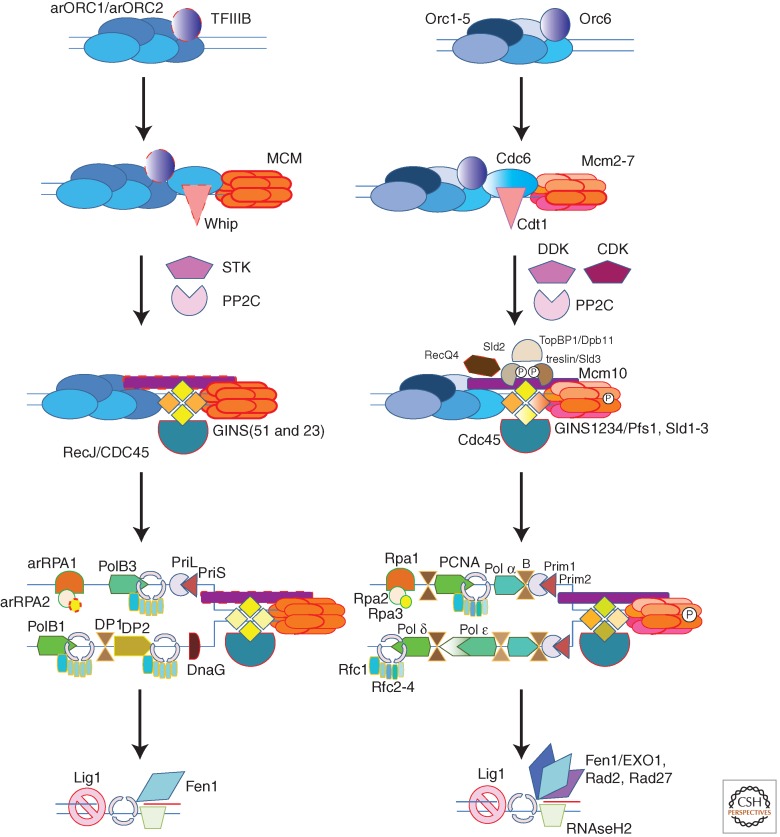
The archaeal DNA replication system shows an unexpected level of complexity. In fact, there is a close correspondence between components of the archaeal and eukaryotic replication systems.
Abstract
Recent advances in the characterization of the archaeal DNA replication system together with comparative genomic analysis have led to the identification of several previously uncharacterized archaeal proteins involved in replication and currently reveal a nearly complete correspondence between the components of the archaeal and eukaryotic replication machineries. It can be inferred that the archaeal ancestor of eukaryotes and even the last common ancestor of all extant archaea possessed replication machineries that were comparable in complexity to the eukaryotic replication system. The eukaryotic replication system encompasses multiple paralogs of ancestral components such that heteromeric complexes in eukaryotes replace archaeal homomeric complexes, apparently along with subfunctionalization of the eukaryotic complex subunits. In the archaea, parallel, lineage-specific duplications of many genes encoding replication machinery components are detectable as well; most of these archaeal paralogs remain to be functionally characterized. The archaeal replication system shows remarkable plasticity whereby even some essential components such as DNA polymerase and single-stranded DNA-binding protein are displaced by unrelated proteins with analogous activities in some lineages.
Double-stranded DNA is the molecule that carries genetic information in all cellular life-forms; thus, replication of this genetic material is a fundamental physiological process that requires high accuracy and efficiency (Kornberg and Baker 2005). The general mechanism and principles of DNA replication are common in all three domains of life—archaea, bacteria, and eukaryotes—and include recognition of defined origins, melting DNA with the aid of dedicated helicases, RNA priming by the dedicated primase, recruitment of DNA polymerases and processivity factors, replication fork formation, and simultaneous replication of leading and lagging strands, the latter via Okazaki fragments (Kornberg and Baker 2005; Barry and Bell 2006; Hamdan and Richardson 2009; Hamdan and van Oijen 2010). Thus, it was a major surprise when it became clear that the protein machineries responsible for this complex process are drastically different, especially in bacteria compared with archaea and eukarya. The core components of the bacterial replication systems, such as DNA polymerase, primase, and replication helicase, are unrelated or only distantly related to their counterparts in the archaeal/eukaryotic replication apparatus (Edgell 1997; Leipe et al. 1999).
The existence of two distinct molecular machines for genome replication has raised obvious questions on the nature of the replication system in the last universal common ancestor (LUCA) of all extant cellular life-forms, and three groups of hypotheses have been proposed (Leipe et al. 1999; Forterre 2002; Koonin 2005, 2006, 2009; Glansdorff 2008; McGeoch and Bell 2008): (1) The replication systems in Bacteria and in the archaeo–eukaryotic lineage originated independently from an RNA-genome LUCA or from a noncellular ancestral state that encompassed a mix of genetic elements with diverse replication strategies and molecular machineries. (2) The LUCA was a typical cellular life-form that possessed either the archaeal or the bacterial replication apparatus in which several key components have been replaced in the other major cellular lineage. (3) The LUCA was a complex cellular life-form that possessed both replication systems, so that the differentiation of the bacterial and the archaeo–eukaryotic replication machineries occurred as a result of genome streamlining in both lines of descent that was accompanied by differential loss of components. With regard to the possible substitution of replication systems, a plausible mechanism could be replicon takeover (Forterre 2006; McGeoch and Bell 2008). Under the replicon takeover hypothesis, mobile elements introduce into cells a new replication system or its components, which can displace the original replication system through one or several instances of integration of the given element into the host genome accompanied by inactivation of the host replication genes and/or origins of replication. This scenario is compatible with the experimental results showing that DNA replication DNA in Escherichia coli with an inactivated DnaAgene or origin of replication can be rescued by the replication apparatus of R1 or F1 plasmids integrated into the bacterial chromosome (Bernander et al. 1991; Koppes 1992). Furthermore, genome analysis suggests frequent replicon fusion in archaea and bacteria (McGeoch and Bell 2008); in particular, such events are implied by the observation that in archaeal genomes, genes encoding multiple paralogs of the replication helicase MCM and origins of replication are associated with mobile elements (Robinson and Bell 2007; Krupovic et al. 2010). Replicon fusion also is a plausible path from a single origin of replication that is typical of bacteria to multiple origins present in archaea and eukaryotes. However, all the evidence in support of frequent replicon fusion and the plausibility of replicon takeover notwithstanding, there is no evidence of displacement of the bacterial replication apparatus with the archaeal version introduced by mobile elements, or vice versa, displacement of the archaeal machinery with the bacterial version, despite the rapid accumulation of diverse bacterial and archaeal genome sequences. Thus, the displacement scenarios of DNA replication machinery evolution are so far not supported by comparative genomic data.
Regardless of the nature of the DNA replication system (if any) in the LUCA and the underlying causes of the archaeo–bacterial dichotomy of replication machineries, the similarity between the archaeal and eukaryotic replication systems is striking (Table 1). Generally, the archaeal replication protein core appears to be the same as the eukaryotic core, but eukaryotes typically possess multiple paralogous subunits within complexes that are homomeric in archaea, many additional components interacting with the core ones and more complex regulation (Leipe et al. 1999; Bell and Dutta 2002; Bohlke et al. 2002; Kelman and White 2005; Barry and Bell 2006). Thus, the archaeal replication system appears to be an ancestral version of the eukaryotic system and hence a good model for functional and structural studies aimed at gaining mechanistic insights into eukaryotic replication.
Table 1.
The relationship between archaeal and eukaryotic replication systems
| Archaea (projection for LACA) | Eukaryotes (projection for LECA) | Comments |
|---|---|---|
| ORC complex | ||
| arORC1 | Orc1, Cdc6 | In LACA the ORC/Cdc6 complex probably consisted of two distinct subunits, and in LECA of six distinct. Both complexes might possess additional Orc6 and Cdt1 components. |
| arORC2 | Orc2, Orc3, Orc4, Orc5 | |
| TFIIB or homologa | Orc6 | |
| WhiP or other wHTH proteina | Cdt1 | |
| CMG complex | ||
| Archaeal Cdc45/RecJ | Cdc45 | In many archaea and eukaryotes, CDC45/RecJ apparently contain inactive DHH phosphoesterase domains. The RecJ family is triplicated in euryarchaea, and some of the paralogs could be involved in repair. MCM is independently duplicated in several lineages of euryarchaea. |
| Mcm | Mcm2, Mcm3, Mcm4, Mcm5, Mcm6, Mcm7 | |
| Gins23 | Gins2, Gins3 | |
| Gins15 | Gins1, Gins5 | |
| Inactivated MCM homologa | Mcm10 | |
| CMG activation factors | ||
| — | RecQ/Sld2 | There is no evidence that kinases and phosphatases in archaea are directly involved in replication, although they probably regulate cell division. |
| — | Treslin/Sld3 | |
| — | TopBP1/Dpb11 | |
| STK | CDK, DDK | |
| PP2C | PP2C | |
| Primases | ||
| Prim1/p48 | PriS | In eukaryotes, Pol α is involved in priming by adding short DNA fragments to RNA primers. In archaea, DnaG might be involved in priming specifically on the lagging strand. |
| Prim2a/p58 | PriL | |
| DnaG | — | |
| Polymerases | ||
| PolB3 | Pol α, Pol δ, Pol ζ | No eukaryotic homologs of DP2 are known, but Zn fingers of Pol ε are apparently derived from DP2. |
| PolB1 | Pol ε | |
| DP1 | B subunits of Pol α, Pol δ, Pol ζ, Pol ε | |
| DP2 | — | |
| DNA polymerase sliding clamp and clamp loader | ||
| RFCL | RFC1 | Eukaryotes have additional duplications of both RFCs and PCNA involved in checkpoint complexes (Rad27 and Rad1, Rad9, Hus1, respectively). |
| RFCS | RFC2, RFC3, RFC4, RFC4 | |
| PCNA | PCNA | |
| Primer removal and gap closure | ||
| RNase H2 | RNase II | There is a triplication of ligases (LigI, LigIII, LigIV) in eukaryotes, but only LigI is directly involved in replication. In a few Halobacteria, ATP-dependent ligase is replaced by NAD-dependent ligase. |
| Fen1 | Fen1/EXO1, Rad2, Rad27 | |
| Lig1 | Lig1 | |
| SSB | ||
| arRPA1_long | Rpa1 | In Thermoproteales, RPA is displaced by the non-homologous ThermoSSB; two short RPA forms in many euryarchaea; expansion of short RPA forms in Halobacteria. |
| arRPA1_short and RPA2 | Rpa2 | |
| arCOG05741a | Rpa3 | |
For eukaryotic genes in Homo sapiens and Saccharomyces cerevisiae, gene names are indicated. Archaeal genes are denoted as in Barry and Bell (2006) or as introduced here.
aNot confidently traced to LACA.
In the last few years, there has been substantial progress in the study of the archaeal replication systems that has led to an apparently complete delineation of all proteins that are essential for replication (Berquist et al. 2007; Beattie and Bell 2011a; MacNeill 2011). The combination of experimental, structural, and bioinformatics studies has led to the discovery of archaeal homologs (orthologs) for several components of the replication system that have been previously deemed specific for eukaryotes (Barry and Bell 2006; MacNeill 2010, 2011; Makarova et al. 2012). Furthermore, complex evolutionary events that involve multiple lineage-specific duplications, domain rearrangements, and gene loss, and in part seem to parallel the evolution of the evolution of the replication system in eukaryotes, have been delineated for a variety of replication proteins in several archaeal lineages (Tahirov et al. 2009; Chia et al. 2010; Krupovic et al. 2010). Here we summarize these findings and present several additional case studies that show the complexity of evolutionary scenarios for the components of the archaeal replication machinery and new aspects of their relationship with the eukaryotic replication system.
PREREPLICATION COMPLEX
DNA replication begins at specific sites in the genome known as the origins of replication. Some archaea possess a single and others possess several replication origins, whereas in all eukaryotes replication starts from numerous, independent origins (Robinson and Bell 2005; Coker et al. 2009). Typically, replication origins encompass a distinct sequence signature, the AT-rich box, that can be used to predict origins in silico (Zhang and Zhang 2005).
The origin recognition complex (ORC) is a hexameric protein complex that binds the origins of DNA replication and recruits additional replication factors resulting in the formation of the prereplicative complex (Fig. 1). Eukaryotic Orc1-5 and the closely related CDC6 are paralogous proteins that belong to the DnaA/ORC clade of AAA+ ATPases (Iyer et al. 2004) and often also contain a carboxy-terminal helix–turn–helix (HTH) DNA-binding domain. All the six families of eukaryotic ORC subunits have been traced back to the last eukaryotic common ancestor (LECA) (Makarova et al. 2005). The Orc2 and Orc3 families are highly diverged and contain inactivated ATPase domains.
Figure 1.
Comparison of the archaeal (reconstructed LACA) and eukaryotic replication systems (reconstructed LECA). Orthologs are shown by shapes of the same color, and paralogs are denoted by similar colors. The gene product names are indicated as follows: for most eukaryotic genes, the Homo sapiens nomenclature is used, but in several cases, both H. sapiens and Saccharomyces cerevisiae gene names are given for clarity. Archaeal genes are named as in Barry and Bell (2006) or as discussed in the text. The dotted outline in some shapes indicates components that could not be confidently traced to LACA. Small circles with “P” inside indicate phosphorylation.
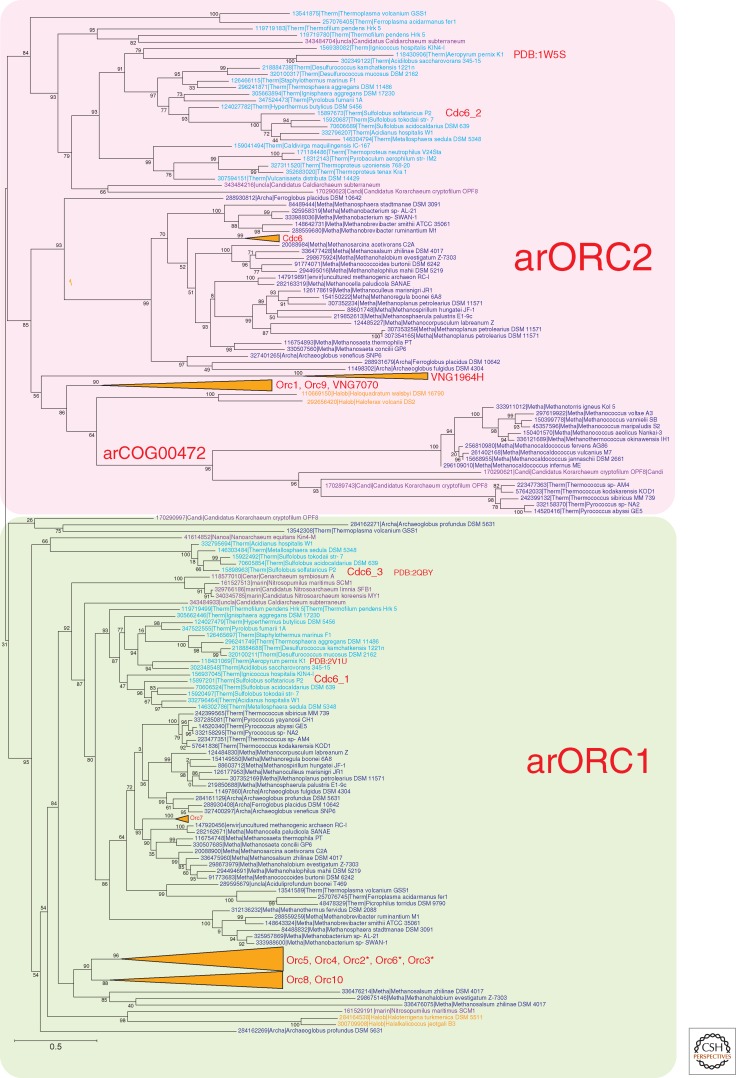
Phylogenetic analysis of archaeal ORC homologs reveals a complex evolutionary scenario of gene duplications, losses, and extreme divergence (Fig. 2). All this complexity notwithstanding, the phylogenetic tree clearly shows the existence of two major groups of ORC proteins, namely, the slow-evolving arORC1 and the fast-evolving arORC2. The vast majority of archaea encode representatives of each of these groups.
Figure 2.
Phylogenetic analysis of the ORC family in archaea and eukaryotes. (Gray) Eukaryotes; (dark blue) Euryarchaeota, with the exception of (orange) Halobacteria; (light blue) Crenarchaeota; (purple) deeply branched archaeal lineages (Thaumarchaeota, Korarchaeota, Nanoarchaeota). The MUSCLE program (Edgar 2004) was used for construction of sequence alignments. The tree was reconstructed using the FastTree program (Price et al. 2010) (293 sequences and 186 aligned positions for the archaeal tree and 84 sequences and 380 aligned positions for the eukaryotic tree). For Halobacteria, the branches are collapsed. For characterized genes of Halobacteria, Sulfolobus, and yeast, the conventional gene name or protein identifies are indicated in red on the right of the corresponding sequence or the corresponding branch. For two archaeal sequences for which the structure is solved, its PDB code is indicated. (*) The corresponding genes are missing in some Halobacteria. (Red) Fast-evolving branches; (green) slow-evolving branches.
Thermoproteales, Nanoarchaeum equitans, and several Thaumarchaeota possess only one ORC homolog that, in the latter two cases, belongs to the arORC1 group. Haloarchaea show evidence of ancestral bursts of duplication events in both arORC1 and arORC2 clades (Fig. 2). The distinction between the two ancestral ORC subfamilies persists, but most of the lineage-specific paralogs show accelerated evolutionary rates, and some have inactivated ATPase domains. Further duplications are detectable in several Halobacterial lineages giving rise to up to 20 ORC paralogs in Haloterrigena turkmenica. It appears that paralogization of ORC genes is driven by the appearance of additional origins of replication including acquisition and integration of extra-chromosomal elements (McGeoch and Bell 2008). In Sulfolobus, it has been shown that the two origins are recognized by distinct monomers or dimers of Orc1/Cdc6 paralogs homologs and that the three Orc1/Cdc6 genes are differentially expressed in different growth phases and cell cycle stages, suggesting a role for these proteins in modulating the activity of the origins (Robinson et al. 2004). However, the functional differences and the causes behind the distinct evolutionary constraints in the two major groups of archaeal ORCs remain poorly understood.
The Euryarchaeota of the order Methanococcales are currently thought to lack ORC proteins, and Thermococcales are assumed to possess only one ORC that belongs to the arORC1 subfamily (Barry and Bell 2006). We identified a subfamily of archaeal AAA+ ATPases (arCOG00472) that are fused to the carboxy-terminal wHTH domain and clusters with the arORC2 branch in the phylogenetic tree. The two major lineages represented in this subfamily, Methanococcales and Thermococcales, are exactly those that were missing from the arORC2 clade. Moreover, the genetic context of these genes in Methanococcales suggests their possible role in replication (Fig. 3). The only archaeon in which we were unable to identify an ORC candidate is Methanopyrus kandleri AV19. Thus, it is most likely that the last common ancestor of the extant archaea and the last archaeo–eukaryotic ancestor (whatever the exact nature of this entity) already encoded at least two paralogous ORC proteins. The nature of the functional distinctions between the slow-evolving and fast-evolving ORC paralogs remains unclear. In S. solfataricus, both the slow-evolving arORC1 and two fast-evolving paralogs from the arORC2 branch all recognize origins of replication albeit with different specificities (Robinson et al. 2004). Furthermore, the loss of ORC1 and the colocalization of the ORC2 gene with replication machinery components in Methanococcales imply that on some occasions in the evolution of archaea, ORC2 could take over the essential function of ORC1 in replication. In addition, the absence of recognizable ORC homologs in M. kandleri suggests that alternative, uncharacterized mechanisms of archaeal replication initiation might exist.
Figure 3.
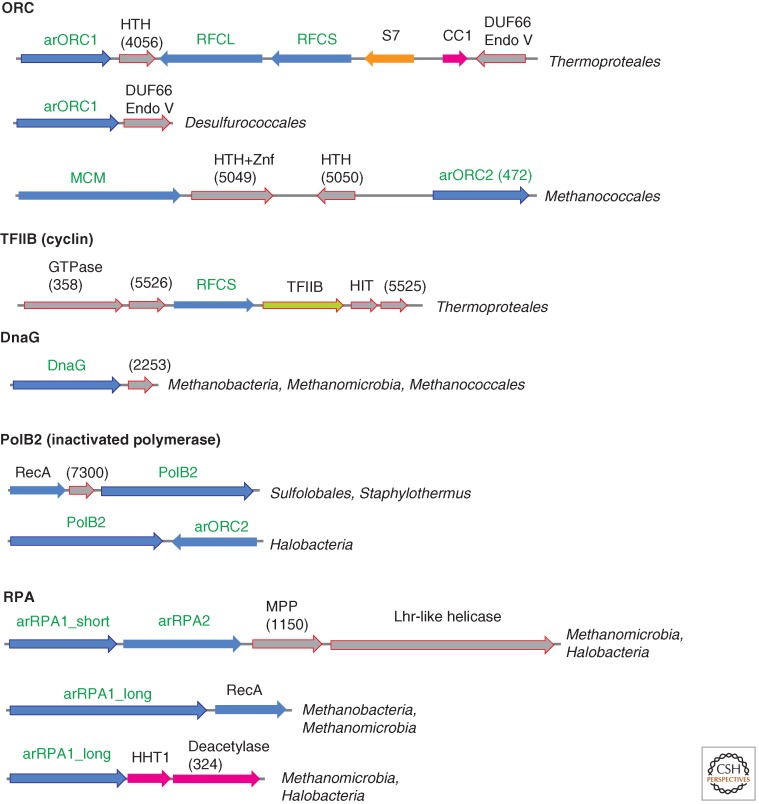
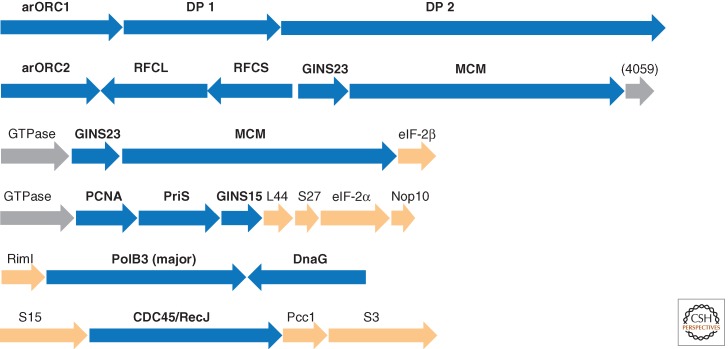
Genomic context of selected genes for proteins involved in replication in archaea. Homologous genes are shown by arrows of the same color; genes are shown approximately to scale. (Blue) Replication genes; (magenta) genes coding for chromatin-binding protein and the respective modification enzymes; (orange) genes for translation system components; (gray) uncharacterized genes. Genes for which the neighborhood is discussed in the text are marked by an outline. The arCOG numbers to which uncharacterized proteins are assigned are shown in parentheses. HTH, helix–turn–helix domain; S7, ribosomal protein S7; CC1, DNA-binding protein CC1; Endo V, homolog of endonuclease V; Znf, Zn finger; TFIIB, transcription initiation factor TFIIB; HIT, HIT family hydrolase; MPP, metallophosphatase superfamily protein; HHT1, histone.
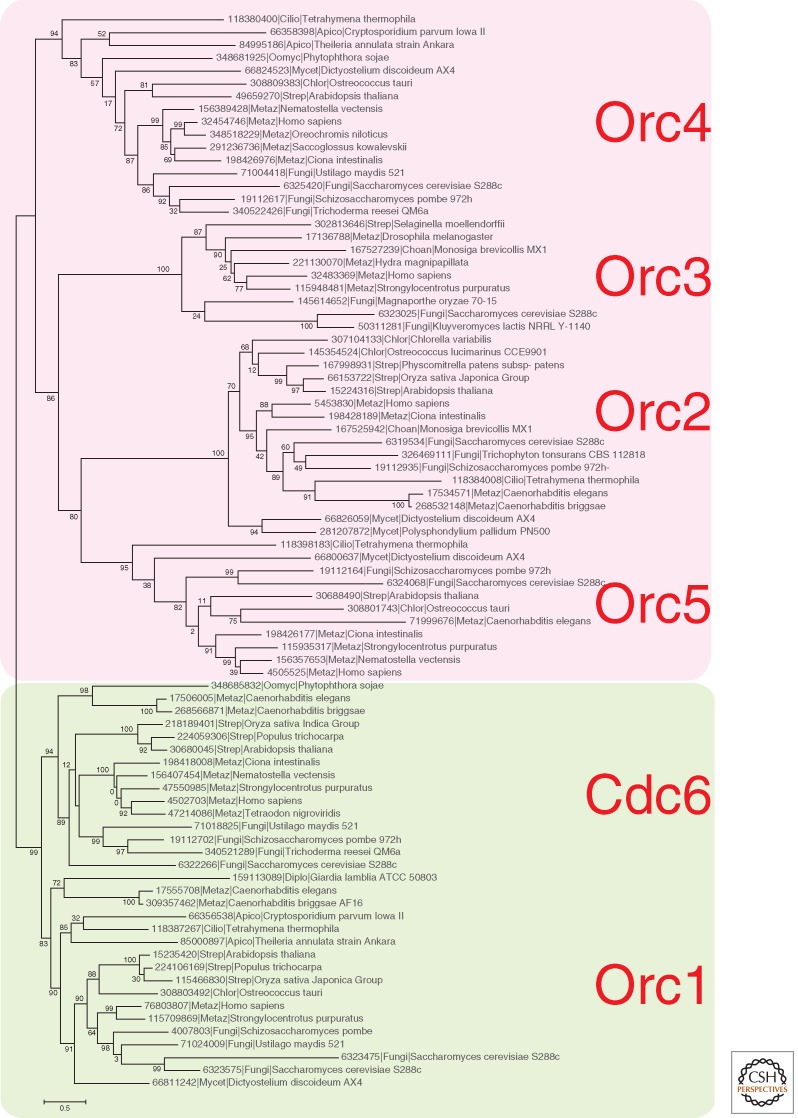
The phylogenetic tree of the eukaryotic ORC/CDC6 family also divides into two branches: the slow-evolving ORC1/CDC6 and fast-evolving ORC2-3-4-5 (Fig. 2). Owing to the extreme sequence divergence, it is difficult to precisely decipher the relationships between archaeal and eukaryotic ORC families, but it seems likely that arORC1 is the ancestor of Orc1/CDC6, whereas arORC2 is the ancestor of the ORC2-3-4-5 branch. However, in eukaryotes, the functional division between the slow-evolving and fast-evolving ORC subunits is different from that in archaea because all of these proteins are components of the heteromeric complex involved in replication.
The sixth component of the ORC complex, Orc6, is shown to be required for DNA replication in eukaryotes, and it is critical for ORC function (Duncker et al. 2009). Orc6 does not display any sequence similarity with ATPases. Structural modeling of the amino-terminal domain of metazoan Orc6 that is essential for replication (Balasov et al. 2009) and the recently resolved structure of the middle portion of human Orc6 both show that Orc6 contains two cyclin-like domains similar to those in the transcription factor TFIIB that belongs to a conserved achaeo–eukaryotic protein family (Liu et al. 2011). Unlike Orc6, the TFIIB family proteins, in addition to cyclin domains, also contain an amino-terminal Zn finger. However, in several archaea, the Zn-binding ligands are lacking or the finger module is lost completely. Interestingly, in several genomes of Thermoproteales, the gene for TFIIB is encoded in a predicted operon with the small subunit of clamp loader, replication factor C (RFC) (Fig. 3). Furthermore, several archaeal lineages encode additional, functionally uncharacterized TFIIB paralogs that typically contain a single cyclin-like domain. Therefore, it cannot be ruled out that TFIIB and/or its paralog perform dual functions in both transcription and replication (Fig. 1).
In eukaryotes, the hexameric ORC recruits two additional components, namely, Cdc6, which is an apparent product of ancestral duplication of Orc1 gene in eukaryotes (Fig. 2), and Cdt1 (Duncker et al. 2009). Both the middle and the carboxy-terminal, MCM-interacting, domains of Cdt1 adopt the winged HTH fold (Lee et al. 2004; De Marco et al. 2009). In the Crenarchaeota Aeropyrum pernix and several Sulfolobus species, a gene adjacent to the origin of replication region (ORI) encodes a winged-helix initiator protein (WhiP) (Robinson and Bell 2007). Given that WhiP contains two winged HTH domains, it has been proposed that this protein is the ortholog and functional analog of Cdt1 (Robinson and Bell 2007). Typical WhiP proteins with two HTH domains so far have been detected only in Desulfurococcales, although many other archaea encode closely related homologs of the amino-terminal HTH domain of WhiP, and some of these proteins might interact with ORI regions. Moreover, in several archaeal genomes, HTH-containing proteins are encoded adjacent to ORC subunits (Fig. 3), suggesting that these proteins also could perform specific roles in replication.
The resulting complete ORC–Cdc6–Cdt1 complex is responsible for the ATP-dependent loading of the replicative helicase, the minichromosome maintenance (MCM) complex, in eukaryotes and probably in some if not most archaea and by inference in LACA (Fig. 1).
THE CMG COMPLEX
The eukaryotic MCM complex consists of six paralogous proteins (Mcm2–7), all of which are essential for cell viability and are required for the initiation of DNA replication and replication fork progression (Bochman and Schwacha 2009). Additional eukaryotic MCM protein families, Mcm8 and Mcm9, might be ancestral but are not present in all species, and their function in replication is unclear (Bochman and Schwacha 2009). Genetic, biochemical, and structural studies have shown that the hexameric MCM complex is the replicative helicase that separates DNA strands during chromosomal replication (Bochman and Schwacha 2009). The conserved core structure of all MCM proteins consists of an α-helical amino-terminal subdomain, an OB fold domain with an inserted Zn finger, an AAA+ ATPase domain, and a carboxy-terminal HTH domain. The HTH domain can be confidently detected only in Mcm6. The structure of this domain has been solved, and it has been shown to bind Cdt1 (Wei et al. 2010). The same core structure is conserved in MCM proteins from archaea including the carboxy-terminal HTH domain. It appears that all eukaryotic MCM paralogs are products of ancestral duplications arising from a single ancestral MCM protein (Chia et al. 2010; Krupovic et al. 2010). All archaea encode at least one MCM protein homolog, and serial duplication of the MCM genes occurred in multiple lineages. At least two paralogs could be traced back to common ancestor of Halobacteriales and independently to the common ancestor of Methanococcales (Chia et al. 2010; Krupovic et al. 2010). Other duplications and losses followed the ancestral duplication in Methanococcales giving rise to up to eight MCM paralogs in Methanococcus maripaludis C6 (Chia et al. 2010; Walters and Chong 2010). Duplications occurred also within a few other narrow archaeal lineages. It has been shown that all four MCM paralogs of M. maripaludis S2 could form a heterohexameric complex (Walters and Chong 2010). In many archaea, some of the paralogous MCM genes are associated with mobile elements or viruses and show accelerated evolution (Chia et al. 2010). To date, a single genetic study has been published showing that only one of the three MCM paralogs in the euryarchaeon T. kodakarensis is essential for cell viability (Pan et al. 2011b). Interestingly, an intein inserted into one of the MCM genes, apparently early in the evolution of euryarchaea, followed by inactivation of the ATPase domain in several intein-containing MCM homologs; all archaea that possess this inactivated MCM protein also encode an intact paralog.
In eukaryotes, in vitro and in vivo experiments have shown that the MCM complex, on its own, is not the active helicase but requires the association with two accessory factors, the tetrameric GINS complex (Sld5, Psf1-3) and the Cdc45 protein (Moyer et al. 2006; Pacek et al. 2006; Labib and Gambus 2007). This complex is referred to as the CMG (Cdc45, MCM, GINS) complex and is thought to be the active replicative helicase unit in vivo (Moyer et al. 2006; Pacek et al. 2006; Labib and Gambus 2007). In addition to binding MCM proteins, the GINS complex also associates with Pol α-primase, the protein complex that synthesizes the primers on the lagging strand, and with the leading and lagging strands polymerases, Pol ε and Pol δ, respectively (MacNeill 2010). In contrast to the eukaryotic Mcm2–7 complex, which does not show helicase activity without the associated GINS and Cd45 proteins, in vitro experiments with the archaeal MCM homohexamers from several organisms have revealed robust helicase activity that did not require additional subunits (Sakakibara et al. 2009).
Highly diverged archaeal GINS homologs have been originally identified in silico (Makarova et al. 2005) and have been subsequently shown to interact with MCM (Marinsek et al. 2006; Bell 2011). Archaea encode two distinct forms of the GINS proteins, one of which appears to have been derived from the other by circular permutation of a small domain (Marinsek et al. 2006). These two forms are also present in eukaryotes, which have two ancestral genes with one domain configuration (Psf2 and Psf3) and two genes with the permuted domain arrangement (Psf1 and Sld5). Although many archaea encode only one GINS protein that belongs to the Psf1/Sld5 subfamily, Crenarchaeota, Thaumarchaeota, Korarchaeaon, and Thermococci encode proteins of both subfamilies, suggesting that eukaryotes inherited both forms from their archaeal ancestor (Marinsek et al. 2006). Recently, the structure of two GINS protein from T. kodakaraensis has been solved, revealing that the backbone structure of each subunit and the tetrameric assembly closely resemble those of the human GINS complex (Oyama et al. 2011).
Until very recently, it was believed that Cdc45, which is present in all eukaryotes in one copy, is specific to eukaryotes (MacNeill 2010). However, an in-depth computational analysis has shown that the amino-terminal region of Cdc45 contains a DHH phosphoestarase domain, suggesting that Cdc45 is the eukaryotic ortholog of archaeal and bacterial RecJ nucleases (Sanchez-Pulido and Ponting 2011). Furthermore, the GINS complex of S. solfataricus, in addition to the association with MCM, binds a protein denoted RecJdbd (RecJ-like DNA-binding domain), which is homologous to the carboxy-terminal domain of bacterial RecJ but lacks the nuclease domain (Marinsek et al. 2006).
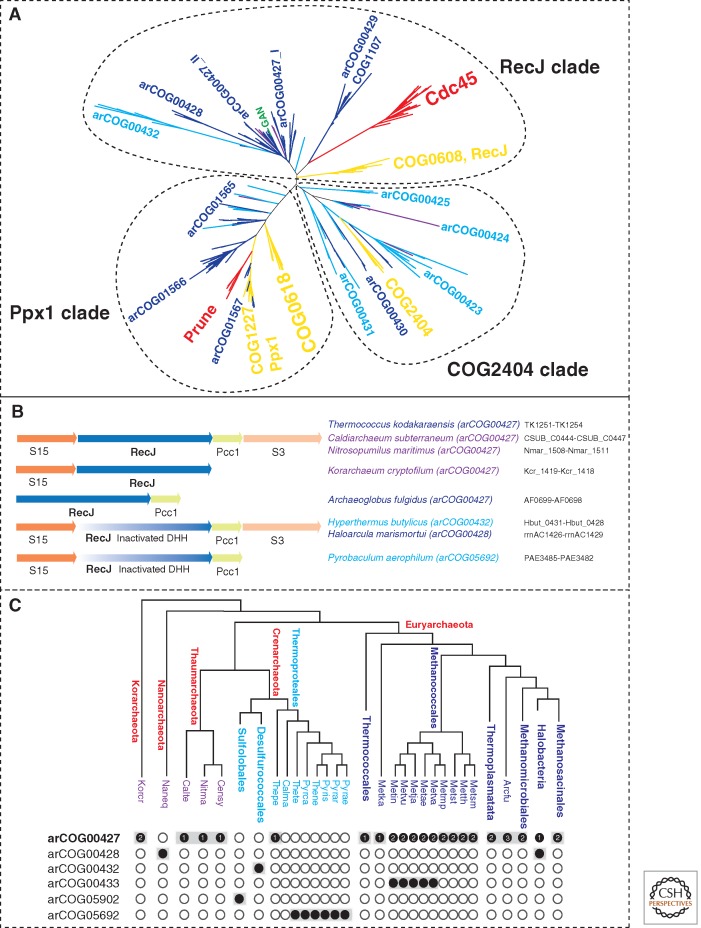
Recently, the GINS complex of the euryarchaeon T. kodakarensis has been shown to interact with primase, MCM, DNA polymerase D, PCNA, and the GINS-associated nuclease (GAN) (Li et al. 2010, 2011). Unlike the RecJdbd of S. solfataricus, GAN is a bona fide ortholog of bacterial RecJ containing a DHH phosphoesterase domain with all the essential catalytic residues. Recent phylogenomic analysis of RecJ homologs in archaea led to the identification of previously unsuspected homologs in Thermoproteales and revealed a complex scenario of RecJ family evolution in archaea and the origin of Cdc45 (Fig. 4A) (Makarova et al. 2012).
Figure 4.
RecJ homologs in archaea. (Red) Eukaryotes; (yellow) bacteria; the color code for archaea is as in Figure 2. (A) Phylogeny of the DHH superfamily of phosphoesterases. The tree was reconstructed using aligned blocks corresponding to the DHH domain (Makarova et al. 2012). The MUSCLE program (Edgar 2004) was used for the construction of sequence alignments (229 sequences in total, 83 aligned positions). The maximum likelihood (ML) phylogenetic tree was constructed by using the MOLPHY program (Adachi and Hasegawa 1992) with the JTT substitution matrix to perform local rearrangement of an original Fitch tree (Fitch and Margoliash 1967). The MOLPHY program was also used to compute RELL bootstrap values. arCOG or COG numbers or family names (for eukaryotes) are indicated for the corresponding branches. (Green) The location of the GAN protein. (B) Genomic context of the RecJ homologs in selected archaea. The designations are as in Figure 3. The arCOGs to which RecJ-like proteins are assigned are indicated in parentheses. The protein IDs for this region in the corresponding genomes are indicated. (C) Phyletic patterns of RecJ-related arCOGs. The phyletic patterns for indicated arCOGs ([filled circles] presence; [empty circles] absence) are superimposed over the phylogenetic tree of archaea. The circles for proteins implicated in replication are shaded. The number of paralogs is indicated inside the circles for arCOG00427 (all other subfamilies have one representative in each genome). The tree is a modified version of the consensus phylogeny of archaea (Makarova et al. 2010) with Caldiarchaeum subterraneum included in the Thaumarchaeota branch and several branches with the same distribution of RecJ-related subfamilies collapsed. Arcfu, Archaeoglobus fulgidus; Metsm, Methanobrevibacter smithii ATCC 35061; Metth, Methanothermobacter thermautotrophicus; Metst, Methanosphaera stadtmanae; Metmp, Methanococcus maripaludis S2; Metva, Methanococcus vannielii SB; Metae, Methanococcus aeolicus Nankai-3; Metja, Methanocaldococcus jannaschii; Metvu, Methanocaldococcus vulcanius M7; Metin, Methanocaldococcus infernus ME; Metka, Methanopyrus kandleri; Pyrae, Pyrobaculum aerophilum; Pyrar, Pyrobaculum arsenaticum DSM 13514; Pyris, Pyrobaculum islandicum DSM 4184; Thene, Thermoproteus neutrophilus V24Sta; Pyrca, Pyrobaculum calidifontis JCM 11548; Thete, Thermoproteus tenax; Calma, Caldivirga maquilingensis IC-167; Thepe, Thermofilum pendens Hrk 5; Censy, Cenarchaeum symbiosum; Nitma, Nitrosopumilus maritimus SCM1; Naneq, Nanoarchaeum equitans; Korcr, Candidatus Korarchaeum cryptofilum OPF8; Calte, Candidatus Caldiarchaeum subterraneum.
Under this scenario, the last common ancestor of all extant archaea possessed a single RecJ ortholog that was encoded in the conserved neighborhood including also the S15, S3, and Pcc1 genes (Fig. 4B). This ancestral protein was an active DHH nuclease and an essential component of the replication machinery. However, the experiments with the S. solfataricus replication system indicate that the nuclease domain is not required for replication; thus, the ancestral RecJ protein might have performed additional functions, for example, in repair, that required the nuclease activity. In Crenarchaeota, the DHH domain partially deteriorated, losing the nuclease activity, and the RecJ homolog apparently became a dedicated replication system component; the subsequent routes of evolution were notably different between the three major crenarchaeal branches—Sulfolobales, Desulfurococcales, and Themrorpoteales—resulting in extreme sequence divergence. The archaeal ancestor of eukaryotes, the exact nature of which remains elusive, also retained the RecJ ortholog (Cdc45) in which some but not all catalytic residues of the DHH domain are conserved; so far, to our knowledge, there are no experimental data showing a nuclease activity of Cdc45. In Euryarchaeota, the RecJ gene seems to have undergone triplication. One clade evolved very fast and developed some specialized function (arCOG00429, COG1107). Two other clades (within arCOG00427) retained significant levels of sequence similarity. Most of these proteins contain an active DHH nuclease domain, suggesting that they are active nucleases. Only one of these paralogs (arCOG00427_II) is often encoded in a conserved neighborhood with the ribosomal proteins S15 and S3, and the Pcc1 subunit of the KEOPS complex (Fig. 4B). Thermococci might have lost one paralog (arCOG00427_I). Methanococcales encode an additional paralog (arCOG00433) that has lost the DHH domain. These genes are located in genomic neighborhoods that encode no other proteins involved in replication; thus, it is unclear whether they are components of replication systems. In Halobacteria, the RecJ orthologs (arCOG00428), which are encoded in the same conserved neighborhood, contain an inactivated DHH domain. Thus, inactivation of the RecJ-like nuclease that apparently became a dedicated replication protein seems to have occurred at least twice independently in different archaeal lineages (Fig. 4C).
In eukaryotes, MCM10 is an essential replication protein that interacts with the assembled preRC and is required for the recruitment of Cdc45 (Wohlschlegel et al. 2002). It stays in the active replisome complex throughout the S phase and appears to be important for both replisome assembly and fork progression. Mcm10 physically interacts with both PCNA and Pol α (Chattopadhyay and Bielinsky 2007). The structure of the conserved middle domain of Mcm10 revealed a unique arrangement of a CCCH-type Zn-binding module and an OB-fold that jointly form a DNA-binding interface (Warren et al. 2008). The carboxy-terminal domain of MCM10 is composed of a second, CCCC-type Zn-binding module that is not involved in DNA binding and a HTH domain. The CCCC-type Zn-binding module of MCM10 is structurally similar to a corresponding module in the amino-terminal oligomerization domain of eukaryotic and archaeal MCM helicases, some of which also possess a carboxy-terminal HTH domain (Robertson et al. 2010). In several fungi, Mcm10 additionally contains an amino-terminal P-loop ATPase domain (Fien et al. 2004; Fien and Hurwitz 2006). The human MCM10 protein forms a hexameric structure similar to the structure formed by other MCM proteins (Okorokov et al. 2007). Taken together, these observations suggest that Mcm10 might be derived from the MCM protein family. Archaea also encode several families of modified MCM homologs. For example, inactivated MCM paralogs originated independently in M. kandleri and in Thermococci (arCOG05761). In both cases, Zn-binding amino acids are also substituted, but the carboxy-terminal HTH domain is present. The function of these MCM homologs in archaea is unknown. Several Methanosarcinales and Halobacteriales possess a protein family with a CCCC-type Zn finger clearly related to that in MCM and an amino-terminal HTH domain (arCOG02259 and arCOG02260, respectively), resembling the configuration of the carboxy-terminal portion of eukaryotic MCM10. Some of these proteins also contain an additional carboxy-terminal RepH/I/J family domain that is involved in replication of plasmid pNRC100 in Haloferax volcanii (Ng and DasSarma 1993). Thus, a homolog of Mcm10 might have been present already in the archaeal ancestor of eukaryotes (Fig. 1).
In eukaryotes, activation of the replicative helicase and loading of the replisome depend on two serine–threonine kinases, CDK (cyclin- dependent kinase) and DDK (Dbf4-dependent kinase) (Tanaka et al. 2007; Araki 2010; Sheu and Stillman 2010). The DDK phosphorylates one of the Mcm2/4/6 proteins (Sheu and Stillman 2010), whereas CDK phosphorylates Mcm5, Sld2, Sld3, and Mcm10 (Tanaka et al. 2007). This phosphorylation results in binding of Cdc45 and Dpb11 proteins and activation of the Mcm2–7 helicase (Sclafani and Holzen 2007). The Sld2, Sld3, and Dpb11 proteins are conserved in most eukaryotes and are essential regulators of replication initiation (Pospiech et al. 2010).
Dpb11 (known as TopBP1 in human) is a member of the BRCT domain superfamily that contains several BRCT repeats (Bork et al. 1997; Garcia et al. 2005). In prokaryotes, the BRCT domain is present only in NAD-dependent DNA ligase (Bork et al. 1997). The NAD-dependent ligase is an essential enzyme of DNA replication and repair that is present in almost all bacteria (except for some intracellular symbionts with the smallest genomes) but only in a few archaea, which probably acquired this gene via horizontal transfer from bacteria. The eukaryotic BRCT domains most likely evolved from the bacterial ones.
The Sld3-like proteins show poor sequence conservation even among eukaryotes. Recently, however, it has been shown that an α-helical conserved domain of Sld3 is present in the majority of eukaryotes (in animals and plants, this protein is called treslin); however, homologs in prokaryotes so far have not been detected (Sanchez-Pulido et al. 2010).
The Sld2 sequence is poorly conserved as well, and this protein so far has been identified only in Fungi. However, the animal RecQ4 helicase contains an amino-terminal Sld2 domain (Capp et al. 2010). We found that the amino-terminal domain of plant RecQ (e.g., GI: 308804427) shows weak sequence similarity and compatible secondary structure with Sld2 (KS Makarova, unpubl.). This observation suggests that the Sld2 domain is involved in the regulation of prereplication complex formation in all eukaryotes. Moreover, the LECA probably contained the Sld2 domain fused with the RecQ-like helicase that might be directly involved in replication (Capp et al. 2010; Pospiech et al. 2010). Most bacteria encode a RecQ helicase, whereas only a few archaea do. In Methanomicrobia, the recQ genes are clearly transferred from bacteria. In contrast, Acidilobus saccharovorans, Aeropyrum pernix, and Korarchaeum cryptofilum possess a small subfamily of RecQ homologs that are highly diverged and contain a long amino-terminal region that in K. cryptofilum encompasses a DEDDh-like 3′–5′ exonuclease domain and in the other two species probably is an inactivated derivative of this nuclease. This domain architecture resembles the animal Werner syndrome RecQ-like helicase (Mushegian et al. 1997; Bernstein et al. 2010). It seems likely that this particular archaeal subfamily of RecQ helicases is ancestral to the eukaryotic RecQ. An interesting question that probably can be addressed only by structure comparison is whether the extended amino-terminal region of these archaeal RecQ-like proteins also contains a domain homologous to Sld2 and if they are involved in replication.
Homologs of CDK and DDK (COG0515) serine–threonine kinases are present in most major archaeal lineages (with the exception of Thaumarchaeota and Nanoarchaeota) and thus can be confidently projected to the archaeal ancestor of eukaryotes as well. Most likely, serine/threonine protein phosphatase(s) might be present in this ancestral form as well. A functional link between kinase and phosphatase and cell division and membrane-remodeling systems in archaea is strongly suggested by conserved genomic contexts (Makarova and Koonin 2010; Makarova et al. 2010), but at this point, there is no evidence of the involvement of these kinases in regulation of archaeal DNA replication.
THE ACTIVE REPLISOME
Once the CMG complex is assembled and activated, it begins separating the two DNA strands and forming a replication “bubble” from which the two replication forks, each containing a leading and a lagging strand, move away (Moyer et al. 2006; Pacek et al. 2006; Labib and Gambus 2007; MacNeill 2010). Once replication is in progress, other proteins required for the bulk DNA synthesis and protection of single-stranded DNA can be recruited to form the full replisome complex at each replication fork (Fig. 1).
Primases
Similarly to the eukaryotic replication system, archaeal DNA replication involves a two-subunit primase (PriS, the catalytic subunit, and PriL, the polymerase-interacting subunit) that synthesizes an 8- to 12-nt RNA primer (Kuchta and Stengel 2010). In eukaryotes, the primer is then elongated by Pol α complexed with the cognate B (small) subunit to form a covalent DNA–RNA hybrid that is required for the replicative polymerase to start elongation (Kuchta and Stengel 2010). In archaea, the RNA primer seems to be sufficient for the activation of the replicative polymerase (Barry and Bell 2006). The evolution of these proteins in both eukaryotes and archaea appears to have involved primarily simple vertical descent because the majority of archaeal and eukaryotic genomes encompass a single gene for each subunit. The only deviation from this simple pattern is found in N. equitans, which encodes a hybrid protein containing regions approximately corresponding to the amino-terminal domain of the small subunit and the carboxy-terminal domain of the large subunit (NEQ395).
In addition to the large and small subunits of the archaeo–eukaryotic primase (PriSL), all archaea possess at least one gene for the bacterial-type DnaG primase, although in contrast to the bacterial homologs, archaeal DnaG lacks the amino-terminal Zn-finger domain and the carboxy-terminal domain that binds the bacterial replicative helicase DnaB (Makarova et al. 1999). The recently characterized DnaG protein of S. solfataricus has been shown to possess similar properties to the Escherichia coli primase and shows a fourfold faster rate of DNA priming than SsoPriSL (Zuo et al. 2010). It has been proposed that DnaG might be involved in the initiation of Okazaki fragment synthesis on the lagging strand (Zuo et al. 2010). The dnaG gene is often colocalized with the gene for the major form of archaeal superfamily B DNA polymerase, although these genes might not be co-regulated given that they are oriented convergently (Fig. 5). In many methanogens, dnaG is encoded in a predicted operon with uncharacterized α-helical, potentially metal-binding protein of arCOG02254 that by inference might have a role in replication (Fig. 3). Most eukaryotes (but not animals in which this gene has been inactivated) encode a DnaG protein that is typically fused to a DnaB-like helicase domain and is not directly related to archaeal DnaG. Instead, this gene appears to have been derived from the mitochondria and has a dedicated function in mitochondrial genome replication (Shutt and Gray 2006).
Figure 5.
Conserved genomic context of selected genes encoding replication proteins. The designations are as in Figure 3. S3, S15, S27, L44, The respective ribosomal proteins; RimI, GNAT family acetyltransferase; eIF-2α and eIF-2β, the respective translation initiation factors; Nop10, an RNA-binding protein; Pcc1, subunit of the KEOPS complex.
DNA Polymerase Sliding Clamp and Clamp Loader
The PCNA (proliferating cell nuclear antigen) functions as the sliding clamp, a trimer that encircles double-stranded DNA and freely slides along the DNA molecule (Beattie and Bell 2011b). The sliding clamp is required for the processivity of the DNA polymerase and coordinates the function of multiple binding partners that are also required for replication and repair processes. The sliding clamp is one of the few universally conserved proteins involved in replication (the bacterial ortholog is known as the DNA polymerase β subunit). In eukaryotes and many archaea, there is a single PCNA gene the product of which forms a homotrimer; this gene can be projected back to the last common ancestor of archaea and the archaeal ancestor of eukaryotes. However, in eukaryotes and independently in Crenarchaea, the PCNA gene is duplicated. Apparently there had been an ancestral duplication in Crenarchaea followed by an additional duplication in the Desulfurococcales lineage (Chia et al. 2010). Some species of Crenarchaea encode multiple copies of PCNA that can form either homotrimers or heterotrimers (Barry and Bell 2006; Pan et al. 2011a). In eukaryotes, the ancestral duplication yielded at least three additional PCNA-like families, all of which are subunits of the checkpoint 9-1-1 complex (components HUS1, RAD1, and RAD9), which is involved in DNA damage checkpoint control (Aravind et al. 1999; Majka and Burgers 2004).
Replication factor C, the clamp loader, is another universally conserved protein that is required for PCNA assembly around a DNA molecule at template–primer junctions (Bloom 2009). In both archaea and eukaryotes, RFC is a pentamer that consists of one large subunit (RFCL) encoded by a single gene and four small subunits (RFCS) that are identical in most archaea but are represented by four distinct paralogs encoded by ancestral genes in eukaryotes (Makarova et al. 2005; Chia et al. 2010). Both large and small subunits are paralogs and belong to the AAA+ superfamily of ATPases (Iyer et al. 2004). In addition, early in eukaryotic evolution, further duplications gave rise to other families, such as Rad24, which were recruited for distinct roles in checkpoint complexes in parallel with the duplication of the corresponding PCNAs and Chl12, a specific chromatid cohesion clamp loader (Majka and Burgers 2004). In archaea, independent duplications of RFCS occurred in Methanomicrobia/Halobacteria, Thermoproteales, and a few smaller lineages independently, often followed by acceleration of the evolutionary rate of the “extra” copies (Chia et al. 2010).
Notably, in archaeal genomes, the “original” paralogs of both PCNA and RFC that retain the ancestral roles in replication are encoded in conserved gene contexts adjacent to other proteins involved in replication, although some additional paralogs are encoded in suggestive contexts as well (Figs. 3 and 5).
DNA Polymerases
In archaea and eukaryotes, the main replicative polymerases belong to the B family of Palm-domain polymerases (Burgers et al. 2001). In addition to the polymerase core domain, all these proteins contain an amino-terminal 3′–5′ exonuclease domain. All eukaryotes possess four paralogous B-family polymerases denoted Pol α, Pol δ, Pol ε, and Pol ζ that are involved in both DNA replication and repair (Makarova et al. 2005; Kunkel and Burgers 2008). Archaea encode at least two paralogous B-family polymerases: the “major” one (present in all archaea), and PolB3 and the minor one (several lineages of methanogens lack this gene), which are both projected to LACA PolB1 (Edgell et al. 1998; Rogozin et al. 2008; Tahirov et al. 2009). A small subset of archaea possesses another, divergent B-family polymerase (arCOG04926), which seems to contain active exonuclease and Palm domains. This family is represented by three paralogs in Methanococcoides burtonii and appears to be prone to duplication and HGT. In addition, several archaea encode a derivative B-family polymerase, PolB2, in which both the exonuclease and the Palm domain appear to be inactivated (Edgell et al. 1998; Rogozin et al. 2008). In Crenarachaeota, this gene often colocalizes with genes encoding a small α-helical protein from arCOG07300 and RadA protein, whereas in Euryarchaeota, it colocalizes with the arORC2 gene, suggestive of involvement in replication (Fig. 3).
In addition to the B-family polymerases, many archaea encode the unique D-family polymerase (Cann et al. 1998), which is absent in Crenarachaea but present in all deeply branching lineages (Thaumarchaea, Nanoarchaeon, Korarchaeon), suggesting that this polymerase was present in LACA. The D-family polymerases consist of two subunits. The large subunit DP2 is a large multidomain protein that forms a homodimer that is responsible for the polymerase activity (Shen et al. 2001; Matsui et al. 2011). The DP2 protein does not display any sequence similarity with other protein families (except for two Zn fingers), but examination of conserved motifs suggests that it might be a highly diverged Palm-domain polymerase (Cann et al. 1998). The small subunit DP1 contains at least two domains, an ssDNA-binding OB-fold and the 3′–5′ exonuclease domain of the metallophosphatase (MPP) family. The DP1 protein is the ancestor of the small B subunit of eukaryotic replicative polymerases of the B family that, however, have lost the catalytic amino acid residues of the 3′–5′ exonuclease (Aravind and Koonin 1998; Klinge et al. 2009). Evidence has been presented that the D-family polymerase specializes in the replication of the lagging strand, whereas the B-family polymerase is involved in the leading-strand replication (Henneke et al. 2005). Interestingly, all Crenarachaea that have no family-D polymerase possess at least one additional active polymerase of the B family, suggesting that the two distinct B-family polymerases specialize on the leading- and lagging-strand replication, respectively, as is the case in eukaryotes.
Phylogenetic analysis of archaeal, eukaryotic, and bacterial B-family DNA polymerases combined with an analysis of domain architectures indicates that eukaryotic B-family polymerases, most likely, originated from two distinct archaeal ancestors, the major archaeal form that gave rise to the catalytically active amino-terminal domain of Pol ε, and the minor form that became the common ancestor of Pol α, Pol δ, and Pol ζ. All eukaryotic B-family polymerases contain two carboxy-terminal Zn-finger modules. Interestingly, the carboxy-terminal module appears to be derived from the Zn finger in the DP2 subunit of archaeal D-family DNA polymerases that are unrelated to the B family, at least as judged by sequence comparison (Tahirov et al. 2009). The Zn finger of Pol ε shows greater similarity to the counterpart in archaeal DP2 than the Zn fingers of other eukaryotic B-family polymerases. The carboxy-terminal portion of eukaryotic Pol ε consists of two additional polymerase and exonuclease domains, both inactivated; there are indications that this module could be of bacterial or bacteriophage origin (Tahirov et al. 2009). The presence of an inactivated exonuclease–polymerase module in Pol ε parallels a similar inactivation of both enzymatic domains in a distinct subfamily of inactivated archaeal B-family polymerases (Rogozin et al. 2008).
The apparent derivation of the large subunits of eukaryotic B-family polymerases from the major (PolB3) and minor (PolB1) forms of archaeal polymerases, and the origin of the small subunit from DP1 and the origin of the carboxy-terminal Zn finger from DP2 emphasize the joint contributions of the B-family and D-family archaeal polymerases to the evolution of the eukaryotic replication machinery. Furthermore, these findings suggest that the archaeal ancestor of eukaryotes possessed a highly complex replication apparatus, possibly more complex than any known extant archaeon.
Primer Removal and Gap Closure
In both eukaryotes and archaea, RNase HII and FEN1 flap endonuclease are responsible for removal of RNA primers during replication (Fig. 1) (Barry and Bell 2006). Both enzymes are present in all archaea with only a few duplications observed. Eukaryotes have a single gene for RNase II, but ancestral duplications have led to at least three FEN1-like families (Rad2, Rad27, EXO1), which, as shown in yeast, possess essentially the same activities and can complement each other in both replication and repair processes (Sun et al. 2003).
Another essential replication enzyme is DNA ligase, which is responsible for joining of the Okazaki fragments during lagging-strand replication. The main ligase involved in DNA replication in archaea is an ATP-dependent ligase (arCOG01347), which is encoded in the vast majority of archaeal genomes and is an apparent ortholog of the three eukaryotic ligases (I, III, and IV) that evolved through ancestral duplications (Ellenberger and Tomkinson 2008; Yutin and Koonin 2009). These enzymes share a common DNA-binding domain, a catalytic nucleotidyltransferase domain, and an OB-fold domain (Martin and MacNeill 2002; Ellenberger and Tomkinson 2008). Only Ligase I in eukaryotes is directly involved in replication, whereas Ligase III and Ligase IV have adopted different roles in DNA repair and homologous recombination. Ligase III and Ligase IV contain carboxy-terminal BRCT domains that are involved in protein–protein interactions via phosphoserine recognition (Martin and MacNeill 2002). As pointed out above, a BRCT domain is also present in bacterial NAD-dependent DNA ligase, which is also found in several archaea. So far only two archaea, Halalkalicoccus jeotgali and Halorhabdus utahensis, lack the ATP-dependent ligase but encode the NAD-dependent ligase, which in these organisms can be predicted to function in replication.
Single-Stranded DNA-Binding Proteins
Single-stranded DNA-binding proteins (SSBs) are essential components of the replication machinery that prevent reannealing of the growing DNA chain with the template and protect ssDNA from degradation. In all three domains of life, the major SSB proteins possess an OB-fold (Murzin 1993). In eukaryotes, the SSB, known as RPA, is a heterotrimer composed of three subunits of 70, 32, and 14 kDa (denoted RPA1, RPA2, and RPA3 in yeast) that contain four, one, and one OB-fold units, respectively. All three subunits are essential for the formation of a stable, functional RPA complex (Bochkarev and Bochkareva 2004). Archaea show multiple variations of RPA domain organization, number of homologs, and modes of interaction. Among the experimentally characterized RPA complexes in archaea, there is a homotetramer containing a single OB-fold in S. solfataricus (Wadsworth and White 2001); single-subunit RPAs containing multiple OB-folds in methanogens Methanosarcina acetivorans, Methanocaldococcus jannaschii, and Methanothermobacter thermautotrophicus (Robbins et al. 2005, and references therein); and a heterotrimeric RPA in Pyrococcus furiosus that consists of three distantly related subunits: RPA41 (COG1599), RPA32 (COG3390), and RPA14 (arCOG05741), each containing an OB-fold; in addition, RPA41 contains a Zn-finger-like motif (Komori and Ishino 2001).
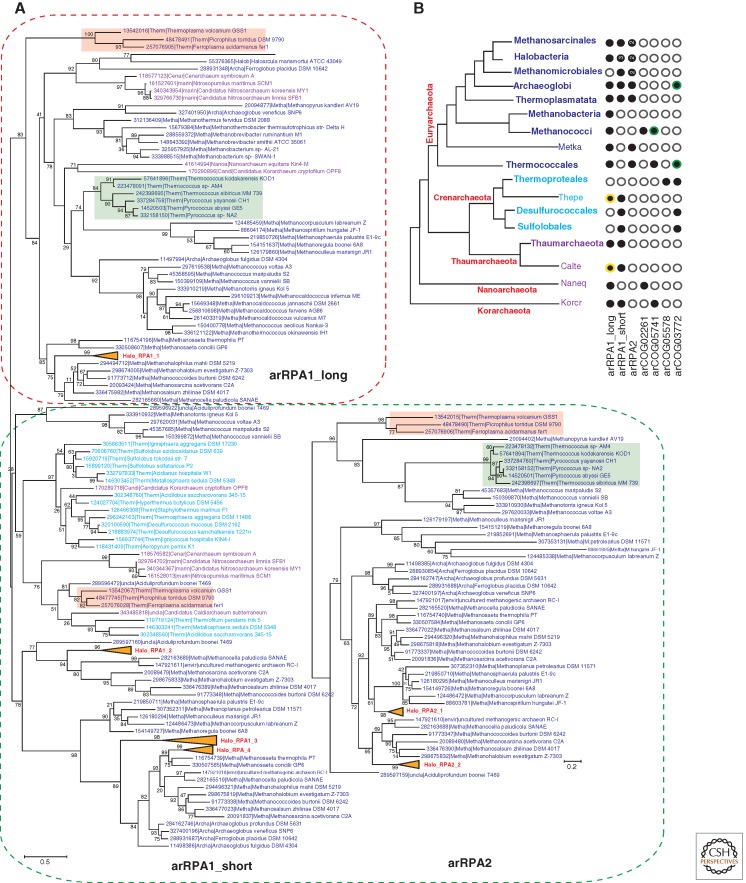
To characterize general trends in the evolution of RPA in archaea, we reconstructed phylogenetic trees for COG1599 and COG3390 (Fig. 6A). The COG1599 subtree (which we here denote arRPA1) divides into two clades: arRPA1_long with several OB-fold domains and often a Zn finger homologous to that in eukaryotic RPA1; and arRPA1_short, typically with a single OB-fold domain.
Figure 6.
Phylogenetic analysis of the RPA family in archaea and eukarya. The designations and the method of tree reconstruction are as in Figure 2. (Green) The Thermococcales branches; (pink) the Thermoplasmata branches. Halobacterial branches are collapsed and numbered as follows: for arRPA1 from Halo_RPA1_1 to Halo_RPA1_4 and for arRPA2 from Halo_RPA2_1 to Halo_RPA2_2. “Long” RPAs are outlined by the red dotted line and “short” RPAs by the green dotted line. (A) Phylogenetic trees of the RPA1 and RPA2 families. RPA1 corresponds to COG1599 (167 sequences in total, 89 aligned positions), and RPA2 corresponds to COG03390 (76 sequences in total, 149 aligned positions). (B) The phyletic patterns of SSB/RPA proteins and their homologs in archaea. The designations are as in Figure 4C. (In panel B, circles with a yellow outline denote) RPA proteins from several organisms that could not be confidently aligned and thus are not present in the corresponding tree but included into the phyletic pattern. (Green outline) Those that do not include all representatives in the corresponding lineage.
The diverged COG3390 (arRPA2) consists of short proteins most of which contain a single OB-fold domain and a carboxy-terminal wHTH domain, the same domain arrangement as in the eukaryotic RPA2 protein (Mer et al. 2000). So far, RPA2 has been detected only in Euryarchaeota (Fig. 6B). It seems likely that arRPA2 evolved by duplication of the short form of arRPA1 followed by accelerated evolution. Similar events have been detected in eukaryotes. For example, fungal CDC13, Stn1, and Ten1, the subunits of a heterotrimeric complex essential for telomere maintenance, are paralogs of the Rpa1–Rpa2–Rpa3 complex subunits but show only remote sequence similarity to RPA1, RPA2, and RPA3, respectively (Sun et al. 2009, 2011).
Thus, most archaea possess at least two RPA genes, one long (arRPA1_long) and one or several short ones (arRPA1_short or arRPA2), which are apparent ancestors of eukaryotic RPA1 and RPA2, respectively, and can be confidently projected to LACA (Fig. 6B). Whether or not LACA encoded a third short RPA (arCOG05741) remains uncertain. but, given the presence of this gene in Korarchaeaon and several recently sequenced genomes of deeply branching archaea (Fig. 6B) (Narasingarao et al. 2012), it seems likely that this RPA is ancestral in archaea as well (Fig. 1). The major exceptions from this pattern of multiple RPA forms are Sulfolobales/Desulfurococcales, which possess only RPA1_short, and Thermoproteales, which are the only known organisms without detectable OB-fold-containing SSBs.
The apparent absence of a canonical RPA/SSB in Thermoproteales stimulated the search for an alternative, which resulted in the identification of a distinct ssDNA-binding protein in T. tenax that is unrelated to RPA and has been denoted ThermoSSB (Paytubi et al. 2012). ThermoSSB (arCOG05578) contains an ssDNA-binding domain with a novel fold and a leucine-zipper domain that mediates dimerization of this protein (Paytubi et al. 2012). ThermoSSB is present in all Thermoproteales with the exception of T. pendens, which encodes two RPA-like proteins. Thus, ThermoSSB perfectly complements the phyletic pattern of RPA such that SSB proteins now have been identified in all archaea.
Some potential archaeal SSBs remain unassigned. In particular, an uncharacterized paralog of ThermoSSB (arCOG03772) shows a broader phyletic distribution being present, in addition to Thermoproteales, in Sulfolobales/Desulfurococcales, Thermococcales, and Archaeoglobi (Paytubi et al. 2012). In addition, there is at least one more protein family containing OB-fold domains and distantly similar to archaeal RPA2 (arCOG02261) that is present in most Methanococcales and Nanoarchaeum equitans.
GENOMIC CONTEXT AND PREDICTION OF NEW COMPONENTS OF THE REPLICATION MACHINERY
The results of comparative genomics outlined here show that despite the fundamental importance of DNA replication, the protein machinery responsible for this process shows remarkable evolutionary plasticity that involves multiple events of gene loss, non-orthologous displacement, and lineage-specific duplication, even among the genes encoding core replication proteins. Examples of such events leading to complex evolutionary scenarios are the substitution a B-family polymerase for the D-family polymerase in Crenarchaeota and substitution of ThermoSSB for RPA in Thermoproteales. Moreover, some components of replication systems evolve rapidly and often lose sequence similarity, which obscures their origin. However, in many such cases, the rapidly evolving gene remains in a conserved and functionally coherent genomic neighborhood (Figs. 3 and 5). One such example is the extremely divergent RecJ homolog in Thermoproteales and another is the numerous GINS proteins that are still annotated as “hypothetical” in sequenced archaeal genomes. These highly divergent proteins are encoded in the same neighborhoods as their better conserved homologs, which facilitates functional prediction. Moreover, using the “guilt by association” principle (Aravind 2000; Galperin and Koonin 2000), conservation of gene neighborhoods can be used for prediction of new genes associated with replication. However, this approach requires caution because some housekeeping genes encoded in the same locus might not be functionally related but rather are highly expressed genes that “hitchhike” with genes that are not directly functional but are expressed similarly (Rogozin et al. 2002). In particular, genes involved in replication are often associated with genes coding for components of the translation system (Fig. 5) (Berthon et al. 2008). Thus, uncharacterized genes in the respective operons might not be involved in replication directly but are nevertheless interesting targets for experimental study (Figs. 3 and 5; Table 1). Recently, for example, analysis of gene neighborhoods led to the prediction that such genes as PACE12, a member of the GPN-loop GTPase family; and NudF, a NUDIX pyrophosphatase family member are involved in DNA replication and/or repair in archaea (Berthon et al. 2008). Here we also identified several uncharacterized genes that could be involved in these processes (Figs. 3 and 5; Table 1). In addition, because replication is a complex, metabolically costly process, many other proteins and cellular systems could be directly or indirectly involved in replication. Recent analysis of proteins interacting with core replication proteins in T. kodakarensis suggests that there could be dozens of such partners, and for many of these, the specific function is not known (Li et al. 2010). Thus, all recent advances notwithstanding, the current understanding of the archaeal replication system is still far from being complete.
GENERAL TRENDS IN THE EVOLUTION OF ARCHAEAL AND EUKARYOTIC REPLICATION SYSTEMS
The study of archaeal replication is currently a dynamic research field that continues to identify previously uncharacterized protein components of the replication machinery, most of which show new connections with the eukaryotic replication machinery. Comparative genomic analysis reveals a (nearly) precise correspondence between the components of the archaeal and eukaryotic replication systems, with a few notable exceptions such as the CMG-activating proteins for which archaeal counterparts have not been detected (Fig. 1; Table 1). Thus, it appears most likely that the archaeal ancestor of eukaryotes possessed a DNA replication apparatus that was as complex in its main features as the eukaryotic replication apparatus. Given this essentially precise correspondence between the archaeal and eukaryotic replication systems, it can be expected that additional replication proteins shared between archaea and eukaryotes eventually will be discovered (Fig. 1; Table 1). A major innovation in eukaryotes could be the regulation of replication by phosphorylation of replisome subunits catalyzed by dedicated kinases. Whether a counterpart to this regulatory circuit exists in archaea remains to be determined.
The close correspondence between the components of the archaeal and eukaryotic replication systems notwithstanding, eukaryotic replication does show increased complexity compared with archaeal replication. The principal modality of this greater complexity is duplication of the replication machinery components followed by subfunctionalization (Lynch and Force 2000; Ward and Durrett 2004) whereby heteromeric complexes in eukaryotes replace homomeric complexes in archaea. The obvious cases in point are the ORC, MCM, GINS, and RFC complexes. This evolutionary trend in the replication machinery is part of a general relationship between eukaryotes and their prokaryotic ancestors as exemplified by the proteasome, the exosome, and many other macromolecular complexes (Makarova et al. 2005).
Although, on the whole, the eukaryotic replication system is more complex than the archaeal counterparts, complicated histories of lineage-specific expansion of gene families accompanied by functional diversification abound in archaea. Unlike eukaryotes, where the main route to increased complexity is serial intragenomic gene duplication, in archaea, replicon fusion and more generally horizontal gene transfer are major factors of evolution, leading in particular to pseudoparalogy (Makarova et al. 2005). Strikingly, in some families of proteins involved in replication, archaea attain far greater diversity than eukaryotes, the RecJ family being the prime example. In other gene families, multiple, independent duplications are traceable in eukaryotes and in different lineages of archaea. The degree of paralogization of the archaeal replication system remains underappreciated, and many divergent paralogs still await functional characterization.
In addition to the diversification via paralogization, archaeal replication systems show less uniformity and a greater plasticity than the eukaryotic counterparts. The striking examples include non-orthologous displacement of DNA polymerases, SSB, and ligases. The actual scope of this plasticity is still unexplored because even among the relatively few archaeal genomes sequenced to date, some gaps within the core of the replication machinery are apparent such as the lack of ORC subunits in M. kandleri, suggestive of additional displacements of essential components.
In summary, the recent experimental and phylogenomic advances in the study of eukaryotic and archaeal replication systems clearly complement each other and jointly are yielding an increasingly complete picture of the organization and evolution of the core replication machinery. This obvious progress notwithstanding, the identity and roles of more peripheral components of the replication apparatus, the functions of paralogs of replicative genes, and especially, the ultimate origins of the complex replication machinery that is inferred to have existed in LACA, remain poorly understood and are targets for future investigation.
ACKNOWLEDGMENTS
The authors' research is supported by the intramural funds of the U.S. Department of Health and Human Services (to the National Library of Medicine).
REFERENCES
- Adachi J, Hasegawa M 1992. MOLPHY: Programs for molecular phylogenetics. In Computer science monographs 27. Institute of Statistical Mathematics, Tokyo [Google Scholar]
- Araki H 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol 22: 766–771 [DOI] [PubMed] [Google Scholar]
- Aravind L 2000. Guilt by association: Contextual information in genome analysis. Genome Res 10: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV 1998. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res 26: 3746–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res 27: 1223–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M, Huijbregts RP, Chesnokov I 2009. Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci 106: 10672–10677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Bell SD 2006. DNA replication in the Archaea. Microbiol Mol Biol Rev 70: 15: 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TR, Bell SD 2011a. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 15: 614–619 [DOI] [PubMed] [Google Scholar]
- Beattie TR, Bell SD 2011b. The role of the DNA sliding clamp in Okazaki fragment maturation in archaea and eukaryotes. Biochem Soc Trans 39: 70–76 [DOI] [PubMed] [Google Scholar]
- Bell SD 2011. DNA replication: Archaeal oriGINS. BMC Biol 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bernander R, Dasgupta S, Nordstrom K 1991. The E. coli cell cycle and the plasmid R1 replication cycle in the absence of the DnaA protein. Cell 64: 1145–1153 [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R 2010. The RecQ DNA helicases in DNA repair. Annu Rev Genet 44: 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquist BR, DasSarma P, DasSarma S 2007. Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1. BMC Genet 8: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon J, Cortez D, Forterre P 2008. Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol 9: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom LB 2009. Loading clamps for DNA replication and repair. DNA Repair (Amst) 8: 570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E 2004. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol 14: 36–42 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A 2009. The Mcm complex: Unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73: 652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlke K, Pisani FM, Rossi M, Antranikian G 2002. Archaeal DNA replication: Spotlight on a rapidly moving field. Extremophiles 6: 1–14 [DOI] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 11: 68–76 [PubMed] [Google Scholar]
- Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, et al. 2001. Eukaryotic DNA polymerases: Proposal for a revised nomenclature. J Biol Chem 276: 43487–43490 [DOI] [PubMed] [Google Scholar]
- Cann IK, Komori K, Toh H, Kanai S, Ishino Y 1998. A heterodimeric DNA polymerase: Evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc Natl Acad Sci 95: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capp C, Wu J, Hsieh TS 2010. RecQ4: The second replicative helicase? Crit Rev Biochem Mol Biol 45: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Bielinsky AK 2007. Human Mcm10 regulates the catalytic subunit of DNA polymerase-α and prevents DNA damage during replication. Mol Biol Cell 18: 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N, Cann I, Olsen GJ 2010. Evolution of DNA replication protein complexes in eukaryotes and Archaea. PLoS ONE 5: e10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JA, DasSarma P, Capes M, Wallace T, McGarrity K, Gessler R, Liu J, Xiang H, Tatusov R, Berquist BR, et al. 2009. Multiple replication origins of Halobacterium sp. strain NRC-1: Properties of the conserved orc7-dependent oriC1. J Bacteriol 191: 5253–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco V, Gillespie PJ, Li A, Karantzelis N, Christodoulou E, Klompmaker R, van Gerwen S, Fish A, Petoukhov MV, Iliou MS, et al. 2009. Quaternary structure of the human Cdt1–Geminin complex regulates DNA replication licensing. Proc Natl Acad Sci 106: 19807–19812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ 2009. The origin recognition complex protein family. Genome Biol 10: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell DR, Doolittle WF 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89: 995–998 [DOI] [PubMed] [Google Scholar]
- Edgell DR, Malik SB, Doolittle WF 1998. Evidence of independent gene duplications during the evolution of archaeal and eukaryotic family B DNA polymerases. Mol Biol Evol 15: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE 2008. Eukaryotic DNA ligases: Structural and functional insights. Annu Rev Biochem 77: 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fien K, Hurwitz J 2006. Fission yeast Mcm10p contains primase activity. J Biol Chem 281: 22248–22260 [DOI] [PubMed] [Google Scholar]
- Fien K, Cho YS, Lee JK, Raychaudhuri S, Tappin I, Hurwitz J 2004. Primer utilization by DNA polymerase α-primase is influenced by its interaction with Mcm10p. J Biol Chem 279: 16144–16153 [DOI] [PubMed] [Google Scholar]
- Fitch WM, Margoliash E 1967. Construction of phylogenetic trees. Science 155: 279–284 [DOI] [PubMed] [Google Scholar]
- Forterre P. 2002. The orgin of DNA genomes and DNA replication proteins. Curr Opin Microbiol 5: 525–532 [DOI] [PubMed] [Google Scholar]
- Forterre P. 2006. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc Natl Acad Sci 103: 3669–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV 2000. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol 18: 609–613 [DOI] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Glansdorff N, Xu Y, Labedan B 2008. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct 3: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan SM, Richardson CC 2009. Motors, switches, and contacts in the replisome. Annu Rev Biochem 78: 205–243 [DOI] [PubMed] [Google Scholar]
- Hamdan SM, van Oijen AM 2010. Timing, coordination, and rhythm: Acrobatics at the DNA replication fork. J Biol Chem 285: 18979–18983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G, Flament D, Hubscher U, Querellou J, Raffin JP 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J Mol Biol 350: 53–64 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146: 11–31 [DOI] [PubMed] [Google Scholar]
- Kelman Z, White MF 2005. Archaeal DNA replication and repair. Curr Opin Microbiol 8: 669–676 [DOI] [PubMed] [Google Scholar]
- Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L 2009. 3D architecture of DNA Pol α reveals the functional core of multi-subunit replicative polymerases. EMBO J 28: 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K, Ishino Y 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J Biol Chem 276: 25654–25660 [DOI] [PubMed] [Google Scholar]
- Koonin EV 2006. Temporal order of evolution of DNA replication systems inferred by comparision of cellular and viral DNA polymerases. Biol Direct 1: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2009. On the origin of cells and viruses: primordial virus world scenario. Ann NY Acad Sci 1178: 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Martin W 2005. On the origin of genomes and cells within inorganic compartments. Trends Genet 21: 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppes LJ 1992. Nonrandom F-plasmid replication in Escherichia coli K-12. J Bacteriol 174: 2121–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker TA 2005. DNA replication, 2nd ed. University Science Books, Sausalito, CA [Google Scholar]
- Krupovic M, Gribaldo S, Bamford DH, Forterre P 2010. The evolutionary history of archaeal MCM helicases: A case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol Biol Evol 27: 2716–2732 [DOI] [PubMed] [Google Scholar]
- Kuchta RD, Stengel G 2010. Mechanism and evolution of DNA primases. Biochim Biophys Acta 1804: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM 2008. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A 2007. A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17: 271–278 [DOI] [PubMed] [Google Scholar]
- Lee C, Hong B, Choi JM, Kim Y, Watanabe S, Ishimi Y, Enomoto T, Tada S, Cho Y 2004. Structural basis for inhibition of the replication licensing factor Cdt1 by geminin. Nature 430: 913–917 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Koonin EV 1999. Did DNA replication evolve twice independently? Nucleic Acids Res 27: 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z 2010. Affinity purification of an archaeal DNA replication protein network. MBio 1: e00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z 2011. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res 39: 6114–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y 2011. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci 108: 7373–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA 2010. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J 425: 489–500 [DOI] [PubMed] [Google Scholar]
- MacNeill SA 2011. Protein–protein interactions in the archaeal core replisome. Biochem Soc Trans 39: 163–168 [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 78: 227–260 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV 2010. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV 1999. Comparative genomics of the Archaea (Euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9: 608–628 [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res 33: 4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV 2010. Evolution of diverse cell division and vesicle formation systems in archaea. Nat Rev Microbiol 8: 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Kelman Z 2012. The archaeal CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep 7: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin IV, MacNeill SA 2002. ATP-dependent DNA ligases. Genome Biol 3: REVIEWS3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui I, Urushibata Y, Shen Y, Matsui E, Yokoyama H 2011. Novel structure of an N-terminal domain that is crucial for the dimeric assembly and DNA-binding of an archaeal DNA polymerase D large subunit from Pyrococcus horikoshii. FEBS Lett 585: 452–458 [DOI] [PubMed] [Google Scholar]
- McGeoch AT, Bell SD 2008. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat Rev Mol Cell Biol 9: 569–574 [DOI] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ 2000. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103: 449–456 [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR 2006. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG 1993. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J 12: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Bassett DE Jr, Boguski MS, Bork P, Koonin EV 1997. Positionally cloned human disease genes: Patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci 94: 5831–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, DasSarma S 1993. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. J Bacteriol 175: 4584–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okorokov AL, Waugh A, Hodgkinson J, Murthy A, Hong HK, Leo E, Sherman MB, Stoeber K, Orlova EV, Williams GH 2007. Hexameric ring structure of human MCM10 DNA replication factor. EMBO Rep 8: 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Ishino S, Fujino S, Ogino H, Shirai T, Mayanagi K, Saito M, Nagasawa N, Ishino Y, Morikawa K 2011. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Pan M, Kelman LM, Kelman Z 2011a. The archaeal PCNA proteins. Biochem Soc Trans 39: 20–24 [DOI] [PubMed] [Google Scholar]
- Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z 2011b. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res 39: 9671–9680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paytubi S, McMahon SA, Graham S, Liu H, Botting CH, Makarova KS, Koonin EV, Naismith JH, White MF 2012. Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales. Proc Natl Acad Sci 109: E398–E405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech H, Grosse F, Pisani FM 2010. The initiation step of eukaryotic DNA replication. Subcell Biochem 50: 79–104 [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP 2010. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JB, McKinney MC, Guzman CE, Sriratana B, Fitz-Gibbon S, Ha T, Cann IK 2005. The Euryarchaeota, nature's medium for engineering of single-stranded DNA-binding proteins. J Biol Chem 280: 15325–15339 [DOI] [PubMed] [Google Scholar]
- Robertson PD, Chagot B, Chazin WJ, Eichman BF 2010. Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif. J Biol Chem 285: 22942–22949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD 2005. Origins of DNA replication in the three domains of life. FEBS J 272: 3757–3766 [DOI] [PubMed] [Google Scholar]
- Robinson NP, Bell SD 2007. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci 104: 5806–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116: 25–38 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, Szekely LA, Koonin EV 2002. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res 30: 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Makarova KS, Pavlov YI, Koonin EV 2008. A highly conserved family of inactivated archaeal B family DNA polymerases. Biol Direct 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara N, Kelman LM, Kelman Z 2009. Unwinding the structure and function of the archaeal MCM helicase. Mol Microbiol 72: 286–296 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Ponting CP 2011. Cdc45: The missing RecJ ortholog in eukaryotes? Bioinformatics 27: 1885–1888 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JF, Ponting CP 2010. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol 20: R509–R510 [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Musti K, Hiramoto M, Kikuchi H, Kawarabayashi Y, Matsui I 2001. Invariant Asp-1122 and Asp-1124 are essential residues for polymerization catalysis of family D DNA polymerase from Pyrococcus horikoshii. J Biol Chem 276: 27376–27383 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt TE, Gray MW 2006. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J Mol Evol 62: 588–599 [DOI] [PubMed] [Google Scholar]
- Sun X, Thrower D, Qiu J, Wu P, Zheng L, Zhou M, Bachant J, Wilson DM 3rd, Shen B 2003. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair (Amst) 2: 925–940 [DOI] [PubMed] [Google Scholar]
- Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M 2009. Stn1–Ten1 is an Rpa2–Rpa3–like complex at telomeres. Genes Dev 23: 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yang Y, Wan K, Mao N, Yu TY, Lin YC, DeZwaan DC, Freeman BC, Lin JJ, Lue NF, et al. 2011. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res 21: 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV 2009. Evolution of DNA polymerases: An inactivated polymerase–exonuclease module in Pol ε and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Wadsworth RI, White MF 2001. Identification and properties of the Crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res 29: 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, Chong JP 2010. An archaeal order with multiple minichromosome maintenance genes. Microbiology 156: 1405–1414 [DOI] [PubMed] [Google Scholar]
- Ward R, Durrett R 2004. Subfunctionalization: How often does it occur? How long does it take? Theor Popul Biol 66: 93–100 [DOI] [PubMed] [Google Scholar]
- Warren EM, Vaithiyalingam S, Haworth J, Greer B, Bielinsky AK, Chazin WJ, Eichman BF 2008. Structural basis for DNA binding by replication initiator Mcm10. Structure 16: 1892–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Liu C, Wu X, Xu N, Zhou B, Liang C, Zhu G 2010. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J Biol Chem 285: 12469–12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC 2002. Xenopus Mcm10 binds to origins of DNA replication after Mcm2–7 and stimulates origin binding of Cdc45. Mol Cell 9: 233–240 [DOI] [PubMed] [Google Scholar]
- Yutin N, Koonin EV 2009. Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: A case of hidden complexity. Biol Direct 4: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang CT 2005. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea 1: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA 2010. Characterization of a functional DnaG-type primase in Archaea: Implications for a dual-primase system. J Mol Biol 397: 664–676 [DOI] [PubMed] [Google Scholar]