e., increase or decrease) as their corresponding double mutant (try versus try cpc, 86/100 match [P = 3.6E-7]; egl3 versus egl3 gl3, 78/100 [P = 7.0E-5]; myb23 versus myb23 wer, 86/100 [P = 3.6E-7]) (Figures 3A to 3C). However, there were significant gene-specific differences in transcript FC in the single mutants relative to the double mutants (Figures 3A to 3C; see Supplemental Data Set 2 online). Remarkably, the expression of some genes is as strongly altered in the single mutant as in their corresponding double mutant (>80% of the double mutant FC; including 11 genes from try, 25 from egl3, and six from myb23; see Supplemental Data Set 2 online), implying that these genes are TRY, EGL3, or MYB23 dependent but apparently are not essential for normal hair/non-hair cell specification.
Figure 3.
Root Epidermal Gene Expression in try, egl3, and myb23 Mutants Relative to the Wild Type and to Their Paralogous Gene Mutants, cpc, gl3, and egl3.
Plots on left side show microarray-derived transcript FC (relative to the wild type [WT]; dashed horizontal line) in each set of single mutants lacking a phenotype (open triangles), single mutants possessing a root epidermis defect (open squares), and double mutants (closed circles) for the top 100 differentially expressed genes in the double mutants (versus the wild type). Summaries on right side show relative abundance of gene expression categories for the 100 loci. Definitions: high, single mutant alters gene’s transcript >80% of the double mutant level; medium, single mutant alters gene’s transcript 20 to 80% of the double mutant level; none, single mutant alters gene’s transcript <20% of the double mutant level. Values in parentheses represent the fraction of genes affected at the given level in the single mutants. Expected gene numbers for each category (based on random distribution) are shown in parentheses. Asterisks indicate categories with significantly greater gene number than expected (P < 0.01).
[See online article for color version of this figure.]
To extend this analysis, we obtained root epidermal transcript profiles (in biological triplicate) from the corresponding paralogous gene mutants (cpc, gl3, and wer), using WER:GFP cell-sorted microarray analysis. Consistent with their stronger morphological phenotypes, these mutants generally exhibited more severe gene expression changes than their paralogous mutants (try, egl3, and myb23, respectively), although this was not uniformly the case (Figure 3; see Supplemental Data Set 2 online). Categorizing the 100 putative gene targets in each set based on their transcriptional response to each single mutant (high response [H], >80% of the double mutant FC; medium [M], 20 to 80% of the double mutant FC; low/none [L], <20% of the double mutant FC), we observed substantial variation in the relative effects of the mutations (Figures 3A to 3C), likely reflecting gene-specific differences in the functional overlap (redundancy) of these pairs of paralogous transcription factor genes. For example, some genes’ transcripts were affected in each single mutant as much as in the corresponding double mutant (the “H-H” category), suggesting that each of the two transcription factors is necessary for the target gene’s expression and so possess no functional overlap with the other (i.e., no redundancy). Other genes exhibit little or no transcript change in either single mutant, relative to the double mutant (the “L-L” category), indicating complete redundancy. Interestingly, genes in these two categories were significantly overrepresented in the try-cpc and egl3-gl3 sets (P value < 0.01; Figure 3), implying biological significance to these types of regulatory relationships.
We reasoned that an analysis of the distribution of these categories among the 208 genes of the core root epidermal network (Figure 1A) might enable dissection of the relative importance of the transcription factors on specific groups of genes in the network. First, as expected, we observed a significant positive correlation in the direction of change in transcript abundance in the single mutants lacking morphological phenotypes versus double mutants (try versus try cpc, P = 7.0E-17; egl3 versus egl3 gl3, P = 5.9E-3; myb23 versus myb23 wer, P = 9.3E-17; see Supplemental Data Set 3 online), further supporting a molecular impact of these single-gene mutations on the root epidermal network. In addition, several statistically significant deviations from random gene distributions among categories were uncovered, including two particularly strong deviations. In one of these cases, the “M-M” category is overpopulated generally in the try-cpc set (P = 7.7E-4); specifically, the “M-M” category is overpopulated by genes in Branch 1 of the epidermal network (P = 1.8E-5; Figure 1A; see Supplemental Data Set 3 online). Correspondingly, Branch 1 genes are underrepresented in categories of the “H” type for either try or cpc. This suggests that additive effects between the TRY and CPC genes dominate their impact on the downstream genes of the root epidermal network, particularly Branch 1. In a second case, a significantly greater fraction of the Branch 3 genes than expected are in the try “H” categories (P = 1.1E-6; Figure 1A; see Supplemental Data Set 3 online), and quantitative RT-PCR (qRT-PCR) analyses with a subset of these genes validated their TRY sensitivity in roots (see Supplemental Figure 1 online). This suggests that regulation of the ROOT HAIR DEFECTIVE 6 (RHD6)-independent Branch 3 of the root hair pathway is dominated by TRY. Interestingly, because try mutants have a normal distribution of cell types, this branch of the network is probably not critical for root epidermal cell specification. It may be that these genes are required under nonlaboratory environmental conditions, or they may reflect TRY’s role in trichome development, though only two of these (At1g16260 and At2g18690) exhibited significant differential expression in a microarray data set of wild-type versus gl3-sst trichomes (Gilding and Marks, 2010).
Defining the Role of TTG2 through Mutant Transcript Profiling
An implication of our results above is that high-resolution cell-specific transcript data from a mutant strain that lacks a visible abnormal phenotype might provide insight into the mutated gene’s function. To test this, we analyzed the root epidermis transcriptome of a mutant affecting TRANSPARENT TESTA GLABRA2 (TTG2), which encodes a WRKY-type transcription factor required for anthocyanin and trichome production (Johnson et al., 2002). TTG2 is presumed to have a role in non-hair epidermis development, due to its preferential expression in differentiating non-hair cells (Johnson et al., 2002; Bruex et al., 2012) (see Supplemental Figure 2A online) and the dependence of this expression on WER, GL3/EGL3, TTG, and CPC/TRY (Ishida et al., 2007; Bruex et al., 2012) (see Supplemental Figure 2B online). However, TTG2’s role in the root epidermis is unclear, principally because ttg2 mutants lack a morphological phenotype (Johnson et al., 2002) (Figures 4A and 4B).
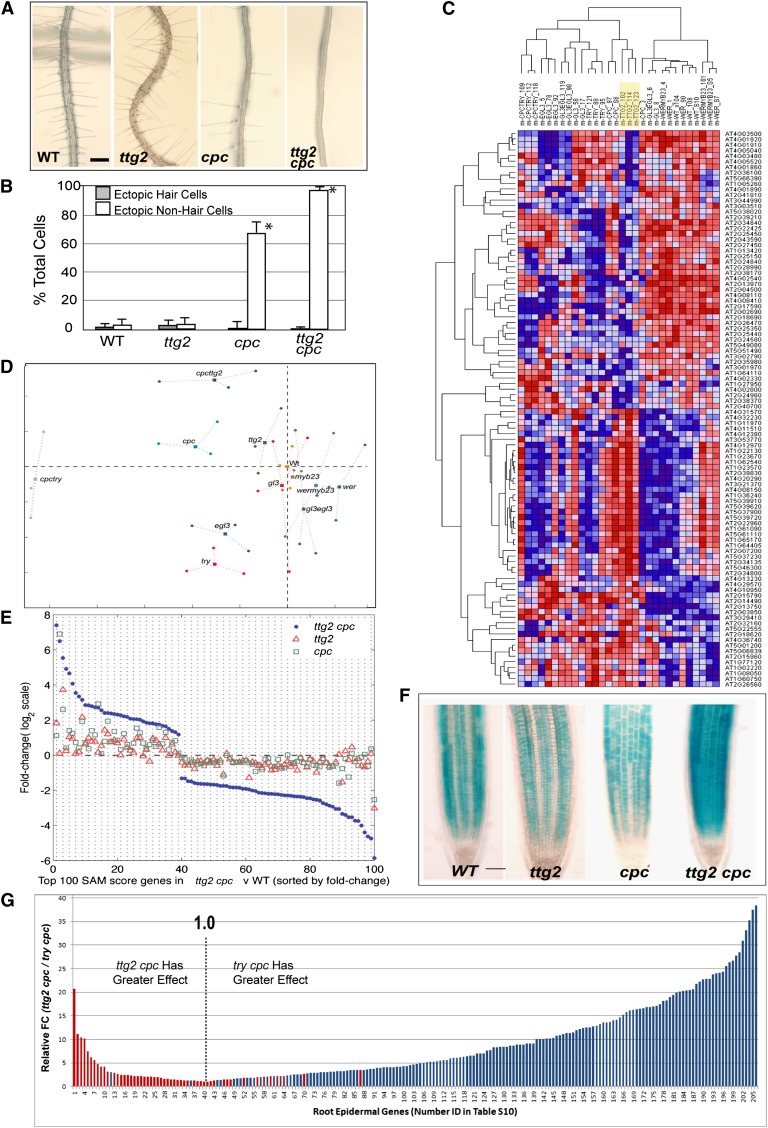
Figure 4.
Global Expression Analysis Links TTG2 and CPC.
(A) Root phenotype of ttg2, cpc, and ttg2 cpc double mutants. WT, the wild type. Bar = 300 µm.
(B) Quantitative analysis of cell type formation in the wild type and ttg2 mutants, showing the mean proportion of cells in the N position that differentiate as hair cells (ectopic hair cells) and cells in the H position that differentiate as non-hair cells (ectopic non-hair cells). Bars indicate sd from three trials. Asterisks indicate significant deviation from the wild type (P < 0.05).
(C) Hierarchical clustering of each microarray replicate, based on transcript abundance in the top 50 upregulated and top 50 downregulated genes by ttg2 (relative to the wild type). The ttg2 replicates (highlighted in yellow) cluster most closely to the cpc replicates. Red = higher transcript accumulation; blue = lower transcript accumulation.
(D) Multidimensional scaling analysis of transcript accumulation from the 208 genes of the core root epidermal gene set using WER:GFP-sorted cells of the indicated wild type, single mutants, and double mutants.
(E) Microarray-derived transcript FC (relative to the wild type; dashed horizontal line) in the ttg2 single mutant (open triangle), cpc single mutant (open squares), and ttg2 cpc double mutants (closed circles) for the top 100 differentially expressed genes in the ttg2 cpc double mutants (versus the wild type).
(F) Expression of the GL2:GUS non-hair cell marker in the wild type, ttg2, cpc, and ttg2 cpc double mutant. Bar = 100 µm.
(G) Relative effect of ttg2 cpc and try cpc on the root epidermal gene set. The bars indicate the absolute value of transcript FC in (ttg2 cpc versus the wild type)/(try cpc versus the wild type) for root hair genes (blue bars) and non-hair genes (red bars) from the core root epidermal gene set (excluding TTG2 and TRY; Bruex et al., 2012). The dotted line (at position 1.0) separates genes exhibiting greater relative FC in ttg2 cpc (left side) from genes exhibiting greater relative FC in try cpc (right side). Numerical data depicted in this graph are available in Supplemental Data Set 7 online.
To assess potential transcriptional effects of ttg2 in the root epidermis, we introduced the WER:GFP marker into the presumed null ttg2-1 mutant and analyzed cell-sorted microarrays, relative to the wild type (three biological replicates each). Among 201 differentially expressed genes (FC > 2.0; SAM t-statistic > 2.5; q-value < 0.05), members of the core root epidermal gene set are overrepresented (P = 1.5E-4; see Supplemental Data Set 4 online). Thus, like try, egl3, and myb23, the ttg2 mutant exhibits an altered transcript profile that preferentially affects root epidermal pathway genes, despite the lack of a detectable morphological change.
Results from comparative expression analyses unexpectedly linked TTG2 to a role in differentiation of root hair cells, rather than non-hair cells as implied by TTG2’s expression/localization pattern. First, the direction of transcript change in the significantly affected root epidermal genes was consistent with TTG2 promoting hair cell transcription (4/4 hair gene transcripts decreased in ttg2; 4/5 non-hair gene transcripts increased; see Supplemental Data Set 4 online). Second, hierarchical clustering with the entire set of mutant transcript profiles shows that, for the 100 genes most affected by ttg2 (50 genes exhibiting the greatest increase and 50 genes exhibiting the greatest decrease in transcript abundance in ttg2 relative to the wild type; see Supplemental Data Set 5 online), the ttg2 transcript accumulation pattern is most similar to cpc mutants (Figure 4C). Third, using multidimensional scaling visually to portray the transcriptional effects of the mutants across the entire 208 members of the core root epidermal gene set, the ttg2 deviates from the wild type in a manner most similar to cpc (Figure 4D).
Reasoning from the above that TTG2’s function in root epidermis development may overlap with CPC, we generated a ttg2 cpc double mutant. Relative to cpc, the double mutant exhibited decreased hair cell production (Figures 4A and 4B) and increased GL2:GUS (for β-glucuronidase) reporter expression throughout the epidermis (Figure 4F), demonstrating that ttg2 enhances the cpc phenotype. To further assess this, we introduced the WER:GFP transgene and conducted tissue-specific transcript profiling of ttg2 cpc. Among the top 100 genes differentially expressed in ttg2 cpc (versus the wild type), only four were categorized as “H” for cpc (>80% of ttg2 cpc) (Figure 4E; see Supplemental Data Set 6 online), showing that ttg2 enhanced the transcriptional effect of cpc in 96 of these genes. Furthermore, among the 208 core root epidermal genes, ttg2 cpc preferentially reduced mean transcript accumulation from hair genes (P = 5.1E-9) and preferentially increased mean transcript accumulation from non-hair genes (P = 4.2E-5), relative to cpc (see Supplemental Data Set 7 online), although in general, the quantitative effect of ttg2 cpc on hair cell gene expression was less than that of try cpc (Figure 4G).
A possible explanation for the similarity in ttg2 cpc and try cpc phenotypes is that TTG2 may help promote transcription of the lateral inhibition gene TRY. Consistent with this, we observed a small reduction in TRY transcripts in ttg2 (67% of the wild type; P < 0.2) and a significant reduction in ttg2 cpc (2.6-fold, relative to cpc; P < 0.01), but the try mutant did not significantly alter TTG2 transcript levels.
DISCUSSION
Molecular Dissection of Duplicate Gene Function
The immediate outcome of gene duplication is genetic redundancy, whereby duplicate genes encode equivalent proteins that carry out a common function. Over time, one of the duplicates may be lost or both duplicates may be retained and diverge in sequence and function, leading to subfunctionalization or neofunctionalization, with corresponding changes in the degree of genetic redundancy (Pickett and Meeks-Wagner, 1995; Briggs et al., 2006; Pérez-Pérez et al., 2009; Chen et al., 2010).
Here, we defined the relative roles of three sets of paralogous transcription factor genes at the resolution of a specific gene regulatory network in Arabidopsis. A fascinating finding of this study is the extensive gene-by-gene variation in the relative transcriptional effects of each paralog on its putative downstream gene targets. For each paralogous gene set, we identified downstream genes exhibiting high, moderate, or low/no sensitivity to the lack of a given paralog or its partner, indicating a diverse array of gene regulatory effects (Figure 3). This illustrates the regulatory complexity that can be generated within a single pathway by the functional divergence of duplicate transcription factor genes. It is tempting to speculate that this imparts greater efficiency and/or fidelity to the system as a whole, enhancing its robustness and phenotypic buffering capacity (Thomas, 1993; Fu et al., 2009).
These data also provide insight into specific aspects of the divergence of these paralogs. First, each of the three paralogous gene sets exhibit evidence of unequal (or asymmetrical) divergence (Wagner, 2002; Ganko et al., 2007) with respect to the root epidermal network because mutants of one member (cpc, gl3, and wer) have a generally greater impact on downstream gene expression than mutants of the other (try, egl3, and myb23), consistent with their morphological effects. Furthermore, we conclude that, among these three paralogous gene sets, the TRY-CPC set appears to have undergone the least diversification and MYB23-WER the most (with respect to the root epidermal network) because (1) TRY-CPC has the smallest frequency (and MYB23-WER the greatest frequency) of target genes exhibiting high sensitivity to the loss of either member (i.e., number of “H” category genes in each set: TRY-CPC [12], EGL3-GL3 [51], and MYB23-WER [76]; Figure 3), and (2) TRY-CPC has the greatest frequency (and MYB23-WER the smallest frequency) of target genes exhibiting synergistic effects (i.e., number of “L-L” genes in each set: TRY-CPC [26], EGL3-GL3 [9], and MYB23-WER [0]; Figure 3).
Redundancy Is Relative
This study highlights the importance of phenotypic characterization for defining genetic redundancy. Because redundancy is assessed by comparing differences in single and multiple mutants, conclusions regarding genetic redundancy are necessarily dependent on the nature of the phenotypic analysis performed (Pickett and Meeks-Wagner, 1995; Pérez-Pérez et al., 2009; Lloyd and Meinke, 2012).
Compared with results from the morphological analysis of these root epidermis mutants, we found that the use of high-resolution transcriptome analyses led to different conclusions regarding the redundancy between the paralogous genes. In particular, the fact that the TRY, EGL3, and MYB23 gene functions are required for normal root epidermal transcript accumulation implies a reduced degree of genetic redundancy than indicated by morphological analyses. Furthermore, the gene-to-gene variation in the transcriptional effects of paralogous gene mutants prevented a simple conclusion for genetic redundancy across the entire root epidermal network. Indeed, different redundancy relationships may be deduced, ranging from complete redundancy to no redundancy, depending on the particular gene(s) or subnetwork chosen for assessment.
Overall, these results emphasize that genetic redundancy is a relative term, strongly dependent on the phenotypic analysis employed. They further suggest that a detailed molecular characterization of a mutant phenotype is necessary to fully assess the nature and impact of genetic redundancy on a biological process.
Using Molecular Phenotypes to Define Gene Function
A current challenge in molecular genetics is the assignment of gene function to loci that lack visible mutant phenotypes. For example, in Arabidopsis, <10% of single-gene insertional mutants exhibit discernable phenotypes, largely due to genetic redundancy originating from duplication events (Bouché and Bouchez, 2001; Hanada et al., 2009a; Pérez-Pérez et al., 2009; Brady et al., 2011).
Here, we discovered that loss-of-function mutations affecting one member of a duplicate gene pair alter tissue-specific transcript profiles in a manner that reflects the known biological role of the gene, despite the lack of a visible mutant phenotype. In addition, we showed that tissue-specific mutant transcriptome information can be used to help define the function of an uncharacterized gene in the root epidermal network (the TTG2 gene). Indeed, our data uncovered a role for TTG2 in hair cell specification, an unexpected finding given that TTG2’s gene expression and protein accumulation is preferentially observed in non-hair cells (Ishida et al., 2007).
The general approach we used here may be applied to the functional characterization of other mutants lacking overt phenotypes. For increased chance of success, several issues should be considered. First, some knowledge of the organ or tissue environment for the likely gene function would be helpful to enrich for molecules of interest and simplify the data set (e.g., root epidermis–specific analysis in this study). Second, a transcriptome-based analysis is necessarily limited to characterizing genes expected to influence transcript accumulation (e.g., transcription factor genes in this study). Other large-scale approaches, such as metabolomics analysis, might be useful to characterize other gene categories (Fu et al., 2009). Lastly, the use of comparative transcript profiling is aided by knowledge of components of the putative gene pathway/network of interest (e.g., the extensively characterized root epidermal network used in this study; Bruex et al., 2012) to enable a focus on an informative gene set.
METHODS
Arabidopsis thaliana Strains and Growth Conditions
The mutant lines used in this study are described in Supplemental Table 1 online. The GUS and GFP reporter lines used here have been previously described, including TTG2:GUS (Johnson et al., 2002), GL2:GUS (Rerie et al., 1994), and WER:GFP (Lee and Schiefelbein, 1999). For seedling growth, Arabidopsis seeds were surface sterilized and grown in a vertical orientation on agarose-solidified nutrient medium as described (Schiefelbein and Somerville, 1990). Multiple mutant or transgene lines were generated by crossing and subsequent PCR testing for mutations or transgenes.
Root Epidermis Phenotype Analysis
The determination of root hair and non-hair epidermal cells in 4-d-old seedlings of the wild-type and mutant lines was conducted using established protocols (Lee and Schiefelbein, 2002; Simon et al., 2007). In these assays, a cell was considered to be of the hair cell type if it possessed a protrusion, irrespective of the length. Each analysis was conducted three independent times.
In Situ RNA hybridization
The in situ RNA hybridization was conducted with intact root tips (∼1 mm of the root terminus, including the meristem, elongation region, and root hair zone) from 4-d-old seedlings, essentially as described (Bernhardt et al., 2005).
GUS Assays
Qualitative histochemical GUS staining of seedling roots was performed as previously described (Hung et al., 1998; Wang et al., 2002).
qRT-PCR Assays
For qRT-PCR, total RNA was extracted from root tips of 5-d-old seedlings using the RNeasy plant mini kit (Qiagen). Following treatment with DNase I (Invitrogen), 1 µg of total RNA was used to synthesize cDNA using the SuperScript II reverse transcriptase kit (Invitrogen) in combination with oligo(dT)12-18 primer. The qRT-PCR amplification was performed using the Mastercycler ep Realplex detection system and Power SYBR Green PCR master mix (Applied Biosystems) with gene-specific primers (see Supplemental Table 3 online). Relative expression of target genes was calculated by the comparative threshold cycle to the wild type using reactions with the ACTIN7 transcript as the internal reference.
Microarray Gene Expression Assays
The WER:GFP marker was crossed into each of the mutant lines to ensure consistency among lines. Protoplasts were prepared and isolated from the root tips of 5-d-old seedlings using FACS as previously described (Birnbaum et al., 2005; Bruex et al., 2012). Briefly, for each sample, ∼1000 root tips were pooled, subjected to cell wall–degrading enzyme solutions, and sorted on a BD FACS unit. RNA was purified from the GFP-positive protoplasts using the Qiagen RNeasy micro kit and amplified using the Affymetrix Small Sample Labeling Protocol VII, with its quality verified by capillary electrophoresis using a Bioanalyzer 2100 (Agilent). Sample preparation for cDNA preparation, hybridization, and detection to the Affymetrix Arabidopsis ATH1 GeneChip was according to Affymetrix protocols. GeneChips were scanned using the Affymetrix 3000 7G GeneChip Scanner with Autoloader. Raw images (CEL format) were generated with Affymetrix GeneChip Operating Software. The microarray results for the wild-type Columbia, try cpc, egl3 gl3, and wer myb23 lines have been described (Bruex et al., 2012), and the microarray analyses of the other eight lines reported here (three biological replicates each acquired at three separate dates) were conducted during the same period and used the same growth conditions.
Statistical Analyses
The probe set summaries from the ATH1 microarrays were computed using the robust multichip average method with quantile normalization (Irizarry et al., 2003). Custom probe set definitions were used for data preprocessing (Dai et al., 2005). All values subsequently analyzed were log2 scale expression levels. SAM (Tusher et al., 2001) was used to define differentially expressed genes employing SAM scores representing the t-statistic value and false discovery rate q-values (Benjamini and Hochberg, 1995).
Fisher's exact test (Fisher, 1922) was used to assess the statistical significance of association between two variables to identify groups with significant deviation from a random distribution. In these tests, genes mutated in a tested line were excluded from the gene group analysis (e.g., the TRY gene was excluded from try mutant analysis).
To evaluate correlation in transcript change direction in the single versus double mutants, the deviation in mean transcript abundance (positive or negative) in each single mutant, relative to the wild type, was compared with the double mutants. For quantitative comparisons, the relative FC (FC in each single mutant to FC in its corresponding double mutant) was determined for the 100 genes exhibiting the greatest (absolute value) SAM score for the double mutant versus the wild type. These genes were categorized as follows: L (low/no difference), single mutant FC < 20% double mutant FC; M (medium difference), single mutant FC > 20% but <80% of double mutant FC; H (high difference), single mutant FC > 80% of double mutant FC. Similar comparisons were conducted using the core root epidermal gene set (Bruex et al., 2012), but limited to those genes exhibiting FC > 2.0 in double mutant versus the wild type and excluding genes with mutations in the tested lines and the regulatory genes ETC1, MYC1, GL2, and RHD6.
Hierarchical clustering of transcript accumulation in the genes and samples was performed using the GenePattern suite of tools (de Hoon et al., 2004; Reich et al., 2006). The gene ontology term enrichment analysis was performed using the online tool at http://amigo.geneontoloty.org/cgi-bin/amigo/term_enrichment with The Arabidopsis Information Resource gene ontology annotations (http://www.Arabidopsis.org/tools/bulk/go/index.jsp) and use of Fisher’s exact test (http://en.wikipedia.org/wiki/Fisher’s_exact_test). Multidimensional scaling of the biological triplicate sets of microarray data from the wild type and single and double mutants was performed using the MATLAB Statistics Toolbox (http://www.mathworks.com/help/toolbox/stats/mdscale.html) using gene data from the core root epidermal gene set.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers listed in Supplemental Table 1 online. The ATH1 microarray data in this study are available from the Gene Expression Omnibus (http://www.ncbi.nim.nih.gov/geo/) under the series records GSE30547 (including accession numbers GSM757837-GSM757839, GSM757846-GSM757848, and GSM757888-GSM757893) and GSE47632 (including accession numbers GSM1153845-GSM1153868).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. qRT-PCR Analysis of TRY-Regulated Genes
Supplemental Figure 2. Regulation of TTG2 Expression by Root Epidermal Transcription Factors.
Supplemental Table 1. Arabidopsis Root Epidermis Mutants Analyzed in This Study.
Supplemental Table 2. Gene Ontology Terms Overrepresented among the Genes Differentially Expressed in try, egl3, and myb23 Mutants.
Supplemental Table 3. Primers Used for qRT-PCR Analysis.
Supplemental Data Set 1. Differentially Expressed Genes in the try, egl3, and myb23 Mutant Lines Relative to the Wild Type.
Supplemental Data Set 2. Lists of 100 Genes Exhibiting the Greatest Differential Expression in the try cpc, egl3 gl3, and myb23 wer Double Mutants Relative to the Wild Type.
Supplemental Data Set 3. Relative Effect of Single and Double Mutants on the 208 Root Epidermal Gene Set.
Supplemental Data Set 4. Differentially Expressed Genes in the ttg2 Mutant Line Relative to the Wild Type.
Supplemental Data Set 5. List of 50 Genes Exhibiting the Greatest Increase and 50 Genes Exhibiting the Greatest Decrease in Transcript Abundance in ttg2 versus the Wild Type.
Supplemental Data Set 6. List of 100 Genes Exhibiting the Greatest Differential Expression in the ttg2 cpc Double Mutant Relative to the Wild Type.
Supplemental Data Set 7. Relative Transcript Accumulation in ttg2 cpc versus try cpc for the 208 Root Epidermal Gene Set.
Acknowledgments
We acknowledge the helpful technical assistance provided by Nichole McCaffrey and the assistance of the University of Michigan Sequencing Core and Microarray Facility. We thank David Smyth (Monash University) for providing the TTG2:GUS transcriptional reporter line. This research was supported by grants from the National Science Foundation (NSF IOS-0723493 and NSF IOS-1121602) to J.S. and from the National Institutes of Health (U54-DA-021519) to P.J.W. and R.M.K.
AUTHOR CONTRIBUTIONS
M.S., A.B., R.M.K., X.Z., L.H., P.J.W., and J.S. designed the research. M.S., A.B., X.Z., L.H., and R.M.K. performed most of the research. R.M.K. and P.J.W. contributed new computational tools and conducted the bioinformatics analyses. Results were evaluated and discussed among all the authors. The article was principally written by M.S. and J.S., and it was read and revised by all of the authors.
Glossary
- FACS
fluorescence-activated cell sorting
- FC
fold change
- SAM
significance analysis of microarrays
- qRT-PCR
quantitative RT-PCR
References
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300 [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Jung J.W., Wang J.Y., Lambert G.M., Hirst J.A., Galbraith D.W., Benfey P.N. (2005). Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Bouché N., Bouchez D. (2001). Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant Biol. 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brady S.M., et al. (2011). A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs G.C., Osmont K.S., Shindo C., Sibout R., Hardtke C.S. (2006). Unequal genetic redundancies in Arabidopsis—A neglected phenomenon? Trends Plant Sci. 11: 492–498 [DOI] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.W., Bandyopadhyay S., Shasha D.E., Birnbaum K.D. (2010). Predicting genome-wide redundancy using machine learning. BMC Evol. Biol. 10: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., Watson S.J., Meng F. (2005). Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., Miyano S. (2004). Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. (1922). On the interpretation of X2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 85: 87–94 [Google Scholar]
- Fu J., et al. (2009). System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat. Genet. 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Ganko E.W., Meyers B.C., Vision T.J. (2007). Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 24: 2298–2309 [DOI] [PubMed] [Google Scholar]
- Gilding E.K., Marks M.D. (2010). Analysis of purified glabra3-shapeshifter trichomes reveals a role for NOECK in regulating early trichome morphogenic events. Plant J. 64: 304–317 [DOI] [PubMed] [Google Scholar]
- Grebe M. (2012). The patterning of epidermal hairs in Arabidopsis—Updated. Curr. Opin. Plant Biol. 15: 31–37 [DOI] [PubMed] [Google Scholar]
- Hanada K., Kuromori T., Myouga F., Toyoda T., Li W.H., Shinozaki K. (2009a). Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol. Evol. 1: 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Kuromori T., Myouga F., Toyoda T., Shinozaki K. (2009b). Increased expression and protein divergence in duplicate genes is associated with morphological diversification. PLoS Genet. 5: e1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.Y., Lin Y., Zhang M., Pollock S., Marks M.D., Schiefelbein J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117: 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Hattori S., Sano R., Inoue K., Shirano Y., Hayashi H., Shibata D., Sato S., Kato T., Tabata S., Okada K., Wada T. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.S., Kolevski B., Smyth D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J.J., Fu J., Terpstra I.R., Garcia J.M., van den Ackerveken G., Snoek L.B., Peeters A.J., Vreugdenhil D., Koornneef M., Jansen R.C. (2007). Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., West M.A., van Leeuwen H., Loudet O., Doerge R.W., St Clair D.A. (2006). Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J., Meinke D. (2012). A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol. 158: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Micol J.L. (2009). Understanding synergy in genetic interactions. Trends Genet. 25: 368–376 [DOI] [PubMed] [Google Scholar]
- Pickett F.B., Meeks-Wagner D.R. (1995). Seeing double: Appreciating genetic redundancy. Plant Cell 7: 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. (2006). GenePattern 2.0. Nat. Genet. 38: 500–501 [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Ryu K.H., Kang Y.H., Park Y.H., Hwang I., Schiefelbein J., Lee M.M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132: 4765–4775 [DOI] [PubMed] [Google Scholar]
- Savage N.S., Walker T., Wieckowski Y., Schiefelbein J., Dolan L., Monk N.A. (2008). A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 6: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J., Kwak S.H., Wieckowski Y., Barron C., Bruex A. (2009). The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W., Somerville C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578 [DOI] [PubMed] [Google Scholar]
- Song S.K., Ryu K.H., Kang Y.H., Song J.H., Cho Y.H., Yoo S.D., Schiefelbein J., Lee M.M. (2011). Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 157: 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra I.R., Snoek L.B., Keurentjes J.J., Peeters A.J., van den Ackerveken G. (2010). Regulatory network identification by genetical genomics: Signaling downstream of the Arabidopsis receptor-like kinase ERECTA. Plant Physiol. 154: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.H. (1993). Thinking about genetic redundancy. Trends Genet. 9: 395–399 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Ishida T., Wada T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286: 67–106 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Nukumizu Y., Sato S., Kato T., Tabata S., Wada T. (2012). Functional divergence of MYB-related genes, WEREWOLF and AtMYB23 in Arabidopsis. Biosci. Biotechnol. Biochem. 76: 883–887 [DOI] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19: 2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wagner A. (2002). Asymmetric functional divergence of duplicate genes in yeast. Mol. Biol. Evol. 19: 1760–1768 [DOI] [PubMed] [Google Scholar]
- Wang H., Lee M.M., Schiefelbein J.W. (2002). Regulation of the cell expansion gene RHD3 during Arabidopsis development. Plant Physiol. 129: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J.G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kwak S.H., Zeng Q., Ellis B.E., Chen X.Y., Schiefelbein J., Chen J.G. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]