An-1 regulates the formation of long awns of seeds, which are important for seed dispersal in wild rice. However, An-1 has been selected in cultivated rice, which leads to loss of awn as well as more grains per panicle and higher yield per plant. Cloning of An-1 provides insight into how artificial selection shaped plant morphology to meet human demands during the long history of domestication.
Abstract
Long awns are important for seed dispersal in wild rice (Oryza rufipogon), but are absent in cultivated rice (Oryza sativa). The genetic mechanism involved in loss-of-awn in cultivated rice remains unknown. We report here the molecular cloning of a major quantitative trait locus, An-1, which regulates long awn formation in O. rufipogon. An-1 encodes a basic helix-loop-helix protein, which regulates cell division. The nearly-isogenic line (NIL-An-1) carrying a wild allele An-1 in the genetic background of the awnless indica Guangluai4 produces long awns and longer grains, but significantly fewer grains per panicle compared with Guangluai4. Transgenic studies confirmed that An-1 positively regulates awn elongation, but negatively regulates grain number per panicle. Genetic variations in the An-1 locus were found to be associated with awn loss in cultivated rice. Population genetic analysis of wild and cultivated rice showed a significant reduction in nucleotide diversity of the An-1 locus in rice cultivars, suggesting that the An-1 locus was a major target for artificial selection. Thus, we propose that awn loss was favored and strongly selected by humans, as genetic variations at the An-1 locus that cause awn loss would increase grain numbers and subsequently improve grain yield in cultivated rice.
INTRODUCTION
Modern genetics and archaeological studies have revealed that the Asian cultivated rice Oryza sativa was domesticated from the ancestor of the wild rice species Oryza rufipogon ∼8000 years ago (Zong et al., 2007; Fuller et al., 2009; Izawa et al., 2009; Huang et al., 2012). Wild rice exhibits a number of traits, such as easy seed shattering, prostrate growth, long awns, black hulls, and few grains per panicle. These unique characteristics have important roles in wild rice and are strongly associated with seed dispersal, dormancy, and survival in harsh environmental conditions. In comparison to its wild progenitor, cultivated rice typically displays reduced seed shattering and dormancy, a reduction of outcrossing rate and awn length, erect growth, and pericarp and hull color conversion (Kovach et al., 2007; Sweeney and McCouch, 2007). All of these traits that distinguish cultivated rice from its wild progenitor are called domestication traits.
To date, the well-characterized rice domestication genes include shattering4 (sh4), QTL of seed shattering in chromosome1 (qSH1), PROSTRATE GROWTH1 (prog1), Black hull4 (Bh4), Red pericarp (Rc), QTL for seed width on chromosome5 (qSW5), the rice ortholog of maize C1 gene, and Waxy. An examination of domestication genes reveals that the molecular mechanisms underlying the evolution of phenotypes are varied. Natural variations have been found to directly disrupt the functions of some genes and to be associated with phenotype changes, such as those in prog1, Bh4, and Rc (Sweeney et al., 2006; Jin et al., 2008; Tan et al., 2008; Zhu et al., 2011). Other domestication genes have been identified as being involved in protein modification, regulatory changes, or both. qSH1 eliminated its expression at the provisional abscission layer to confer nonshattering to japonica rice (Konishi et al., 2006), while sh4 differs both in the coding region and 5′ regulatory sequence between wild rice and cultivated rice (Li et al., 2006a). Furthermore, domestication genes often have pleiotropic effects on multiple traits. Prog1 resulted in erect growth and an increased number of grains per panicle (Jin et al., 2008; Tan et al., 2008). Another aspect that increases the intricacy of rice domestication is that O. sativa is divided into japonica and indica subspecies. Previous research showed that the domestication alleles of sh4, Prog1, qSW5, and Bh4 were fixed in both japonica and indica, while qSH1, Waxy, and Rc were fixed only in the japonica population (Huang et al., 2012).
A recent study revealed the origins of O. sativa ssp japonica and ssp indica by massive analyses of the 55 regions of selective sweeps and genome-wide patterns (Huang et al., 2012). This study demonstrated that japonica rice was first domesticated from the wild populations of O. rufipogon in the middle Pearl River regions in southern China and that indica rice was subsequently domesticated through introgression of domestication genes from japonica into wild populations toward southeastern and southern Asia (Huang et al., 2012). This research provides an important resource and an effective genomics approach for identifying genes that regulate key differences in plant structure and physiology that distinguish cultivated rice from its wild progenitors.
The awn is one of the morphological characteristics of rice seeds and is also found in other species, such as wheat (Triticum aestivum), barley (Hordeum vulgare), oats (Avena sativa), and sorghum (Sorghum bicolor). Awn is an extension of apex of the lemma of spikelet. As a characteristic of seeds in wild plants, long awns are reported to aid seed dispersal and seed burial and protect cereal grains from animal predation (Elbaum et al., 2007). However, long awns are not favorable during harvest and storage; hence, this trait was artificially selected during domestication. Even so, long awns are retained in some cereal crops, such as wheat and barley, because long awns contribute significantly to photosynthesis and yield (Abebe et al., 2010). On the contrary, most cultivated rice bear no awns or very short awns because the round rice awn contains only one vascular bundle and may not contribute to photosynthesis (Toriba et al., 2010).
Genetic analysis has shown that the awn is a complex trait and many awn-related quantitative trait loci (QTLs) have been identified in rice (Cai and Morishima, 2002; Thomson et al., 2003; Gu et al., 2005a, 2005b; Wang et al., 2011), but no genes have been molecularly identified to date. Therefore, the molecular mechanism that transforms the long awns of wild rice to the awnless trait of cultivated rice is still unknown.
In this study, we investigated genetic variations involved in awn loss in domesticated rice. We cloned the major QTL Awn-1 (An-1) that encodes a basic helix-loop-helix (bHLH) protein and regulates the long-awn trait in wild rice. An-1 is intensely expressed at the apex of the lemma primordia, specifically causing continuous cell division to form a long awn. Upregulation of An-1 expression could induce long awns and grain elongation as well as reduce grain number per panicle in awnless cultivated rice, whereas RNA interference (RNAi) of An-1 in a long-awn indica variety Kasalath generated shorter awns and shorter grains but produced more grains per panicle compared with the control plants. Our population genetic analysis of wild and cultivated rice showed a significant reduction in nucleotide diversity of the An-1 locus and revealed that the An-1 locus was a major target for artificial selection. We thus propose that awn loss was also favored and selected by humans, in addition to easy harvest and storage, as the genetic variation causing awn loss would increase grain number and subsequently improve yield in cultivated rice during the long course of domestication.
RESULTS
Cloning and Characterization of the Wild Rice Allele of An-1, Which Regulates Long-Awn Development
We previously constructed a wild rice chromosome 4 substitution line (SL4) derived from a cross between cultivated rice variety O. sativa ssp indica cv Guangluai4 (GLA4) (awnless) as the recurrent parent and wild rice accession O. rufipogon Griff W1943 (W1943) (long awn) as the donor parent (Figures 1A and 1B) (Zhu et al., 2011; Zhou et al., 2012). SL4 had long awns of 36.45 ± 11.32-mm long in Sanya (N 18.2°, E 109.5° in China) and 24.81 ± 4.17-mm long in Shanghai (N 31.2°, E 121.4° in China) (see Supplemental Table 1 online). The awn length was affected to a certain degree by some environmental factors, such as photoperiod and temperature during the floral differentiation stage, which was consistent with a previous study (Aspinall, 1969). However, the proportion of the awned plants (awn rate) of SL4 remained stable and ranged from ∼70 to 90% in Sanya and Shanghai, whereas GLA4 had no awns in either area (see Supplemental Table 1 online).
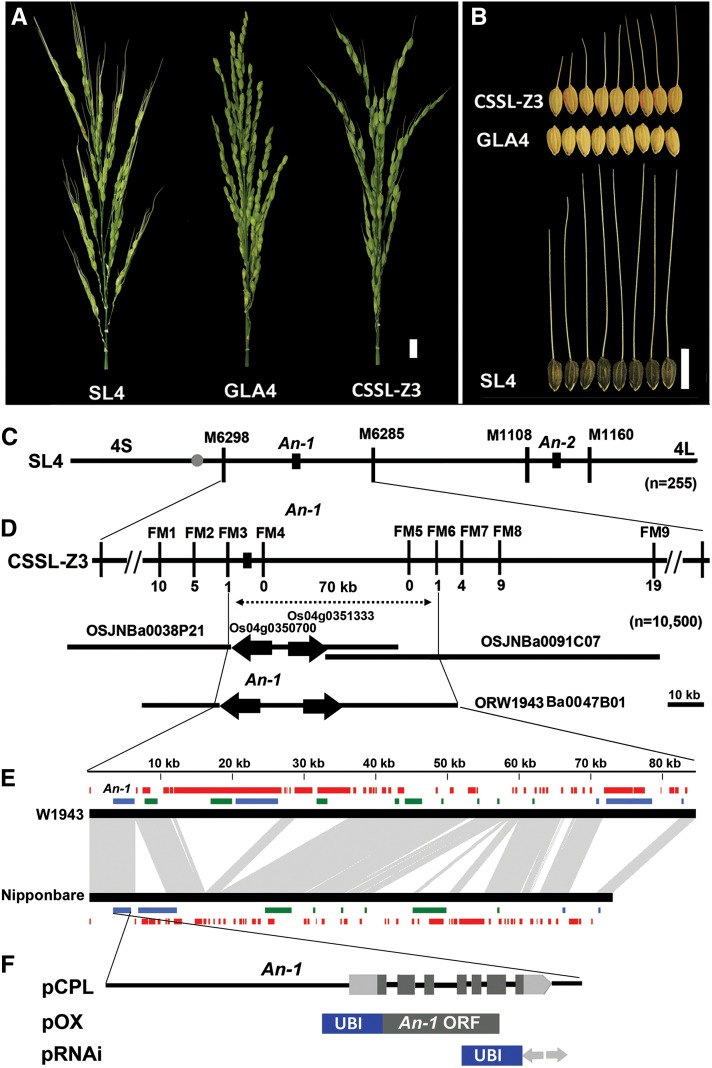
Figure 1.
Map-Based Cloning and Identification of An-1.
(A) Panicle comparison among SL4, GLA4, and CSSL-Z3. Bar = 10 mm.
(B) Awn length comparison among CSSL-Z3, GLA4, and SL4. Bar = 10 mm.
(C) Two QTLs for awn length were identified on chromosome 4 in SL4, and An-1 was first mapped to the interval between the markers M6298 and RM6285, while An-2 localized between the markers M1108 and M1160.
(D) An-1 was further delimited to a 70-kb genomic region between the markers FM3 and FM6, which corresponded to two Nipponbare BACs and one W1943 BAC. FM1 to FM9 are primers used for fine mapping. The numbers underneath the bars indicate the number of recombinants between An-1 and the molecular markers. The black arrows indicate gene direction. Bar = 10 kb.
(E) Comparison of BAC sequence and annotation between W1943 and Nipponbare. Black bars represent genomic sequences of W1943 and Nipponbare. Red bars represent transposon-like and repeat sequences. Green/blue bars represent predicated ORFs. Green bars represent ORFs with a forward direction. Blue bars represent ORFs with a reverse direction. Gray represents regions sharing sequence collinearity between W1943 BAC and Nipponbare BAC. Genes annotated on the BACs are listed in Supplemental Table 2 online.
(F) Gene structure of An-1 and constructs used in An-1 function investigation. pCPL represents the 10-kb W1943 genomic fragment used for the complementation test; pOX contains W1943 An-1 ORF used for ectopic expression and overexpression; pRNAi denotes the RNA interference construct. UBI is a maize Ubiquitin promoter.
Using the F2 population derived from the cross between SL4 and GLA4, two QTLs for awn length, designated as Awn-1 (An-1) and Awn-2 (An-2), were mapped between the markers M6298 and M6285 (∼5.88 Mb) and the markers M1108 and M1160 on the long arm of chromosome 4, respectively (Figure 1C). The wild alleles of An-1 and An-2 showed positive effects and accounted for 52.5 and 12% of the phenotypic variations, respectively. To further map the loci and clone the genes for awn length, we constructed a set of single chromosome segment substitution lines (CSSLs). Among the CSSLs, CSSL-Z3 (Z3) contained the major QTL, the An-1 locus (see Supplemental Figure 1 online), and displayed a stable awn phenotype in both Sanya and Shanghai. The awn length of Z3 was 18.82 ± 3.25 mm in Sanya (Figures 1A and 1B) and 15.74 ± 2.16 mm in Shanghai. The awn rate of Z3 was 67.16% ± 9.12% in Sanya and 60.55% ± 8.30% in Shanghai (see Supplemental Table 1 online). The F1 plants from the cross between Z3 and GLA4 were awned. An examination of awn phenotypes in 126 F2 plants revealed that 87 plants were awned but 39 plants were awnless, which fitted a 3:1 segregation ratio. This indicated that An-1 is a single dominant gene that regulates the awn phenotype in this population. Then, a larger F2 population containing 10,500 plants was genotyped using the flanking markers M6298 and M6285. By progressively examining the insertion or deletion (indel) and single nucleotide polymorphism (SNP) markers between M6298 and M6285, An-1 was finally delimited between FM3 and FM6, representing a 70-kb genomic region on chromosome 4 (Figure 1D) according to the Build 4.0 pseudomolecules of the Nipponbare genome (Feng et al., 2002; International Rice Genome Sequencing Project, 2005).
To clone the wild allele An-1, a BAC library of W1943 was screened using the FM3 and FM6 probes. The BAC (ORW1943Ba0047B01) containing the An-1 locus was identified, sequenced, and annotated. Meanwhile, the BAC (OSIGBa0144C23) corresponding to the collinear FM3-FM6 regions from GLA4 had been sequenced before, and the same gene content was identified within this region between GLA4 and Nipponbare. However, a sequence comparison between W1943 and Nipponbare revealed that multiple reciprocal insertions and deletions existed within this region (Figure 1E). Most of those indels were repeat sequences (Figure 1E), which might make further recombination difficult. Within the collinear FM3-FM6 regions between W1943 and Nipponbare, we found that there were only two unique genes, Os04g0350700 and Os04g0351333, that were likely candidates for awn length according to the Rice Annotation Project Database (Figure 1E) (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/build4/). Other predicted open reading frames (ORFs) within this region were all transposon element–like sequences or highly repetitive sequence-related genes (see Supplemental Table 2 online). Furthermore, Os04g0350700 encodes a bHLH transcription factor with a few polymorphisms in the coding region and 5′ upstream region (see Supplemental Table 3 online), while Os04g0351333 encodes an RF1-like gene and contains polymorphisms in the promoter region between W1943 and GLA 4.
To determine which one is the casual gene, we generated two complementary constructs, pCPL and pCPL-RF. pCPL contained a 10,244-bp W1943 genomic sequence covering the entire Os04g0350700 gene region and 6-kb 5′ upstream and 500-bp 3′ downstream sequences (Figure 1F). The pCPL-RF contained a 10,501-bp W1943 genomic sequence covering the entire Os04g0351333 gene region and 4-kb 5′ upstream and 2-kb 3′ downstream sequences (see Supplemental Figure 2A online). Since we were unable to regenerate shoots from the callus of awnless indica variety GLA4, we transformed an awnless japonica Nipponbare to determine the function of An-1. The vector pCAMBIA1301 was used as a control. Eighty-two percent of the T0 plants transformed with pCPL produced long awns, whereas none of T0 plants transformed with pCPL-RF produced any awns (see Supplemental Table 4 and Supplemental Figures 2B and 2C online). The long awn phenotypes were stably inherited in the T1 and T2 progeny of different pCPL transgenic lines, which indicated that Os04g0350700 indeed regulates awn length.
We further transformed Nipponbare with an ectopic and overexpression construct of pOX and the indica Kasalath, which produces long awns, with the pRNAi construct (Figure 1F). The pTCK303 vector was used as the control for RNAi plants. The phenotypes of T0 transgenic plants revealed that the pOX construct could also produce long awns, whereas the pRNAi construct could induce shortened awns in transgenic plants. Control plants transformed with either pCAMBIA1301 or pTCK303 did not show any morphological changes during both vegetative and reproductive stages (see Supplemental Table 4 online). Therefore, the results demonstrate that Os04g0350700 is An-1 and that this gene regulates awn development in SL4.
Analysis of Functional Allelic Variations in the An-1 Locus Associated with Awn Loss in Cultivated Rice
A 1978-bp An-1 cDNA, which encodes a 262–amino acid protein containing a typical bHLH domain, was obtained in W1943 by 5′ and 3′ rapid amplification of cDNA ends (RACE) (see Supplemental Figure 3 online). The 1979-bp cDNA cloned in GLA4 was almost identical to An-1 cDNA but for a 3-bp indel causing an insertion of Ala and an SNP causing a substitution of Gly by Ala in the first exon, and two 1-bp deletions and a SNP in the 3′ untranslated region (see Supplemental Figure 4 online). The C terminus of An-1 was found to contain transcriptional activation activity in a yeast one-hybrid activation assay (see Supplemental Figure 5A online). The green fluorescent protein–An-1 fusion was predominantly located in the nucleus (see Supplemental Figures 5B and 5C online). Thus, An-1 is a transcription factor with transactivation activity.
Based on a BLAST search of a public database using the An-1 protein sequence (http://blast.ncbi.nlm.nih.gov/Blast.cgi), we constructed a phylogenetic tree of An-1 and its homologs. An-1 clustered with three grass bHLH proteins in a small branch with 100% bootstrap support (see Supplemental Figure 6 and Supplemental Data Set 1 online). This group contained bHLH proteins from Brachypodium distachyon, Aegilops tauschii, and barley, all of which have grains with long awns, indicating that those proteins might also be responsible for awn formation in those species.
As the An-1 allele from W1943 could induce long-awn formation, the corresponding allele in awnless O. sativa was denoted as an-1. We compared sequence variations between An-1 of W1943 and an-1 of Nipponbare or GLA4. Except for two SNPs in the 5′ upstream region, the sequence of an-1 in Nipponbare was nearly identical to that in GLA4. Sequence analysis of An-1 and an-1 revealed the existence of a 3-bp indel, one SNP in the coding region, and several variants in the promoter region (see Supplemental Table 3 online).
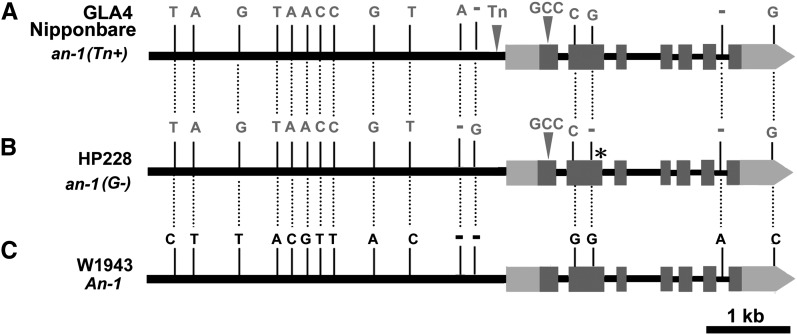
To identify possible functional variations, we sequenced about an 8-kb genomic region of An-1 covering the 4.5-kb promoter region and the 3.5-kb gene region in 27 accessions of wild rice and 43 cultivars, including 21 japonica varieties and 22 indica varieties. By comparative sequence analysis of An-1 and an-1 alleles, 12 common SNPs, four 1-bp indels, and one 4.4-kb mutator-like transposon polymorphism were detected. According to those common variants, we identified two major haplotypes in cultivars (Figures 2A and 2B) and wild rice (Figure 2C). The cultivar haplotypes could be further divided into two subhaplotypes based on a transposon-like indel in the promoter region and a 1-bp deletion in the second exon (Figures 2A and 2B). The Nipponbare subhaplotype an-1(Tn+) harbored the transposon-like indel in the promoter region (Figure 2A), while another typical subhaplotype an-1(G-) (Figure 2B) contained a 1-bp nucleotide-G deletion in the second exon of An-1 that led to a frame shift and generated a premature stop codon. The truncated protein only consists of 97 amino acids, which does not contain the bHLH domain and probably loses its functions (see Supplemental Figure 7 online). Twelve of 21 japonica varieties displayed the an-1(Tn+) subhaplotype, while 17 of 22 indica varieties displayed the an-1(G-) subhaplotype (see Supplemental Table 5 online). Among those 43 cultivars, awn length was examined in 30 cultivars; 25 were found to be awnless and five awned. Among the 25 awnless cultivars, nine japonica varieties displayed the an-1(Tn+) subhaplotype, while 13 of 16 indica cultivars displayed the an-1(G-) subhaplotype, but GLA4 displayed the an-1(Tn+) subhaplotype. The two exceptions, HP219 and HP188, displayed neither the an-1(Tn+) nor the an-1(G-) subhaplotype (see Supplemental Table 5 online). Except for two major subhaplotypes, another haplotype was also observed in a small proportion of cultivars (see Supplemental Tables 3 and 5 online). However, sequence variations were more diverse in wild rice. Although some accessions displayed the indica genotype (see Supplemental Table 5 online), ∼70% of wild rice accessions shared the same haplotype with W1943 (see Supplemental Table 3 online). Thus, sequence variations between the An-1 and an-1 haplotypes might generate awn differences between wild and cultivated rice.
Figure 2.
Typical Haplotypes of An-1/an-1 in O. sativa and O. rufipogon.
(A) The an-1(Tn+) subhaplotype in Nipponbare, GLA4, and most japonica varieties.
(B) The an-1(G-) subhaplotype in HP228 and most indica varieties.
(C) The An-1 haplotype in W1943 and most accessions of wild rice.
The common variations between cultivated rice and wild rice are indicated in this figure. Black bars represent 5′ upstream regions and introns. Light-gray bars represent 5′ and 3′ untranslated regions. Dark-gray bars represent coding regions. The triangles represent insertions. The short dashes represent single base pair deletions. The star in (B) represents the premature stop codon site in an-1(G-). Bar = 1 kb.
An-1 Promotes Awn Development and Grain Length
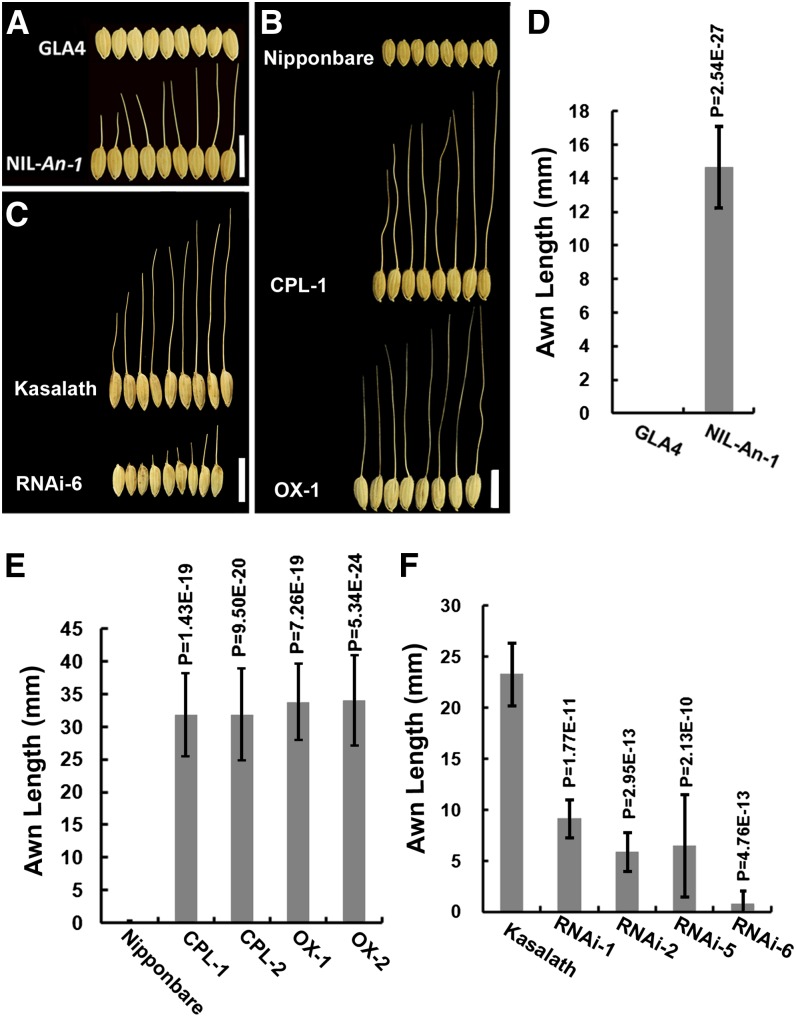
To investigate An-1 functions, we developed a near-isogenic line, NIL-An-1, that contained only a 120-kb W1943 genomic fragment of the An-1 locus in GLA4 and exhibited a stable and similar awn length and awn rate as Z3 (Figure 3A; see Supplemental Figure 1 online). The awn length of NIL-An-1 was 14.68 ± 2.45 mm, while the awn rate was 52.81% ± 12.8% (Figure 3D; see Supplemental Table 6 online).
Figure 3.
Awn Length Comparison.
(A) Photograph of apical grains of primary branches in GLA4 and NIL-An-1.
(B) Photograph of apical grains of primary branches in Nipponbare, CPL-1, and OX-1.
(C) Photograph of apical grains of primary branches in Kasalath and RNAi-6.
(D) Awn length comparison between GLA4 and NIL-An-1. Whereas GLA4 lacked awns, the awns of NIL-An-1 were 10 to 25 mm long.
(E) Awn length comparison among Nipponbare, CPL, and OX. There was no visible awn in Nipponbare, but the awns of both the An-1 complementation plants and An-1–overexpressing plants were 30 to 50 mm long.
(F) Awn length comparison among RNAi lines and Kasalath. The awns of Kasalath were 15 to 40 mm long and shorter in the RNAi plants.
CPL, An-1 complementation plants; OX, An-1–overexpressing plants; RNAi, An-1 suppression plants. Bars = 10 mm. In (D) to (F), for GLA4, NIL-An-1, Nipponbare, and Kasalath, the sample size was n = 60. For transgenic lines, the sample size was n = 30. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
[See online article for color version of this figure.]
For complementation and RNAi experiments, we performed the analysis on single-copy T2 homozygotes of CPL and RNAi transgenic lines. For the overexpression study, we just analyzed the T0 OX transgenic plants. The awn length and awn rate were greatly increased in CPL and OX plants compared with those in Nipponbare. The awn length was 31.84 ± 6.36 mm and 31.90 ± 6.98 mm, while the awn rate was 54.48% ± 14.41% and 53.21% ± 15.14%, respectively, in CPL-1 and CPL-2 plants. The awn length was 33.82 ± 5.81 mm and 34.04 ± 6.99 mm, while the awn rate was 63.41% ± 14.19% and 65.74% ± 16.82%, respectively, in OX-1 and OX-5 plants (Figures 3B and 3E; see Supplemental Table 6 online).
Kasalath harboring the pRNAi construct exhibited shortened awns and a reduced awn rate (Figure 3C). The awn length of RNAi plants ranged from 0.82 ± 1.29 mm to 9.14 ± 1.87 mm, while the awn length of Kasalath was 23.30 ± 3.05 mm (Figure 3F). The awn rate of RNAi plants ranged from 4.25% ± 3.44% to 23.84% ± 9.49%, which was lower than the 56.32% ± 4.11% awn rate in Kasalath (see Supplemental Table 6 online).
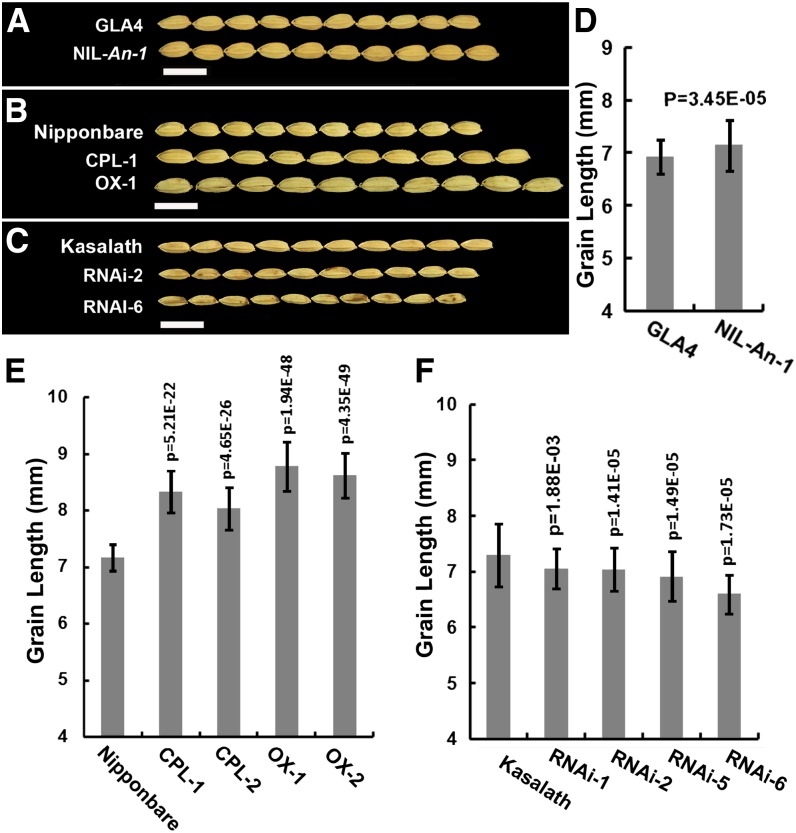
In addition to awn elongation, An-1 also regulated grain length. The grains of NIL-An-1 were 3.19% longer than those of GLA4 (Figures 4A and 4D). The grain length in CPL-1 and CPL-2 plants was 13.90 and 11.97% longer, respectively, than that in Nipponbare. The grain length in OX-1 and OX-5 plants increased 19.99 and 16.74%, respectively, compared with that in Nipponbare (Figures 4B and 4E). By contrast, the grain length in RNAi lines was decreased, being 3.30 to 9.60% shorter than those of Kasalath (Figures 4C and 4F). Therefore, An-1 is a major regulator of awn length and awn rate and also plays a role in grain elongation.
Figure 4.
Grain Length Comparison.
(A) Photograph of 10 grains of GLA4 and NIL-An-1.
(B) Photograph of 10 grains of Nipponbare, CPL-1, and OX-1.
(C) Photograph of 10 grains of Kasalath, RNAi-2, and RNAi-6.
(D) Grain length comparison between GLA4 and NIL-An-1; grains in NIL-An-1 are slightly longer than those in GLA4.
(E) Grain length comparison among CPL, OX, and Nipponbare; grains in CPL and OX are much longer than those in Nipponbare.
(F) Grain length comparison among RNAi lines and Kasalath; grains in RNAi lines are shorter than those in Kasalath.
CPL, An-1 complementation plants; OX, An-1–overexpressing plants; RNAi, An-1 suppression plants. Bars = 10 mm. In (D) to (F), the sample size for all analyses was n = 100. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
[See online article for color version of this figure.]
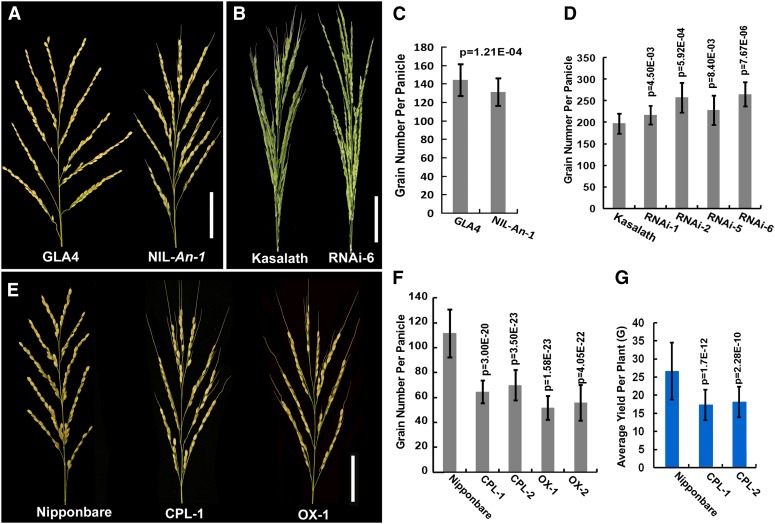
An-1 Negatively Regulates Grain Number per Panicle and, Thus, an-1 Increases Yield per Plant
Except for awn elongation and grain elongation, An-1 plays an additional role in regulating grain number per panicle. NIL-An-1 generated 10.58% fewer grains per panicle than GLA4 (Figures 5A and 5C). However, the RNAi plants generated more grains per panicle. The different RNAi lines produced 13.60 to 38.4% more grains per panicle than Kasalath (Figures 5B and 5D). The examination of complementary and overexpressing plants revealed similar changes in grain number per panicle as found in NIL-An-1 (Figure 5E). Nipponbare generated 9.67 ± 1.57 primary branches, 19.76 ± 3.95 secondary branches, and 111.56 ± 19.29 grains per panicle. Both primary branches and secondary branches were reduced in CPL plants and overexpressing plants (see Supplemental Table 6 online). The CPL-1 and CPL-2 plants produced 63.61 ± 9.17 and 68.47 ± 12.19 grains per panicle separately, while OX-1 and OX-5 plants produced 51.79 ± 9.57 and 55.82 ± 14.34 grains per panicle separately (Figure 5F). The grain number per panicle decreased by 42.08, 38.62, 54.58, and 49.96% in CPL-1, CPL-2, OX-1, and OX-5 plants, respectively, compared with that in Nipponbare. The panicle phenotypes of CPL plants were more severe than those of NIL-An-1, which might be due to the different genetic backgrounds.
Figure 5.
Grain Number per Panicle and Yield per Plant Comparison.
(A) Photograph of the panicles in GLA4 and NIL-An-1.
(B) Photograph of the panicles in Kasalath and RNAi-6.
(C) Grain number per panicle comparison between GLA4 and NIL-An-1; the grain number per panicle in NIL-An-1 is slightly lower than that in GLA4.
(D) Grain number per panicle comparison among RNAi lines and Kasalath; grain number per panicle was greater in RNAi lines than in Kasalath.
(E) Photograph of the panicles in Nipponbare, CPL-1, and OX-1.
(F) Grain number per panicle comparison among Nipponbare, CPL, and OX lines; grain number was less in CPL and OX than in Nipponbare.
(G) Yield per plant comparison among Nipponbare, CPL-1, and CPL-2.
CPL, An-1 complementation plants; OX, An-1–overexpressing plants; RNAi, An-1 suppression plants. Bars = 50 mm. In (C), (D), and (F), for GLA4, NIL-An-1, Nipponbare, and Kasalath, sample size was n = 60. For transgenic lines, sample size was n = 30. In (G), sample size was n = 48. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
[See online article for color version of this figure.]
An analysis of yield per plant was performed on Nipponbare and CPL plants. The analysis showed that yield per plant decreased by 34.83 and 30.80% in CPL-1 and CPL-2 plants, respectively, compared with that in Nipponbare (Figure 5G). Thus, introducing the W1943 allele An-1 into cultivated rice reduced grain number per panicle and yield per plant.
An-1 Expression Level Changes in NIL-An-1 and Transgenic Plants
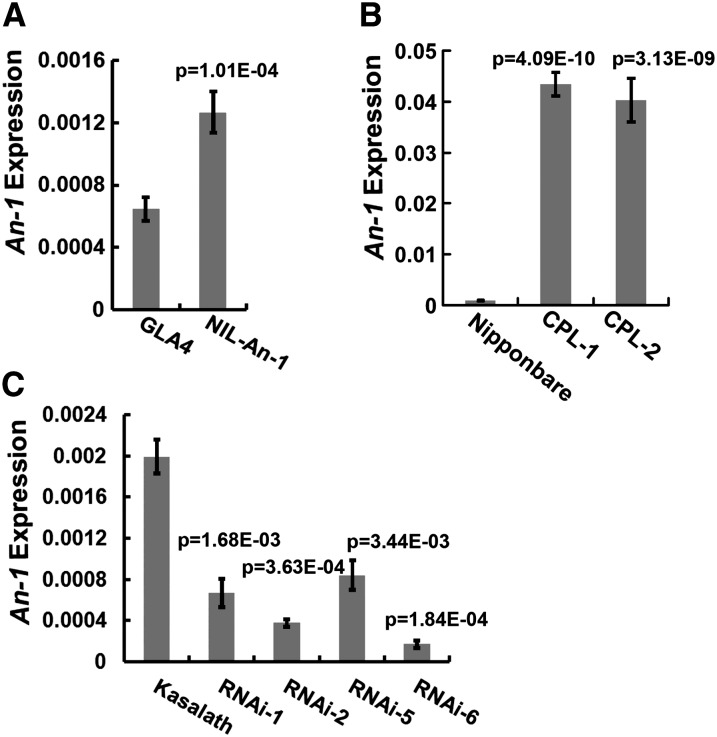
To characterize the An-1 expression level changes underlying the panicle phenotypes, we collected young panicles <4 cm and analyzed An-1 expression using real-time quantitative RT-PCR (qRT-PCR). The results showed that An-1 expression of young panicles was increased twofold in NIL-An-1 compared with that in GLA4 (Figure 6A). For complementary plants, An-1 expression increased nearly 40- to 45-fold in CPL-1 and CPL-2 plants compared with that in Nipponbare (Figure 6B). Considering both CPL lines in analysis contained only a single-copy transgene, expression level having been raised thus high might be mainly due to insertion of W1943 An-1 allele in different genetic background. In RNAi plants, the expression of endogenous An-1 gene decreased (Figure 6C). The extent of An-1 downregulation was highly correlated with panicle phenotypes in RNAi plants. The qRT-PCR results and panicle phenotypes suggested An-1 expression level was directly proportional to awn length and grain length but inversely proportional to grain number per panicle.
Figure 6.
An-1 Expression Level Comparison.
(A) The An-1 transcript level comparison between GLA4 and NIL-An-1 panicles.
(B) The An-1 transcript level comparison among Nipponbare, CPL-1, and CPL-2 panicles.
(C) The An-1 transcript level comparison between Kasalath and RNAi panicles.
Young panicles <4 cm high were collected and subjected to qRT-PCR. The data represent the average of three independent biological replicates and were normalized to the EF1α gene as a reference. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd. CPL, An-1 complementation plants; RNAi, An-1 suppression plants.
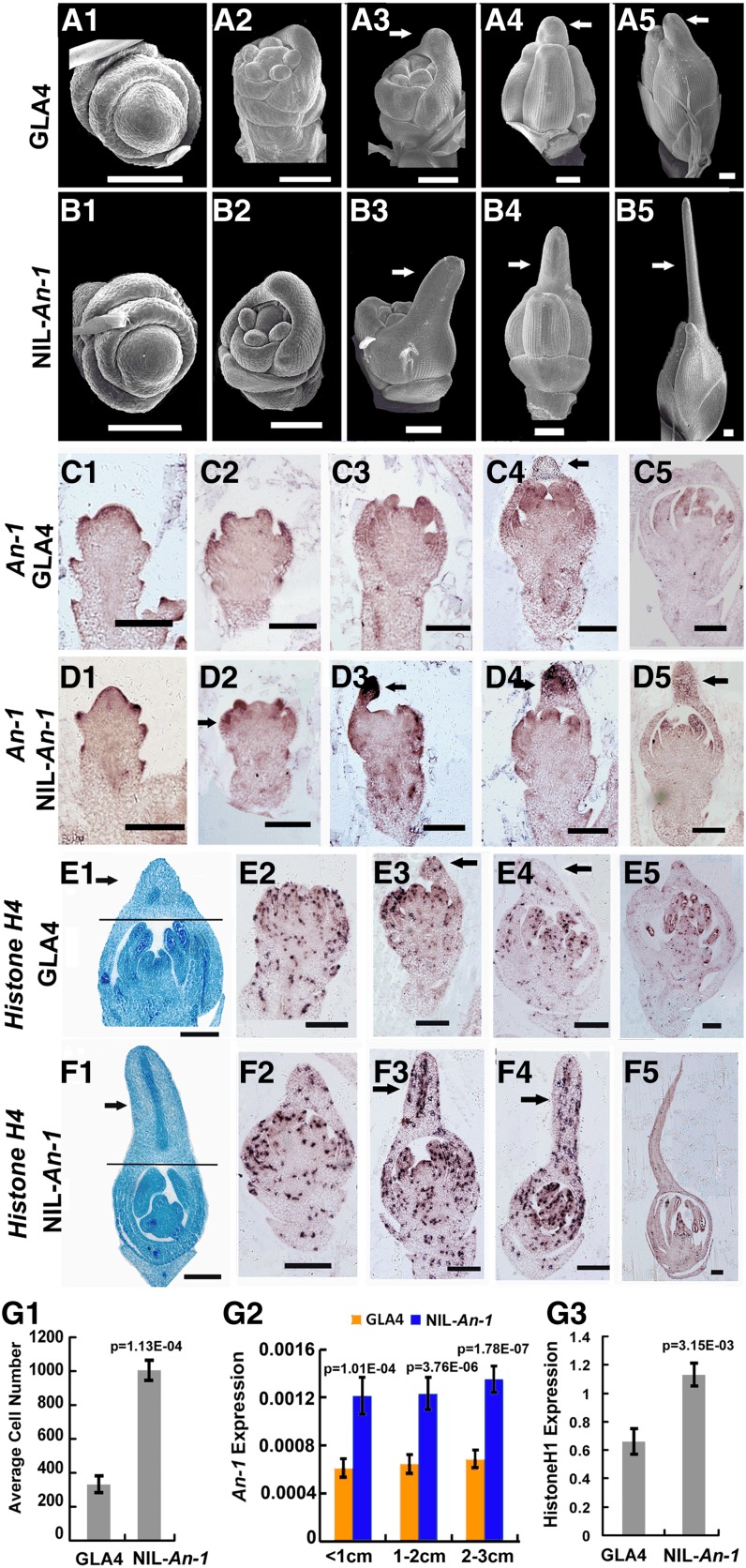
Specific Expression of An-1 Induces Cell Division and Awn Development
To identify the specific stage when the awn differentiated, we compared the spikelet development between GLA4 and NIL-An-1 using scanning electron microscopy. According to the rice spikelet development (Sp) stages defined before (Itoh et al., 2005), lemma primordia initiated at the Sp3 stage and palea primordia formed at the Sp4 stage. No difference was found on spikelet primordia until the Sp6 stage between NIL-An-1 and GLA4 (Figures 7A1 and 7B1). At the Sp6 stage, the lemma primordia in NIL-An-1 grew faster than that in GLA4 (Figures 7A2 and 7B2). At the Sp7 stage, the apex of the lemma in NIL-An-1 protruded and the awn primordia formed, whereas no awn primordia formed in GLA4 (Figures 7A3 and 7B3). At the Sp8 early stage (Sp8e) when lemma and palea were gradually closed, awn primordia extended further in NIL-An-1, while the apex of lemma formed a round tip in GLA4 (Figures 7A4 and 7B4). In the Sp8 late stage (Sp8l), the awn primordia kept extending, but the apex of lemma stopped growth in GL A4 (Figures 7A5 and 7B5).
Figure 7.
Awn Development and Expression Analysis of An-1 and Histone Hs in GLA4 and NIL-An-1.
(A1) to (A5) Scanning electron microscopy images of spikelets at different developmental stages in GLA4. Arrows point to the apex of lemma primordia.
(B1) to (B5) Scanning electron microscopy images of spikelets at different developmental stages in NIL-An-1. Arrows point to awn primordia.
(C1) to (C5) Expression patterns of An-1 during spikelet development in GLA4. Arrow points to the apex of lemma primordia.
(D1) to (D5) Expression patterns of An-1 during spikelet development in NIL-An-1. Arrows point to awn primordia.
(E1) A longitudinal histological section of a spikelet at the Sp8e stage in GLA4. The arrow points to the apex of the lemma primordium. The cells above the black line were quantified.
(E2) to (E5) Expression patterns of Histone H4 during spikelet development in GLA4. Arrows point to the apex of lemma primordia.
(F1) A longitudinal histological section of a spikelet at the Sp8e stage in NIL-An-1. The arrow points to awn primordium. The cells above the black line were quantified.
(F2) to (F5) Expression patterns of Histone H4 during spikelet development in NIL-An-1. Arrows point to awn primordia.
(G1) Cell number comparison between the apices of spikelets in GLA4 and awn primordia in NIL-An-1.
(G2) A comparison of An-1 expression in the 1- to 3-cm young panicles of NIL-An-1 and GLA4.
(G3) A comparison of Histone H1 expression in the young panicles of NIL-An-1 and GLA4.
(A1), (B1), (C1), and (D1) Sp4-Sp5, formation of lemma and palea primordia stage.
(A2), (B2) (C2), (D2), (E2), and (F2) Sp6, formation of stamen primordia stage.
(A3), (B3), (C3), (D3), (E3), and (F3) Sp7, formation of carpel primordia stage.
(A4), (B4), (C4), (D4), (E4), and (F4) Sp8e (Sp8 early), differentiation of ovule and pollen stage.
(A5), (B5), (C5), (D5), (E5), and (F5) Sp8l (Sp8 late), differentiation of ovule and pollen stage.
In (G1), sample size was as follows: GLA4 (n = 8) and NIL-An-1 (n = 8). In (G2) to (G3), the data represent the average of three independent biological replicates and were normalized to the EF1α gene as a reference. The statistical significance was at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd. Bars = 100 μm.
To determine how An-1 regulates awn development, we examined the expression pattern of An-1 by RNA in situ hybridization. During spikelet development, An-1 transcripts were first detected at the two rudimentary glume primordia and two empty glume primordia, next at the lemma and palea primordia (Figures 7C1 and 7D1), and then at the stamen (Figures 7C2and 7D2) and carpel primordia (Figures 7C3 and 7D3). During the early stages of spikelet development, the An-1 expression pattern in NIL-An-1 was the same as that in GLA4. Differences emerged in the Sp6 stage, when the lemma and palea elongated; An-1 expression increased gradually at the apices of lemma primordia, which would subsequently form awn primordia in NIL-An-1 (Figure 7D2). Then, An-1 expression strongly increased in awn primordia of Sp6 (Figure 7D3), was maintained until Sp8e (Figure 7D4), and gradually faded during the Sp8l stage (Figure 7D5). By contrast, An-1 was consistently weakly expressed in all floral organ primordia in GLA4 (Figures 7C2 and 7C3) but not expressed in the apex of the lemma (Figure 7C4). Finally, An-1 expression almost ceased in the lemma and palea during the Sp8l stage in GLA4 (Figure 7C5). The specific expression of An-1 at the apices of lemma and awn primordia in NIL-An-1 demonstrated its role in awn initiation and formation.
We further examined An-1 expression level in young panicles at different developmental stages using qRT-PCR. The results showed that within 1- to 3-cm young panicles, An-1 expression was about 2 times higher in NIL-An-1 than in GLA4 in each pair of samples (Figure 7G2). In summary, both the pattern and level of An-1 expression contributed to the awn phenotypes in NIL-An-1.
On historical sections of spikelets at the Sp8e stage, we quantified the number of cells in awn primordia and spikelet apices (Figures 7E1 and 7F1, above the black lines). The awn primordia in NIL-An-1 contained 3 times more cells than the apex of lemma in GLA4 (Figures 7E1, 7F1, and 7G1), indicating that cell division plays a crucial role in awn formation. As HISTONE Hs are expressed during the G1-S phase, their expression is usually used as a marker for cell division (Marzluff and Duronio, 2002). RNA in situ hybridization indicated that Histone H4 was evenly expressed at the early floral development stages, and no difference was found between NIL-An-1 and GLA4 (Figures 7E2 and 7F2). However, from the Sp7 stage, when awn primordia formed, Histone H4 was highly expressed in the awn primordia, lemma, and palea primordia of young spikelets in NIL-An-1 (Figures 7F3 and 7F4), whereas Histone H4 expression at the lemma and palea primordia gradually weakened in GLA4 (Figures 7E3 and 7E4). In Sp8l spikelets, Histone H4 transcript was almost absent from the lemma and palea and only stayed in pollen and ovule tissues in both NIL-An-1 and GLA4 (Figures 7E5 and 7F5). A qRT-PCR analysis further revealed that Histone H1 expression in NIL-An-1 young panicles was almost 2 times higher than that in GLA4 (Figure 7G3), which indicated that cell division was more active in NIL-An-1 primordia than in the apices of GLA4 spikelets.
Therefore, the specific expression of An-1 could induce continually rapid cell division at the apex of the lemma to lead to awn primordia formation, which was confirmed by Histone Hs expression. By contrast, genetic variations of an-1(Tn+) eliminated its expression at the apex of lemma, resulting in loss of awns in GLA4.
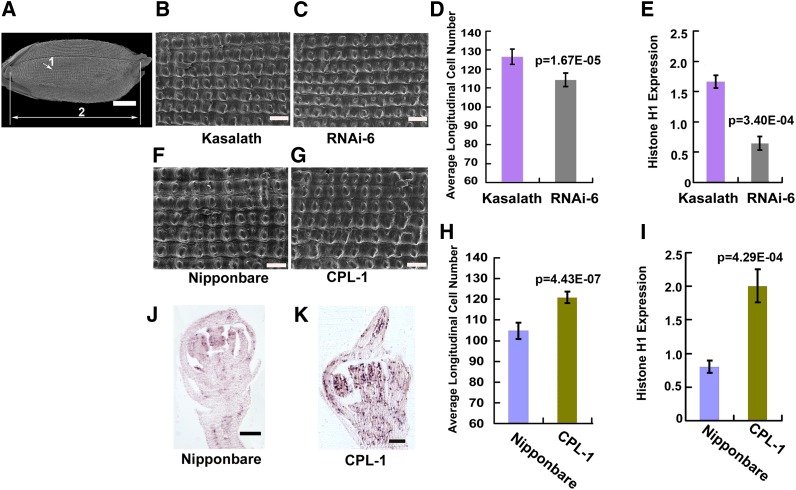
An-1 Drives Grain Elongation through Cell Division
The cell number and cell size of the outer epidermis of grains were examined to determine what caused the changes in grain length in transgenic plants. The cell number of a single line was quantified along the long axis of the lemma using scanning electron microscopy images (Figure 8A). Scanning electron microscopy images revealed that the outer epidermal cell size at the lemma did not change obviously in RNAi-6 grains compared with that in Kasalath grains (Figures 8B and 8C), whereas quantification of epidermal cells in the lemma showed that cell number decreased by 9.68% in RNAi-6 grains compared with that in Kasalath grains (Figure 8D). The qRT-PCR results confirmed that Histone H1 expression in the young panicles in RNAi plants decreased to about half of that in Kasalath (Figure 8E).
Figure 8.
Epidermis Cell Number Comparison and Histone Hs Expression Analysis.
(A) Scanning electron microscopy photograph of a whole grain. Arrow 1 indicates the magnified area on the outer epidermis of the lemma, and arrow 2 represents the longitudinal axis along which cell number was quantified. Bar = 1 mm.
(B) and (C) Scanning electron microscopy photographs of lemma outer epidermis cells in Kasalath and RNAi-6 grains. Bars = 100 μm.
(D) Comparison of average cell number along the longitudinal axis between Kasalath and RNAi-6 grains.
(E) qRT-PCR analysis of Histone H1 expression in 3- to 4-cm young panicles of Kasalath and RNAi-6.
(F) and (G) Scanning electron microscopy photographs of lemma outer epidermis cells in Nipponbare and CPL-1 grains. Bars = 100 μm.
(H) Comparison of the average cell number along the longitudinal axis between Nipponbare and CPL-1 grains.
(I) qRT-PCR analysis of Histone H1 expression in the 3-4 cm young panicles of Nipponbare and CPL-1.
(J) Expression pattern of Histone H4 in spikelets at the Sp8l stage in Nipponbare.
(K) Expression pattern of Histone H4 in spikelets at the Sp8l stage in CPL-1. Bars = 100 μm.
CPL, An-1 complementation plants; RNAi, An-1 suppression plants. In (D) and (H), sample size was n = 8 for all analyses. In (E) and (I), the data represented the average of three independent biological replicates and were normalized to the EF1α gene as a reference. The statistical significance was at P < 0.05 based on two-tailed Student’s t test. Error bars represent the sd.
The epidermal cell size in the lemma was nearly the same in CPL-1 grains as in Nipponbare grains (Figures 8F and 8G). However, the epidermal cell number in CPL-1 was 14.16% more than that in Nipponbare (Figure 8H). Accordingly, Histone H1 expression in CPL young panicles was more than twofold upregulated compared with that in Nipponbare (Figure 8I). RNA in situ hybridization also revealed that Histone H4 was expressed in the lemma and palea primordia until the Sp8l stage in the CPL spikelet, whereas no Histone H4 transcript was detected in the Nipponbare spikelet at the same stage (Figures 8J and 8K). We conclude that prolonged cell division led to an increase in both cell number and grain length in CPL plants.
Upregulation of An-1 in the Inflorescence Reduces Meristem Activity and Grain Number per Panicle
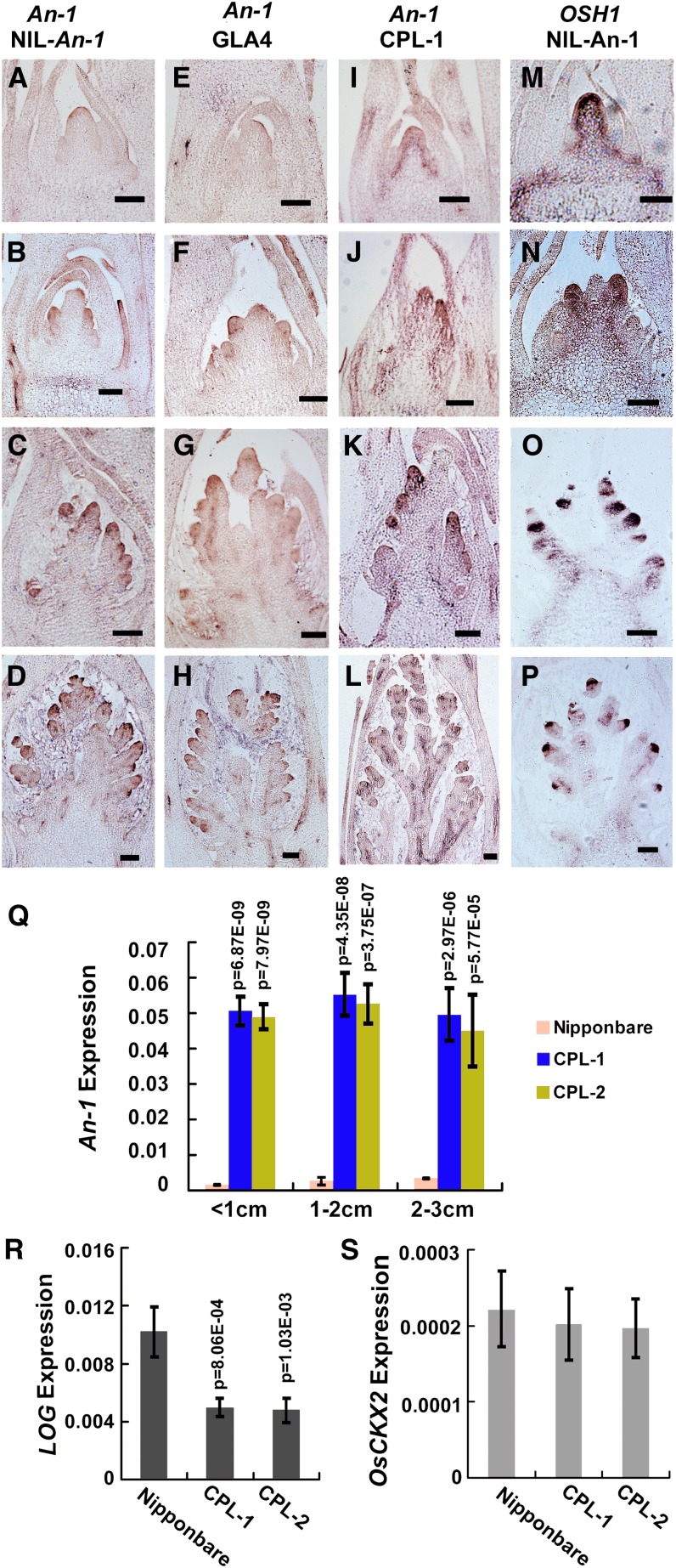
To investigate how An-1 regulates grain number per panicle, we characterized the pattern and level of An-1 expression during inflorescence development. As defined before, rice inflorescence development was divided into nine stages (Itoh et al., 2005). During the early stages of inflorescence development, An-1 shared a similar expression pattern in GLA4, NIL-An-1, and CPL-1. At the In1 stage, when bract primordia formed, An-1 began to be weakly expressed at the surface layers of the rachis meristem in both NIL-An-1 and GLA4 (Figures 9A and 9E), while An-1 expression in CPL-1 increased and expanded to the pith and procambium of the rachis meristem (Figure 9I). At the In2-3 stage, An-1 was moderately expressed at the primary branch meristem in NIL-An-1 and GLA4, but strongly expressed in CPL-1 (Figures 9B, 9F, and 9J). Once the secondary branch meristem initiated at the In5 stage, An-1 transcript was detected at the secondary branch meristem in NIL-An-1, GLA4, and CPL-1 (Figures 9C, 9G, and 9K). From stage In6, when spikelet development started, An-1 expression appeared at the spikelet primordia (Figures 9D, 9H, and 9L). No specific signal was detected at any stage of inflorescence development on An-1 sense control sections (see Supplemental Figures 8A to 8C online). Then, we quantified the An-1 expression level in young panicles at different developmental stages. Within 1- to 3-cm young panicles, the An-1 expression was highly increased in CPL transgenic plants compared with in Nipponbare in each group of samples (Figure 9Q).
Figure 9.
Expression Analysis of An-1, OSH1, LOG, and Os CKX2 during Panicle Development in NIL-An-1, GLA4, and CPL-1.
(A) to (D) Expression patterns of An-1 in young NIL-An-1 panicles.
(E) to (H) Expression patterns of An-1 in young GLA4 panicles.
(I) to (L) Expression patterns of An-1 in young CPL-1 panicles.
(M) to (P) Expression patterns of OSH1 in young NIL-An-1 panicles.
(A), (E), (I), and (M) Rachis meristem stage.
(B), (F), (J), and (N) Primary branch formation stage.
(C), (G), (K), and (O) Secondary branch formation stage.
(D), (H), (L), and (P) Spikelet primordia formation stage. Bars = 100 μm.
(Q) A comparison of An-1 expression in the 1- to 3-cm young panicles of Nipponbare, CPL-1, and CPL-2 plants.
(R) A comparison of LOG expression in young panicles <1 cm of Nipponbare, CPL-1, and CPL-2.
(S) A comparison of OsCKX2 expression in young panicles <1 cm of Nipponbare, CPL-1, and CPL-2.
In (Q) to (S), the data represent the average of three independent biological replicates and were normalized to the EF1α gene as a reference. The statistical significance was at P < 0.05 based on two-tailed Student’s t test. Error bars represent the sd.
We further compared the expression patterns between OSH1 and An-1 in NIL-An-1. At the In1-5 stages, OSH1 was highly expressed in the rachis meristem, primary branch meristem, and secondary branch meristem (Figures 9M to 9O), where An-1 expression was relatively low. The differences in expression pattern between An-1 and OSH1 began at In6, when spikelet primordia initiated. OSH1 was only expressed in the floral meristem, but An-1 started its expression at floral organ primordia (Figures 9D and 9P). Although both genes were coexpressed during the early stages of inflorescence development, phenotypes in NIL-An-1 and transgenic plants indicated that An-1 might have an opposite role in meristem maintenance compared with OSH1.
Considering the expression of An-1 in the rachis meristem, the primary branch meristem, and the secondary branch meristem, we predicted that the increased An-1 expression level in NIL-An-1 and CPL might influence the expression of genes that are involved in meristem maintenance and grain number per panicle. LONELY GUY (LOG) and Grain number1a (Gn1a) are known to regulate cytokinin concentration and grain number per panicle in rice. LOG encodes a key enzyme that converts inactive cytokinin to the biologically active form, whereas Gn1a encodes cytokinin oxidase (Os CKX2), which degrades cytokinin (Ashikari et al., 2005; Kurakawa et al., 2007). The two genes directly determined the concentration of active cytokinin, which plays an important role in meristem maintenance (Leibfried et al., 2005; Chickarmane et al., 2012). We analyzed the expression level of LOG and Os CKX2 in CPL plants, which exhibit a significant reduction in grain number per panicle. The qRT-PCR results showed that LOG expression in the young panicles (<1 cm) of both CPL-1 and CPL-2 plants was downregulated to about half of that in Nipponbare (Figure 9R), while Os CKX2 expression did not change markedly between CPL and Nipponbare (Figure 9S). Therefore, upregulation of An-1 expression during the early stages of inflorescence formation could downregulate LOG expression to reduce meristem activity and then reduce grain number per panicle and yield per plant in CPL plants.
Neutrality Test and Phylogenetic Tree of An-1
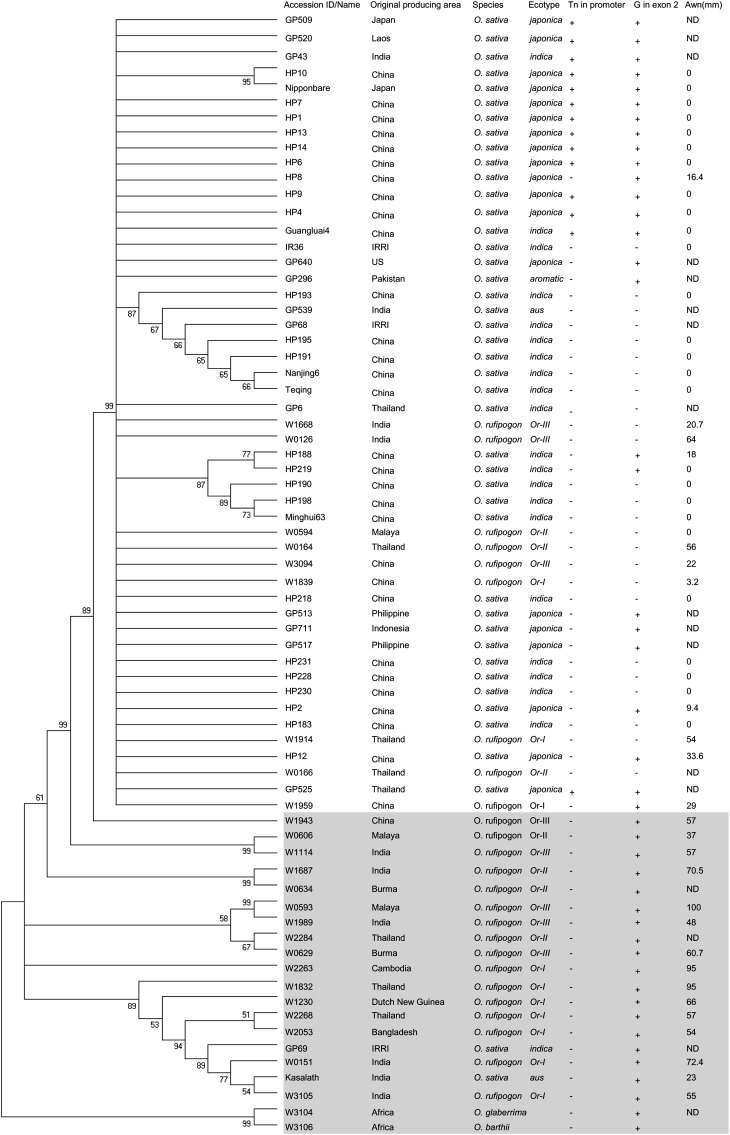
To investigate whether An-1 was subjected to artificial selection, nucleotide diversity was calculated and a neutrality test was performed based on total polymorphic sites of An-1 sequences (∼7 kb, including its flanking genomic region). The nucleotide diversity of An-1 in cultivated rice (π = 0.00073; θw = 0.00229) was significantly reduced compared with that of An-1 in wild rice (π = 0.01012; θw = 0.01174) (Table 1). The Tajima’ D test results revealed that only An-1 in cultivated rice deviated significantly from the neutral expectation (P < 0.01), which indicated that An-1 might have been subjected to artificial directional selection (Table 1). We also noticed that An-1 was located around a major domestication sweep (see Supplemental Figure 9 online) that showed the third strongest selection signal across the rice genome (Huang et al., 2012). A combination of the above analysis of An-1 and its chromosome location suggest that An-1 was probably an important target during rice domestication. Moreover, we constructed a phylogenetic tree based on the An-1 sequence from 43 cultivated rice varieties and 27 wild rice accessions. Two accessions, W3106 (Oryza barthii) and W3104 (Oryza glaberrima), were selected as outgroup. A phylogenetic analysis of multiple An-1 sequences showed that nearly all of the cultivars grouped together tightly, which revealed that these cultivars might have originated from a common progenitor in a single domestication event (Figure 10).
Table 1. Nucleotide Diversity and Tajima’s D Test.
| Taxon | N | L | H | S | π | θw | Tajima's D |
|---|---|---|---|---|---|---|---|
| O. sativa | 82 | 6791 | 18 | 78 | 0.00073 | 0.00229 | −2.28439 P < 0.01 |
| O. rufipogon | 50 | 6789 | 17 | 363 | 0.01012 | 0.01174 | −0.55367 P > 0.10 |
N, total number of sequences; L, average length (bp) of the sequences per taxon; H, total number of haplotypes per taxon; S, total number of polymorphic sites per taxon; π, average number of pairwise nucleotide differences per site calculated based on the total number of polymorphic sites; θw, Watterson’s estimator of per base pair calculated based on the total number of polymorphic sites. Kasalath and GP69 were not included in the analysis.
Figure 10.
Phylogenetic Tree of An-1 in Rice.
Maximum parsimony phylogenetic tree of An-1 in rice cultivars and wild relatives. Bootstrap values were estimated (with 1000 replicates) to assess the relative support for each branch. The bootstrap values of 50% and above are indicated on the tree. ND, not detected. Alignments used to generate the phylogeny are presented in Supplemental Data Set 2 online.
DISCUSSION
An-1 Has Pleiotropic Effects on Awn Development, Grain Length, and Grain Number per Panicle in Rice
Since long awns are a vital trait for seed dispersal and implantation in wild rice, many important genes are involved in awn development. Twenty QTLs for awn length were identified on nine chromosomes by different mapping populations (http://www.gramene.org). However, genes involved in awn development have not hitherto been molecularly identified in rice. Recently, Lks2, which regulates awn length in barley, was cloned and natural variation in the conserved domain of Lks2 led to short awns in some varieties. The putative ortholog of Lks2 in rice is Os06g0712600, but no QTL for awn length has been mapped within this region in rice (Yuo et al., 2012).
In our study, two QTLs, An-1 and An-2, were mapped on chromosome 4. We cloned and functionally characterized the major QTL, An-1. When a single W1943 An-1 allele was introduced into an awnless variety, GLA4 or Nipponbare, long awns were induced in NIL-An-1 and the transgenic plants. RNAi of An-1 in long-awned Kasalath led to shorter and fewer awns.
Scanning electron microscopy revealed that awn primordia formed in NIL-An-1 from the Sp6 stage. Specific expression of An-1 at the apex of lemma at the Sp6 stage promoted continuous and rapid cell division at the apex of lemma, which induced awn primordia formation. Conversely, the an-1(Tn+) allele eliminated its expression at the apex of lemma and caused loss of awn initiation in GLA4. Unlike Lks2 in barley that only regulates awn length (Yuo et al., 2012), An-1 regulated both awn initiation and awn length. Therefore, An-1 is a major regulator of awn development in rice.
Furthermore, we found long awn induction accompanied by grain elongation and reduction of grain number per panicle in both NIL-An-1 and transgenic plants. The An-1 expression level was highly correlated with grain length and grain number per panicle. Therefore, An-1 is a multifunctional gene with pleiotropic roles in rice development.
An-1 Promotes Cell Division
An-1 encodes a typical bHLH transcription factor, which contains a bHLH DNA binding domain and transcriptional activation activity. The bHLH transcription factors belong to a large gene family, which contains 147 members in Arabidopsis thaliana and 167 members in O. sativa (Li et al., 2006b). The bHLH genes in O. sativa have been reported to be involved in grain size, leaf and root hair development, pericarp color, and axillary meristem formation (Sweeney et al., 2006; Ding et al., 2009; Zhang et al., 2009; Heang and Sassa, 2012; Yang et al., 2012). As a member of the bHLH gene family in rice, An-1 regulates awn development, seed elongation, and grain number.
Awn formation is dependent on cell division, which was confirmed by RNA in situ hybridization of Histone H4 expression. Grain elongation is also promoted by enhancing cell division. The upregulation of An-1 in CPL plants resulted in a corresponding increase in Histone H1 expression, whereas suppression of An-1 downregulated Histone H1 expression in RNAi plants. Therefore, An-1 might directly or indirectly promote cell division in rice. SPATULA (SPT) and BIGPETALp (BPEp) are two members of the bHLH family in Arabidopsis. SPT plays pleiotropic roles in carpel development, root growth, and leaf size (Heisler et al., 2001; Ichihashi et al., 2010; Makkena and Lamb, 2013). In the spt loss-of-function mutant, root meristem size and leaf size increased due to increased cell number (Ichihashi et al., 2010; Makkena and Lamb, 2013). BPEp interacts with AUXIN RESPONSE FACTOR8 to limit cell division during the early stages of petal developmental and limit cell expansion during later stages (Varaud et al., 2011). Both SPT and BPEp are negative regulators of cell division, whereas An-1 is a positive regulator of cell division.
An-1 Downregulates Meristematic Activity and Grain Number per Panicle
Except for regulating awn development and grain elongation, An-1 also affects grain number per panicle. Cytokinin concentration has been reported to affect grain number per panicle and yield in rice (Ashikari et al., 2005; Kurakawa et al., 2007; Li et al., 2013). Grain number per panicle depends on the meristematic activity of the inflorescence. Both OSH1 and cytokinin are key players in maintaining meristem identity (Sato et al., 1996; Leibfried et al., 2005; Tsuda et al., 2011; Chickarmane et al., 2012). There is reciprocal positive regulation between OSH1 and cytokinin (Rupp et al., 1999; Frank et al., 2000; Yanai et al., 2005). RNA in situ hybridization analysis showed that An-1 is coexpressed with OSH1 in the rachis meristem and primary and secondary branch meristems during earlier stages of inflorescence development. Based on the panicle phenotypes in NIL-An-1 and transgenic plants, we considered that An-1 functions as a negative regulator in maintaining meristematic activity. Upregulation of An-1 led to downregulation of LOG expression, which might reduce the active cytokinin level as well as meristem identity. Although in the vivo cytokinin concentration was not measured in our study, the reduction in grain number per panicle and yield per plant coincided with the downregulation of LOG in transgenic plants. Therefore, we propose that An-1 is a negative regulator of meristem identity. In cultivated rice, an-1(Tn+) reduced An-1 expression and an-1(G-) abolished An-1 function, which increased meristematic activity of the inflorescence and subsequently increased grain number per panicle and yield per plant.
An-1 Is a Major Target of Artificial Selection
The results in this study revealed that formation of long awns was accompanied by a reduction of grain number per panicle and yield per plant. In contrast with long awns increasing yield in barley and wheat (Abebe et al., 2010), long-awn formation reduced yield per plant in rice, which might partly explain why the long awn trait was under strong artificial selection during rice domestication.
Tajima’s D statistic is widely used as a neutrality test. Positive values of Tajima’s D arise from an excess of intermediate frequency alleles, indicating a decrease in population size and/or balancing selection. Negative values of Tajima’s D arise from an excess of low-frequency alleles and indicate population expansions (e.g., after a bottleneck or a selective sweep) and/or purifying selection (Tajima, 1989). The reduced nucleotide diversity of an-1 in cultivated rice and negative Tajima’ D statistics indicated that An-1 in cultivated rice might have been subjected to artificial selection. According to a recent large genomic investigation of cultivated rice and wild rice, 55 selective sweeps occurred during rice domestication (Huang et al., 2012). Like all well-characterized domestication genes, An-1 locates in one of the selective sweeps on chromosome 4, which is the third strongest selective sweep. This indicates that An-1 is a major regulator of awn and grain number and is a main target of artificial selection with strong selection pressure.
The phylogenic tree revealed that nearly all cultivars of O. sativa grouped together. This suggested that An-1 might originate from a common progenitor with a single domestication event. Although phylogenic analysis revealed that an-1 of cultivated rice originated from a single domestication event, we also detected two major haplotypes in cultivated rice. The flanking sequence comparison between An-1 and an-1 loci revealed that this gene is located in a complicated genomic context, comprised of multiple reciprocal indels of transposon-like or repeat sequence. This demonstrates that the evolution process of the An-1 locus during domestication might have undergone multiple recombination events or genetic drifts.
METHODS
Plant Materials
In QTL mapping, map-based cloning, and subsequent analysis of An-1, we used an awnless cultivar GLA4 and Nipponbare, awned Kasalath, and awned SL4, Z3, and NIL-An-1. Z3 was screened from the BC5F3 population of SL4 and GLA4. NIL-An-1 was identified in a BC1F3 population derived from the cross between Z3 and GLA4. The 43 varieties of cultivated rice (Oryza sativa) and the 28 accessions of wild rice used in the phylogenetic analysis are listed in Supplemental Table 7 online.
Primers
The primers used in this study are listed in Supplemental Table 8 online.
QTL Mapping and Fine Mapping of An-1
An F2 population, containing 255 plants, from the cross between SL4 and GLA4 was constructed for QTL mapping. The total 28 indel primers were used to construct a genetic linkage map of chromosome 4 by Joinmap4.0 (Stam, 1993). QTL Cartographer V2.5 software was used to detect two QTLs of An-1 and An-2 on chromosome 4 (Wang et al., 2007). An F2 population containing 10,500 plants was raised from the cross between Z3 and GLA4 for fine mapping of An-1. The molecular markers used in QTL mapping and fine mapping are listed in Supplemental Table 8 online. The An-1 locus was finally narrowed down to an interval between SNP markers FM3 and FM6.
BAC Screening, Sequencing, Assembly, and Annotation
The BAC clone ORW9143Ba0047B01, containing An-1, was identified from the W1943 BAC library and sequenced by shotgun sequencing. The BAC sequence was assembled by Phred/Phrap and the Gap4 software package (Ewing and Green, 1998; Ewing et al., 1998) and was annotated by Genescan (Burge and Karlin, 1997). The annotated genes were compared with gene annotations on RAP-DB (http://rapdb.dna.affrc.go.jp/viewer/gbrowse/build4).
Constructs and Transformation
A 10244-kb fragment from W1943 BAC harboring the entire An-1 gene was inserted into the binary vector pCAMBIA1301 to form the pCPL construct. A 10,501-kb fragment from W1943 BAC harboring the entire Os04g0351333 gene was inserted into the binary vector pCAMBIA1301 to form the pCPL-RF construct. The pOX construct contained an ORF of W1943 An-1 under the control of the maize Ubiquitin promoter with pCAMBIA1300 as the backbone. The RNAi construct contained an inverted repeat harboring the 282-bp An-1 cDNA fragment in vector pTCK303 (Wang et al., 2004). All plasmid constructs were introduced to Agrobacterium tumefaciens strain EHA105 and subsequently transferred into the japonica cultivar Nipponbare or Aus variety Kasalath.
Phenotypic Evaluation
For the near-isogenic line NIL-An-1 and the recipient parent GLA4, we used 20 plants to measure awn length and the number of primary branches, secondary branches, and grains per panicle on the main panicles. Three main panicles of each plant were collected for analysis. The awn length of apical spikelets of all primary branches was measured, which was used to represent the awn length of the whole panicle. To avoid gene dosage effects caused by multiple copies of transgenes, T2 homozygotes with a single-copy transgene were identified by a hygromycin resistance test. Phenotypic measurements were performed using two independent complementary T2 homozygous lines (CPL-1 and CPL-2), two independent overexpressing T0 lines (OX-1 and OX-5), four RNAi T2 homozygous lines (RNAi-1, -2, -5, and -6), and 10 plants from each line. Twenty Nipponbare and Kasalath plants were used as the control. The 100 fully matured seeds from NIL-An-1, GLA4, Nipponbare, Kasalath, and independent transgenic lines were randomly selected for grain length measurement. The seeds with their awns removed were mounted on a scanner for scanning. Scanning images were analyzed using software to calculate the average grain length and width.
Yield Analysis
The average yield per CPL-1, CPL-2, and Nippponbare plant was determined at the experimental farm in Shanghai during the growing season (May to September) of 2012. The germinated seeds were sown in a seedling bed, and seedlings were transplanted to a paddy field 30 d later with a single plant per hill spaced at 20 × 25 cm. Each plot included eight rows with 12 plants per row. The 48 plants in the middle of each plot were selected to investigate grain yield per plant. After harvest and awn removal, all fully mature seeds of each plant were collected and weighed. The average weight of 48 plants was used in the data analysis.
Quantification of Cell Number
The cell number in the awn primordia or apex of the spikelet was quantified in histological sections. The quantification was performed on three serial sections of three samples, and average cell numbers were compared. The outer epidermal cells of lemmas were quantified on scanning electron microscopy images. The quantification was performed using eight grains, and the average cell numbers were compared.
5′ and 3′ RACE and qRT-PCR
Total RNA was extracted using TRIzol reagent, treated with DNaseI, and reverse transcribed with oligo(dT20) primer using SuperScript II reverse transcriptase (Life Technologies). First-strand cDNAs were used as templates in qRT-PCR using SYBR Green PCR Master Mix (Takara). The qRT-PCR was performed and analyzed using the ABI Prism 7500 sequence detection system and software (PE Applied Biosystems). The qRT-PCR was performed in triplicate, and the rice gene eEF-1α (AK061464) was used as a control to normalize all data. The 5′ and 3′ RACE reactions were performed using the GeneRacer kit (Life Technologies), following the manufacturer’s instructions.
Scanning Electron Microscopy
The young panicles were fixed in formalin\x{2013}acetic acid\x{2013}alcohol fixative solution, dehydrated through an ethanol series, and dried using a carbon dioxide critical-point dryer. Mature seeds were cleaned with 1% Tween 20 and dried at 45°C. The dry panicles and seeds were gold plated and observed using a Hitachi S-2460 scanning electron microscope at 15 kV.
RNA in Situ Hybridization
Young panicles of GLA4, NIL-An-1, and CPL-1 were fixed in 4% paraformaldehyde, dehydrated through an ethanol series, and embedded in Paraplast. A gene-specific region of An-1, OSH1, and OsHistone H4 cDNA was used to generate digoxigenin-labeled RNA probes (Roche). RNA in situ hybridization with sense and antisense probes was performed on 8-μm sections of young panicles, as described by Luo et al. (1996).
Neutrality Test and Phylogenetic Tree
Multiple sequences were aligned with ClustalX (Thompson et al., 1997). The program DnaSP, version 5.0 (Rozas et al., 2003), was used to calculate nucleotide diversity and perform Tajima's D test. These values were calculated after first excluding all regions of alignment gaps and missing data. A phylogenetic tree of the sequenced accessions was reconstructed by the Maximum parsimony method. MEGA version 5.1 was used to perform the phylogenetic reconstruction (Tamura et al., 2011). Bootstrap values were estimated (with 1000 replicates) to assess the relative support for each branch, and bootstrap values were labeled with cutoff = 50.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: An-1 cDNA from W1943 (FO681395), an-1 cDNA from GLA4 (FO681475), ORW1943Ba0047B01 (FO681399), and OSIGBa0144C23 (CR855121). The accession numbers of An-1/an-1 genomic sequences in 43 cultivated and 27 wild rice varieties are listed in Supplemental Table 7 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genotypes of Guangluai-4, SL4, CSSL-Z3, and NIL-An-1 Lines.
Supplemental Figure 2. The T0 Transgenic Phenotypes in Complementation Tests.
Supplemental Figure 3. Full-Length cDNA of An-1 in W1943.
Supplemental Figure 4. Full-Length cDNA of an-1 in GLA4.
Supplemental Figure 5. Transcription Activation Activity Assay and Nuclear Localization of An-1.
Supplemental Figure 6. Phylogenetic Tree of An-1 and Its Homologs.
Supplemental Figure 7. Full-Length cDNA of an-1 in HP228.
Supplemental Figure 8. In Situ Hybridization of An-1 Sense Probe at Different Developmental Stages in NIL-An-1.
Supplemental Figure 9. Screening of Domestication Sweeps on Chromosome 4.
Supplemental Table 1. Comparison of Awn Length and Awn Rate among SL4, CSSSL-Z3, and GLA4.
Supplemental Table 2. Gene Annotation Summary of Genomic Region between FM3 and FM6 in W1943 and Nipponbare.
Supplemental Table 3. Summary of An-1 Genotypes and Sequence Variations in Cultivated and Wild Rice.
Supplemental Table 4. Phenotype Summary of T0 Transgenic Plants.
Supplemental Table 5. Summary of An-1 Haplotypes and Phenotypes in Cultivated and Wild Rice.
Supplemental Table 6. Panicle Phenotype Comparison in NIL-An-1/CPL /OX/RNAi Plants.
Supplemental Table 7. The List of Accession Numbers and Names in 43 Cultivars and 28 Wild Rice Varieties Used in This Study.
Supplemental Table 8. Summary of Primers Used in This Study.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny in Supplemental Figure 6 Online.
Supplemental Data Set 2. Alignments Used to Generate the Phylogeny in Figure 10.
Acknowledgments
We thank Jiqin Li and Xiaoyan Gao for scanning electron microscopy experiments, Xiaoshu Gao for the confocal microscopy experiment, and Lizhen Si for the neutrality test and transcriptional activity analysis in yeast. We thank Kang Chong for providing the vector pTCK 303 and Hongquan Yang for the vector pA7. This work was supported by the grants from the Ministry of Science and Technology of China (2011CB100205, 2012AA10A302, and 2012AA10A304), the Ministry of Agriculture of China (2011ZX08009-002 and 2011ZX08001-004), and the National Natural Science Foundation of China (31121063).
AUTHOR CONTRIBUTIONS
B.H. conceived the project and its components. H.L., T.Z., B.G., T.S., and Zixuan W. performed map-based cloning and construction of the introgression lines. J.L., T.Z., Y.S., and J.Z. performed transgenic, cell biological, and other functional analyses. Y.W., Ziqun W., A.W., and B.G. performed fieldwork. X.H., Y.Z., Q.Z., and Y.L. performed evolutionary study. J.L., H.L., and T.Z analyzed the data. J.L. and B.H. wrote the article.
Glossary
- QTL
quantitative trait locus
- bHLH
basic helix-loop-helix
- RNAi
RNA interference
- SNP
single nucleotide polymorphism
- indel
insertion or deletion
- ORF
open reading frame
- RACE
rapid amplification of cDNA ends
- qRT-PCR
real-time quantitative RT-PCR
References
- Abebe T., Melmaiee K., Berg V., Wise R.P. (2010). Drought response in the spikes of barley: Gene expression in the lemma, palea, awn, and seed. Funct. Integr. Genomics 10: 191–205 [DOI] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Aspinall D. (1969). The effects of day length and light intensity on the growth of barley. VI. Interactions between the effects of temperature, photoperiod, and the spectral composition of the light source. Aust. J. Biol. Sci. 22: 53–67 [Google Scholar]
- Burge C., Karlin S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Cai W., Morishima H. (2002). QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., Yu Z., Tong Y., Huang W., Chen H., Wu P. (2009). A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res. 19: 1309–1311 [DOI] [PubMed] [Google Scholar]
- Elbaum R., Zaltzman L., Burgert I., Fratzl P. (2007). The role of wheat awns in the seed dispersal unit. Science 316: 884–886 [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feng Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Frank M., Rupp H.M., Prinsen E., Motyka V., Van Onckelen H., Schmülling T. (2000). Hormone autotrophic growth and differentiation identifies mutant lines of Arabidopsis with altered cytokinin and auxin content or signaling. Plant Physiol. 122: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller D.Q., Qin L., Zheng Y., Zhao Z., Chen X., Hosoya L.A., Sun G.P. (2009). The domestication process and domestication rate in rice: Spikelet bases from the Lower Yangtze. Science 323: 1607–1610 [DOI] [PubMed] [Google Scholar]
- Gu X.Y., Kianian S.F., Foley M.E. (2005a). Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice. Genetics 171: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.Y., Kianian S.F., Hareland G.A., Hoffer B.L., Foley M.E. (2005b). Genetic analysis of adaptive syndromes interrelated with seed dormancy in weedy rice (Oryza sativa). Theor. Appl. Genet. 110: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Heang D., Sassa H. (2012). Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS ONE 7: e31325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Atkinson A., Bylstra Y.H., Walsh R., Smyth D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Horiguchi G., Gleissberg S., Tsukaya H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51: 252–261 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Izawa T., Konishi S., Shomura A., Yano M. (2009). DNA changes tell us about rice domestication. Curr. Opin. Plant Biol. 12: 185–192 [DOI] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J.P., Yang J., Shi M., Zhu M.Z., Luo D., Lin H.X. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369 [DOI] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Kovach M.J., Sweeney M.T., McCouch S.R. (2007). New insights into the history of rice domestication. Trends Genet. 23: 578–587 [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006a). Rice domestication by reducing shattering. Science 311: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Li S., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 110: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2006b). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Makkena S., Lamb R.S. (2013). The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. BMC Plant Biol. 13: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Duronio R.J. (2002). Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14: 692–699 [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J.C., Messeguer X., Rozas R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497 [DOI] [PubMed] [Google Scholar]
- Rupp H.M., Frank M., Werner T., Strnad M., Schmülling T. (1999). Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 18: 557–563 [DOI] [PubMed] [Google Scholar]
- Sato Y., Hong S.K., Tagiri A., Kitano H., Yamamoto N., Nagato Y., Matsuoka M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93: 8117–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P. (1993). Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J. 3: 739–744 [Google Scholar]
- Sweeney M., McCouch S. (2007). The complex history of the domestication of rice. Ann. Bot. (Lond.) 100: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., Fu Y., Cai H., Wang X., Xie D., Sun C. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364 [DOI] [PubMed] [Google Scholar]
- Thomson M.J., Tai T.H., McClung A.M., Lai X.H., Hinga M.E., Lobos K.B., Xu Y., Martinez C.P., McCouch S.R. (2003). Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T., Suzaki T., Yamaguchi T., Ohmori Y., Tsukaya H., Hirano H.Y. (2010). Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22: 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Ito Y., Sato Y., Kurata N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varaud E., Brioudes F., Szécsi J., Leroux J., Brown S., Perrot-Rechenmann C., Bendahmane M. (2011). AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang A., Huang X., Zhao Q., Dong G., Qian Q., Sang T., Han B. (2011). Mapping 49 quantitative trait loci at high resolution through sequencing-based genotyping of rice recombinant inbred lines. Theor. Appl. Genet. 122: 327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Basten, C., and Zeng, Z. (2007). Windows QTL Cartographer 2.5. (Raleigh, NC: North Carolina State University). [Google Scholar]
- Wang Z., Chen C., Xu Y., Jiang R., Han Y., Xu Z., Chong K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22: 409–417 [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Yuo T., Yamashita Y., Kanamori H., Matsumoto T., Lundqvist U., Sato K., Ichii M., Jobling S.A., Taketa S. (2012). A SHORT INTERNODES (SHI) family transcription factor gene regulates awn elongation and pistil morphology in barley. J. Exp. Bot. 63: 5223–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lu D., Li C., Luo J., Zhu B.F., Zhu J., Shangguan Y., Wang Z., Sang T., Zhou B., Han B. (2012). Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B.F., et al. (2011). Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155: 1301–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y., Chen Z., Innes J.B., Chen C., Wang Z., Wang H. (2007). Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449: 459–462 [DOI] [PubMed] [Google Scholar]