Abstract
Engineered tissues require enhanced organization of cells and extracellular matrix (ECM) for proper function. To promote cell organization, substrates with controlled micro- and nanopatterns have been developed as supports for cell growth, and to induce cellular elongation and orientation via contact guidance. Micropatterned ultra-thin biodegradable substrates are desirable for implantation in the host tissue. These substrates, however, need to be mechanically robust to provide substantial support for the generation of new tissues, to be easily retrievable, and to maintain proper handling characteristics. Here, we introduce ultra–thin (<10 μm), self-assembled chitin nanofiber substrates micropatterned with replica molding for engineering cell sheets. These substrates are biodegradable, mechanically strong, yet flexible, and easily manipulated into the desired shape. As a proof-of-concept, fibroblast cell proliferation, elongation, and alignment were studied on the developed substrates with different pattern dimensions. On the optimized substrates, the majority of the cells aligned (<10°) along the major axis of micropatterned features. With the ease of fabrication and mechanical robustness, the substrates presented herein can be utilized as versatile system for the engineering and delivery of ordered tissue in applications such as myocardial repair.
Introduction
Engineered tissues require enhanced organization of cells and extracellular matrix (ECM) for proper function.1-3 The cells reorganize according to the interaction with the ECM based on topography and mechanical properties such as matrix stiffness, elasticity, and viscosity.4 Alignment of ECM molecules and concentration gradients of immobilized growth factors also play a crucial role in cellular organization.3 To promote cell organization, substrates with controlled micro- and nanopatterns have been developed as supports for cell growth, and to induce cellular elongation and orientation via contact guidance in engineered tissues.5-11 These substrates produce highly ordered and functional cell-sheets that may be detached from the substrate for delivery to the host tissue via enzymatic degradation or thermal stimulus.12-17 Delivery of detached engineered cell-sheets often occurs with a support platform because free-standing cell sheets are mechanically weak and difficult to handle.18 An alternate approach is to create cell-sheets on ultra-thin biodegradable substrates that are implanted in the host tissue. These substrates need to be thin and flexible for conformal contact to the tissue of choice and robust for ease of handling.19 Robustness of the substrate is particularly important in myocardial tissue repairs.16, 17 In these repairs, the substrate is also required to restore the mechanical properties of the damaged tissue while the new tissue is growing. To fabricate robust biodegradable substrates, structural biopolymers such as collagen, chitin, and chitosan are particularly appealing for their biocompatibility and mechanical strength.20-22 Specifically, chitin and its deacetylated derivative, chitosan, possess multiple advantages as tissue engineering substrates including nontoxicity, cytocompatibility, and tunable biodegradability.20, 23-28 In addition, the 3-D assembly of nanofibrous structures with chitin and chitosan is known to mimic the natural ECM and promote cell attachment and spreading ability. 29 While chitin nanofibers already exist in nature, chitosan nanofibers are typically produced by electrospinning.30-32 Electrospinning is difficult to couple with micro- and nanofabrication to yield micropatterned substrates. However, chitosan nanofibers have found broader use than chitin due to chitin's intractability and insolubility in common organic solvents. We have previously demonstrated the self-assembly of ultrathin (3 nm) α-chitin nanofibers from solution33 and the micro- and nanofabrication of self-assembled chitin nanofiber structures with soft lithography strategies.34 Here, we utilize this facile and versatile approach to produce transparent, ultra–thin(<10 μm), mechanically robust, yet flexible self-assembled chitin nanofiber micropatterned substrates for tissue engineering (Fig. 1). With these micropatterned substrates, we demonstrated cell sheets of aligned fibroblasts as a proof of concept. The supported cell sheets made in this manner were mechanically robust and easy to handle, yet flexible and bendable in the shape of choice.
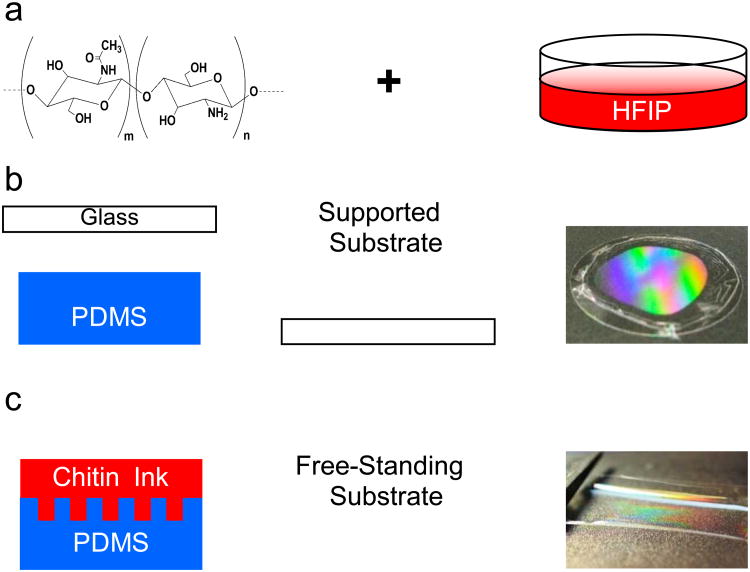
Fig. 1.
Chitin nanofiber-based micropatterned substrate fabrication process. (a) chitin/HFIP solution (0.1 % w/w) was poured on the top of the (b) mold covered with a glass slide to create a supported substrate after drying overnight and its optical image with diffraction pattern on it (c) Thicker films were obtained from more concentrated solutions (0.2 % w/w) to create free-standing substrates, which were robust and easy to handle. The optical image demonstrated the diffraction pattern on the free-standing chitin film.
Experimental
Fabrication of Chitin Substrates
Substrates with micropatterns were fabricated using replica molding from a “chitin nanofiber ink” as previously described.34 The chitin nanofiber ink was prepared by stirring appropriate amounts of β-chitin squid extract (Industrial Research Ltd-New Zealand) in hexafluoro-2-propanol (Oakwood Products, Inc.) for one week in ambient conditions. The starting material had a degree of acetylation defined as n/ (m+n) (Figure 1a) of 84%. Two optical gratings (THORLABS) were used as masters: GE2550-3263 Echelle Grating with3.16 μm spacing and 63° blaze angle (G1) and a GE 2550-0875 Echelle Grating, 12.66 μm, 75° blaze angle (G2). Replica molds were made with PDMS from SYLGRAD 184 (10% curing agent) and cured at 50°C for 8 hours. The chitin nanofiber ink was drop cast onto the mold (50 μl) to create the micropatterned features. Alternatively, control substrates (no micropatterns) were prepared using a flat PDMS mold. Micropatterned and control substrate were supported on plasma-cleaned glass cover slips (Ted Pella, Inc.). For supported micropatterned and control substrate, a chitin solution of 0.1 % w/w was used. Free-standing chitin substrates were fabricated with a 0.2 % w/w chitin solution. Solutions of higher concentrations resulted in thicker films that were easier to handle. Both types of films were dried overnight in ambient and peeled off from the PDMS mold for further experiments.
Characterization of Chitin Substrates
A Veeco Multimode V (Nanoscope IV controller) and Veecoprobes Sb-doped Si cantilevers (ρ = 0.01-0.025 Ω.cm, k = 40 N/m, υ ∼ 300 kHz) were used for atomic force microscopy (AFM). Fourier Transform Infrared (FTIR) spectra were recorded on free-standing substrates with a Bruker vector 33 FTIR spectrophotometer (4000 to 400 cm−1, 4 cm−1 resolution). The degree of deacetylation (chitin vs. chitosan) was evaluated using the ratio of A1560/A1030 according to a previously reported procedure.28, 35 Stress vs. strain data of the free-standing substrates were recorded with a Shimadzu AGS-X. The substrates were tested dry before and after deacetylation as well as after immersion in Dulbecco's Phosphate Buffered Saline (DPBS) (GIBCO) for 1 and 5 days. Wet substrates were immersed in PBS for 1 day and tested immediately after removing from the solution.
Chemical modification of chitin substrate
Chitin substrates were partially deacetylated into and coated with fibronectin (FN) to improve cell attachment and proliferation. Deacetylation was performed with 5mM NaOH at 40°C for 1 hour. The substrates were then washed with distilled water three times and air-dried. FN (from bovine plasma, 0.1% sterile solution, Sigma-Aldrich) was coated on the substrate by incubating the chitin substrates in 200 μn of FN solution (50 μl ml−1 in DPBS) for two hours (T=37 °C). After incubation, the substrates were rinsed thoroughly in DPBS to remove the non-adsorbed and excessive aggregates of FN. The substrates were then dried in a desiccator overnight for further biological experiments.
Cell culture preparation
NIH/3T3 fibroblast cells were used in this work. The cells were maintained in T-75 flasks at 37 °C and 5 % CO2 and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS and 1% penicillin-streptomycin. Prior to seeding cells on chitin substrates, the samples were rinsed in NIH/3T3 cell culture medium twice. 80% confluent 3T3 cells were trypsinized, washed and suspended in fresh culture medium and subsequently the cell suspension was diluted with cell growth medium to reach the desired cell concentration. A cell suspension containing 1000 cells/500supof medium was then added to each sample for further testing.
Cell proliferation and immunofluorescence for cytoskeletal organization
Cell attachment and proliferation on the micropatterned and control (without pattern) chitin substrates were evaluated with direct cell counting at days 0 and 5 of culture. Immunostaining assesses the actin cytoskeletal organization (F-actin) of the cells attached to different samples. The samples were washed for three times in DPBS and are fixed in 4% paraformaldehyde (PF) solution in DPBS for 20 min at room temperature. The substrates were then submerged in 0.1% Triton X-100 solution in DPBS for 30 minutes in order to permeabilize the cells' membrane and blocked in 1% Bovine Serum albumin (BSA) for 1 h. The actin cytoskeleton was stained using 1:40 dilution of Alexa Fluor-594 phalloidin (Invitrogen) in 1% BSA. Following immunostaining, the cell nuclei were stained with 1:1000 dilution of 4′,6-diamidino-2-phenyl indole dihydrochloride (DAPI) stain (Invitrogen) in DPBS for 5 min. Upon staining, the samples were imaged using an inverted fluorescence microscope (Nikon TE 2000-U, Nikon instruments Inc., USA) and the fluorescence images were analyzed using NIH image J software (version 1.4).
Quantification of cellular alignment and shape index
To quantify cellular alignment within micropatterned and control chitin substrates, fluorescence images were obtained at 5-6 different locations of each sample after 5 days of culture. The shape of each individual nucleus was first fitted with an ellipse and the normalized nuclei alignment were defined according to previously published procedure36. The normalized cellular alignment angles were finally grouped in 10 degree increments to compare the alignment of the nuclei on patterned and control substrates. Cell elongation within each sample was evaluated using fluorescence images of the cells cytoskeleton stained for F-actin filaments (5-6 regions). Cell elongation was determined as the ratio of cell length to cell width. The cell length was defined as its longest cord and the cell width was defined as the longest chord perpendicular to its length. The angle between the cells longest cord and the direction of the micropattern grating was defined as the cell orientation angle.
Results and Discussion
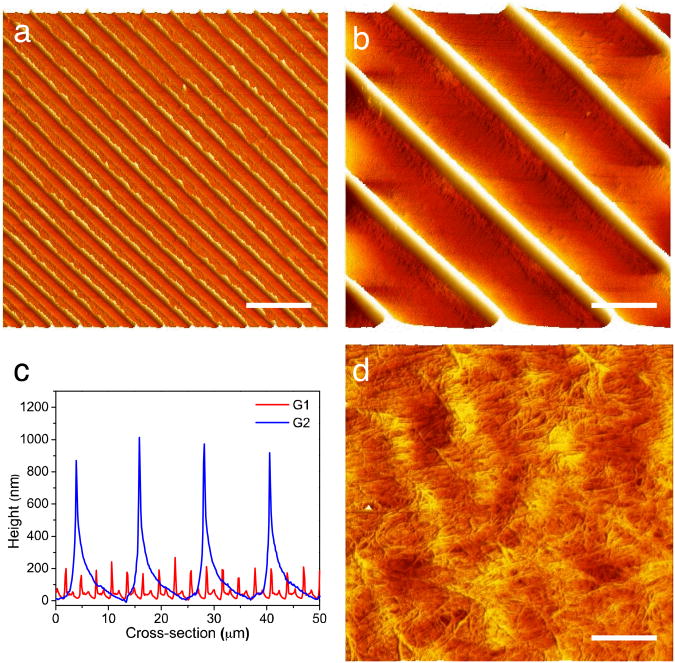
We prepared the chitin micropatterned substrates with replica molding of a “chitin nanofiber ink” (Fig. 1). The chitin nanofiber ink is a solution of squid pen β-chitin that self-assembles into 3 nm diameter α-chitin nanofibers upon drying.33, 34 To create the desired substrates, an appropriate amount of chitin nanofiber ink was drop cast on the top of the mold and allowed to dry overnight in ambient conditions. In this work, we used PDMS replica of different gratings as molds to create the substrates. We chose micropatterns with 3.2 μm spacing (G1) and 11.2 μm spacing (G2) to evaluate the effects of groove spacing on cell alignment, G1 being smaller than the average cell diameter, and G2 being slightly larger than the average cell diameter. Supported chitin micropatterned substrates were obtained by placing a glass slide on the top of the solution during drying (Fig. 1b). This simple strategy afforded the attachment of chitin nanofiber substrates to an arbitrary support without requiring additional adhesive, which may be toxic for the cells. Free-standing substrates were typically thicker than the supported substrate and were peeled off directly from the mold (Fig. 1c). The free-standing substrates were robust and could be easily lifted out from the molds with a pair of lab-tweezers. Both supported and free-standing were easy to handle and stable in cell culture media for extended periods of time. The microfabricated substrates prepared in this fashion exhibited the desired microstructure, which is inferred from the diffraction patterns in the optical images (Fig. 1b and 1c) and AFM micrographs (Fig. 2 a and b). Micropatterned substrates G1 and G2 closely replicated the spacing and height of the original gratings (Fig. 2c). Both G1 and G2 substrates had saw tooth cross-section that derived from the cross section of the diffraction grating. The features in G1 were on average 200 nm tall, while the features on the G2 substrates were 900 nm tall. For both substrates, uniform features were found in extended areas up to several cm2 only limited by the size of the original master and mold. With appropriate masters and molds, features of arbitrary shape and size ranging from a few nanometers to hundreds of microns were also available with the “chitin nanofiber ink” expanding the kind of substrates available for future studies. Over fifty substrates were reliably reproduced from the same master without observing any degradation of the replicated master. Each PDMS mold was reused at most ten times before noticing some degradation of the patterns due to mold contamination with the chitin material. As previously reported, chitin nanofiber random morphology was maintained intact in the micropatterns with the replica molding process (Fig. 2d).34 In the future, substrates with chitin nanofibers aligned along the micropattern direction may be fabricated with directional drying.33 This replica molding process was low cost, easy to use, high throughput, and did not require expensive clean room equipment. With this simple solution processing micropatterned supported and free standing substrates were created in one step for further experiments.
Fig. 2.
AFM imaging of supported chitin substrates (a, b) AFM height images of chitin substrates G1 and G2 respectively (Scale bar: 10 μm), (c) Cross-sectional height profiles of (a) and (b). G1 pitch= 3.0 ± 0.09 μ , height= 193.4 ± 30.4 nm and G2 pitch = 12.2 ± 0.2 μm and height 943.3 ± 62.6, (d) Zoom in of (b) showing chitin nanofiber morphology (scale bar = 200nm).
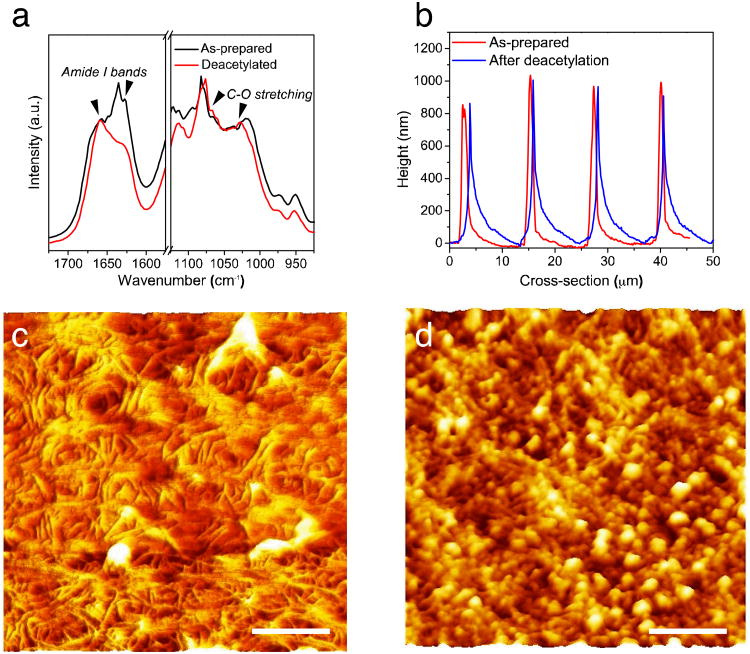
To employ the micropatterned nanofiber substrates for tissue engineering, NIH-3T3 fibroblast cells were seeded on the substrates with different size of groove and their behavior was subsequently investigated. We chose fibroblasts for proof-of-concept because the organization of fibroblasts within the ECM of native myocardial tissue is critical to cell alignment, which influences the electrical and mechanical properties of the heart. 37 Fibroblast cell attachment on the as-prepared chitin substrates was low, as previously observed for neuronal cells, possibly due to the lack of reactive species and positive charges on the chitin surface.28 To improve cell attachment, the chitin substrates were partially deacetylated and coated with a thin layer of FN. FN is an ECM protein and plays a major role in cell adhesion, growth, migration, and differentiation.38 Deacetylation replaced the acetyl group in chitin (Fig. 1a, n block) with an amine (Fig. 1b, m block) resulting in a more hydrophilic and positively charged polymer. When the ratio between acetyl groups and amines is lower than 1:1 (n<m), the polymer is typically referred to as chitosan. This deacetylation process of the chitin nanofibers allowed fine-tuning the chemistry of the substrates. The degree of deacetylation (m/n+m) of the chitin nanofiber substrates was determined with FTIR analysis (Fig. 3a) using the ratios of the C-O (amide II) peak at 1560 cm−1 and the C–O peak at 1030 cm−1.28, 35 The “as prepared” film was ca. 14% deacetylated, while after treatment the degree of deacetylation decreased to ca. 30%. Importantly, 30% deacetylation did not significantly affect the quality of the micropatterns. (Fig. 3b).The deacetylation process to form 30% deacetylated chitin starting from 16% deacetylated chitin was easier than increasing the degree of acetylation of highly deacetylated chitosan as previously reported.20 Highly deacetylated chitosan does not self-assemble into nanofibers from solution because the nanofiber self-assembly process is driven by the intramolecular hydrogen bonding of the acetyl groups in chitin.39 However, reducing the number of acetyl groups from 84% to 70% in the self-assembled chitin nanofibers did not disrupt the nanofiber morphology (Fig. 3 c) indicating that enough acetyl groups were still present to maintain the hydrogen bonding of the nanofiber structures. Nanofibers with high surface area provide abundant adhesion sites and enhance the overall cell-substrate interaction. However, further studies with similarly deacetylated chitin without a nanofiber structure are required to pinpoint the exact effects of the nanofiber morphology on cell proliferation, growth, and alignment. Chitin substrates with 30% degree of deacetylation were typically more stable in culture media than the easier to process highly deacetylated chitosan. Highly deacetylated chitosan is soluble in slightly acidic water and often requires additional chemical modification and crosslinking to retain structural integrity in aqueous environments.40 Furthermore, coating the samples with FN retained the quality of the micropatterns for cell-culture experiments. While deacetylation did not affect nanofiber morphology, FN coating was present on top of the nanofibers (Fig. 3c and d). In particular, FN formed globules that are approximately 20 nm in diameter on the top of the nanofiber substrates. It is conceivable that these FN globules may assemble along the chitin nanofibers as it is suggested by the topography Figure 3d. However, further studies are required to determine the exact FN-chitin interaction and its effects on the FN structure.
Fig. 3.
Effect of post-treatment of chitin substrates for biological testing (deacetylation in NaOH, fibronectin treatment). (a) FT-IR spectra, (b) Cross-sectional height profiles of the chitin substrates obtained from the G2 grating mold, (c, d) Morphology of chitin nanofiber films after deacetylation in NaOH and fibronectin treatment, respectively (All scale bars = 200 nm).
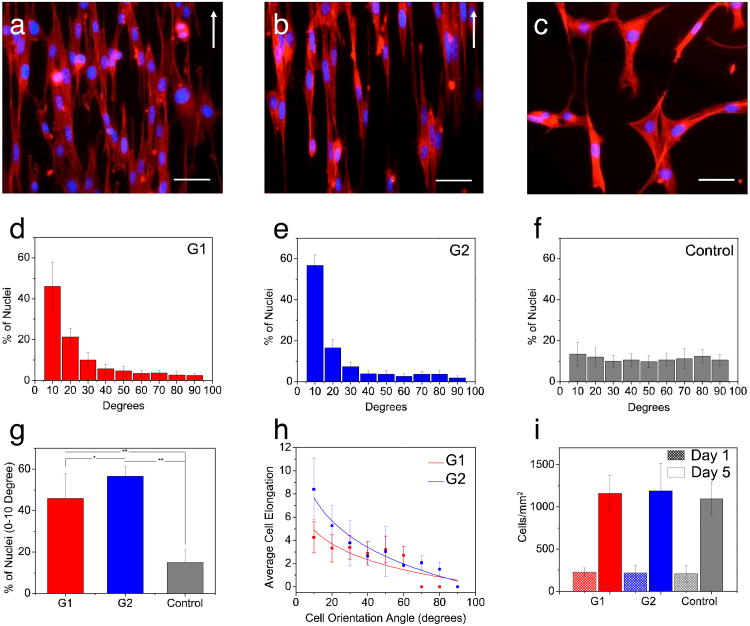
To evaluate the chitin substrates for tissue engineering, we cultured NIH-3T3 cells on the micropatterned and control chitin substrates and measured cell attachment, alignment, and proliferation. On the chitin micropatterned substrates, G1 and G2 (Fig. 4a and b) cells with a spindle-like morphology aligned their cytoskeletal structure along the major axis of the micropatterned features (contact guidance). In contrast, the cells grown on the control chitin substrates did not have any preferred orientation (Fig. 4c). We further quantified cells nuclei alignment on G1 and G2 micropatterns after 5 days of cell culture (Fig. 4d, e, and f). In G1 and G2, a larger proportion of the cells aligned within the 0-10° preferred angle as opposed to control substrate (Fig 4f). The degree of cells nuclei alignment on G1 (∼45%) and G2 (∼55%) samples was significantly higher (p<0.05) than the degree of alignment for the control sample (15%) (Fig. 4g).In addition, on G1 and G2, cell elongation increased with cell alignment. This increase indicated that the substrate topography affected cell morphology along with cellular alignment (Fig. 4h). Cell attachment and proliferation was also evaluated with the direct cell counting method at days 1 and 5 of culture. NIH-3T3 fibroblast cells proliferated at day 5 of culture compared to day 1, indicating that the chitin substrates were non cytotoxic. In addition, there was no significant difference in cell proliferation on different micropatterned samples (G1, G2) compared to the control substrate (Fig. 4i). Our observations are consistent with earlier work in terms of cellular alignment within micropatterns.41 In general, decreasing groove width and increasing groove depth enhances cellular alignment.41 We observed more pronounced alignment on the G2 micropatterns, which are wider, but also deeper than the G1 micropatterns. In G1 micropatterned features, where the cells size is larger than the groove width and the width is decreased beyond a threshold, the cells easily bridge the neighboring ridges. Such behavior ultimately results in overall decreased cellular alignment. On the other hand, on G2 micropatterns, where the groove width was within the range of a single cell size (10 μm), the cells were laterally confined within the grooves in between the taller ridges. This confinement resulted in improved cellular alignment (Supplementary Fig. S1). Having the groove width in the range of a single cell size, the effect of the width on G2 patterns is expected to be more pronounced on cellular alignment compared to the height. However, some effect on cellular alignment from the increased height in the G2 patterns respect to G1 cannot be completely ruled out at this stage.
Fig. 4.
Fluorescence images of the actin cytoskeleton of the cells on (a) G1, (b), G2 substrates and (c) control sample after 5 days of culture (Scale bar 50 μ(S. The white arrow shows the longitudinal direction of the patterns. (d-f) Distribution of cells nuclei alignment angles on the patterned and control samples, (g) Percentage of cells that have orientation angles within 0-10 degree angle, (h) Cellular elongation function of cell orientation angle within the patterned substrates (i) Proliferation of the cells on the patterned and control substrates.
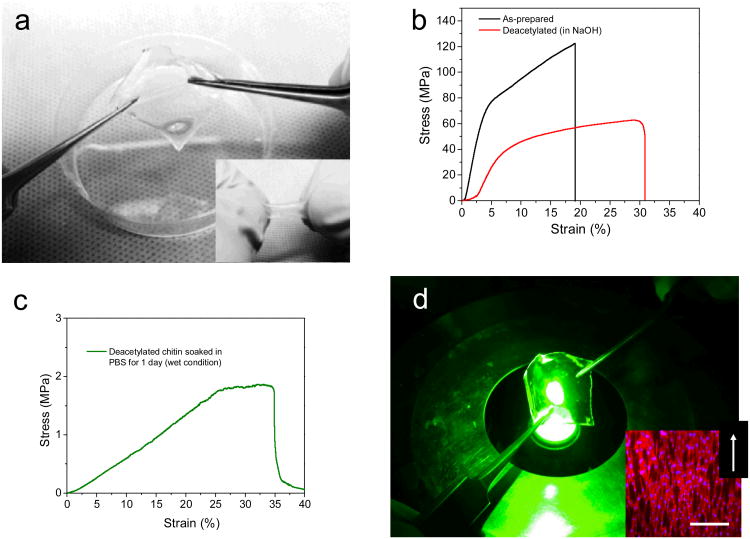
The NIH-3T3-seeded free-standing films were sturdy and easily manipulated as well as flexible (Fig 5a) noting that the substrates absorb water and expand up to five fold their initial volume. Absorption of water is not surprising because the chitin nanofiber substrates themselves are hydrophilic and porous with a density of ∼1 g/cm3, which is lower than the density of chitin (1.44 g/cm3). Water absorption and porosity are desirable properties to afford tunable optimal flow of nutrients to, and waste from the growing tissue.1 To gather further insights on the mechanical properties of the substrates, we performed tensile test of the pristine chitin substrates, as well as the 30% deacetylated chitin nanofiber substrates before and after immersion in cell culture media for 1 and 5 days. The elastic modulus (E) of the pristine dry chitin substrates was 2.5 GPa with a tensile strength exceeding 100 MPa (Fig. 5b). As expected from partial removal of the acetyl groups and corresponding lower degree of hydrogen bonding, the elastic modulus of the chitin substrates after deacetylation to 30% decreased to 926 MPa still retaining a tensile strength of 60 MPa (Fig. 5b). These values for elastic modulus and tensile strength are significantly higher than the values previously reported for chitosan electrospun nanofibers (150 MPa).42 Even after deacetylation to 30%, the self-assembled chitin nanofibers most likely still have a higher degree of crystallinity and stronger intramolecular hydrogen bonding from the remaining acetyl groups.39 Notably, only a small decrease in elastic modulus was observed for the 70% acetylated chitin nanofiber substrates after immersion in PBS for 1 day (E=875 MPa) and 5 days (E=674 MPa) when measured after drying off the excess water from the substrates for 30 minutes in ambient conditions (Fig S2). These measurements confirmed the stability of the 30% deacetylated chitin nanofiber substrates in cell culture media. The tensile strength of the substrates immersed in PBS for 1 and 5 days decreased, however, to ∼ 35 MPa. This lower value for tensile strength may be attributed to a higher water content (∼ 25-30%) of the PBS immersed substrates respect to the 30% deacetylated dry chitin nanofiber substrates (∼ 10%). However, some degradation of the substrate material cannot be completely ruled out. To investigate the mechanical properties of the substrates during cell culture, we measured substrates after immersion in PBS for 1 day immediately after removal from the solution while the substrates were still in the swollen state. The elastic modulus was 5 MPa with a tensile strength of 1 MPa (Fig 5c). Incorporation of water made the substrates more flexible and better suited for tissue engineering applications.19, 43 In these applications it is desirable for the substrate to have similar mechanical properties of the host tissue.44-46 The substrates formed in this way were transparent, which allows the optical characterizations of cell interaction with substrate (Fig 5d).In addition, the cells spread and covered the entire film and aligned along the micropattern major axis as already observed on the glass-supported chitin substrates (Fig. 5d). With these favorable properties, the cell-seeded chitin substrates were still mechanically robust but more flexible than the dry chitin and withstood bending and rolling to produce more complex 3D structures or transfer to the tissue of choice. These transparent, robust, ultra-thin, free-stranding chitin substrates covered with aligned fibroblasts with tunable and superior mechanical properties could make very a promising candidate to form 3D functional tissue and mimic the complex hierarchical of the ECM.
Fig. 5.
Cell seeded free-standing flexible chitin substrates, (a) Stretching and Rolling (insert) flexibility the free-standing micropatterned chitin substrates (G2) seeded with 3T3 fibroblast (b) Mechanical properties of micropatterned free standing chitin substrates before and after deacetylation to 30%, (c) mechanical properties of the 30% deacetylated chitin nanofiber substrates after immersion in PBS for 1 day (measured wet) (d) Chitin nanofiber substrates are transparent and afford optical inspection, insert Fluorescence images of actin cytoskeleton of the cells on G2 showing the entire covering and alignment of cells within the direction of the patterned features. White arrow on the right corner of the images indicates the direction of the patterns. Scale bar 100 μm.
Conclusions
In this work, we have developed supported and free-standing micropatterned substrates made of self-assembled chitin nanofibers. These ultrathin micropatterned substrates are biodegradable, mechanically robust, yet flexible and easily manipulated. The substrates were seeded with NIH-3T3 cells that aligned along the major micropattern axis creating ultra-thin (<10 μm) and free-standing ordered cell sheets. These sheets are sturdy in cell culture media yet flexible and easily manipulated to create more complex tissue structures for regenerative medicine and tissue engineesring applications. These applications include myocardial repair were the damaged tissue could be mechanically supported by the chitin substrate while the new tissue is growing. Additionally, these chitin substrates are optically transparent and may find use in retinal regeneration.
Supplementary Material
Supplementary Fig. S1: Phase contrast Images of the cells attached on (a) G1 and (b) G2 micropatterned substrates after 5 days of culture. Scale bars represent 50(a)
Supplementary Fig. S2: Tensile testing on 30% deacetylated chitin substrates dry and after immersion in PBS for 1 day and for 5 days.
Acknowledgments
A.K. acknowledges funding from the Presidential Early Career Award for Scientists and Engineers (PECASE), the Office of Naval Research Young National Investigator Award, the National Science Foundation CAREER Award (DMR 0847287), and the National Institutes of Health (HL092836, AR057837, DE021468, DE019024, EB012597, HL099073, EB008392). M.R. acknowledges funding from the Washington Research Foundation, the University of Washington Center for Commercialization, the Coulter Foundation, and a 3M Untenured Faculty Award.
References
- 1.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Nat. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zorlutuna P, Annabi N, Camci-Unal G, Nikkhah M, Cha JM, Nichol JW, Manbachi A, Bae H, Chen S, Khademhosseini A. Adv Mater. 2012;24:1782–1804. doi: 10.1002/adma.201104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvir T, Timko BP, Kohane DS, Langer R. Nature Nanotech. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 5.Kaihara S, Borenstein J, Koka R, Lalan S, Ochoa ER, Ravens M, Pien H, Cunningham B, Vacanti JP. Tissue Eng. 2000;6:105–117. doi: 10.1089/107632700320739. [DOI] [PubMed] [Google Scholar]
- 6.Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR, Vacanti JP. Biomed Microdevices. 2002;4:167–175. [Google Scholar]
- 7.Vozzi G, Flaim C, Ahluwalia A, Bhatia S. Biomaterials. 2003;24:2533–2540. doi: 10.1016/s0142-9612(03)00052-8. [DOI] [PubMed] [Google Scholar]
- 8.King K, Wang C, Kaazempur-Mofrad M, Vacanti J, Borenstein J. Adv Mater. 2004;16:2007. [Google Scholar]
- 9.Fidkowski C, Kaazempur-Mofrad MR, Borenstein J, Vacanti JP, Langer R, Wang Y. Tissue Eng. 2005;11:302–309. doi: 10.1089/ten.2005.11.302. [DOI] [PubMed] [Google Scholar]
- 10.Bettinger CJ, Langer R, Borenstein JT. Angew Chem. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao F, Veldhuis JJ, Duan Y, Yang Y, Christoforou N, Ma T, Leong KW. Mol Ther. 2010;18:1010–1018. doi: 10.1038/mt.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamato M, Kushida A, Okano T. Tanpakushitsu Kakusan Koso. 2000;45:1766–1772. [PubMed] [Google Scholar]
- 13.Shimizu T, Yamoto M, Kikuchi A, Okano T. Tissue Eng. 2001;7:141–151. doi: 10.1089/107632701300062732. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu T, Yamato M, Isoi Y, Akutsu T, Setomaru T, Abe K, Kikuchi A, Umezu M, Okano T. Circ Res. 2002;90:E40–E48. doi: 10.1161/hh0302.105722. [DOI] [PubMed] [Google Scholar]
- 15.Miyahara Y, Nagaya N, Kataoka M, Yanagawa B, Tanaka K, Hao H, Ishino K, Ishida H, Shimizu T, Kangawa K, Sano S, Okano T, Kitamura S, Mori H. Nat Med. 2006;12:459–465. doi: 10.1038/nm1391. [DOI] [PubMed] [Google Scholar]
- 16.Ke Q, Wang X, Gao Q, Wu Z, Wan P, Zhan W, Ge J, Wang Z. J Tissue Eng Regen Med. 2011;5:138–145. doi: 10.1002/term.298. [DOI] [PubMed] [Google Scholar]
- 17.Haraguchi Y, Shimizu T, Yamato M, Okano T. Rsc Adv. 2012;2:2184–2190. [Google Scholar]
- 18.Obokata H, Yamato M, Tsuneda S, Okano T. Nat Protoc. 2011;6:1053–1059. doi: 10.1038/nprot.2011.356. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg AW, Feigel A, Shevkoplyas SS, Sheehy S, Whitesides GM, Parker KK. Science. 2007;317:1366–1370. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- 20.Francesko A, Tzanov T. Adv Biochem Eng Biot. 2011;125:1–27. doi: 10.1007/10_2010_93. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Hu X, Lin W, Dong C, Wu H. In vitro Cell Dev-An. 2011;47:234–240. doi: 10.1007/s11626-010-9381-4. [DOI] [PubMed] [Google Scholar]
- 22.Timnak A, Gharebaghi FY, Shariati RP, Bahrami SH, Javadian S, Emami Sh H, Shokrgozar MA. J Mater Sci-Mater M. 2011;22:1555–1567. doi: 10.1007/s10856-011-4316-5. [DOI] [PubMed] [Google Scholar]
- 23.Yang TL. Int J Mol Sci. 2011;12:1936–1963. doi: 10.3390/ijms12031936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayakumar R, Chennazhi KP, Srinivasan S, Nair SV, Furuike T, Tamura H. Int J Mol Sci. 2011;12:1876–1887. doi: 10.3390/ijms12031876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji C, Annabi N, Khademhosseini A, Dehghani F. Acta Biomater. 2011;7:1653–1664. doi: 10.1016/j.actbio.2010.11.043. [DOI] [PubMed] [Google Scholar]
- 26.Jeong SI, Krebs MD, Bonino CA, Samorezov JE, Khan SA, Alsberg E. Tissue Eng Part A. 2011;17:59–70. doi: 10.1089/ten.TEA.2010.0086. [DOI] [PubMed] [Google Scholar]
- 27.Reis LA, Chiu LL, Liang Y, Hyunh K, Momen A, Radisic M. Acta Biomater. 2012;8:1022–1036. doi: 10.1016/j.actbio.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 28.Cooper A, Zhong C, Kinoshita Y, Morrison RS, Rolandi M, Zhang M. J Mater Chem. 2012;22 [Google Scholar]
- 29.Jayakumar R, Menon D, Manzoor K, Nair SV, Tamura H. Carbohyd Polym. 2010;82:227–232. [Google Scholar]
- 30.Ji W, Sun Y, Yang F, van den Beucken JJ, Fan M, Chen Z, Jansen JA. Pharm Res. 2011;28:1259–1272. doi: 10.1007/s11095-010-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladd MR, Lee SJ, Stitzel JD, Atala A, Yoo JJ. Biomaterials. 2011;32:1549–1559. doi: 10.1016/j.biomaterials.2010.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Beachley V, Wen X. Mater Sci Eng: C. 2009;29:663–668. doi: 10.1016/j.msec.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong C, Cooper A, Kapetanovic A, Fang ZH, Zhang MQ, Rolandi M. Soft Matter. 2010;6:5298–5301. [Google Scholar]
- 34.Zhong C, Kapetanovic A, Deng Y, Rolandi M. Adv Mater. 2011;23:4720–4720. doi: 10.1002/adma.201102639. [DOI] [PubMed] [Google Scholar]
- 35.Shigemasa Y, Matsuura H, Sashiwa H, Saimoto H. Int J Biol Macromol. 1996;18:237–242. doi: 10.1016/0141-8130(95)01079-3. [DOI] [PubMed] [Google Scholar]
- 36.Nikkhah M, Eshak N, Zorlutuna P, Annabi N, Castello M, Kim K, Dolatshahi-Pirouz A, Edalat F, Bae H, Yang Y, Khademhosseini A. Biomaterials. 2012;33:9009–9018. doi: 10.1016/j.biomaterials.2012.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papadaki M, Bursac N, Langer R, Merok J, Vunjak-Novakovic G, Freed LE. Am J Physiol Heart C. 2001;280:H168–178. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]
- 38.Couchman JR, Austria MR, Woods A. J Invest Dermatol. 1990;94:7S–14S. doi: 10.1111/1523-1747.ep12874973. [DOI] [PubMed] [Google Scholar]
- 39.Zhang K, Geissler A, Fischer S, Brendler E, Baucker E. J Phys Chem B. 2012;116:4584–4592. doi: 10.1021/jp210469x. [DOI] [PubMed] [Google Scholar]
- 40.Cooper A, Bhattarai N, Kievit FM, Rossol M, Zhang MQ. Phys Chem Chem Phys. 2011;13:9969–9972. doi: 10.1039/c0cp02909b. [DOI] [PubMed] [Google Scholar]
- 41.Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A. Biomaterials. 2012;33:5230–5246. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiffman JD, Schauer CL. Biomacromolecules. 2007;8:594–601. doi: 10.1021/bm060804s. [DOI] [PubMed] [Google Scholar]
- 43.Lovett M, Lee K, Edwards A, Kaplan DL. Tissue Eng Part B Rev. 2009;15:353–370. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Nat Mater. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dvir T, Timko BP, Brigham MD, Naik SR, Karajanagi SS, Levy O, Jin H, Parker KK, Langer R, Kohane DS. Nat Nano. 2011;6:720–725. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS. Adv Funct Mater. 2013 doi: 10.1002/adfm.201300570. 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1: Phase contrast Images of the cells attached on (a) G1 and (b) G2 micropatterned substrates after 5 days of culture. Scale bars represent 50(a)
Supplementary Fig. S2: Tensile testing on 30% deacetylated chitin substrates dry and after immersion in PBS for 1 day and for 5 days.