Abstract
Synexpression groups are genetic modules composed of genes that share both a complex expression pattern and the biological process in which they function. Here we investigate the regulation of BMP4 synexpression by studying the enhancers of bambi, smad7 and vent2 in Xenopus. We find that a BMP4 synexpression promoter module is compact and (i) requires direct BMP responsiveness through Smad and Smad-cofactor binding motifs, (ii) may contain an evolutionary conserved BMP-responsive element, bre7 (TGGCGCC), that is crucial for expression of bambi and smad7 and is highly prognostic for novel BMP-responsive enhancers (BREs); and (iii) requires a narrow window of BMP inducibility, because minor enhancement or reduction of BMP responsiveness abolishes synexpression. Furthermore, we used a bioinformatic model to predict in silico 13 novel BREs, and tested five of them that were found in the id1-4 genes. The results highlight that in vivo analysis is required to reveal the physiological, spatio-temporal regulation of BMP-responsive genes.
Keywords: bambi, bmp, smad7, synexpression, Xvent2
Introduction
Recent large-scale analyses have revealed coordinate expression of genes functioning in the same biological process. Such clusters of coexpressed genes were found by microarray experiments in yeast and in mammalian cultured cells, and in a systematic gene expression screen by RNA in situ hybridization, we have previously discovered four clusters of about 50 coexpressed genes during early Xenopus development (Gawantka et al, 1998). Such clusters, or synexpression groups (Niehrs and Pollet, 1999), were linked to distinct molecular processes, such as protein secretion, chromatin assembly and growth factor signaling. More recently, synexpression groups were also identified in mouse and zebrafish embryos, indicating that they are a widespread phenomenon (Tsang et al, 2000; Grotewold et al, 2001; Niehrs and Meinhardt, 2002). Member genes of synexpression groups show tight spatio-temporal RNA coexpression over various stages of embryogenesis and, wherever investigated, these genes cofunction in the respective molecular pathway (Onichtchouk et al, 1996, 1999). The tight coexpression of member genes of a synexpression group raises the question of how coregulation is achieved. A likely possibility is that genes of a synexpression group share similar enhancers, which in its simplest case may be regulated by one master control gene. A well-characterized example is the BMP4 synexpression group (Figure 1A), members of which are expressed like this growth factor—dorsally in the eye, heart and proctodeum of tailbud stage Xenopus embryos (Figure 1B). This group consists of eight members, which all encode components of the BMP signaling cascade as studied in early development, including ligands, receptors and downstream components of the pathway.
Figure 1.
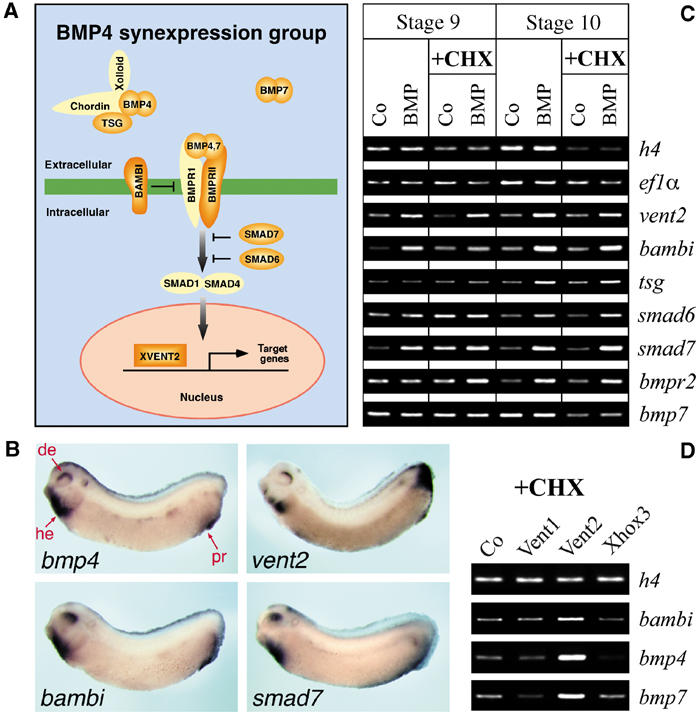
BMP4 synexpression group. (A) Simplified scheme of the BMP signaling pathway with coexpressed components (synexpression group member genes) indicated in orange. (B) Whole-mount in situ hybridization for bmp4, vent2, bambi and smad7 in Xenopus tailbud embryos reveals their highly similar expression pattern in the dorsal eye (de), heart region (he) and proctodeum (pr). (C) RT–PCR analysis of ectodermal explants (animal caps) from embryos injected animally with 2 ng bmp4 RNA and either treated or untreated with cycloheximide (CHX) from stage 8 onwards. Animal caps were collected at stage 9 (late blastula) or 10 (early gastrula) and analyzed for expression of the indicated RNAs. The samples were normalized by ef1α and histone h4 expression. (D) RT–PCR analysis of animal caps from embryos injected animally with 2 ng vent1, vent2 or xhox3 RNA and treated with CHX from stage 8 for 3 h.
BMPs belong to the large family of TGF-β signaling molecules and are important regulators of early embryonic patterning, organogenesis and tissue homeostasis. BMP ligands bind and induce heterodimerization of two classes of receptors with Ser/Thr kinase activity that transmit the signal mainly via the Smad pathway (Heldin et al, 1997; Miyazawa et al, 2002; Shi and Massagué, 2003). Smad proteins belong to three distinct functional classes. The receptor activated Smads (R-Smads) are phosphorylated by the activated receptors and transduce the BMP (Smad1, 5 and 8) or the TGF-β/activin type of signals (Smad2 and 3). In the cytoplasm, R-Smads form heteromeric complexes with the Co-Smad (Smad4) and translocate to the nucleus, where they regulate target genes' expression in complexes with other transcription factors. Inhibitory Smads (Smad6 and 7) attenuate the signaling cascade by inhibition of R-Smad phosphorylation and oligomerization or by targeting the receptors to degradation (Shi and Massagué, 2003).
Smads can bind directly to certain DNA motifs, but with relatively low affinity and specificity (ten Dijke et al, 2000; Shi and Massagué, 2003; Zwijsen et al, 2003). R-Smads and Smad4 recognize the so-called Smad binding element (SBE) motif GTCT (Shi et al, 1998) optimally: GTCTG, which is found in the promoters of most TGF-β and BMP target genes. Another motif, GCAT, was described as a Smad1 binding element in the promoter of Xenopus vent2 (Henningfeld et al, 2000). Furthermore, Drosophila R-Smad (Mad) was shown to bind GC-rich sequences with a GCCG core consensus (Kim et al, 1997). Recently, GC-rich elements were described as Smad binding motifs in several vertebrate BMP-responsive promoters, for example, smad6 (Ishida et al, 2000) and id1 (Korchynskyi and ten Dijke, 2002; Lopez-Rovira et al, 2002). Multimerized GCCG elements were used as BMP-specific reporters in cell culture assays (Kusanagi et al, 2000).
Smads alone bind to DNA with low affinity and selectivity and therefore need to associate with various transcription factors (TF) and it is these R-Smad/Co-Smad/Co-TF complexes that ensure promoter and cell-type specific activation of TGF-β target genes (Miyazawa et al, 2002; Zwijsen et al, 2003). Many transcription factors (Fast, AP1, ATF2, TFE3, Mixer, Lef/TCF and others) were found as Smad partners in the activin/TGF-β pathway (Shi and Massagué, 2003) and a common Smad interacting motif (SIM), which could be utilized for bioinformatic prediction of other Smad cofactors, was identified in some of them (Germain et al, 2000). Unlike TGF-β-responsive enhancers, little is known at present about how Smad/Co-TF complexes form and regulate BMP-responsive enhancers (BREs). The best-studied case is the Xenopus vent2 gene. OAZ (Hata et al, 2000) and Vent2 itself (Henningfeld et al, 2002) were shown to interact physically with Smad complexes and DNA elements on the vent2 promoter.
The coordinate transcriptional regulation of members of the BMP4 synexpression group raises the question of its molecular basis. Some of the genes, such as vent2 and smad6, are direct targets of the BMP/Smad signaling pathway. It is unknown whether other BMP synexpression group genes are also directly regulated by Smads, thus making BMP4 the master regulator of the entire synexpression group. Are there common elements in the enhancers of member genes and if so which? If the enhancers are composed of different modules, what is their individual contribution to the domains of expression? Is it possible to derive a consensus structure for the regulatory elements of this synexpression group? Importantly, it is unclear whether BMP responsiveness alone is sufficient to confer synexpression: despite the many studies on BREs there is little information regarding their physiological relevance for in vivo expression.
To address these questions, we have cloned and characterized three BREs from Xenopus bambi and smad7 and performed comparative analyses of these BREs with that of vent2 in transgenic frogs. Combining experimental and bioinformatic methods, we identified a GC-rich phylogenetically conserved motif as a critical BMP-responsive element that can be used for in silico identification of novel BREs.
Results
Synexpression group members are immediate early BMP4 target genes
The BMP4 synexpression group consists of eight genes that encode ligands (BMP4 and 7), receptors (BMPR2, BAMBI), extra- and intracellular modulators (Tsg, Smad6, Smad7), and transcription factors (Vent2) involved in BMP signaling (Figure 1A). The similar expression pattern of these genes (Figure 1B) suggests that they may be under common transcriptional regulation. Indeed, Xenopus vent2 and mouse smad6 genes are direct targets of BMP signaling (Hata et al, 2000; Ishida et al, 2000). To address whether other members of the synexpression group are also immediate BMP targets, Xenopus embryos were injected with bmp4 RNA and analyzed by RT–PCR for target gene expression at the late blastula (st. 9) and early gastrula stage (st. 10). In order to distinguish between direct and indirect induction, we treated embryos with or without the protein synthesis inhibitor cycloheximide (CHX). In Xenopus embryos, RNA starts being translated immediately after injection, whereas zygotic transcription begins at the mid-blastula stage when embryos were transferred to CHX. RT–PCR analysis (Figure 1C) revealed that vent2, bambi, tsg, bmpr2, smad6 and smad7 are induced by bmp4 in the presence of CHX, albeit with subtle kinetic differences, and hence are direct targets of BMP signaling. In contrast, bmp7 is not induced by bmp4. Similar results were obtained when embryos were injected with RNA for downstream components of the pathway, namely constitutively active BMP receptor—CABR (Candia et al, 1997)—or Smad1 plus Smad4, and CHX treated (data not shown). To test whether downstream effectors of the BMP signaling cascade can also induce BMP4 synexpression group genes, vent1, vent2 and xhox3 RNAs were injected and the embryos were treated with CHX (Figure 1D). Only Vent2 was capable of directly inducing the tested genes bambi, bmp4 and bmp7. bmp4 contains Vent2 binding sites that are required for its autoregulation (Schuler-Metz et al, 2000). As bambi is induced both by BMP4 as well as its transcriptional target Vent2 in the presence of CHX, it is both a direct as well as an indirect (via Vent2) BMP target gene. These results suggest that developmental synexpression results from transcriptional coregulation by the BMP signaling cascade.
Identification of the BRE of bambi
Xenopus bambi encodes a pseudoreceptor that negatively regulates TGF-β signaling (Onichtchouk et al, 1999) and is tightly coexpressed with BMP4 during development in Xenopus (Figure 1B), mouse (Grotewold et al, 2001) and zebrafish (Tsang et al, 2000). To analyze its enhancer and to compare it with already known BREs, a 13 kb clone, containing around 6 kb from the 5′ flanking region of bambi, was isolated from a Xenopus genomic library (Figure 2A). When the upstream region of bambi was sequenced and compared with the corresponding genomic segment of the human ortholog nma, a single highly conserved fragment of around 200 bp was found 2 kb upstream of bambi and 0.5 kb upstream of nma (Figure 2A). Conserved noncoding sequences (CNCS) are reliable indicators of regulatory elements, especially when found in distant species, due to the selective pressure exerted on functionally important DNA regions (exons, promoters, enhancers, etc.).
Figure 2.
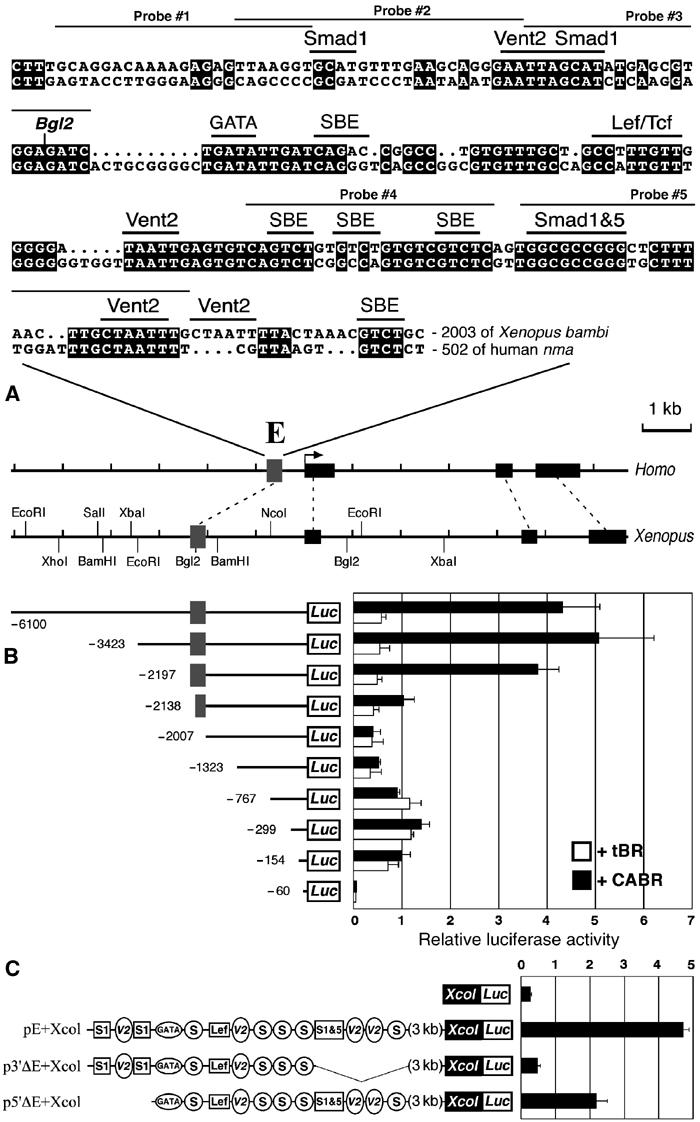
Isolation of the BRE of bambi. (A) Comparative map of Xenopus bambi and human nma genomic regions. The exons are shown as black bars, a conserved 200 bp fragment (putative enhancer) is labeled ‘E' (gray box) and its Xenopus/Homo sequence alignment is shown. The putative transcription factor binding sites are shown above the alignment. Also highlighted are sequences used to derive probes for EMSA assays. (B) Deletion analysis of the 5′ flanking region of bambi confirms that E is a BRE. Xenopus embryos were coinjected with the indicated promoter constructs and either tBR or CABR mRNA and harvested at the gastrula stage for luciferase reporter assay. (C) The bambi enhancer confers BMP inducibility to a heterologous (collagen X) minimal promoter. Xenopus embryos were coinjected with the indicated promoter constructs and CABR. Putative TF binding sites from panel A are represented schematically: S, SBE; S1, Smad1; V2, Vent2; S1&5, Smad1 and 5 motifs.
We further analyzed this putative enhancer for potential TF binding sites that are linked to BMP signaling. Interestingly, eight Smad binding sites were detected, five of them fully conserved between the frog and the human DNA sequence. Most represent the common SBE motif GTCT (or AGAC in reverse orientation). In addition to SBE, two Smad1 binding motifs (GCAT) and one fully conserved GC-rich element (potential Smad1/5 motif) were found in the putative bambi enhancer (Figure 2A). Consensus binding sites were also found for GATA, Lef/TCF and Vent2 transcription factors. Of special interest is the presence of four putative Vent2 binding motifs (C)TAATT, because we observed direct induction of bambi by Vent2 in RT–PCR experiments and since Vent2 interacts with Smad1 to activate its own promoter (Henningfeld et al, 2002).
To confirm that this 200 bp fragment represents a functional enhancer, we carried out a systematic deletion analysis of the bambi upstream genomic region via luciferase reporter assays (Figure 2B). Deletion constructs were coinjected in Xenopus embryos either with constitutively active (CABR) or truncated (dominant-negative) BMP receptor (tBR), to enhance or suppress embryonic BMP signaling. This analysis showed that the evolutionary conserved 200 bp segment was indeed the BMP-responsive enhancer of bambi (compare constructs −2197 and −2007). The −154 construct harbors BMP-independent, basal promoter activity driven by two evolutionary conserved, overlapping Sp1 sites, deletion of which eliminates this activity (data not shown). In addition, a silencer element is located around −1 kb (compare −1323 to −767 constructs) that reduces the basal activity of the proximal promoter only. The bambi enhancer is active not only on its own proximal promoter but also on a heterologous minimal promoter from the collagen X gene (Harada et al, 1997). Strong stimulation of pE+Xcol was observed by CABR mRNA coinjection, which was significantly reduced when 5′ and, especially, 3′ deletions in the enhancer were introduced (Figure 2C).
Characterization of the BRE of bambi
We next dissected the bambi enhancer through deletions (Figure 3A) and point mutations (Figure 3B) of the predicted TF binding sites. Both 5′ and 3′ deletions of the enhancer confirmed the importance of most Smad and Vent2 consensus motifs. Multimerization of the 5′ 30 bp segment containing such motifs led to strong BMP responsiveness (p11). However, BMP inducibility is partially retained when deleting the 5′ Smad/Vent2 motifs (e.g. p3), but almost abolished by deleting the 3′ motifs (e.g. p9, p15 and Figure 2C). The greater importance of the 3′ enhancer part is supported by multi-species sequence alignment (see below).
Figure 3.
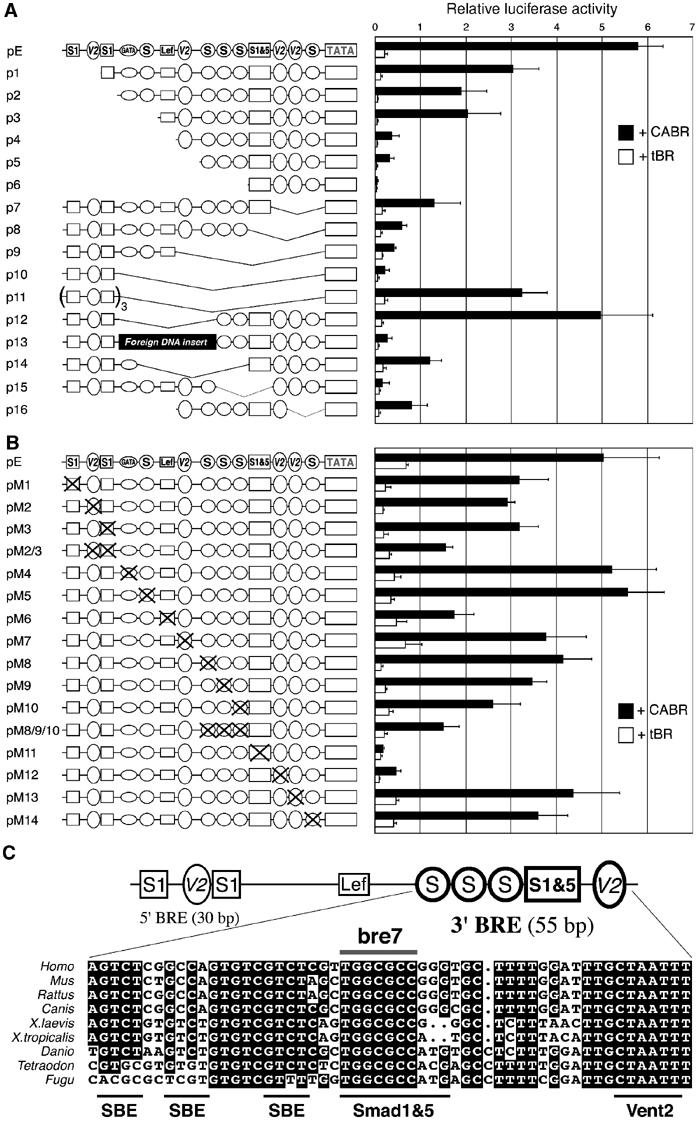
Functional dissection of the bambi enhancer. (A) Deletion and (B) point-mutation analyses of the 200 bp enhancer. Frog embryos were injected with the indicated constructs together with tBR or CABR mRNA and collected at gastrula stage for luciferase reporter assays. Putative TF binding sites are represented schematically: S, SBE; S1, Smad1; V2, Vent2; S1&5, Smad1 and 5 motifs. ‘TATA' represents the heterologous 13 bp core adenovirus E1b promoter. (C) Structure of the bambi enhancer with functionally relevant TF binding sites. The major 3′ BMP-responsive element (3′ BRE) is highlighted and its phylogenetic conservation in nine vertebrate species is shown. Putative TF binding sites are underlined. Sequences were extracted from the assembled genomic data for Homo (human), Mus (mouse), Rattus (rat), Danio (zebrafish) and Fugu (pufferfish), or from the whole-genome shotgun trace archives for Canis (dog), X. tropicalis (frog) and Tetraodon (pufferfish).
While deletions of the putative GATA and the 5′ SBE motifs did not affect reporter activity, elimination of the consensus Lef/TCF binding site strongly reduced reporter levels (compare p3 and p4 constructs). Lef/TCF are HMG box transcription factors that mediate Wnt/β-catenin signaling, but may also serve an architectural function in the assembly of TF complexes by DNA bending. In the case of the bambi enhancer regulation, Wnt signaling is not involved, since neither overexpression of XWnt8 (by plasmid injection) nor the Wnt inhibitor Dkk1 affects reporter activity (data not shown). This suggests that Lef/TCF or related proteins may have architectural rather than signaling function in this case. This is further supported by substituting a 60 bp fragment containing this element with irrelevant DNA, which strongly reduced expression (p13). In contrast, its deletion did not affect significantly reporter activity (p12), possibly because proximity of the 5′ and 3′ enhancer parts can compensate for lack of DNA bending.
While point mutations in the 5′ sites only reduce enhancer activity, mutating the GC-rich box (potential Smad1/5 binding sequence, pM11) completely abolishes BMP inducibility (Figure 3B). This is in agreement with the deletion experiments (p15, Figure 3A) and points to the critical importance of this putative Smad1/5 binding motif. Mutating the three adjacent SBEs (pM8/9/10) also significantly reduces reporter activity. One of the two consensus Vent2 elements adjacent to the Smad1/5 motif is also essential (pM12 and p7, Figure 3A).
Consistent with the greater importance of the 55 bp 3′ BMP-responsive element (3′ BRE), it is phylogenetically highly conserved (Figure 3C). A heptanucleotide TGGCGCC in the critical Smad 1/5 motif is fully conserved, but the overlapping GCCG core, believed to be essential for Smad1/5 binding, is not. There is no consensus SBE in Fugu, one in Tetraodon, and two or three copies are found in the higher vertebrates. The region upstream of the 3′ BRE is partially conserved in Xenopus tropicalis, zebrafish and mammals, but not in the pufferfish species. Thus, there is a trend toward more Smad motifs and presumably greater BMP responsiveness in higher vertebrates.
We next tested the ability of recombinant Smad and Vent2 proteins to bind to the predicted motifs in EMSA assays. Smad1 interacts with the probe containing the putative Smad1/5 binding site (#5), as well as with the 3 × SBE probe (#4, Figure 4A). However, no Smad1 binding to the GCAT-containing probes was observed (#2 and #3), which may reflect the reported importance of flanking sequences within the vent2 promoter (Henningfeld et al, 2000). Expectedly, recombinant Smad4 interacted only with the 3 × SBE probe (#4) and Vent2 with the probe containing the CTAATT motif (Figure 4B and C). The in vivo role of Vent2 in the regulation of bambi was confirmed in a reporter assay where dominant-negative Vent2 mRNA was co-injected with CABR, which significantly reduced reporter activity (Figure 4D).
Figure 4.
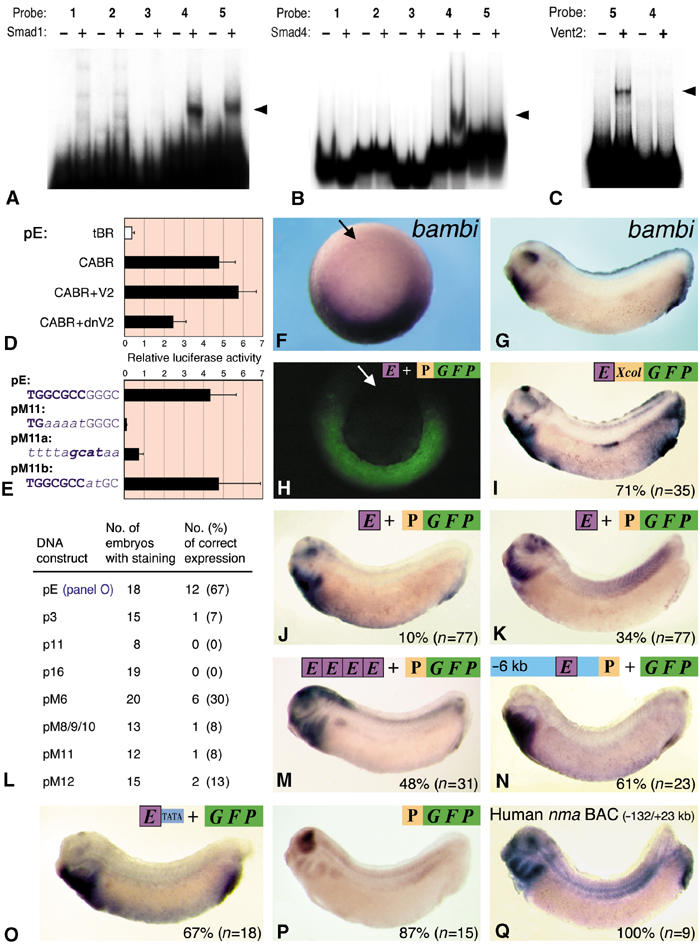
In vitro and transgenic characterization of the bambi enhancer. EMSA assays with recombinant Smad1 (A), Smad4 (B) and Vent2 proteins (C) of probes 1–5 (see Figure 2A). Specific bandshifts are indicated with arrowheads. (D) Xenopus embryos were injected with the 200 bp enhancer construct pE together with the indicated RNAs. V2, Vent2; dnV2, dominant-negative Vent2. (E) Luciferase reporter assay in Xenopus embryos of enhancer constructs with mutations in the crucial GC-rich element. All constructs were coinjected in embryos with CABR mRNA. Constructs pE (wild-type) and pM11 are shown in Figure 3B. The replaced nucleotides and the alternative Smad1 motif GCAT are highlighted. (F, G) In situ hybridization of bambi at gastrula (vegetal view) and tailbud stage embryos. The arrows in panels F and H point to the blastopore. (H–Q) Transgenic Xenopus assays with the indicated DNA constructs. Transgene expression was visualized by fluorescence (H) or in situ hybridization for gfp (I–P) and nma mRNA (Q). The percentage of embryos with the depicted pattern is indicated. E, 200 bp bambi enhancer; Xcol, collagen X minimal promoter; P, bambi minimal promoter (−154/+60). (L) Results from transgenic experiments with constructs from Figure 3. The correct expression pattern referred to (heart+proctodeum) is the one shown in panel O.
Next we asked whether the crucial Smad1/5 GC-rich element is functionally interchangeable with the other known Smad1 motif GCAT, found in the 5′ BRE and in the vent2 promoter. When the GC element was replaced by a GCAT motif, enhancer activity was only partially rescued (compare pM11 and pM11a; Figure 4E), consistent with the EMSA data showing that GCAT has a lower Smad binding affinity than the GC-rich element (Figure 4A), at least in vitro. In line with the poor evolutionary conservation of the GCCG core within the GC-rich element (Figure 3C), we found that its integrity was not important (pM11b) but that the conserved TGGCGCC motif, referred to as bre7 is required for enhancer activity (pM11, Figure 4E). As it turns out this motif is found in many other BMP-responsive genes (see below).
Transgenic analysis of the bambi enhancer
An important question is whether the 200 bp enhancer is capable of reproducing the characteristic expression pattern of the endogenous bambi gene. We therefore performed transgenic experiments in Xenopus (Kroll and Amaya, 1996) with GFP-based reporter constructs and monitored the expression by GFP fluorescence or in situ hybridization. In most experiments, we mixed enhancer DNA fragments and promoter/GFP constructs (‘cotransgenesis'; Hartley et al, 2001).
In gastrula stage embryos, the 200 bp enhancer is sufficient to drive reproducibly GFP transgene expression similar to bambi in the ventral and lateral marginal zone (Figure 4F and H). For later embryonic stages, GFP expression was monitored not by fluorescence but by in situ hybridization, because the stability of the GFP protein caused an overlap of the early and late expression. At the tailbud stage, the characteristic eye–heart–proctodeum synexpression pattern (Figure 4G) was seen fully reproduced in around 10% (n=77) of the transgenic embryos (Figure 4J), suggesting that the 200 bp enhancer is in principle capable of successfully driving synexpression. However, likely due to position effect variegation (PEV), the majority of embryos showed a combination of correct and ectopic expression (example in Figure 4K), also when using a heterologous minimal promoter from the collagen X gene (Figure 4I). Multimerization of the enhancer to reduce PEV indeed leads to a 5-fold more reproducible expression pattern, but adds ectopic expression in the brain (Figure 4M) where low-level BMP signaling is present. This indicates that synexpression requires a certain threshold of BMP inducibility.
Expression in the heart and proctodeum was present in the majority of transgenic embryos, whereas expression in the dorsal eye was rarely seen and only when the bambi enhancer was combined with its own proximal promoter (Figure 4J). Using the entire 6 kb bambi promoter improved the reproducibility of pattern except for dorsal eye expression, as was the case for a 155 kb bacterial artificial chromosome containing the human bambi ortholog nma (Figure 4Q). We therefore conclude that the bambi enhancer is sensitive to PEV, especially concerning expression in the dorsal eye.
We also tested some of the deletion and point-mutation constructs from Figure 3 in transgenic frogs. The intact enhancer in front of a TATA box drives reproducible, albeit weak expression in the heart and proctodeum (Figure 4O). Mutation in the consensus Lef/TCF site (pM6) reduced the percentage of correct expression twofold, while mutations in the 3′ BRE elements SBE (pM8/9/10), bre7 (pM11) and Vent2 (pM12) strongly affected it (Figure 4L), in agreement with the reporter assay data (Figure 3B). Interestingly, deletion of the 5′ BRE (p3), which only moderately reduced overall luciferase reporter activity and retained BMP inducibility (Figure 3A), had a dramatic effect on transgene expression (Figure 4L). Likewise, neither 3′ BRE alone (p16) nor the multimerized 5′ BRE (p11) was able to drive correct expression, although they display BMP inducibility (Figure 3A). We conclude that (i) the full-length bambi enhancer is required for proper developmental expression and (ii) a minor change in BMP responsiveness has a profound effect on synexpression.
Transgenic analysis of the vent2 promoter
The transcriptional regulation of the Xenopus vent2 gene was extensively studied by several groups and is a paradigm for a BMP-responsive promoter. It contains a core 50 bp BMP-responsive element (BRE), composed of one SBE and an adjacent motif for the cofactor OAZ, which are critically important for BMP stimulation in cell culture assays (Hata et al, 2000). Furthermore, two GCAT Smad1 binding motifs upstream of the BRE are necessary for full promoter activity in Xenopus embryos (Henningfeld et al, 2000). In addition, Vent2 autoregulates its own expression in a complex with Smad1 (Henningfeld et al, 2002). However, the role and importance of these elements for the synexpression pattern of vent2 (Figure 5B) have not been previously analyzed. We tested the 300 bp vent2 promoter (Figure 5A) in transgenic frogs and found it sufficient to reproduce robustly the BMP4-like pattern of the endogenous vent2 gene except for a relatively weaker expression in the tailbud (Figure 5C). Point mutations in the Smad1 motifs (Figure 5D,E) and even deletion of the entire region upstream from the BRE (Figure 5I), while weakening the expression, did not significantly affect the pattern. However, mutating the SBE or OAZ motifs resulted in a complete loss of correct expression and in a weak ectopic expression in the somites (Figure 5F and G). Likewise, a 3′ deletion including the proximal Vent2 binding site also failed to produce the wild-type pattern (data not shown). Sequence comparison of the promoters of vent2 and vent1 (a closely related homeobox gene) revealed a conserved consensus Lef/TCF binding sequence and its mutation in the vent2 promoter decreased the strength and reproducibility of transgene expression (Figure 5H). We conclude that, similar to bambi, binding sites for Smads and transcription cofactors are required in the vent2 promoter for synexpression.
Figure 5.
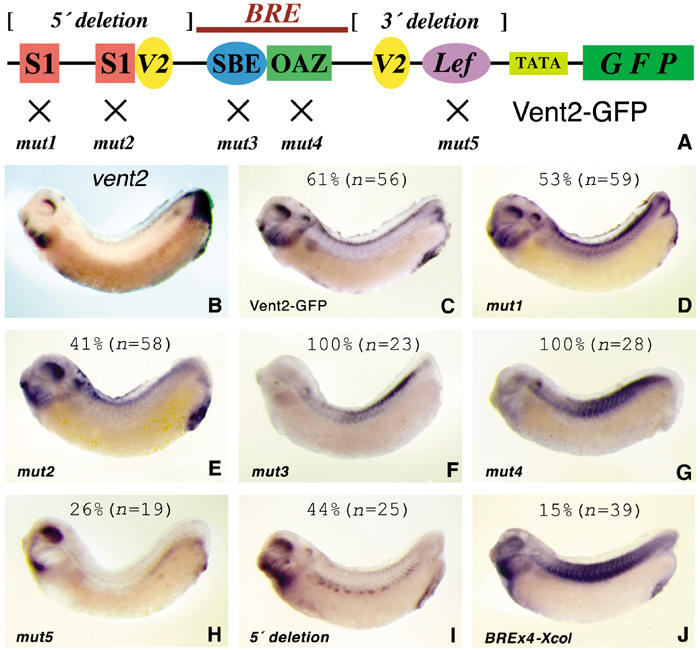
Transgenic analysis of the vent2 promoter. (A) Scheme of the vent2 promoter (Hata et al, 2000; Henningfeld et al, 2000, 2002). S1, Smad1 motif (GCAT); OAZ, binding site (TGGAGC) for the Smad cofactor OAZ; Lef, putative Lef/TCF motif (CCTTTGAT); V2, Vent2 motifs. (B) In situ hybridization of vent2 at the tailbud stage. (C–J) gfp in situ hybridization of tailbud stage embryos transgenic for the indicated constructs (see panel A). BREx4-Xcol, four copies of the vent2 BRE in front of the collagen X minimal promoter. The percentage of embryos with the depicted expression pattern is shown.
We also tested three artificial, multimerized and in vitro strongly BMP-inducible BREs for their ability to confer BMP4 synexpression. BRE × 4 (Hata et al, 2000) transgenic embryos displayed variable expression and even the few embryos with correct pattern showed additional ectopic expression in the somites (Figure 5J). Likewise, 3GC2 (Ishida et al, 2000) and 12 × GCCG (Kusanagi et al, 2000) transgenic frogs failed to reproduce the BMP4 synexpression pattern (data not shown). These results further highlight that different BREs drive very different expression patterns.
Characterization of BMP-responsive enhancers in the Xenopus smad7 gene
To study a third member of the synexpression group, we chose smad7, a negative feedback regulator of BMP and qTGF-β signaling (Nakao et al, 1997), which is coexpressed with BMP4 during Xenopus development (Figure 1B; Casellas and Hemmati-Brivanlou, 1998). smad7 expression is induced by both TGF-β and BMP signaling in various cell types and organisms (Zwijsen et al, 2000; Locklin et al, 2001; Ota et al, 2002) and our RT–PCR experiments showed that it is a direct transcriptional target of BMP signaling in Xenopus (Figure 1C). We isolated, sequenced and compared a Xenopus smad7 genomic clone with the corresponding human and Fugu DNA regions. Two highly conserved noncoding segments were detected, one in the 5′ flanking region (E1) and one near the 3′ end of intron1 (E2) (Figure 6A). The mouse smad7 promoter region containing E1 was previously described as only activin/TGF-β-responsive (Nagarajan et al, 1999). Phylogenetic footprinting of E1 shows perfect conservation of the octanucletide palindrome composed of two SBEs and adjacent putative binding sites for the transcription factors EKLF, USF and AP1. A GC-rich sequence at the 3′ end of E1 is another potential Smad1/5 binding site. The predicted intronic enhancer E2 contains conserved binding motifs for Smads (SBE and GC-rich element), GATA and NFY transcription factors. Remarkably, the GC-rich element contains the same bre7 motif that we found to be critically important in the bambi enhancer. In E2, bre7 is present on both strands, forming a perfect palindrome (Figure 6A, red arrows). Similar to bambi, there is a GC-rich stretch 3′ of the bre7 motif.
Figure 6.
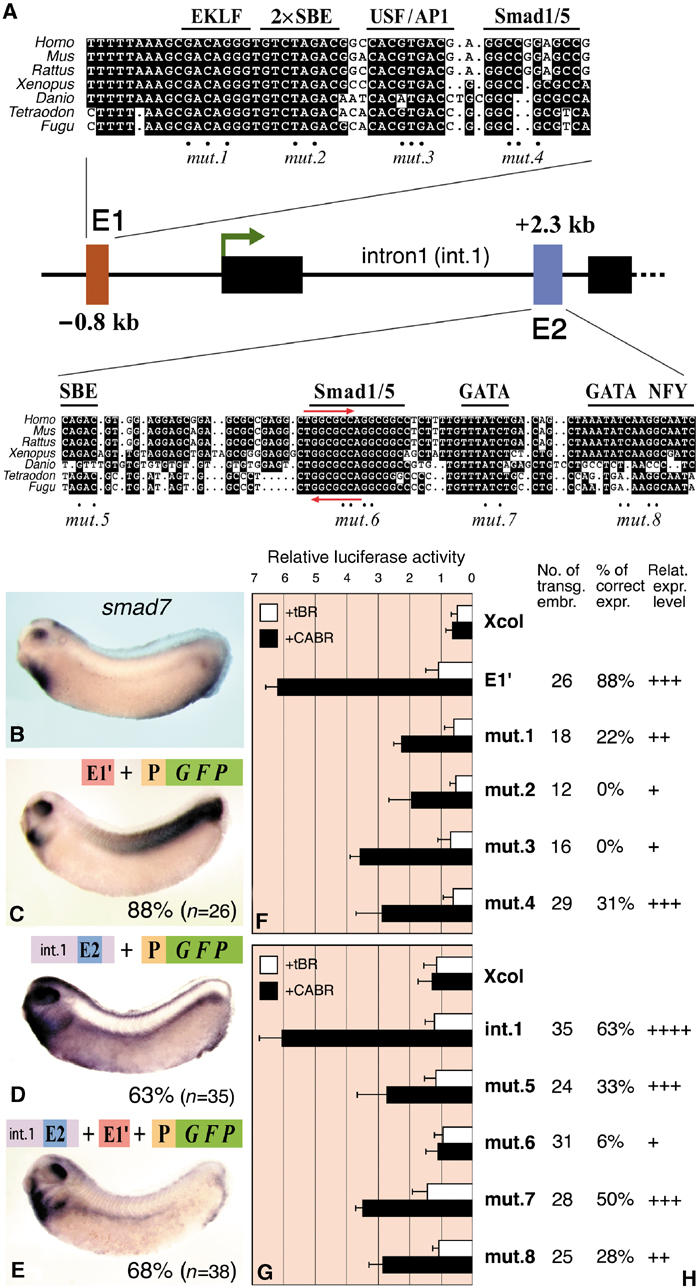
Characterization of the BREs of smad7. (A) Map of the Xenopus smad7 gene with the phylogenetically conserved putative enhancers E1 and E2. Multi-species sequence alignment with putative TF binding sites is shown for each enhancer. Red arrows in E2 indicate the bre7 motif also found in the bambi enhancer. Mutated nucleotides are indicated with dots. For transgenic and luciferase reporter assays, E1 was cloned with 24 bp upstream and 58 bp downstream nonconserved flanking sequences (E1′). (B) In situ hybridization of smad7 mRNA at the tailbud stage. (C–E) gfp in situ hybridization of transgenic embryos with the depicted constructs. P is the minimal promoter of bambi (−154/+60). The percentage of embryos with the depicted expression pattern is shown. (F, G) Data from luciferase reporter assays of the smad7 enhancers. E1′, intron1 or their mutated versions (see panel A) were cloned upstream of the Xcol minimal promoter and injected in embryos together with tBR or CABR mRNA as indicated. (H) Tabular results of embryos transgenic for the intact or mutated constructs (see panel A). The pattern referred to as correct is the one depicted in panel C (for E1′) and D (for intron1). The number of transgenic embryos analyzed, the percentage of embryos with correct pattern and the relative strength of transgene expression for each construct are shown.
Transgenic Xenopus assays demonstrated that either E1 (Figure 6C) or intron1 (Figure 6D) could drive an expression pattern reminiscent to that of smad7, but with ectopic expression in somites (E1) or the lateral plate (intron1). Only when both enhancers were combined, ectopic expression was suppressed and the majority of transgenic embryos showed a pattern very similar to that of the endogenous gene (Figure 6E). We conclude that smad7 synexpression requires both the upstream and intronic enhancers, even though both are individually BMP-responsive in luciferase assay (Figure 6F,G).
To further analyze the two enhancers, reporter and transgenic assays were performed with intact and mutated constructs (Figure 6F–H). Mutations of all predicted TF binding sites in E1 and E2 affected luciferase reporter and GFP transgene expressions. For E1, complete loss of correct transgene expression was observed when the 2 × SBE or the putative USF/AP1 motifs were disrupted (mut.2 and 3, Figure 6H), although BMP responsiveness in luciferase assay was only partially reduced (Figure 6F). Mutating the GC-rich (bre7) motif in E2 resulted in a complete loss of enhancer activity in the luciferase assay and in only very few transgenic embryos with correct, but much weaker expression (mut.6, Figure 6G and H). These data again demonstrate (i) the critical importance of the bre7 motif for BMP responsiveness and (ii) the nonequivalence of BMP inducibility and synexpression.
In silico prediction of novel BREs
The conservation of the bre7 motif in the BREs of bambi and smad7 encouraged us to search in silico for other putative BREs in known BMP target genes. We specifically looked for adjacent SBE and bre7 motifs in phylogenetically conserved regions that are close to the transcription start sites of the genes of interest. This resulted in 15 hits in 13 BMP target genes (Table I and Supplementary Figure 2).
Table 1.
BMP target genes with conserved clusters of SBE and bre7 motifs
| Gene (BRE) | CNCS (kb) | SBE | Spacing (bp) | bre7 |
|---|---|---|---|---|
| bambi (nma) | −0.5 | GTCT | 3 | TGGCGCC |
| smad7 | +2.1 | GTCTG | 28 | TGGCGCCA |
| id1 | −1.0 | GTCTG | 58 | TGGCGCC |
| id1 | +3.3 | GTCTG | 5 | AGGCGCC |
| id2 | −3.2 | GTCTG | 4 | TGGCGCCA |
| id3 | −2.9 | GTCTG | 4 | TGGCGCCA |
| id3 | +1.3 | GTCTG | 5 | TGGCGCC |
| id4 | +1.7 | GTCTG | 4 | TGGCGCCA |
| msx1 | −2.8 | GTCTG | 16 | TGGCGCC |
| msx2 | −3.3 | GTCT | 18 | TGGCGCC |
| gata2 | +5.5 | GTCTG | 16 | TGGCGCCA |
| gata3 | −0.1 | GTCTG | 16 | TGGCGCC |
| bmpr2 | −1.8 | GTCTG | 9 | TGGCGCCA |
| tlx2 (hox11L1) | −1.2 | GTCT | 12 | TGGCGCC |
| vox (vega1) | −0.2 | GTCTG | 5 | TGGCGCC |
| vent (vega2) | −0.3 | GTCTG | 4 | TGGCGCC |
|
tbx2 |
−2.9 |
GTCTG |
60 |
TGGCGCCA |
| The position of the conserved noncoding segments (CNCS) is shown according to the annotation of the NCBI human genomic browser, except for vox and vent (zebrafish). BREs experimentally analyzed in this study are in bold. Previously known BREs are underlined. | ||||
One class of genes identified are id (inhibitors of differentiation) genes (Figure 7), which are immediate targets of BMP signaling (Hollnagel et al, 1999). A bre7 motif was recognized previously as a 6 bp GGCGCC palindrome in the id1 promoter (Korchynskyi and ten Dijke, 2002; Supplementary Figure 1A). In addition to this known BRE, we identified a second putative BRE downstream of the id1 gene (Figure 7B), containing a GGCGCC palindrome and an SBE motif 5 bp apart, which are fully conserved from Fugu to Homo. Analysis of id2, 3 and 4 genomic regions also revealed a single highly conserved noncoding segment in each gene (Figure 7A). These regions are located upstream of the genes (id2, id3) or inside an intron (id4). Remarkably, all of them contain conserved SBE and bre7 motifs in the same orientation and with the same spacing, despite a lack of overall homology. A second putative BRE for id3 is located inside an intron (Figure 7B). When cloned and tested, all five in silico predicted enhancers proved to be BMP-responsive in frog embryos, with inductions between 3- and 6-fold (Figure 7C). We conclude that clusters of evolutionary conserved bre7 and SBE motifs are highly prognostic for BREs.
Figure 7.
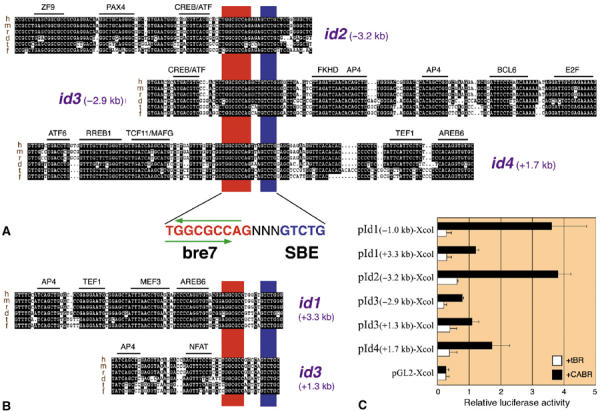
Bioinformatic prediction and experimental verification of five novel BREs for id1-4 genes. (A, B) Multi-species sequence alignment of the in silico identified novel id enhancers. The SBE and bre7 motifs are highlighted in blue and red, respectively. Putative TF binding sites are indicated. The currently available genomic sequences of Homo (h), Mus (m), Rattus (r), Danio (d), Tetraodon (t) and Fugu (f) are aligned. (C) Luciferase reporter assay of the five novel and the known (id1 −1.0 kb) BREs, cloned upstream of the Xcol minimal promoter (see Supplementary Figure 1C for fragments cloned). Frog embryos were injected with the indicated constructs together with tBR or CABR mRNA and harvested at the gastrula stage for luciferase assay.
Discussion
One shortcoming of the many studies dealing with BREs is the lack of in vivo analysis to characterize promoters in whole animals. Thus, prior to this study it was unknown whether the identified BREs are sufficient to drive faithful expression of the gene in question in transgenic animals. Here we have addressed this issue.
Common architecture of the BREs of bambi, smad7 and vent2
bmp4 and -7 are subject to complex regulation and integrate a variety of inputs, which control their developmental expression pattern (Schuler-Metz et al, 2000). In contrast, all other member genes of the BMP4 synexpression group proved to be direct transcriptional targets of the BMP/Smad signaling pathway in Xenopus (Figure 1). Therefore, developmental synexpression in the BMP4 group seems to result from the common, BMP/Smad-dependent transcriptional regulation of the member genes where the BMP ligands are the master regulators.
The 200 bp enhancer of bambi has a complex structure, consisting of 5′ BRE and 3′ BRE that contain multiple Smad and Vent2 binding sites. Both BREs and the linker region between them synergize in eliciting BMP-induced activation. Consistent with the leading importance of the 3′ BRE, it is highly conserved in nine vertebrate species from fish to mammals and is the only conserved region in a Fugu/Homo sequence comparison. However, neither of the two BREs alone is capable of driving strong reporter activity or conferring the correct pattern of expression; only the intact enhancer can mimic the expression pattern of bambi in transgenic assays.
The smad7 expression pattern is driven by two BREs (upstream and intronic). Crucial for expression are the 2 × SBE and putative USF/AP1 motifs in the upstream enhancer, and the bre7 element in the intronic enhancer. While this manuscript was in preparation, an independent study described the mouse smad7 intronic enhancer as GATA and Smad dependent, but the role of bre7 was not examined there (Benchabane and Wrana, 2003).
For the vent2 promoter, both the SBE and OAZ motifs are strictly required for correct developmental expression, but sequences outside the SBE and OAZ sites also affect expression, since the related gene vent1 has the same motifs (Hata et al, 2000) and is not coexpressed with vent2 at the tailbud stage (Gawantka et al, 1998).
Thus, all enhancers studied are relatively compact and contain at least one functional SBE motif. In addition, the evolutionary conserved GC-rich element bre7 was found to be critically important in the bambi and the smad7 intronic enhancers. This element is located close to conserved SBE and cofactor binding motifs. Smads can bind to certain GC-rich sequences (Shi and Massagué, 2003; Zwijsen et al, 2003) and bre7-containing probes specifically bind recombinant Smad5 (Korchynskyi and ten Dijke, 2002). Substitution of the bre7 element with a GCAT Smad1 motif partially rescues the bambi enhancer activity. Collectively, these data suggest that bre7 may function in vivo as a binding site for the BMP-specific R-Smads and, in combination with SBE motifs, can be utilized to predict novel BREs. Interestingly, bre7 (TGGCGCC) has similarity to the proposed OAZ motif TGGAGC (Hata et al, 2000), raising the possibility that Smad complexes may bind to bre7 through or synergistically with OAZ. On the other hand, id genes, which contain bre7 in their enhancers, are rapidly induced by BMP stimulation in C2C12 myoblast cells, which lack OAZ (Korchynskyi and ten Dijke, 2002), and the vent2 reporter is unresponsive to BMPs in these cells, unless OAZ is exogenously supplied (Hata et al, 2000). Furthermore, we find a bre7 motif in the tlx2 BMP-responsive promoter, reported to be OAZ independent (Hata et al, 2000). The possible interaction of OAZ with bre7 therefore needs further clarification.
Enhancer model for in silico prediction of novel BREs
Based on the phylogenetic conservation of bre7 and SBE motifs in the bambi and smad7 enhancers, we built an enhancer model and predicted a dozen novel BREs in other BMP target genes (Table I). A prominent case are msx genes, which are co-expressed with BMPs in many tissues and are immediate targets of BMP signaling (Hollnagel et al, 1999). msx1 also participates in a positive feedback loop, regulating bmp4 expression during mammalian palatogenesis and tooth development (Zhang et al, 2002). Another important hit were id family genes, which are BMP-responsive (Hollnagel et al, 1999; Locklin et al, 2001; Ota et al, 2002) and crucial for many developmental processes and for the maintenance of ES cell pluripotency. One of the six BREs predicted in id1-4, that upstream of id1, was previously characterized (Korchynskyi and ten Dijke, 2002; Lopez-Rovira et al, 2002). It harbors functional bre7 and SBE motifs, which are essential for BMP inducibility and Smad binding. For the five newly predicted id1-4 enhancers, we experimentally confirmed that they are also functional BREs, proving the usefulness of our bioinformatic search. The results highlight the utility of combining experimentally based enhancer modeling with phylogenetic footprinting to predict in silico novel pathway-specific regulatory elements.
BMP inducibility and synexpression
While there is no simple BMP4 synexpression promoter module shared by all genes, they have common features which are crucial for BMP inducibility and synexpression: (i) one or more functional SBE and/or alternative Smad motifs (bre7), (ii) adjacent functional binding sites for transcription factors that can physically interact with Smad proteins (Vent2, OAZ, AP1), and (iii) a restricted window of BMP inducibility. The member genes of the BMP4 synexpression group are a subset of all BMP target genes. In order to be coexpressed with BMP4, they should have similar thresholds of BMP inducibility. This is important because BMP4 in Xenopus like Dpp in Drosophila acts as a morphogen in a graded fashion, inducing target genes at different signaling thresholds. For example, vent1 is a BMP-responsive gene whose expression requires higher levels of BMP signaling than the expression of vent2 (Dosch et al, 1997). While both share a similar BRE, vent1 is not synexpressed but has a more restricted pattern in the marginal zone of the blastula embryo, as well as in later stages. Conversely, artificial, multimerized BREs, which are highly sensitive to BMP signaling, typically show ectopic sites of transgene expression. Collectively, the results indicate that synexpression promoter modules respond to similar levels of BMP signaling.
In general, we conclude that different BREs that show BMP inducibility in vitro confer different spatiotemporal expression in vivo, highlighting the importance of transgenic analysis for understanding developmental regulation in general and synexpression in particular.
Materials and methods
Embryo manipulation and RT–PCR
In vitro fertilization, embryo culture, staging, microinjection and explantation were carried out as described (Gawantka et al, 1995). Cycloheximide (CHX) treatment was performed from stage 8 onwards with 30 μg/ml until control embryos reached stage 9 or 10. CHX treatment was effective since cell division was arrested and H4 mRNA levels were reduced. RT–PCR assays on animal cap explants were carried out as described previously (Gawantka et al, 1995). Primers for h4, vent1, vent2 and bmp4 were as described (Gawantka et al, 1995; Onichtchouk et al, 1996). Other primers are listed in Supplement 3.
Enhancer cloning
A total of 2 × 106 clones from the Lambda FIX II Xenopus laevis genomic library (Stratagene) were screened with 5′ cDNA probes for bambi and smad7. Several genomic clones were isolated, partially sequenced (GenBank accession numbers: AY499615—bambi, AY499616—smad7) and subcloned in the promoter or enhancer cloning sites of the luciferase reporter vectors pGL2-Basic (Promega), pGL2-Xcol (Harada et al, 1997) and pE1b-luc (Song et al, 1998). Deletion and point mutation constructs were generated by PCR and verified by sequencing. The point-mutations in the bambi and vent2 enhancers are listed in Supplement 3. The 51 bp insert in construct p13 (Figure 3A) was cut from the MCS of the pTimer vector (Clontech) with BglII and BamHI. The six id BREs were cloned from human gDNA by PCR (see Supplementary Figure 1C), sequence verified and subcloned in the enhancer cloning site of pGL2-Xcol.
Xenopus transgenesis and in situ hybridization
Transgenesis was performed using the REMI nuclear transplantation method as described (Kroll and Amaya, 1996; Sive et al, 2000). When cointegration of two or three DNA pieces was performed (Hartley et al, 2001), they were produced with the same NotI overhangs and added in equimolar concentrations to sperm nuclei. A total of 200 ng of the BAC RP11-48B24 (RZPD), containing the human nma gene and linearized with NotI, was used as a transgene in Figure 4Q. The frequency of BAC transgenics was around 6% (9/140 embryos). Xenopus whole-mount in situ hybridization and bleaching of pigment was performed as described (Sive et al, 2000).
Luciferase reporter and EMSA assays
Embryos were injected at the four-cell stage in each blastomere with luciferase reporter constructs (50 pg/blastomere) plus synthetic mRNAs for CABR (300 pg/blastomere), tBR (600 pg/blastomere), Vent2 (1 ng/blastomere) or dominant-negative Vent2 (1 ng/blastomere). CABR is an ALK3 (Q228D) constitutively active variant (Candia et al, 1997); tBR is a kinase domain-deficient dominant-negative BMP receptor (Graff et al, 1994); dnVent2 harbors a point mutation in the homeodomain and may sequester wild-type Vent2 protein (Onichtchouk et al, 1998). For each sample, three pools of three embryos were collected at gastrula stage 11, homogenized in passive lysis buffer (Promega) and assayed for luciferase activity. Data were normalized per embryo. Electrophoretic mobility shift assay (EMSA) and the production of recombinant proteins were carried out as described previously (Henningfeld et al, 2000). All DNA oligonucleotides used as probes for EMSA (shown in Figure 2A) had additional eight-nucleotide PCR primer sequences at the ends: 5′-CCGAGCTC and GTCGACGC-3′.
Bioinformatic tools
Described in Supplementary Figure 1A legend.
Supplementary Material
Supplemental Figure 1
Supplemental Figure 2
Supplemental Table I
Acknowledgments
We are indebted to Enrique Amaya for a practical training on Xenopus transgenesis. We thank Nicolas Pollet, Joachim Niedermeyer and Henner Friedle for helpful discussions and Joan Massagué, Ken Cho, Naoto Ueno, Mitsuyasu Kato and Takeshi Imamura for sharing plasmids. We thank Sandra Kneissel for the Xenopus genomic library, Katrin Arnold for human gDNA and Doris Weber for recombinant proteins.
References
- Benchabane H, Wrana JL (2003) GATA- and Smad1-dependent enhancers in the Smad7 gene differentially interpret bone morphogenetic protein concentrations. Mol Cell Biol 23: 6646–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW (1997) Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development 124: 4467–4480 [DOI] [PubMed] [Google Scholar]
- Casellas R, Hemmati-Brivanlou A (1998) Xenopus Smad7 inhibits both the activin and BMP pathways and acts as a neural inducer. Dev Biol 198: 1–12 [DOI] [PubMed] [Google Scholar]
- Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C (1997) Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development 124: 2325–2334 [DOI] [PubMed] [Google Scholar]
- Gawantka V, Delius H, Hirschfeld K, Blumenstock C, Niehrs C (1995) Antagonizing the Spemann organizer: role of the homeobox gene Xvent-1. EMBO J 14: 6268–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Pfister R, Vingron M, Nitsch R, Blumenstock C, Niehrs C (1998) Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev 77: 95–141 [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS (2000) Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14: 435–451 [PMC free article] [PubMed] [Google Scholar]
- Graff JM, Thies RS, Song JJ, Celeste AJ, Melton DA (1994) Studies with a Xenopus BMP receptor suggest that ventral mesoderm-inducing signals override dorsal signals in vivo. Cell 79: 169–179 [DOI] [PubMed] [Google Scholar]
- Grotewold L, Plum M, Dildrop R, Peters T, Rüther U (2001) Bambi is coexpressed with Bmp-4 during mouse embryogenesis. Mech Dev 100: 327–330 [DOI] [PubMed] [Google Scholar]
- Harada S, Sampath TK, Aubin JE, Rodan GA (1997) Osteogenic protein-1 up-regulation of the collagen X promoter activity is mediated by a MEF-2-like sequence and requires an adjacent AP-1 sequence. Mol Endocrinol 11: 1832–1845 [DOI] [PubMed] [Google Scholar]
- Hartley KO, Hardcastle Z, Friday RV, Amaya E, Papalopulu N (2001) Transgenic Xenopus embryos reveal that anterior neural development requires continued suppression of BMP signaling after gastrulation. Dev Biol 238: 168–184 [DOI] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massagué J (2000) OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100: 229–240 [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P (1997) TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390: 465–471 [DOI] [PubMed] [Google Scholar]
- Henningfeld KA, Friedle H, Rastegar S, Knöchel W (2002) Autoregulation of Xvent-2B; direct interaction and functional cooperation of Xvent-2 and Smad1. J Biol Chem 277: 2097–2103 [DOI] [PubMed] [Google Scholar]
- Henningfeld KA, Rastegar S, Adler G, Knöchel W (2000) Smad1 and Smad4 are components of the BMP-4 induced transcription complex of the Xvent-2B promoter. J Biol Chem 275: 21827–21835 [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 274: 19838–19845 [DOI] [PubMed] [Google Scholar]
- Ishida W, Hamamoto T, Kusanagi K, Yagi K, Kawabata M, Takehara K, Sampath TK, Kato M, Miyazono K (2000) Smad6 is a Smad1/5-induced Smad inhibitor. Characterization of bone morphogenetic protein-responsive element in the mouse Smad6 promoter. J Biol Chem 275: 6075–6079 [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A (1997) Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388: 304–308 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem 277: 4883–4891 [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E (1996) Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122: 3173–3183 [DOI] [PubMed] [Google Scholar]
- Kusanagi K, Inoue H, Ishidou Y, Mishima HK, Kawabata M, Miyazono K (2000) Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol Biol Cell 11: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locklin RM, Riggs BL, Hicok KC, Horton HF, Byrne MC, Khosla S (2001) Assessment of gene regulation by bone morphogenetic protein 2 in human marrow stromal cells using gene array technology. J Bone Miner Res 16: 2192–2204 [DOI] [PubMed] [Google Scholar]
- Lopez-Rovira T, Chalaux E, Massagué J, Rosa JL, Ventura F (2002) Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J Biol Chem 277: 3176–3185 [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K (2002) Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells 7: 1191–1204 [DOI] [PubMed] [Google Scholar]
- Nagarajan RP, Zhang J, Li W, Chen Y (1999) Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J Biol Chem 274: 33412–33418 [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P (1997) Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 389: 631–635 [DOI] [PubMed] [Google Scholar]
- Niehrs C, Meinhardt H (2002) Modular feedback. Nature 417: 35–36 [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N (1999) Synexpression groups in eukaryotes. Nature 402: 483–487 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massagué J, Niehrs C (1999) Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature 401: 480–485 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C (1996) The Xvent-2 homeobox gene is part of the BMP-4 signaling pathway controlling dorsoventral patterning of Xenopus mesoderm. Development 122: 3045–3053 [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Glinka A, Niehrs C (1998) Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development 125: 1447–1456 [DOI] [PubMed] [Google Scholar]
- Ota T, Fujii M, Sugizaki T, Ishii M, Miyazawa K, Aburatani H, Miyazono K (2002) Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-beta in human umbilical vein endothelial cells. J Cell Physiol 193: 299–318 [DOI] [PubMed] [Google Scholar]
- Schuler-Metz A, Knöchel S, Kaufmann E, Knöchel W (2000) The homeodomain transcription factor Xvent-2 mediates autocatalytic regulation of BMP-4 expression in Xenopus embryos. J Biol Chem 275: 34365–34374 [DOI] [PubMed] [Google Scholar]
- Shi Y, Massagué J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP (1998) Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 94: 585–594 [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM (2000) Early Development of Xenopus laevis, A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Song CZ, Siok TE, Gelehrter TD (1998) Smad4/DPC4 and Smad3 mediate transforming growth factor-beta (TGF-beta) signaling through direct binding to a novel TGF-beta-responsive element in the human plasminogen activator inhibitor-1 promoter. J Biol Chem 273: 29287–29290 [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin CH (2000) Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci 25: 64–70 [DOI] [PubMed] [Google Scholar]
- Tsang M, Kim R, de Caestecker MP, Kudoh T, Roberts AB, Dawid IB (2000) Zebrafish nma is involved in TGFbeta family signaling. Genesis 28: 47–57 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y (2002) Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129: 4135–4146 [DOI] [PubMed] [Google Scholar]
- Zwijsen A, van Rooijen MA, Goumans MJ, Dewulf N, Bosman EA, ten Dijke P, Mummery CL, Huylebroeck D (2000) Expression of the inhibitory Smad7 in early mouse development and upregulation during embryonic vasculogenesis. Dev Dyn 218: 663–670 [DOI] [PubMed] [Google Scholar]
- Zwijsen A, Verschueren K, Huylebroeck D (2003) New intracellular components of bone morphogenetic protein/Smad signaling cascades. FEBS Lett 546: 133–139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Supplemental Figure 2
Supplemental Table I


