Abstract
Enveloped viruses enter cells via a membrane fusion reaction driven by conformational changes of specific viral envelope proteins. We report here the structure of the ectodomain of the tick-borne encephalitis virus envelope glycoprotein, E, a prototypical class II fusion protein, in its trimeric low-pH-induced conformation. We show that, in the conformational transition, the three domains of the neutral-pH form are maintained but their relative orientation is altered. Similar to the postfusion class I proteins, the subunits rearrange such that the fusion peptide loops cluster at one end of an elongated molecule and the C-terminal segments, connecting to the viral transmembrane region, run along the sides of the trimer pointing toward the fusion peptide loops. Comparison with the low-pH-induced form of the alphavirus class II fusion protein reveals striking differences at the end of the molecule bearing the fusion peptides, suggesting an important conformational effect of the missing membrane connecting segment.
Keywords: emerging viruses, envelope glycoproteins, enveloped viruses, flavivirus, membrane fusion, structure/function relations
Introduction
Controlled fusion between different membrane-bound compartments at the right time and space is central to the organization of all living organisms. This process is tightly controlled by specific proteins that interact with each other and with lipids, as well as with other proteins, to form a complex at the fusion site (reviewed in Jahn et al, 2003). Enveloped viruses also use membrane fusion for cell entry, and have evolved virus-specific fusion proteins that mediate the merging of viral and cellular membranes. These fusion proteins contain a special segment of their polypeptide chain, termed ‘fusion peptide', that becomes exposed during the conformational change and is inserted into the target membrane. At least two different structural classes of viral membrane fusion proteins, displaying a completely different molecular architecture, have been described. Pioneering structural studies on the influenza virus hemagglutinin (Wilson et al, 1981) and on other functionally and structurally related viral envelope proteins, collectively termed as class I fusion proteins (reviewed in Weissenhorn et al, 1999; Colman and Lawrence, 2003), have provided major breakthroughs for understanding the mechanisms used to drive the membrane fusion reaction.
Class I fusion proteins form trimeric spikes in their native metastable conformation at the surface of infectious virions. Their postfusion conformation is a hairpin-like structure in which both the fusion peptide (near the N-terminus) and the membrane anchor (near the C-terminus) are juxtaposed at the same end of a very stable protein rod. The class II fusion proteins, E of flaviviruses (Rey et al, 1995; Modis et al, 2003) and E1 of alphaviruses (Lescar et al, 2001), have been shown to be homologous in spite of the absence of sequence conservation, and display a molecular architecture completely different from that of class I proteins. The fusion peptide is internal, contained in a loop between two β-strands. The structural alignment of the flavivirus tick-borne encephalitis virus (TBEV) envelope protein E and the alphavirus Semliki Forest virus fusion protein E1 (SFV E1) provided in Figure 1 summarizes the domain organization as well as the secondary structure nomenclature used for the class II proteins. These proteins are synthesized and folded in a metastable form as a complex with a second, ‘chaperone' viral envelope protein—prM (precursor of M) in the case of flavi and pE2 (precursor of E2) in the case of alphaviruses. Proteolytic cleavage of the chaperone (instead of cleavage of the fusion protein itself, as in the case of class I viral fusion proteins) lowers the energy barrier between the metastable state and the final, lowest energy conformation of the fusion protein, thus priming them to trigger membrane fusion. Both flavivirus and alphavirus virions are icosahedral assemblies, the surface of which is a continuous protein lattice of E homodimers (Kuhn et al, 2002) and E1/E2 heterodimers (Lescar et al, 2001; Zhang et al, 2002), respectively, with the fusion peptide loop buried in the respective dimer interfaces. Both viruses enter target cells by receptor-mediated endocytosis (Wahlberg and Garoff, 1992; Allison et al, 1995), with the receptor recognition function carried by the fusion protein itself in flaviviruses and by the companion E2 protein in alphaviruses. Exposure to the acidic pH of the endosomes triggers a major conformational change of the virion's surface, involving dissociation of the native protein complexes (which destroys the surface icosahedral lattice), and the formation of homotrimers of the fusion proteins (reviewed in Heinz and Allison, 2000; Kielian et al, 2000). The energy released during the transition from the metastable dimers at the viral surface to the very stable target-membrane-inserted homotrimers is used to drive the merging of the viral and cellular membranes. This conversion also occurs in solution with solubilized E dimers (Stiasny et al, 1996).
Figure 1.
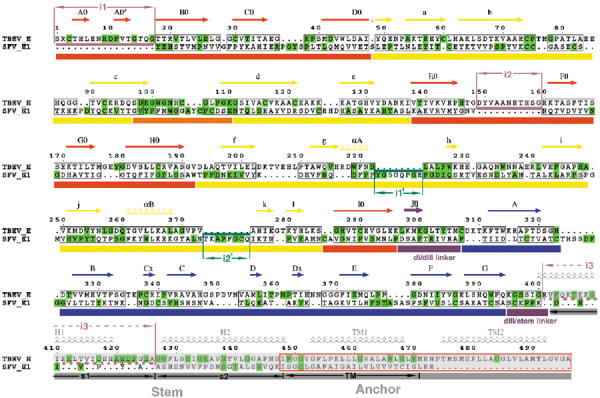
Structural alignment of the class II membrane fusion proteins TBEV E and SFV E1. The secondary structure, domain nomenclature and color coding follow the definitions used for the TBEV sE dimer (Rey et al, 1995). A color-coded bar below the sequence indicates domains dI, dII, and dIII in red, yellow, and blue, respectively. The fusion peptide loop is colored orange and segments in between domains are purple (the dI/dIII and dIII/stem linkers). Highly conserved residues are given on a green background (note the similar pattern of conservation within the two viruses). A gray semitransparent bar overlays the C-terminal segments that are not part of the crystal structures. The secondary structure elements are indicated above the sequence, with arrows and helices for β-strands and α-helices, respectively. In the stem region, the predicted α-helices (H1, H2, TM1, and TM2) are indicated in gray above the sequence, and only apply to the top TBEV sequence. The corresponding segments in alphaviruses are indicated by s1, s2, and TM below the sequence. The actual transmembrane regions of the proteins are framed in red. The structural alignment was obtained using the DALI server with the three domains separately, using the low-pH-induced trimeric conformation of the two proteins. Structurally equivalent amino acids in the two proteins are boxed. A few important insertions are indicated above the sequence in TBEV E (i1, i2, and i3, in pale red) and below for SFV E1 (i1′ and i2′, in dark green). The insertion in between residues 204 and 209 in TBEV (in the fg loop) is also an insertion in tick-borne with respect to mosquito-borne flaviviruses, and therefore is not marked. The i3 insertion in TBEV E is indicated by broken lines since this segment is α helical in flaviviruses and extended in alphaviruses (shown by stretching the residues so that only every third or fourth amino acid in E1 has its counterpart in E in this region of the alignment). The i1′ and i2′ insertions in SFV E1 occur downstream of the two α-helices observed in dII, at regions of contact with s1, and are postulated to compensate for the less bulky s1 region compared to H1.
TBEV as well as several mosquito-borne flaviviruses, like dengue, yellow fever, Japanese encephalitis, and West Nile viruses, are important human pathogenic viruses. The flavivirus E polypeptide chain contains about 500 amino acids. The structure of the TBEV E ectodomain (termed sE and containing about 400 residues) has shown that the E subunits are elongated rods that associate in an antiparallel fashion, leading to a brick-shaped dimer with the C-termini at either end (Rey et al, 1995). Infectious virions contain 90 dimers at their surface, forming a smooth protein shell completely covering the viral membrane (Kuhn et al, 2002). The sE ectodomain is responsible for the lateral contacts between dimers at the viral surface. The viral membrane-interacting segment of the protein contains roughly the 100 C-terminal amino acids that immediately follow sE in the amino-acid sequence. This segment is folded as four α-helices that have been visualized by electron cryomicroscopy (cryo EM) and image reconstruction of mature dengue virus particles (Zhang et al, 2003a). The first two helices are amphipathic and lie flat on the viral membrane, interacting with the lipid heads of the outer leaflet of the lipid bilayer. This region of the protein is called the ‘stem' (Allison et al, 1999). The third and fourth α-helices traverse the membrane as an antiparallel coiled-coil.
Exposure of the soluble sE fragment to low pH leads to a reversible dissociation of the dimer, resulting in exposure of the fusion peptide loops but not to trimerization as on the virion envelope (Stiasny et al, 1996). However, as initially shown for the soluble ectodomain of SFV E1 (Klimjack et al, 1994), when the low-pH treatment is carried out in the presence of liposomes, the interaction of the fusion peptide loops with lipids triggers an irreversible trimerization of sE, which remains stably membrane associated (Stiasny et al, 2002). Solubilization from the liposomes with nonionic detergents allowed the isolation, biochemical characterization, and crystallization of this sE trimer (Stiasny et al, 2004). We describe here the crystal structure of the ectodomain of the TBEV E protein in its low-pH-induced trimeric form.
Results and discussion
Structural transition from dimer to trimer
The crystal structure determination is described in the Materials and methods section, and the relevant statistics are given in Table I. The structure of the trimeric low-pH form of sE shows that the conformational change reorients the molecule from a horizontal, antiparallel dimeric conformation to a vertical trimer in which the subunits display a parallel arrangement (Figure 2). The fusion peptide loops (in orange) are exposed at one end of the trimer, but retain a conformation similar to that observed in the structure of the dimer. Their main chains expose a number of polar groups to solvent, indicating that they are not likely to insert deeply into the lipid bilayer, but would instead interact mostly with the lipid heads of the outer leaflet, as diagrammed in Figure 2B, lower panel. This is consistent with the measurements of the molecule made in electron micrographs of liposomes containing TBEV sE trimers at their surface (Stiasny et al, 2004). The aromatic residues W101 and F108 of the fusion peptide loop, which in the neutral-pH form are buried in the dI/dIII pocket at the dimer interface (Rey et al, 1995), are exposed in the trimer, presumably interacting with disordered detergent molecules in the crystal, and with the aliphatic moiety of the lipid bilayer on membranes.
Table 1.
Summary of crystallographic dataa
| Crystal form | O1 | O2 | T |
|---|---|---|---|
| Space group | P212121 | P21212 | P4222 |
| Cell dimensions (Å) | a=121.5 | a=120.4 | a=b=127.6 |
| b=142.9 | b=180.6 | c=224.6 | |
| c=173.6 | c=68.5 | ||
| Number of molecules per asymmetric unit | 6 | 3 | 3 |
| Maximum resolution (Å) | 2.7 (2.8–2.7) | 3.2 (3.1–3.2) | 3.5b (3.6–3.5) |
| Number of observations | 306 381 | 105 296 | 113 305 |
| Rsym (%) | 11.1 (45.5) | 11.2 (39.0) | 13.1 (42.9) |
| I/σ(I) | 5.4 (2.0) | 9.7 (3.7) | 9.5 (4.0) |
| Resolution range for refinement (Å)c | 40–2.7 | 30–3.2 | 30–3.5 |
| Number of reflections | 80 896 | 25 236 | 24 110 |
| Completeness (%) | 96.9 (83.9) | 99.8 (100) | 99.8 (99.7) |
| Rfact (%) | 20.6 (31.6) | 29.7 | 27.8 |
| Rfree (%) | 24.2 (34.0) | 32.8 | 29.0 |
| rms deviation, bond lengths (Å) | 0.007 | 0.009 | 0.010 |
| rms deviation, bond angles (deg) |
1.4 |
1.5 |
1.6 |
| Rsym=∑h∑j∣Ih, j−〈Ih〉∣/∑h∑jIh,j, where 〈Ih〉 is the average intensity of the j observations of the reflection of unique index h. | |||
| Rfact=∑h∣∣Fobs∣−k∣Fcalc∣∣/∑h∣Fobs∣ for the 76 806 reflections used in refinement; Rfree was calculated with the same formula for a random 4090 reflections not used in refinement. | |||
| b Anisotropic, 3.8 Å along a*, 3 Å along c*. | |||
| c For crystal forms O2 and T, a single round of positional refinement and group B factor refinement was performed after molecular replacement with the refined structure determined with form O1. Noncrystallographic symmetry constraints were enforced. | |||
aValues for the outermost resolution shell are shown in parentheses.
Figure 2.
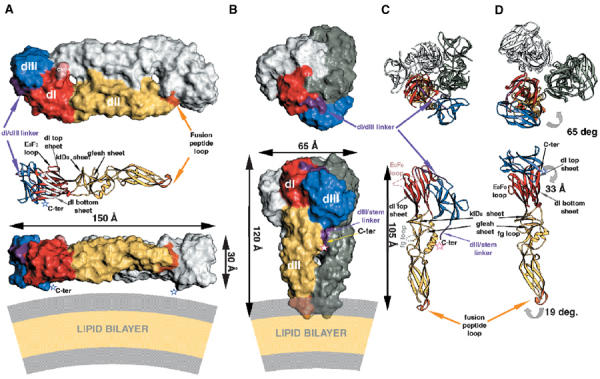
Conformational rearrangement of protein E. Comparison of the overall organization of the protein in the neutral- and acid-pH forms. The ‘top' and ‘side' views are indicated in the top and bottom rows, respectively. The three domains of sE are labeled dI, dII, and dIII. The color coding is defined in the legend to Figure 1. (A) Neutral–pH, dimeric conformation of sE in a surface representation. The carbohydrate residues (labeled CHO) are indicated in pink. A ribbon diagram is intercalated between the top and side views, at the same scale and orientation as the foreground subunit in the side view. Several β-sheets that are referred to in the text are indicated (i.e., the top and bottom β-sheets of dI, the klD0 and gfeah sheets of dII). In the ribbon diagram, the arrows representing the β-strands in the bottom sheet of dI are white. The last amino acid observed in the crystal structure (K395; Rey et al, 1995) is indicated by an open blue star, labeled C-term. The lipid bilayer is diagrammed at the same scale underneath the dimer in the side view, with the aliphatic region in pale yellow and the lipid head regions in gray. (B) Low-pH conformation of sE. As in panel A, only one subunit is colored and the others are shown in white and gray. The arrows show the dimensions of the molecule, including all atoms with a Van der Waals radius of 2 Å. In the side view, the purple region indicates the dIII/stem linker, which ends at the last amino acid visible in the electron density map (R401) indicated by an open red star (labeled C-ter). Note the vertical groove that follows the C-terminus along the interface between neighboring dIIs in the trimer. The lipid bilayer is diagrammed as in (A), indicating the postulated interaction of the fusion peptide loops with the lipid heads. (C) Ribbon diagram of the polypeptide chain of sE in the trimeric conformation. In the top view, note the extended conformation of the dI/dIII linker (purple). In the side view, only the colored subunit displayed in (B) is shown for clarity. The disordered segments (the E0F0 loop in dI and the fg loop in dII) are indicated by broken lines and labeled. The C-terminus is indicated as in (B). It shows that the predicted α-helix H1 of the stem would interact with the two short helices of dII. (D) Conformational rearrangement of sE. dI of the sE subunit in the conformation observed in the dimer (Figure 1A) was superposed on dI of the colored subunit in the trimer shown in (C), as explained in the text. Curved gray arrows show the movement of the domains to reach the conformation indicated in (C). In the side view, the two β-sheets of dII that change their relative orientation are labeled (klD0 and gfeah).
Importantly, in the conformational transition, the individual domains of sE rearrange such that the C-terminal segment, including dIII, runs along the sides of the trimer toward the fusion peptide loops. The sE C-terminus (indicated by an open red star in Figures 2B and C, lower panel, at the end of the dIII/stem linker) is located about mid-way. The conformation of the molecule suggests that in the postfusion form of full-length E, the stem will continue to reach the lipid bilayer such that the TM helices and the fusion peptide loops will be juxtaposed. This overall arrangement is analogous to that observed in the postfusion form of class I fusion proteins (Weissenhorn et al, 1999), suggesting a common mechanism to appose the two different lipid bilayers in the first step of membrane fusion. In contrast to class I, however, no major refolding of the protein takes place during the fusogenic rearrangement, with the individual domains retaining their original fold, as summarized in Table II. For an easier visualization of the change in conformation within the sE polypeptide, we have compared it to the neutral-pH form by artificially holding dI constant and referring all the domain movements to it, as shown in Figure 2D. For this purpose, the sE subunit in the conformation observed in the dimer was superimposed on dI in the trimer, and the trimer axis was used to assemble artificially a trimer of sE subunits in the neutral-pH conformation through similar dI/dI contacts. Figure 2 shows that the relative arrangement of dII with respect to dI is similar in the two forms (compare the lower panels of Figures 2C and D), except for a rotation of dII of 19° about the dI/dII junction, which results in a straight 105 Å long rod formed by dI and dII (measured between Cα atoms at the two ends as indicated in Figure 2C). In contrast, the relative dI/dIII orientation is radically different, with dIII now lying on a side of the trimer, with its C-terminal end and the dIII/stem linker projecting toward dII. In the trimeric form, the β-strands of the Ig-like dIII run roughly parallel to the strands of dI, whereas they were nearly orthogonal in the neutral-pH form. The center of mass of dIII is displaced by 33 Å. In spite of this dramatic change in the environment of dIII, its internal structure changes the least among the three domains in both conformations, as indicated by the small rms deviation (0.6 Å) obtained when directly superposing dIII from the two forms (Table II). In contrast, the dI/dIII linker, which comprises residues 296–310 (in purple), stretches considerably to reach the new position of dIII in the acid form.
Table 2.
Pairwise superposition of domainsa
| Structures compared domains superposed | TBEa/TBEnb |
TBEa/SFVa |
TBEn/SFVn |
||||||
|---|---|---|---|---|---|---|---|---|---|
| dI | dII | dIII | DI | dII | dIII | dI | dII | dIII | |
| No. of equivalent Cα | 90 | 82 | 91 | 78 | 157 | 77 | 75 | 158 | 75 |
| rmsdc (Å) |
2.1 |
1.4 |
0.6 |
2.3 |
3.7 |
3.3 |
2.5 |
3.5 |
2.9 |
| ba, acidic-pH form; n, neutral-pH form. | |||||||||
| crmsd=root mean squared deviation. | |||||||||
aThe superposition was made using the Dali server, http://www.ebi.ac.uk/dali/. The PDB accession numbers of the coordinates used are TBEa, 1URZ; TBEn, 1SVB; SFVa, 1RER; SFVn, 1I9W, except for dIII the coordinates of which will be deposited along with submission of a manuscript (J Lescar and FA Rey, in preparation), describing the refined model of the SFV E1 protein in its neutral-pH form and its interactions in the viral particle.
In addition to the important change in the orientation of dIII and the accompanying stretching of the dI/dIII linker, several changes take place in dI and at the dI/dII interface. Two loops are disordered: the E0F0 loop of dI, which carries the single N-linked glycosylation site of TBEV E, and the fg loop in dII, which contains a six-residue insertion in all tick-borne flaviviruses with respect to the mosquito-borne flaviviruses. These two loops are indicated in Figure 2A, C and D. The rms deviation of the individual superpositions (Table II) shows that in the conformational change the internal structure of dI changes most, and that of dII is intermediate. The changes in dI are best described by comparing to the corresponding changes in dI from SFV E1 and will be dealt with in a later section. In dII, the 19° rotation with respect to dI results from a sliding of the two apposed β-sheets near the dI/dII junction, the klDO and gfeah sheets (labeled in Figure 2). This region overlaps with the area of alternative conformation observed in the dengue virus sE protein structure at neutral pH in the presence of detergent (Modis et al, 2003), where the kl β-hairpin switches to the gfeah sheet to create a hydrophobic pocket that accommodates the aliphatic chain of the detergent.
Interactions that must break in the neutral-pH form to allow the dIII rearrangement
Examination of the dimer contacts shows that only a few of the amino acids involved are conserved among flaviviruses, except for those in the fusion peptide loop, as shown in Figure 3A. In contrast, the region of contacts between dI and dIII within the subunit in the dimer does exhibit a conserved patch across the domain interface, displayed in Figure 3B. The surface buried by dIII in the dimer is 970 Å2, of which 750 Å2 belongs to contacts with dI in the same polypeptide chain and the remainder to the interaction with the fusion peptide loop across the dimer interface. The dI/dIII interaction involves a number of Van der Waals contacts, salt bridges, and direct and water-mediated hydrogen bonds that have to be broken in the transition from dimer to trimer. Figure 3C shows a stereo view of the details of these interactions. It shows, in particular, a salt bridge between the strictly conserved residues R9 (from dI) and E373 (from dIII). It also shows the presence in the contact area of two strictly conserved histidine residues, H323 from dIII and H146 from dI, which are involved in hydrogen bonds with the main chain of the opposite domain across the interface. Protonation of these histidine residues is likely to be an important factor in the destabilization of this interface upon lowering the pH, since the transition is triggered at a pH of about 6.5, near the pK of this amino acid.
Figure 3.
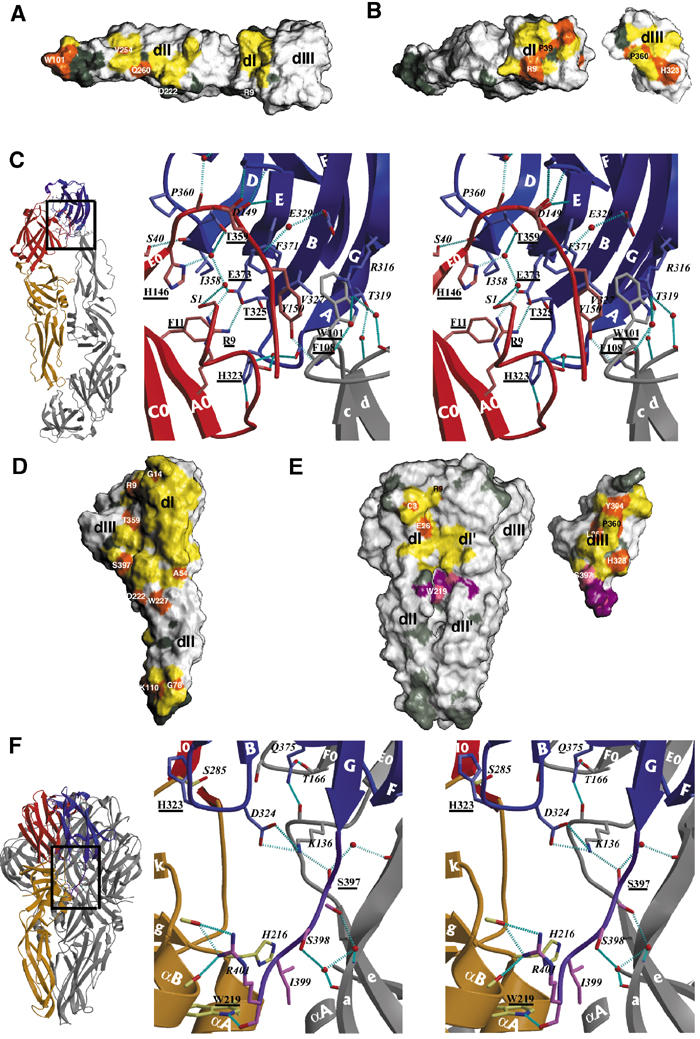
Comparison of the intra-oligomeric and intra-subunit contacts of E before and after the low-pH-induced conformational change. The interactions within the dimeric form are indicated in panels A–C and those in the trimeric form in D–F. In the surface representations of panels A, B, D, and E, the contact regions are indicated in yellow, with the patches formed by conserved residues in orange. Conserved residues outside the contact area are shown in dark gray. The domains are labeled as in Figure 2. (A) sE dimer contacts. The foreground subunit of the dimer, in the side orientation shown in Figure 2A, bottom panel, was removed to show its imprint on the subunit in the background. Except for the fusion peptide loop (labeled with W101), the dimer contact area is essentially made by nonconserved residues. (B) dI/dIII contact surface within the sE dimer. The subunit in (A) was rotated by about 45° about a vertical axis passing through the dI/dIII interface. dIII was then cut out and rotated by 180° about the same axis to show the interaction surface. Note the strong orange patch of conserved residues on both interacting surfaces. Some residues are labeled to allow comparison with the area involved in contacts in the trimeric form. (C) Stereo diagram showing the interactions between dI and dIII across the interface shown in (B). On the left, a square superposed on the ribbon diagram of the sE dimer indicates the region corresponding to the enlargement shown on the right. The polypeptide backbone is shown as ribbons colored according to domains, and gray for the neighboring subunit. The side chains are shown as ball and stick, colored according to atom type (nitrogen: blue; oxygen: red; carbon: light red for dI, light blue for dIII, gray for the adjacent subunit). Water molecules trapped at the interface are shown as red spheres. Hydrogen bonds are indicated as broken cyan lines. The amino acids involved in the contact are labeled, with conserved residues underlined. The secondary structure elements are labeled in white, following Figure 1. (D) sE trimer contacts. The orientation corresponds to that of the subunit in the foreground in Figure 2B, lower panel, which has been rotated 180° about the vertical axis, to show the regions involved in contacts (thus, dIII is on the left). Note the discontinuity of contacts in the central part of dII. Most of the conserved residues have been labeled. Note their distribution at the periphery of the contact area. (E) Contacts made by dIII and the dIII/stem linker in the trimer. The orientation of the trimer is the same as that shown in Figure 2B, lower panel. dIII and the dIII/stem linker were cut from the trimer and rotated by 180° about a vertical axis, shown on the right. Note that the face of dIII involved in the contacts is the same one that faces dI in the dimer (compare the location of H323 and P360, labeled in both panels B and D). The surface of contact of the dIII/stem linker is shown in purple (as well as the residues in the body of the trimer that contact it) with conserved residues colored pink. dI′ and dII′ label the contacted domains from the adjacent subunit (in gray in Figure 2). (F) The ribbons diagram of the left indicates the region of the enlargement shown on the stereo diagram of the right, which depicts the interactions at the interface displayed in (E), the lower half. The color coding is as in (C), with the main chain of the dIII/stem linker in purple, and its side chains as ball and sticks with carbon atoms colored pink. Note the same conserved histidine residue seen in (C) (H323) as being part of the new set of interactions with dI. Note the proximity of helices αA and αB of dII to the C-terminus (R401) of sE, which in the intact molecule would bring them into contact with helix H1.
Trimer contacts
The total buried surface of each subunit in the trimer is roughly 4000 Å2. Figure 3D shows a discontinuous contact surface, divided into two patches at either end of the subunit. The most important patch at the top corresponds to the bottom sheet of the dI β-barrel and parts of the gfeah β-sheet in dII (both β-sheets are labeled in Figure 2). The smaller patch at the tip of dII involves residues from the fusion peptide loops. The residues of dI and dII that are involved in contacts at the trimer interface are also not exposed in the neutral-pH form, where they either face the viral membrane or are buried in contacts between adjacent dimers at the viral surface (Kuhn et al, 2002; Zhang et al, 2003a). The exposed residues in the trimer are also exposed in the dimer, lying on its outward facing side, which carries the antigenic determinants. The fact that these exposed residues in the dimer do not participate in trimer contacts is likely to allow the virus a certain degree of antigenic drift without affecting the trimerization step needed for membrane fusion.
In the trimer, dIII packs against the interface between two subunits, contacting both dI and the dI/dII junction. The surface buried by dIII in the trimer is 1050 Å2, of which 600 Å2 belongs to contacts within the same subunit and 450 Å2 with the adjacent subunit in the trimer. dIII therefore contributes to the stability of the trimeric form, despite the absence of direct dIII/dIII contacts. The contact area of dIII in the trimer is displayed in Figure 3E, showing that the conserved H323 also plays a central role in this interaction. Again, the dIII residues that participate in trimer contacts belong to the buried face of dIII in the dimer. Figure 3E also shows contacts made by the dIII–stem linker (in purple). The details of these interactions are depicted in Figure 3F, which shows that the last amino acid of the crystallized fragment, R401, is involved in extensive Van der Waals contacts and a bidentate hydrogen bond to the main chain of dII in the same subunit. This arginine caps the C-terminus of helix αB in dII while also hydrogen bonding a main-chain carbonyl two residues downstream the αB helix. Up to residue 398, the polypeptide chain makes a number of hydrogen bonds with the adjacent subunit (in gray) and switches to interact with dII in the same chain (in yellow) at I399 (Figure 3F).
The stem region
As indicated in Figures 3E and F (as well as in Figures 2B and C, lower panels), the dIII/stem linker (purple) follows a path that fills a gap at the interface between dII domains in the trimer. The distance from the observed C-terminus in the trimer and the outer leaflet of the bilayer (as diagrammed in Figure 2B) is about 50 Å. In the final postfusion conformation of the full-length molecule, the stem region (which contains about 50 amino acids, see Figure 1) is expected to span this distance in order to place the TM coiled-coil juxtaposed to the fusion peptide loops. The observed discontinuity in trimer contacts displayed in Figure 3D begins precisely at the location of the sE C-terminus. Biochemical studies have shown that, in contrast to the sE fragment, longer fragments of TBEV E, containing helix H1 of the stem, trimerize upon low-pH treatment even in the absence of lipids (Allison et al, 1999), suggesting a stabilizing effect of H1 in the trimer. As indicated in Figure 1, helix H1 is predicted to begin at residue 400 and displays a clear conservation pattern, with every third residue being strictly conserved.
There is a vertical groove at the trimer interface (under the red open star in Figure 2B, lower panel) in between dII domains directly under the C-terminal R401. Modeling the H1 helix as a continuation of the polypeptide chain within this groove, as shown in Figure 4A, indicates that the best visual fit is obtained when the conserved residues are directed toward dII from the same polypeptide chain. This brings the conserved F403 to lie in a pocket made by the strictly conserved W229 and L223, from the conserved patch in between helix αA and β-strand h of dII (see Figure 1). One turn of the helix further down, T406 is in position to hydrogen bond the strictly conserved Q260, at the beginning of the α-B helix of dII. Thus, the H1 helix placed in the observed groove will contact both the dII αA and αB helices and generate a clustering of nine α helices (counting the six symmetry-related helices). In addition, the body of helix H1 would bridge the two contact patches displayed in Figure 3D. In Figure 4A, the exposed residues of H1 are labeled, and correspond essentially to the nonconserved residues indicated in Figure 1, since the conserved ones are buried. Toward the C-terminal end of H1, however, the groove closes underneath the helix and the predicted interactions with dII are less satisfactory. The strictly conserved R412 projects away from the trimer, and the hydrophobic residues V415 and I416 (which are not conserved but are always hydrophobic) are exposed to solvent. This suggests that some rearrangement may take place at the tips of dII in the intact molecule to better accommodate the H1 helix. No attempt was made therefore to optimize the fit of H1 by energy minimization since it would involve altering the conformation of dII as well.
Figure 4.
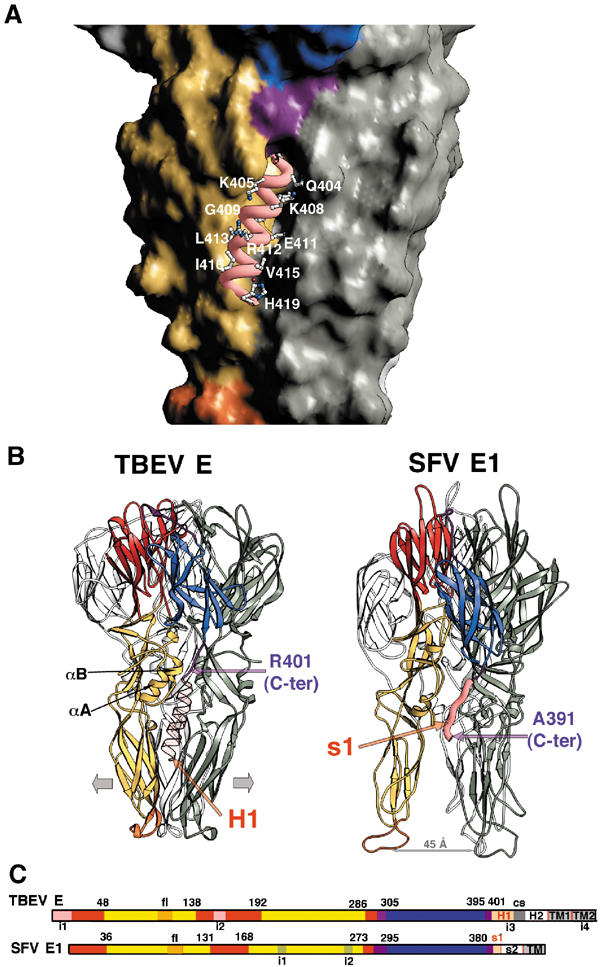
H1 helix of the stem. Comparison of the closed (TBEV) and open (SFV) conformations of the low-pH-induced trimers. As in Figure 2, the subunit in the foreground is colored and those in the background are gray. (A) Close-up view of the side of the trimer in a surface representation, with the dIII/stem linker, colored purple, ending at a groove at the interface between dIIs in the trimer. Helix H1 was modeled within the groove as explained in the text, and its backbone is displayed as a pink thick ribbon. The exposed side chains are displayed as ball and stick, colored according to atom type (carbon: white; nitrogen: blue; oxygen: red). The exposed side chains are labeled. The conserved residues at the N-terminal side of the helix (see Figure 1) are buried and interact with conserved residues from dII, as explained in the text. (B) Side-to-side comparison of the structure of the sE trimer with the modeled H1 helix (left) and the SFV E1 trimer (Gibbons et al, 2004) (right) in a ribbons representation. The molecules were superposed on dI, and are oriented as in Figure 2B, bottom panel. The αA and αB helices of dII are labeled. The modeled H1 helix is indicated in a semitransparent pink ribbon, showing that its path in between dII is similar to that of the s1 segment in SFV E1 (shown as a thick pink ribbon). In the latter, the fusion peptide loops are about 45 Å apart (indicated by the gray arrow). We postulate that the presence of the H1 helix will force the tips of dII to move apart (indicated by the two large gray arrows at either side of the sE trimer). (C) Linear representation of the alignment between the two proteins represented in panel B. Each domain is indicated by color bars according to the color code defined in Figure 1, roughly to scale. The s1 and H1 segments, in pink (labeled in red), are necessary for the molecule to adopt a conformation that can lead to a hemifusion intermediate, as explained in the text. In TBEV, cs indicates a conserved sequence (in gray) in between the H1 and H2 helices. The insertions in each of the molecules (i1–i4) defined in Figure 1 are also indicated.
Comparison to the alphavirus fusion protein E1
The structure of the SFV E1 ectodomain in its trimeric low-pH-induced form (Gibbons et al, 2004) shows a very similar arrangement of domains as that observed in TBEV E (Figure 4B). However, the structures show a striking difference in conformation at the tips of dII, which are kept 45 Å apart, and the fusion peptide loops do not interact with each other within the trimer, as in the TBEV sE structure. The SFV E1 fragment contains all of the ectodomain of the protein except for the last 21 amino acids that precede the viral TM region. The missing residues thus roughly correspond to the H2 helix of flaviviruses (as indicated in the alignment of Figure 1, where this segment has been labeled s2). The s2 segment has been observed in cryo EM reconstructions of Sindbis virus particles as also adopting an α-helical conformation, although this helix does not lie flat on the viral membrane (Zhang et al, 2002). The C-terminal segment of the crystallized fragment of SFV E1 (labeled s1 in Figure 1) has an extended conformation, but its path is very similar to the path followed by the modeled H1 helix in TBEV E, and leads to a similar distance from the fusion peptide loops. The presence of the s1 segment, colored pink in Figure 4, is likely to be responsible for the open conformation observed in the low-pH-induced trimeric form of SFV E1. This open conformation of the trimer allows two-fold related lateral interactions between fusion peptides from adjacent molecules, as described in Gibbons et al (2004), leading to a model for an intermediate state in the fusion reaction that involves a closed ring of trimers in the open conformation that interact with each other and with lipids exclusively via the fusion peptide loops (Gibbons et al, 2004). However, the closed conformation observed in the flavivirus low-pH-induced trimer is incompatible with the type of contacts that led to this model. We propose therefore that in the full-length trimer, the H1 helix will fit in between the dII ‘legs', giving it a ‘tripod' appearance like the SFV E1 trimer, maintaining the fusion peptide loops apart so that they can take part in interactions similar to those observed between SFV E1 trimers. The SFV E1 ectodomain, when inserted into membranes via its fusion loop, forms more ordered arrays than does the TBEV sE fragment, as indicated by electron micrographs of liposomes containing these proteins at their surface. Liposomes coated with the SFV E1 fragment featured projections that were about 70 Å long as measured from the lipid bilayer, with an in register density within the membrane (Gibbons et al, 2003). The TBEV sE-containing liposomes displayed, in contrast, no density within the membrane and the projections were measured to be longer, about 90 Å (Stiasny et al, 2004). These data suggest that the ‘open' and ‘closed' conformations of the fusion proteins may interact differently with the lipid bilayer.
Evolutionary considerations: comparison of the conformational rearrangement of TBEV E and SFV E1
The flavivirus E protein in its neutral-pH form carries two additional functions compared to the alphavirus E1 protein, that is, receptor binding and burying the fusion peptide loop within homodimeric interactions. Two relatively small insertions (i1 and i2, see Figure 1) in the dI region of flaviviruses appear to be related to these additional functions. These i1 and i2 insertions (highlighted in white in Figure 5) occur in regions of dI that contact dIII and allow the use of the latter as a receptor binding module. As illustrated in Figure 5, the interactions provided by i1 and i2 maintain dIII in a vertical orientation so that the FG loop, which has been implicated in receptor interactions (Rey et al, 1995), is exposed at the surface of the virion (see Figure 5C). In addition, the modified dI/dIII interaction also serves the purpose of creating a pocket for burying the fusion peptide loop of the adjacent subunit at the dimer interface (see Figure 5A). In alphaviruses, both these functions, that is, binding the cellular receptor and masking the E1 fusion loop, are carried by glycoprotein E2 (Kielian et al, 2000; Lescar et al, 2001; Zhang et al, 2002).
Figure 5.
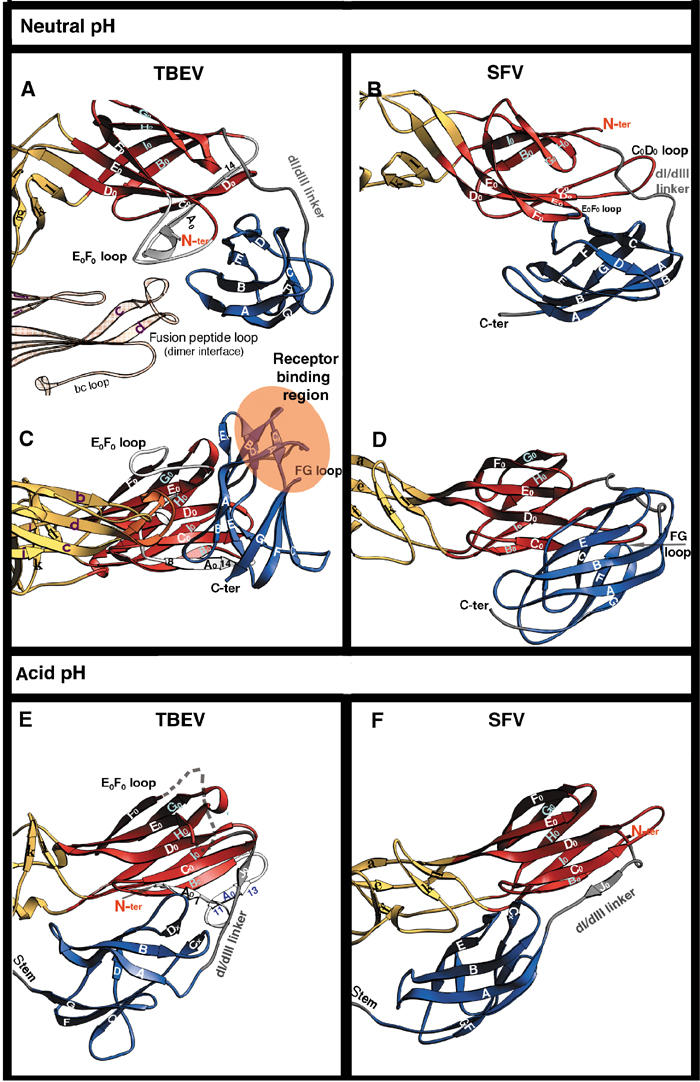
Comparison of the fusogenic structural transition of TBEV E and SFV E1. Ribbon diagram showing the changes that take place in the dI/dIII area of the molecule. The two molecules were superposed on dI (see Table II for rms deviation values). TBEV E is displayed on the left and SFV E1 on the right. The top panels (A, B) show a view from the top, looking down the flavivirus particle. All the other panels (C, D, E, F) show a side view, with dI always in the same orientation. The i1 and i2 insertions in TBEV E dI (from Figure 1) are colored white within the red dI ribbon. Residues from the dI/dIII linker are shown in gray. The β-strands are labeled; note the dI bottom sheet indicated in Figure 2 formed by strands G0H0I0B0 and the dI top sheet by F0E0D0C0A0. The residues at the beginning and end of the A0 strand are labeled to facilitate the description in the text. Strand A0′ is labeled in blue (panel E).
These insertions correspond to 18 amino acids at the N-terminus, including β-strand A0 (i1) and 12 residues within the E0F0 loop (i2). Figure 5 shows that while the relative orientation of dIII with respect to dI is very different in the neutral-pH forms (compare panels C and D), it is very similar in the trimeric forms (compare panels E and F). This is also true for the conformation of the dI/dIII linker in the two proteins. In the conformational change of TBEV E, the A0 strand has to ratchet forward in sequence, while its C-terminal end swaps to the opposite β-sheet of the dI β-barrel, to allow room for the extended dI/dIII linker. Thus, in the sE dimer the A0 strand is formed by amino acids 8–14, while in the sE trimer it is formed by residues 2–5. After the rearrangement, residues 8–14 end up at the other side of the β-barrel, in the new strand A0′ (see Figures 5C and D). The dI/dIII linker makes a short β-strand along its path, J0, which runs antiparallel to strand C0 at its C-terminal end, and would collide with strand A0 if it had not moved out of the way (compare panels C and E in Figure 5).
An additional important change in dI is that the E0F0 loop (i2), which contacts both dIII and the fusion peptide loop of the adjacent subunit across the dimer interface, is disordered in the trimer. The absence of these segments in SFV E1 makes the conformational rearrangement much simpler. The region of dIII that is believed to be responsible for receptor binding in flaviviruses (including the dIII FG loop) is involved instead in contacts with dI (see Figures 5B and D). The change in dIII is a simple translation toward the E1 dI/dII interface, without the important rotation observed in sE. The main difference in the low-pH form of the two molecules, besides the open (SFV) and closed (TBEV) conformation of the tips of dII, is that E1-dIII moves further down toward dII than does sE-dIII (compare panels E and F in Figure 5).
The fact that SFV E1 displays a simpler conformational rearrangement suggests that the common precursor to alpha- and flaviviruses may have had an E1-like structure. The insertions in dI described above would then have been acquired during evolution to endow protein E with additional functions, while still retaining its membrane fusion activity. This is in keeping with the very similar architecture of the flavivirus immature particles and alphavirus virions. The spikes formed by E/prM have a similar arrangement as those of E1/E2, with the fusion peptide loop masked by the companion protein in both cases (Zhang et al, 2003b). The observed evolutionary divergence is likely to relate to the different budding sites of the two viruses. Because the budding competent form is the heterodimer, budding of alphaviruses at the plasma membrane implies that the maturation process, which cleaves pE2 into E2 and E3, must occur without E1/E2 heterodimer dissociation. Flaviviruses, in contrast, are budded at the ER membrane (or at the membrane of nearby internal compartments), before particle maturation in the trans-Golgi network. The proteolytic cleavage of prM therefore causes heterodimer dissociation without interfering with the budding process and, furthermore, leads to a loss of most of the second glycoprotein from the viral surface. In this context, the loss of the functions carried by the companion protein seems to have been rescued during evolution by the i1 and i2 insertions that appeared in E.
Implications for membrane fusion
Figure 6A shows a diagram of the full-length flavivirus glycoprotein E in its neutral-pH form (left panel), and in its proposed final, three-fold symmetric postfusion form, anchored to the fused membrane (right panel). As mentioned above, to reach this final state the protein has to bring its viral membrane-anchored TM segment close to the fusion peptide loops, which are inserted into the target membrane. The supramolecular assembly of five trimers, interacting via the fusion peptide loops and forming a volcano-shaped ring, identified for SFV E1 (Gibbons et al, 2004), was interpreted as corresponding to an intermediate state of the membrane fusion reaction. The ring is such that the 15 participating fusion loops form a crater for interaction with the lipid heads, which induces a nipple-like deformation of the target membrane to cause hemifusion (i.e., fusion of the outer leaflets of the target and viral membranes). This state precedes the opening of the actual fusion pore, which is likely to require the overcoming of an additional energy barrier. This resulting model for the membrane fusion process explains many of the biochemical and biophysical properties of the membrane fusion reaction driven by class I fusion proteins (Chernomordik et al, 1998; Markosyan et al, 2001; Markosyan et al, 2003), suggesting that it is likely to be valid for viral fusion proteins of both structural classes, including flaviviruses. The postulated existence of the same type of intermediate for the latter (Figure 6B) leads us to predict that the presence of the stem in the E protein should affect the trimer conformation such that the fusion peptide loops are spaced appropriately for the relevant interactions to take place. The H2 segment (or s2 in SFV E1) is likely to play the role of an adaptor to allow for the symmetry break proposed for this intermediate, which has the TM segments still anchored in the viral membrane, and therefore does not allow the entire molecule to obey the three-fold molecular symmetry.
Figure 6.
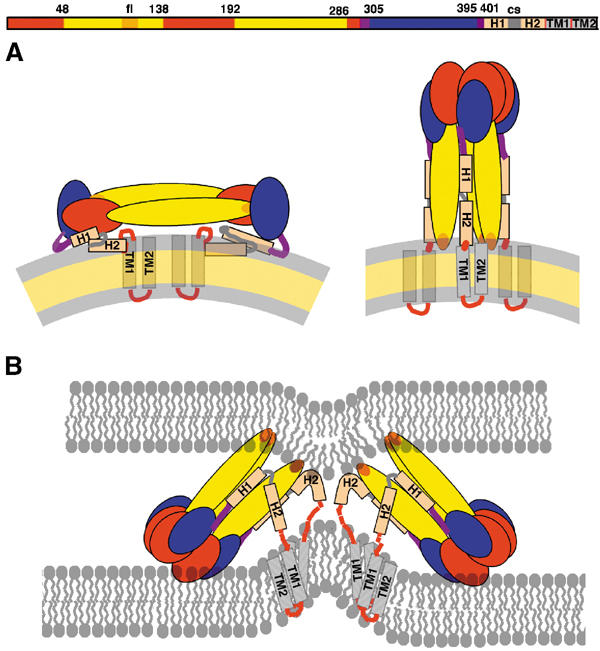
Diagram of the full-length E protein in its different conformations. Organization of the flavivirus E protein in three conformations: on the mature viral particle, in its postfusion form, and in the asymmetric, low-pH-induced intermediate conformation, responsible for the hemifusion step. The linear diagram at the top summarizes the arrangement of domains polypeptide segments, defining the color code used in (A, B). (A) Cartoon of TBEV E as lying on the viral membrane at neutral pH, as observed in the dengue virus particles (Zhang et al, 2003a) (left panel), and in its final, postfusion conformation change (right panel). (B) Proposed structural intermediate responsible for causing fusion of the outer leaflets of the target and viral membranes (hemifusion step). Helix H1 maintains the tips of dII in an open conformation, allowing two-fold related lateral interaction between adjacent trimers via the fusion peptide loops. This arrangement leads to the formation of a ring of five trimers, each interacting identically with its neighbors, which destabilizes the target membrane by creating a lipid nipple. We propose that the H2 segment of the polypeptide chain is used to accommodate the temporary symmetry violation during this intermediate, acting as a tether to the TM segments. Zipping up of the H2 (or s2 segment in SFV) will force juxtaposition of the fusion peptide loops and the TM segments, forcing the opening of an initial fusion pore, as proposed for SFV E1 (Gibbons et al, 2004). For clarity, only two trimers are drawn (out of five proposed to form a closed ring).
Most of the kinetic studies of membrane fusion driven by viral proteins indicate that the initial steps leading to the hemifusion state are fast, and that there is still an important energy barrier to be overcome, from the hemifusion state, to force the opening of the initial fusion pore (Markosyan et al, 2001). It is possible that the remaining, ‘bystander', fusion proteins that coat the viral particle, but are away from the fusion site, may play a role in this process by providing additional viral membrane-destabilizing energy through lateral interactions at the virus surface, as postulated in the ‘fusion protein coat' hypothesis (Kozlov and Chernomordik, 2002). The data presented in this and in the paper describing the dengue virus sE protein in its low-pH-induced conformation (Modis et al, 2004) suggest that the interactions between the stem region and dII, required to complete the fusion process, would be potential targets for antiviral fusion inhibitors analogous to those developed for the class I viral fusion protein of HIV (Eckert and Kim, 2001). It is likely that the best flavivirus inhibitors of fusion would be molecules that mimic H2 (or s2 segment in alphaviruses), since upstream segments would have to inhibit formation of the intermediate, which may not provide a long enough time window for the inhibitor to act efficiently.
Materials and methods
Production, crystallization, and structure determination of the sE trimer
The production (Stiasny et al, 2002), characterization, and crystallization (Stiasny et al, 2004) of the sE trimer have been described separately. Briefly, dimeric sE was cleaved from virions by limited trypsin digestion and purified. Membrane-bound trimers were produced by exposure of sE to mildly acidic pH in the presence of liposomes. The resulting trimers were solubilized with detergent and purified. Crystals were obtained from purified preparations of sE trimers in 0.1 M ammonium acetate (pH 5) and 15 mM N, N-dimethyldecylamine-N-oxide (DDAO). Several crystal forms were characterized (Table I). The crystals were transferred to cryoprotecting solutions containing 25% glycerol, and diffraction data were collected at cryogenic temperatures on synchrotron sources. Diffraction data were processed with the HKL package (Otwinowski and Minor, 1997) or XDS (Kabsch, 2001). The structure was determined by molecular replacement using the available coordinates of the neutral pH, dimeric form of TBEV sE (Rey et al, 1995) and data from crystal forms O2 and T, using a combination of the ‘dyad search' procedure implemented in Molrep (Vagin and Teplyakov, 2000) and the AMoRe package (Navaza, 1994). Refinement and rebuilding were carried out using the high-resolution data from crystal form O1 (Table I). Initial electron density maps were computed with BUSTER (Bricogne, 1993; Roversi et al, 2000). Rebuilding was carried out with TURBO-FRODO (Roussel and Cambillaud, 1991) and refinement with CNS (Brünger et al, 1998). Temperature factors were refined as groups (two groups per residue) during rebuilding. Tight noncrystallographic symmetry restraints were kept throughout for dI and dIII (with the exception of a few surface loops). The final model contains all amino acids except for the N-terminal residue and loops 148–159 (E0F0) and 200–204 (fg).
Coordinates. The atomic coordinates of the TBEV sE trimer have been deposited in the PDB, accession number 1URZ.
Illustrations. Figures 2, 3A, B, D, E, 4A, B and 5 were made with program Ribbons (Carson, 1987). Figures 3C and F were prepared with Molscript (Kraulis, 1991).
Acknowledgments
We thank K Diederichs for writing a specific program to rescue our best diffraction data set, which suffered from a desynchronization between spindle and shutter during data collection; MC Vaney, G Squires, F Coulibaly, DL Gibbons, J Lepault, and J Navaza for help and discussion; A Vigouroux, W Holzer, and S Röhnke for technical assistance; C Schulze-Briese and T Tomikazi for help during diffraction data collection; and M Kielian for comments on the manuscript. Diffraction data were collected at the synchrotron beam line X06SA of the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland. Beam line ID14 of the European Synchrotron Radiation Facility, Grenoble, France, was used for the initial characterization of the crystals. This work was supported in part by the Human Frontiers Science Program Grant RC0509/96 to FAR and FXH. FAR acknowledges support from the CNRS and INRA, the SESAME Program of the ‘Région Ile-de-France', the French ‘Fondation pour la Recherche Médicale', the ‘Association pour la Recherche contre le Cancer', CNRS program ‘Physique et Chimie du Vivant', and the EU grant ENHCV: QLK2-CT-2001-0112. SB has a long-term EMBO fellowship.
References
- Allison SL, Schalish J, Stiasny K, Mandl CW, Kunz C, Heinz FX (1995) Oligomeric rearrangement of tick-borne encephatitis virus envelope proteins induced by an acidic pH. J Virol 69: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX (1999) Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol 73: 5605–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G (1993) Buster. Acta Crystallogr D 49: 37–60 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Carson M (1987) Ribbon models of macromolecules. J Mol Graphics 5: 103–106 [Google Scholar]
- Chernomordik L, Frolov VA, Leikina E, Bronk P, Zimmerberg J (1998) The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol 140: 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman PM, Lawrence MC (2003) The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol 4: 309–319 [DOI] [PubMed] [Google Scholar]
- Eckert DM, Kim PS (2001) Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci USA 98: 11187–11192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Erk I, Reilly B, Navaza J, Kielian M, Rey FA, Lepault J (2003) Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114: 573–584 [DOI] [PubMed] [Google Scholar]
- Gibbons DL, Vaney M-C, Roussel A, Vigouroux A, Reilly B, Kielian M, Rey FA (2004) Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature 327: 320–325 [DOI] [PubMed] [Google Scholar]
- Heinz FX, Allison SL (2000) Structures and mechanisms in flavivirus fusion. Adv Vir Res 55: 231–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Lang T, Sudhof TC (2003) Membrane fusion. Cell 112: 519–533 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2001) XDS. In International Tables for Crystallography, Arnold E (ed) Dordrecht: Kluwer Academic Press [Google Scholar]
- Kielian M, Chatterjee PK, Gibbons DL, Lu YE (2000) Specific roles for lipids in virus fusion and exit: examples from the alphaviruses. Subcell Biochem 34: 409–455 [DOI] [PubMed] [Google Scholar]
- Klimjack MR, Jeffrey S, Kielian M (1994) Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J Virol 68: 6940–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, Chernomordik LV (2002) The protein coat in membrane fusion: lessons from fission. Traffic 3: 256–267 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Kuhn RJ, Zhang W, Rossman MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG et al. (2002) Structure of Dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA (2001) The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105: 137–148 [DOI] [PubMed] [Google Scholar]
- Markosyan RM, Cohen FS, Melikyan GB (2003) HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol Biol Cell 14: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan RM, Melikyan GB, Cohen FS (2001) Evolution of intermediates of influenza virus hemagglutinin-mediated fusion revealed by kinetic measurements of pore formation. Biophys J 80: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA 100: 6899–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 327: 313–319 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Macromolecular Crystallography–Part A, Sweet RM (ed) pp 307–326. London: Academic Press [DOI] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375: 291–298 [DOI] [PubMed] [Google Scholar]
- Roussel A, Cambillaud C (1991) Turbo Frodo. In Silicon Graphics GeometryPatners Directory. Mountain View, CA: Silicon Graphics [Google Scholar]
- Roversi P, Blanc E, Vonrhein C, Evans G, Bricogne G (2000) Modelling prior distributions of atoms for macromolecular refinement and completion. Acta Crystallogr D 56: 1316–1323 [DOI] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX (1996) Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol 70: 8142–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Allison SL, Schalich J, Heinz FX (2002) Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J Virol 76: 3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny K, Bressanelli S, Lepault J, Rey FA, Heinz FX (2004) Characterization and crystallization of a trimeric low-pH form of the class II viral fusion protein E from tick-borne encephalitis virus. J Virol, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (2000) An approach to multi-copy search in molecular replacement. Acta Crystallogr D 56: 1622–1624 [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Garoff H (1992) Membrane fusion process of Semliki Forest virus I: low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol 116: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Calder LJ, Harrison SC, Skehel JJ, Wiley DC (1999) Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol 16: 3–9 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC (1981) Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289: 366–378 [DOI] [PubMed] [Google Scholar]
- Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn R (2003a) Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol 10: 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG (2003b) Structures of immature flavivirus particles. EMBO J 22: 2604–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG (2002) Placement of the structural proteins in sindbis virus. J Virol 76: 11645–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]


