Abstract
Recent studies, mainly in yeast, have identified various cofactors that associate with the 26S proteasome and appear to influence its function. To identify these proteins in different cells and physiological states, we developed a method to gently and rapidly isolate 26S proteasomes and associated proteins without the need for genetic modifications of the proteasome. This method is based on the affinity of this complex for the ubiquitin-like (UBL) domain of hHR23B and elution with a competing polypeptide containing a ubiquitin-interacting motif. Associated with 26S proteasomes from rat muscle were a variety of known proteasome-interacting proteins, activators, and ubiquitin conjugates. In addition, we identified over 40 proteins not previously known to associate with the 26S proteasome, some of which were tightly associated with the proteasome in a substoichiometric fashion, e.g., the deubiquitinating enzymes USP5/isopeptidase T and USP7/HAUSP and the ubiquitin ligases ARF-BP1/HUWE1 and p600/UBR4. By altering buffer conditions, we also purified by this approach complexes of the ATPase p97/VCP associated with its adaptor proteins Ufd1-Npl4, p47, SAKS1, and FAF1, all of which contain ubiquitin-binding motifs. These complexes were isolated with ubiquitin conjugates bound and were not previously known to bind to the UBL domain of hHR23B. These various UBL-interacting proteins, dubbed the UBL interactome, represent a network of proteins that function together in ubiquitin-dependent proteolysis, and the UBL method offers many advantages for studies of the diversity, functions, and regulation of 26S proteasomes and p97 complexes under different conditions.
The ubiquitin proteasome system catalyzes the bulk of protein degradation in the eukaryotic cell. Most of these proteins are initially linked to a chain of ubiquitin molecules, which targets them for degradation by the 26S proteasome. This 2.4 MDa ATP-dependent proteolytic complex is composed of two smaller particles with distinct functions. The 20S particle is a barrel-shaped hollow structure composed of four stacked rings, each containing seven homologous subunits. They enclose a central chamber, where its six proteolytic active sites are located. The 20S proteasome is flanked by either one or two 19S regulatory particles (PA700), which bind ubiquitinated substrates and disassemble ubiquitin chains. This complex contains six ATPases that unfold protein substrates, open the gated entry channel in the 20S, and thus facilitate translocation of the unfolded substrate into the 20S particle (1, 2). In addition, the 20S proteasome associates with other activating complexes that also open the gate and stimulate peptide entry such as the HEAT-repeat protein PA200 (Blm10 in yeast) or the heptameric adaptor complexes PA28 α, β, and γ (3). These ATP-independent proteasome activators may replace the 19S complex or form hybrid structures (e.g., 19S-20S-PA200). In addition, various proteins have been found to associate with the 19S particle though less tightly than its core subunits. Some of them appear to promote delivery of substrates, catalyze the disassembly of ubiquitin chains, or may even regulate proteasomal activity under specific conditions (4). These various cofactors and activators mean that proteasomes are heterogeneous, dynamic structures, which differ in properties and probably in their specialized functions (5).
The various proteasome-associated proteins have been studied most extensively in yeast. The characterization of subunit heterogeneity and functional plasticity in mammalian tissues faces major technical challenges and has not been systematically studied. Most proteasome-associated proteins are removed by the lengthy multistep chromatographic procedures commonly used to isolate proteasomes. Therefore, several groups have genetically altered proteasomes and added affinity tags to one of the core subunits to allow their one-step isolation from yeast (6–8) and mammalian cell cultures (9–11). Unfortunately, this approach limits the isolation and characterization of proteasomes and their associated proteins to organisms that are accessible to genetic modifications, which is time-consuming or impossible for certain studies. For example, analyses of proteasomes derived from animal models of diseases or human patients are of major interest in conditions where the capacity of cells to degrade proteins is accelerated (e.g., in muscle during atrophy (12)) or is decreased as is believed to occur with aging and in brain during several neurodegenerative diseases (13, 14).
To facilitate such studies and to better understand proteasome function in vivo, we developed a method that allows rapid and gentle isolation of 26S proteasomes from diverse cells without the need for genetic manipulation. This method is based on the affinity of 26S proteasomes for the ubiquitin-like (UBL)1 domain of human Rad23 (15), which binds to the Rpn1 and/or Rpn10 subunit of the 19S particle (16, 17). After binding to the UBL domain, these particles are eluted with an excess of a recombinant His-tagged ubiquitin-interacting motif (UIM)1 derived from human S5a. The UIM domain competes with the 26S proteasome for UBL binding, and the His tag allows its subsequent removal. Using this method, we can rapidly isolate 26S proteasomes with associated proteins from any cell type. To separate 26S proteasomes from other UBL-bound proteins, we used glycerol gradients and native gels. Mass spectrometric analysis of these samples identified 62 proteasome-associated proteins, 43 of which have not previously been shown to interact with the proteasome, including several deubiquitinating enzymes, E3 ligases, and others, whose functional significance on the 26S will be important to understand.
Among the nonproteasomal proteins found to directly associate with the UBL domain were large amounts of the p97/VCP/CDC48 complex. This ubiquitous ATPase complex is a member of the AAA family of hexameric ATPases and is implicated in diverse processes ranging from ER-associated protein degradation (ERAD) to membrane fusion events (18), myofibril biogenesis (19), and chromatin dynamics (20). Mutations in p97 cause inclusion body myopathy associated with Paget’s disease of bone and frontotemporal dementia (IBMPFD), an autosomal adult-onset disease marked by the occurrence of protein inclusions in muscle and brain (21). p97’s N-terminal domain has a weak affinity for ubiquitin (22), but it also binds tightly to the UBD/UBX domain family of adaptor proteins, which appear to mediate the interaction of p97 with its diverse substrates (23). Among these adaptor proteins, six contain ubiquitin-binding motifs, four of which, namely, Ufd1-Npl4, p47, SAKS1, and FAF1, were purified with the UBL method. Interestingly, these complexes were isolated with ubiquitin conjugates bound to them. Also a small fraction of p97 complexes appeared to directly interact with the proteasome. Ufd1-Npl4 together with p97 plays a well-established role in quality control in the secretory pathway, where it removes ubiquitinated proteins from the ER membrane prior to proteasomal degradation. Subsequently, these substrates are delivered to the proteasome by a mechanism, which seems to involve Rad23 and Dsk2 in yeast (24). Our findings suggest that, besides Ufd1-Npl4, also these other p97-associated UBX1 proteins containing ubiquitin-binding domains may play a role in protein degradation.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Reagents were obtained from Sigma if not noted otherwise. Anti-Rpt5, anti-20S, anti-α3, and anti-polyUb conjugates were purchased from Biomol. Anti-USP14, anti-Ecm29, and anti-p97 were purchased from Abcam. Anti-Rpn11 (GenWay), anti-PA200 (Affinity Bio-Reagents), and anti-USP5 (ProteinTech Group, Inc.) were purchased as indicated in parentheses.
Cloning GST-UBL
The UBL domain of hHR23B (aa 1–82) was amplified by PCR from a plasmid encoding full-length hHR23B (pGEX-4T2-HHR23B, obtained from Peter Howley, Harvard Medical School, Boston) and cloned into pDEST15 using the Gateway system (Invitrogen). The plasmid encoding His10-UIM2 (from human S5a) was kindly provided by Kylie Walters (University of Minnesota, Minneapolis).
Expression and Purification of GST-UBL
pDEST15-HHR23BUBL was transformed into BL21AI (Invitrogen) according to the manufacturer’s instructions. Cells were grown at 37 °C to an OD600nm of 0.6 and induced for 3 h with 0.001% l-arabinose. Cell pellets corresponding to 2 L of culture were lysed in 50 mL of PBS using a French press and ultracentrifuged for 1 h at 100000g. The supernatant was loaded onto a 5 mL GSTTrap column (GE Healthcare). After being washed with 10 column volumes of PBS, GST-UBL was eluted with a gradient of 1–20 mM reduced GSH in elution buffer (100 mM Tris, 100 mM NaCl, 1 mM DTT). Fractions containing GST-UBL were combined, dialyzed against the UBL buffer (25 mM Hepes, pH 7.4, 10% glycerol, 5 mM MgCl2, 1 mM DTT), and stored at −20 °C (~4 mg/mL).
Expression and Purification of His10-UIM
His10-UIM2 was expressed as described (25). Cell pellets corresponding to 2 L of culture were lysed with the French press in 50 mL of the binding buffer (25 mM Hepes, pH 7.4, 500 mM NaCl, 1 mM DTT, 0.0025% NP40 substitute, 20 mM imidazole) and centrifuged for 1 h at 100000g. The supernatant was loaded onto a 5 mL HisTrap column (GE Healthcare). After being washed with 10 column volumes of binding buffer, His10-UIM2 was eluted with a gradient of 20–500 mM imidazole. Fractions containing His10-UIM2 were combined, dialyzed against UBL buffer, and stored at −20 °C (~2 mg/mL).
Polyacrylamide Gel Eletrophoresis
Native gels and overlay assay with Suc-LLVY-amc (Bachem) were conducted as described (26). For mass spectrometric analysis, native 3–8% Tris-acetate gels from Invitrogen were used as instructed by the manufacturer. The running buffer was modified to contain 5 mM MgCl2, 0.5 mM ATP, and 0.5 mM DTT. SDS gels were prepared using 4–12% BisTris gels (Invitrogen). Silver stains were performed with the SilverSnap stain kit II (Thermo).
UBL Purification Method
Two grams of rat skeletal muscle were homogenized in 12 mL of purification buffer: 25 mM Hepes, pH 7.4, 10% glycerol, 5 mM MgCl2, 1 mM ATP, and 1 mM DTT in the presence or absence of 150 mM NaCl and/or 25 mM β-glycerophosphate/1 mM Na3VO4. Cell debris was removed by low speed centrifugation (15 min 1500g at 4 °C). The supernatant was ultracentrifuged for 1 h at 100000g to remove the microsomal fraction. The cleared cell lysate was supplemented with 0.1–0.2 mg/mL GST-UBL and 0.25 mL of GSH-Sepharose (GE Healthcare) per milligram of GST-UBL. The suspension nutated for 2 h at 4 °C and then was poured into a 20 mL empty column body (Bio-Rad). The flow through was collected for analysis, and the Sepharose resin was washed with 3 × 25 mL of purification buffer. For elution, the resin was carefully agitated in 1 bed volume of 2 mg/mL His10-UIM in UBL buffer supplemented with 1 mM ATP and incubated for 15 min. The buffer was removed and the elution step repeated. The combined eluates were incubated for 20 min with preequilibrated Ni-NTA1 (100 µL/mg of His10-UIM) (Quiagen). The Ni-NTA was removed by spinning the eluate through a 0.22 µm filter (Millipore). His10-UIM2 and associated proteins were washed in 500 µL of buffer and eluted from the Ni-NTA by agitation of the resin in twice the bed volume of purification buffer supplemented with 500 mM imidazole. After 10 min at 4 °C the Ni-NTA suspension was spun again through the 0.22 µm filter, and the UIM-bound proteins were stored at −20 °C until further analysis. The UBL eluates containing 26S proteasomes and p97 complexes were supplemented with 40% glycerol and stored at −20 °C. The GST-UBL was recovered by eluting the GSH-Sepharose in twice the bed volume of purification buffer supplemented with 20 mM reduced GSH.1
Glycerol Gradients
Glycerol gradients were prepared in the purification buffer containing 25–50% glycerol. UBL-bound proteins (100 µg) were separated on a 36 mL gradient for 22 h at 100000g (4 °C). After centrifugation, 1 mL fractions were removed with a pipet from the top of the gradient. About 60 µL of each fraction was used for ATPase and peptidase activity assays. The remaining 940 µL were subjected to TCA precipitation.
ATPase Activity
ATPase activity was determined by measuring the amount of organic phosphate in a 50 µL gradient fraction after 1 h at 37 °C. Organic phosphate was detected as described elsewhere (27).
Proteasomal Peptidase Activity
Proteasomal peptidase activity was measured using the small fluorogenic peptide Suc-LLVY-amc (Bachem) in the absence or presence of 1 µM proteasome inhibitor bortezomib as described elsewhere (28).
Mass Spectrometry
UBL eluates were digested by adding 10% acetonitrile (ACN) (v/v) and trypsin (Promega) in an enzyme-to-substrate ratio of 1:40 (w/w). TCA precipitates were solubilized in 50 mM NH4HCO3/10% acetonitrile before adding trypsin as described above. Gel bands were digested as described elsewhere (29). The generated peptides were analyzed by microcapillary liquid chromatography mass spectrometry (LC-MS/MS) on a hybrid ion trap/FT-ICR mass spectrometer (LTQ FT, Thermo Electron) essentially as described previously (30). MS/MS data were searched against the IPI rat protein sequence database using the SEQUEST algorithm (31) and applying the target-decoy database search strategy (32). Assignments to unique peptides, defined as peptides with different amino acid sequences, modification states, or charge state, were subjected to strict filtering so that the protein false discovery rate in the reported data sets was below 1%. Each sample was analyzed twice. Only proteins identified with at least two unique peptides in two combined runs were regarded as valid hits and listed in the Supporting Information.
Number of Biological Preparations Analyzed by Mass Spectrometry
The in-gel digest presented in Figure 3C was carried out once. For the UBL interactome as referred to in this study three biological preparations were analyzed under salt-free conditions and their results combined. Proteasomes purified in the presence of 150 mM NaCl were analyzed once. For analysis of TCA-precipitated fractions from glycerol gradient as presented in Figure 6B three fractions were analyzed containing p97 complexes and four fractions containing 26S proteasomes. The bands from the native gel presented in Figure 6A were analyzed once.
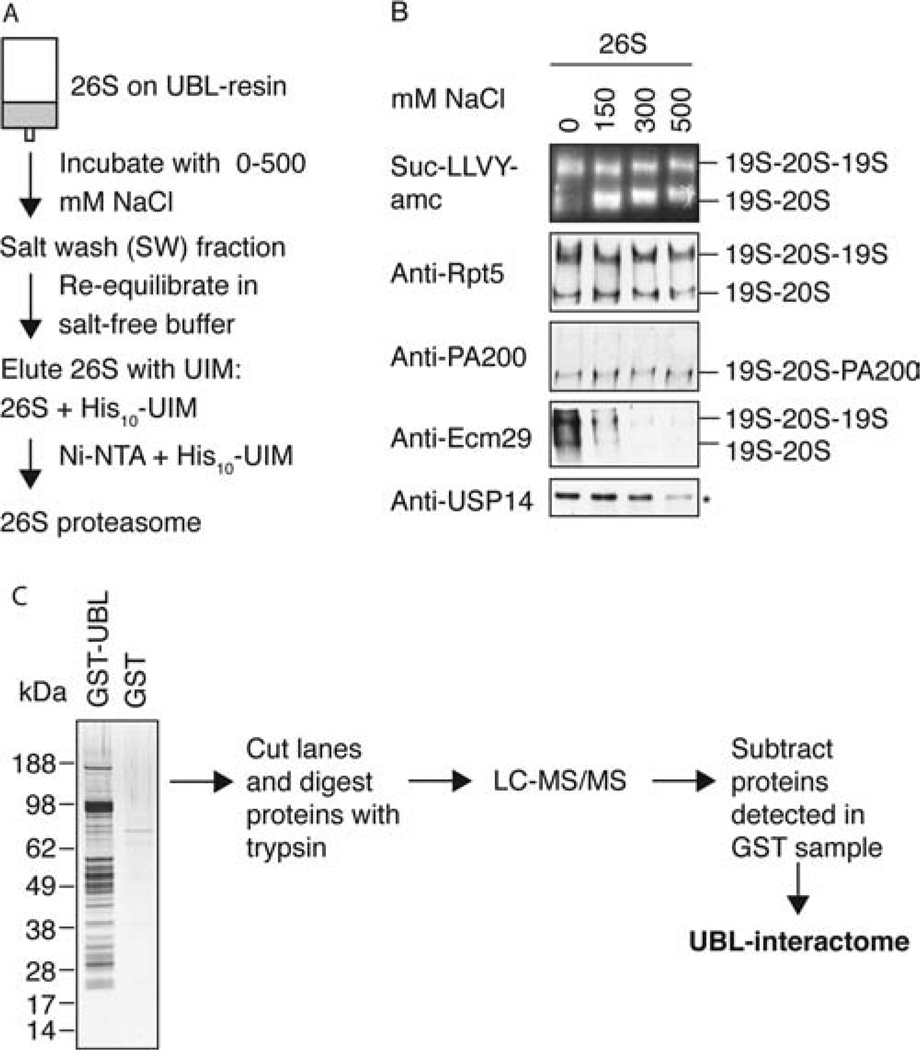
Figure 3.
The 26S proteasome is purified together with known proteasome-associated proteins. (A) 26S proteasomes were purified from rat skeletal muscle as described in Figure 2A but in the absence of salt. Before elution, the UBL-bound proteins were incubated in 500 µL of buffer with different salt concentrations (0, 150, 300, 500 mM NaCl). The salt extract (SW) was collected, and after reequilibration in salt-free buffer, the 26S proteasomes were eluted in the 500 µL buffer containing His10-UIM. (B) Equal volumes of 26S eluates were loaded onto 3.5% native gels and analyzed by LLVY-overlay assay and Western blot as indicated. The Western blot with anti-USP14 was performed on a SDS gel (*). Doubly capped, singly capped, and PA200 hybrid proteasomes were identified as indicated. No 19S nor free 20S was detected (data not shown). (C) Mass spectrometric analysis of proteins isolated with the GST-UBL in the absence of NaCl. For a complete list of all mass spectrometric data, see Supporting Information Table 2.
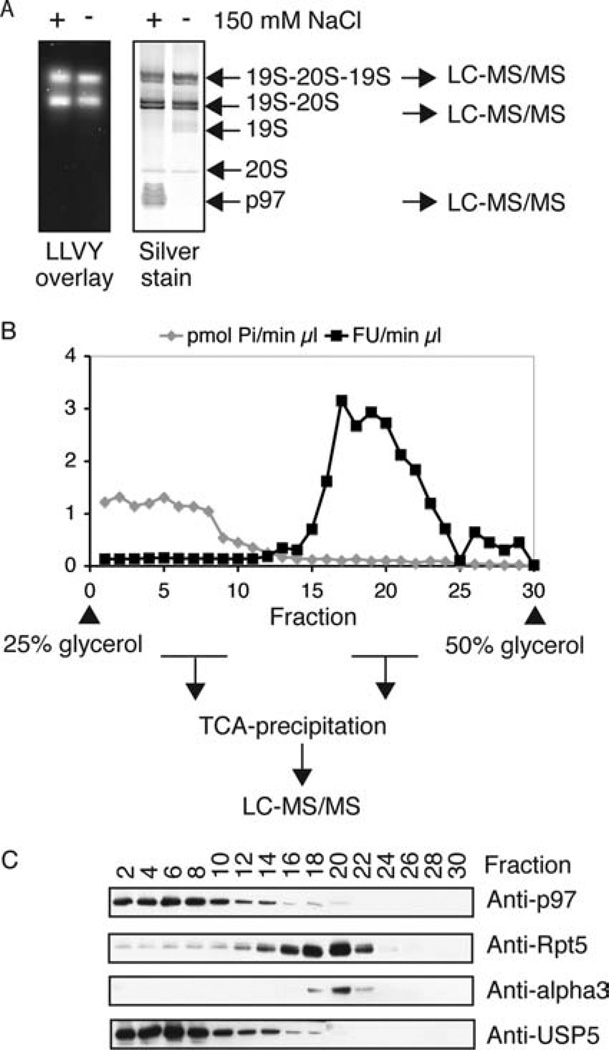
Figure 6.
Identification of proteins associated with the 26S proteasome. (A) UBL purifications from rat muscle were carried out in the presence or absence of 150 mM NaCl. Samples containing equal amounts of proteasomal peptidase activity were separated on native gels. Proteasomes were detected by Suc-LLVY-amc overlay assay (left panel). The right panel presents a silver stain of the respective gel. The silver-stained bands of the indicated complexes were analyzed by LC-MS/MS. (B) About 100 µg of protein after UBL purification were separated on a 25–50% glycerol gradient by ultracentrifugation (22 h, 100000g). Individual fractions were analyzed for proteasomal peptidase activity (Suc-LLVY-amc) and ATPase activity (Malachite green assay). Proteins in the fractions were TCA-precipitated and subjected to mass spectrometric analysis or (C) Western blot. For a complete list of all mass spectrometric data, see Supporting Information Table 2.
RESULTS
The UBL-Affinity Isolation of Proteasomes
The ubiquitin-like domain of Rad23 binds to the Rpn1 and/or Rpn10 subunit of the 26S proteasome (15–17). We decided to utilize this interaction to establish a novel method for purification of proteasomes from mammalian cells that unlike prior approaches would not require genetic manipulation to affinity tag these particles (Figure 1). We cloned the UBL domain of human Rad23 (hHR23B) as a recombinant GST-fusion protein and expressed it in Escherichia coli. Cell extracts were derived from rat skeletal muscle and cleared from microsomes by ultracentrifugation. Incubation of the purified recombinant GST-UBL with these extracts and subsequent isolation of the GST-UBL with GSH-Sepharose led to copurification of 26S proteasomes (Figure 2A), which were identified by measuring their peptidase activity using Suc-LLVY-amc (Supporting Information Table 1). No such activity was detected in similar experiments with GST alone (Figure 2B). In initial experiments, we compared the capacity of UBL domains from several proteins, including those from hHR23A and USP14, to bind proteasomes (data not shown). Because the UBL domain of hHR23B proved most efficient, it was used in subsequent studies.
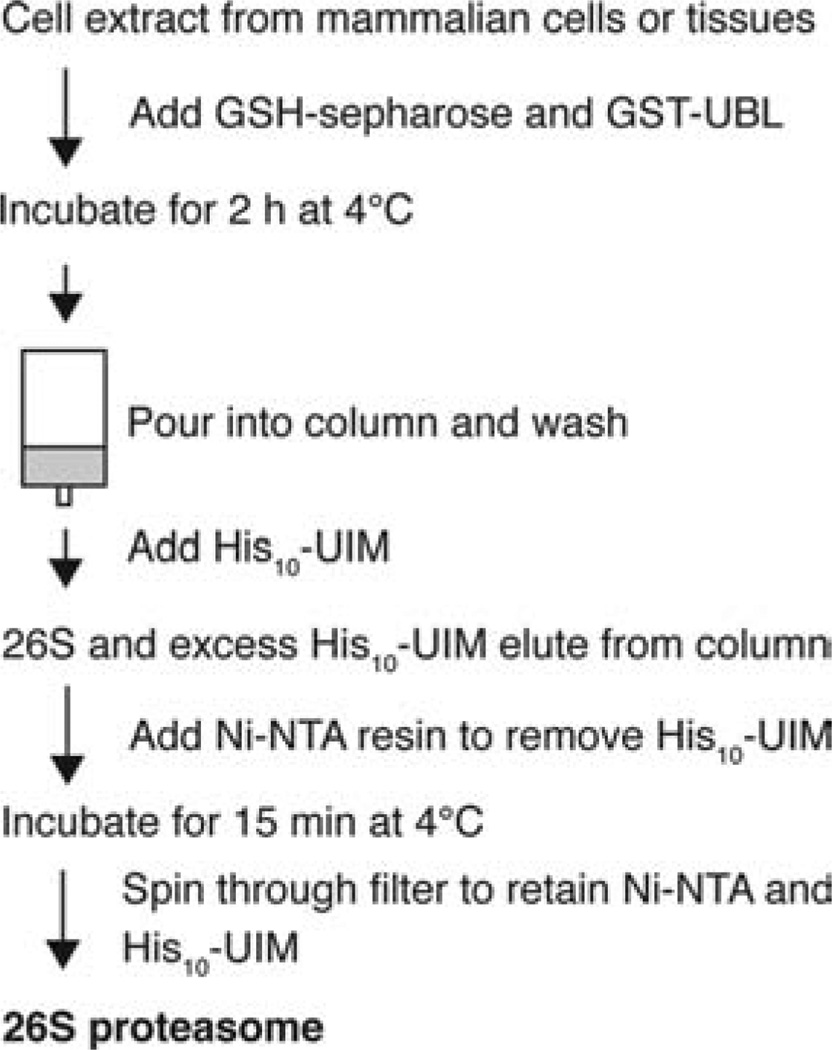
Figure 1.
Flow chart of affinity method to isolate 26S proteasomes from mammalian cells and tissues. GST-UBL, recombinant fusion protein of glutathione S-transferase (GST) and the ubiquitin-like domain (UBL) of human HR23B; His10-UIM, recombinant protein containing the second ubiquitin-interacting motif of human S5a; GSH-Sepharose, glutathione-coupled resin with affinity for GST; Ni-NTA, nickel-charged nitrilotriacetic acid (NTA) agarose that binds to His tags.
Figure 2.
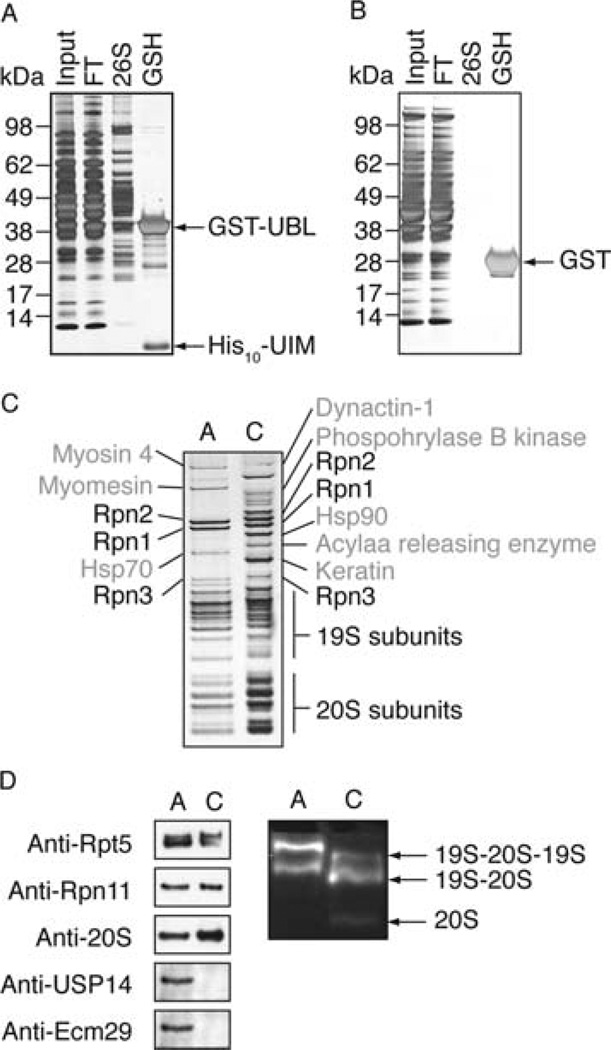
The UBL-affinity purification yields pure 26S proteasomes. (A) Muscle extract (2 g of rat skeletal muscle/10 mL of buffer) was cleared by 1 h 100000g ultracentrifugation (input) and incubated with 1 mg of GST-Ubl and 250 µL of GSH-Sepharose for 2 h. The suspension was poured into an empty column, and the flow through (FT) was collected. After washing the proteasome was eluted with 500 µL of buffer containing 2 mg/mL His10-UIM. The remaining GST-UBL was eluted with 500 µL of buffer plus 20 mM GSH (GSH). Ten micrograms of total protein input and FT, 1 µg of 26S proteasome, and 5 µg of GST-UBL from the GSH fraction were separated by SDS–PAGE and silver stained. (B) As in (A) with GST instead of GST-UBL. (C) UBL-affinity-purified 26S proteasome (A) and 26S proteasome conventionally purified using multistep chromatography (C) from rabbit muscle ((34)) were separated by SDS–PAGE (1.5 µg each) and silver stained. Individual bands were cut and identified by mass spectrometry as indicated. (D) Conventionally and affinity-purified proteasomes were compared by Western blot and native PAGE (4%). Proteasome activity was detected by cleavage of the small fluorogenic peptide Suc-LLVY-amc.
The second UIM domain of human S5a binds the UBL domain of hHR23B with high affinity (33). Therefore, we expressed and purified His10-UIM2 of human S5a from E. coli. A 5-fold molar excess of His10-UIM over GST-UBL was found to efficiently elute the UBL-bound 26S proteasomes from the resin without releasing the GST-UBL. Subsequently, the excess His10-UIM was removed by incubation with Ni-NTA resin, and in the released 26S proteasomes, there was no contaminating trace of GST-UBL or His10-UIM (Supporting Information Figure 1). The UBL-affinity-purified 26S proteasomes showed high ATP-stimulated activity against the small fluorgenic peptide Suc-LLVY-amc (Supporting Information Table 1) and ubiquitinated proteins (data not shown). Because of its rapidity, this approach is now routinely used in our laboratory in place of conventional chromatographic methods used previously that require several days to complete.
Purity of Isolated 26S Proteasomes
In order to analyze the purity of these preparations, we isolated 26S proteasomes from rabbit skeletal muscle by the UBL approach and compared equal amounts with conventionally purified 26S preparations from this tissue (34) by SDS–PAGE. The UBL-affinity-purified 26S proteasomes displayed the characteristic band patterns of 19S and 20S subunits (Figure 2C). They typically contained less 20S particles than conventionally purified proteasomes and a higher content of doubly capped proteasomes (19S-20S-19S). Also some free 20S particles were generally copurified with conventionally purified 26S proteasomes (Figure 2D). Since the UBL domain binds to the 19S particle (16, 17), this method specifically yields doubly and singly capped proteasomes. Mass spectrometric analysis of the individual bands also revealed that the UBL-affinity-purified 26S contained fewer contaminating bands than conventionally purified 26S (Figure 2C).
As described in the Expeerimental Procedures, the ratios of GST-UBL, GSH-Sepharose, and muscle cell lysate were optimized to isolate routinely about ~50 µg of 26S proteasomes from 2 g of rat skeletal muscle. In a typical purification about 80% of the proteasome activity in the cell extract bound to the UBL column (Supporting Information Table 1). When the flow through was subsequently supplemented with fresh GST-UBL and GSH-Sepharose, 26S proteasomes were depleted from the cell extract, indicating that the method could isolate all 26S proteasomes and not just a subfraction (data not shown). Usually we worked with an excess of extract over the column’s capacity to ensure saturation of binding sites. However, depletion of 26S proteasomes from cell extracts is possible by this approach and could be useful for certain types of experiments.
The 26S Preparation Contains Known Proteasome-Associated Proteins
Most of the proteins reported to be associated with the 26S proteasome in yeast and mammals are removed by the multistep purification methods that involve high salt concentrations. To test if the UBL method copurified several reported 26S-associated proteins, we isolated proteasomes from rat skeletal muscle in buffer lacking salt to preserve as many associated proteins as possible and then washed the proteasomes on the UBL column with different concentrations of NaCl. After reequilibration in salt-free buffer, the proteasomes were then eluted with the His10-UIM and analyzed by native gel electrophoresis (Figure 3A). The bands corresponding to doubly and singly capped proteasomes were detected by their activity toward Suc-LLVY-amc and antibodies against Rpt5 (an ATPase subunit in the 19S). These bands did not significantly vary after exposure to different salt concentrations. Thus, the structural integrity of the 26S proteasome was preserved during this experiment (Figure 3B). We then analyzed the abundance of three known proteasome-associated proteins by Western blot analysis of native gels.
PA200 (the homologue of yeast Blm10) is a proteasomal activator that interacts with the 20S particle and stimulates its peptidase activity (35, 36). PA200 was present within hybrid complexes (19S-20S-PA200), and its association with the 20S was not affected by this brief exposure to NaCl.
Ecm29 is a proteasome-associated protein that has been proposed to confer stability to the 19S-20S interaction in yeast (37) and to localize proteasomes to membranes in mammals (38). Unlike PA200, it was removed almost completely by 150 mM NaCl, as reported previously in yeast (6).
The deubiquitinating enzyme, USP14 (the homologue of yeast Ubp6), which reversibly associates with the proteasome via its N-terminal UBL domain in mammals and yeast (6, 39), was extracted from the proteasome with higher salt concentrations than was Ecm29 (Figure 3B).
Neither Ecm29 nor USP14 was detected in conventionally purified 26S from rabbit muscle (Figure 2D). Thus, the preparation obtained with the UBL method appears to resemble more closely 26S particles as found in vivo than conventionally purified ones.
The p97/VCP Complex and Other Proteins That Directly Bind to the UBL Domain
To identify any novel proteasome-associated proteins, we used the UBL method to purify 26S from muscle using a salt-free buffer, consisting of 25 mM Hepes, pH 7.4, 10% glycerol, 5 mM MgCl2, 1 mM ATP, and 1 mM DTT. Proteins isolated with GST-UBL (or GST) were separated on a silver-stained SDS gel, and the whole lane was subjected to mass spectrometry (Figure 3C). This analysis identified all standard subunits of the 26S proteasome along with several proteins previously shown to associate with the proteasome (Supporting Information Table 2). In addition, we identified a variety of proteins not known to be associated with the proteasome, including several deubiquitinating enzymes (DUBs) and E3 ligases (see below).
Most strikingly, we found large amounts of the p97/VCP ATPase complex in our preparations. p97/VCP is a homohexameric complex, whose specific functions depend on its association with adaptor proteins that contain a UBD/UBX domain (23). Four of these adaptors were detected in our purifications, namely, p47, FAF-1, SAKS1, and the heterodimer UFD1-NPL4, which together with p97 has been implicated in degradation of misfolded proteins in the ER, ubiquitin–protein fusions by the UFD pathway (23), and heat and oxidative damaged proteins (40). Although the p97 complex is not known to associate with the UBL domain of hHR23B, these adaptors contain ubiquitin-binding domains such as UBA domains in p47, FAF-1, and SAKS1 and a ubiquitin-binding zinc finger in Npl4 (23) which presumably account for the binding of these complexes to the column.
Salt Prevents Association of p97 Complexes with the UBL Domain but Not of Proteasomes
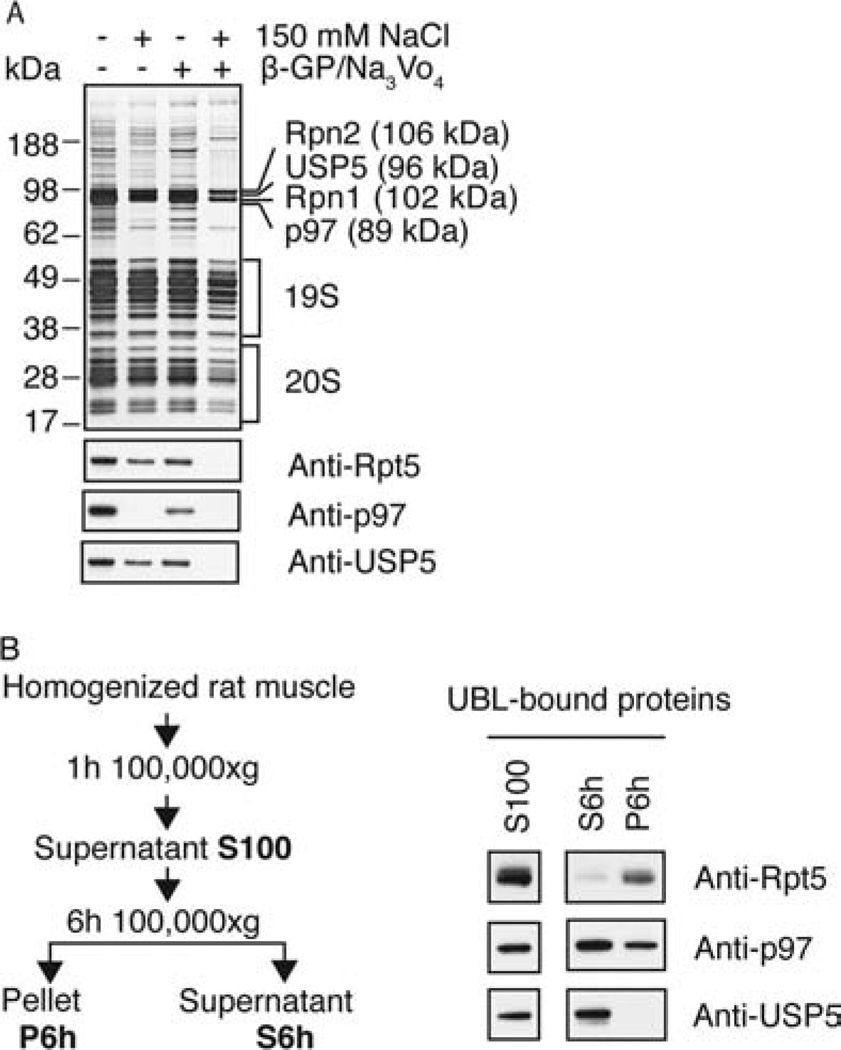
These findings contrasted with our initial studies, where we used buffers containing 150 mM NaCl (together with β-glycerophosphate and sodium orthovanadate) and obtained only pure 26S proteasomes (Figure 2C). The presence of p97 and adaptors in the preparations obtained in the absence of NaCl suggested that the lower ionic strength of the buffer had led to binding to the UBL domain of these additive proteins. We therefore compared the influence of 150 mM NaCl on the purity of the isolated 26S proteasomes. This analysis revealed that, in addition to the 26S proteasome, the deubiquitinating enzyme USP5/isopeptidase T (homologue of Ubp14 in yeast) and the p97 ATPase constituted the most abundant proteins in the preparations lacking salt, as shown by SDS gel analysis (Figure 4A) and native gel (Figure 6A). The presence of 150 mM salt in the purification buffer removed p97 from the preparation and reduced the content of USP5. The addition of β-glycerophosphate and sodium orthovanadate with 150 mM NaCl completely also removed USP5 (Figure 4A), in accordance with our initial results (Figure 2C).
Figure 4.
In the absence of salt, USP5 and p97 complexes are purified in large amounts together with the 26S and bind to the UBL domain directly. (A) UBL-affinity preparations were carried out in the presence or absence of 150 mM NaCl and 25 mM β-glycerophosphate (β-GP)/1 mM Na3VO4. Samples containing equal amounts of proteasomal peptidase activity were analyzed by SDS–PAGE (upper panel) and Western blot (lower panels). The single bands corresponding to Rpn1, Rpn2, USP5, and p97 were identified by mass spectrometry. (B) A crude cell extract derived from 2 g of rat skeletal muscle was centrifuged for 1 h at 100000g. The pellet was discarded and the supernatant (S100) ultracentrifuged for another 6 h at 100000g. S100, S6h, and P6h were each subjected to UBL-affinity purifications and analyzed by Western blot with the indicated antibodies.
p97 Complexes and USP5 Bind to the UBL Domain in the Absence of Proteasomes
To test if p97-containing complexes and USP5 bound to the UBL domain independently from the proteasome, we depleted muscle cell extracts of proteasomes by ultracentrifugation for 6 h at 100000g, as measured by assay of peptidase activity (data not shown), and analyzed what components in the supernatant and resuspended pellet could bind to the UBL domain (Figure 4B). p97 and USP5 were purified by this approach from the proteasome-depleted supernatant. Thus, they bound to the UBL resin independently of the proteasome, although more weakly than the 26S, since their binding was reduced or abolished in presence of NaCl.
The UBL domain of hHR23B is known to interact with ubiquitin-binding motifs, e.g., the UIM domain of S5a utilized in our purification (17). Recently, a heterodimeric complex of hHR23A with hPLIC2 formed by reciprocal UBL–UBA interactions has been observed (41), and Rad23 protects itself from rapid proteasomal degradation by intramolecular interaction of the N-terminal UBL domain with its C-terminal UBA domain (42). USP5 contains two UBA domains that could mediate the interaction with the GST-UBL moiety. p97 by itself has very weak affinity for monoubiquitin (22), and no direct interaction with a ubiquitin-like domain has been reported. Thus, it seems likely that p97 binds to the UBL domain via the ubiquitin-binding domains of its adaptor proteins.
UBL Interactome
To analyze further the proteins isolated with the UBL domain, we performed mass spectrometric analysis of the proteins purified from rat muscle in the presence or absence of salt. The samples were analyzed by LC-MS/MS in a gel-free manner, which increased the sensitivity compared to our previous data set from gel slices (Supporting Information Table 2). In total, we identified more than 100 proteins in the salt-free sample, 38 of which corresponded to proteasome subunits. Twelve known proteasome-associated proteins were detected, among them the deubiquitinating enzymes (DUBs), Uch-L5/Uch37 (43) and USP14/Ubp6 (39), and the HECT ubiquitin ligase KIAA0010/E3C/Hul5 (6), all of which have a defined binding site at the 26S, as well as E6-AP/E3A, another HECT ubiquitin ligase recently reported on the 26S isolated from 293 cells (10). The other 57 proteins identified included more DUBs and E3s, p97 and adaptors, the signalosome (a regulator of SCF ligases (44)) and several other proteins. Whether any of these proteins associated with the proteasome or the UBL domain will be discussed below. Neither p97 nor its adaptors were detected in the presence of 150 mM NaCl with the exception of FAF-1. We termed the sum of proteins interacting with the UBL domain of Rad23 the “UBL interactome”.
Coisolation of Ubiquitin Conjugates with 26S and p97 Complexes
A primary function of the proteasome is to bind ubiquitin-conjugated proteins. To determine whether ubiquitin conjugates were isolated with the 26S and on p97 complexes, the particles were bound to the UBL matrix in the absence of NaCl and washed with different salt concentrations (see Figure 3A). Western blot analysis with antipoly ubiquitin antibody revealed that ubiquitin conjugates were washed off the UBL-bound complexes with increasing salt concentration (salt wash fractions (SW), Figure 5A). Surprisingly, no ubiquitin conjugates were detected on the UBL-bound proteins that had not been exposed to NaCl (Figure 5A). This apparent contradiction was resolved when the proteins isolated with the His10-UIM (in the second step) were analyzed (Figure 5B). The UIM2 domain of S5a (besides binding the UBL domain of hHR23B) is a ubiquitin interacting motif (25). To test if this step removed ubiquitin conjugates, we released the His10-UIM with imidazole from the Ni-NTA resin and by Western blot compared the ubiquitin conjugate levels in the UBL eluate with the respective imidazole-extracted UIM fraction. Indeed, the UIM partially removed ubiquitin conjugates from the UBL-bound proteins during the last step of the purification (Figure 5B).
Figure 5.
Ubiquitin conjugates copurify with 26S and p97 complexes and are partially removed with the His10-UIM in the last purification step. (A) Proteins were isolated with the UBL domain from rat muscle as illustrated in Figure 3A. The same amount of UBL and salt wash (SW) fractions were separated by SDS–PAGE and analyzed by Western blot with anti-ubiquitin conjugate antibody. (B) To retrieve ubiquitin conjugates, His10-UIM was eluted from the Ni-NTA in binding buffer supplemented with 500 mM imidazole as illustrated by the scheme. One microgram of UBL-bound proteins from three different purifications and 1/10th of the corresponding UIM eluate were separated on SDS–PAGE and analyzed by Western blot with an anti-ubiquitin conjugate antibody. (C) UBL purifications from rat muscle were carried out in the presence or absence of 150 mM NaCl. Equal amounts of UBL- and UIM-bound proteins were analyzed by Western blot as indicated.
We then tested whether both p97 and 26S complexes were purified together with associated ubiquitin conjugates and analyzed the UBL- and UIM-bound proteins purified in the presence and absence of 150 mM salt. Western blot analysis with an anti-ubiquitin conjugate antibody revealed that the presence of p97 correlated with a higher amount of ubiquitin conjugates in the UBL- and UIM-bound fractions, and some p97 was removed together with the conjugates by the His10-UIM (Figure 5C). Since 150 mM NaCl did not seem to remove ubiquitin conjugates from the UBL-bound proteins in our earlier studies (Figure 5A), we concluded that the higher amount of conjugates detected in the absence of salt was due to the presence of p97 and its ubiquitin-binding adaptors. Thus this approach should also be useful for the characterization of the ubiquitin conjugates associated with p97 and with 26S complexes under different conditions.
Novel Proteasome-Associated Proteins
In order to identify the proteasome-associated proteins and to make sure, that these proteins did not bind directly to the UBL domain, we used two approaches.
Purification by the UBL method in the absence of NaCl was performed, and then the eluted proteasomes and p97 complexes were separated by glycerol gradient centrifugation. Fractions containing the p97 complexes and the 26S proteasomes were precipitated with TCA and analyzed by mass spectrometry (Figure 6B and Supporting Information Table 2).
Proteins purified with the UBL method in the presence or absence of 150 mM NaCl were separated on native gels (Figure 6A). The bands corresponding to the singly and doubly capped 26S proteasomes and p97-complexes were also analyzed by mass spectrometry (Supporting Information Table 2).
Table 1 presents a summary of all proteins that were proteasome-associated and the approaches used to support this conclusion.
Table 1.
Proteasome-Associated Proteinsa
| unigene | protein | peptides | methods |
|---|---|---|---|
| Proteasome Activators | |||
| Rn.163607 | PA200 | 37 | GG |
| Rn.2742 | PA28α | 14 | GG |
| Rn.198325 | PA28γ | 3 | GG |
| Proteasome-Associated Proteins | |||
| Rn.103325 | PI31 | 14 | GG |
| Rn.154631 | Ecm29 | 164 | GG |
| Rn.9320 | Rpn13 | 10 | GG |
| Rn.40424 | Uch37 | 51 | NP/GG |
| Rn.11790 | Usp14 | 32 | NP/GG |
| Rn.198497 | E6-AP/E3A | 20 | GG |
| UBL-UBA Proteins | |||
| Rn.107103 | Sequestosome/p62 | 6 | GG |
| E3 Ligases | |||
| Rn.37755 | p600/UBR4 | 159 | NP/GG |
| Rn.12130 | HUWE1/ARF-BP | 116 | NP/GG |
| IPI00196914 | HECT E3b | 7 | GG |
| p97 and N-Terminal Adaptors | |||
| Rn.98891 | p97 | 83 | GG |
| Rn.144645 | NPL4 | 6 | GG |
| Rn.11946 | UFD1 | 8 | NP/GG |
| Rn.2771 | p47 | 14 | GG |
| Rn.198539 | SAKS1 | 6 | NP/GG |
| Deubiquitinating Enzymes | |||
| Rn.44078 | USP5 | 138 | NP/GG |
| Rn.72721 | USP7 | 17 | GG |
| Ubiquitin | |||
| Rn.106034 | ubiquitin | 18 | NP/GG |
| Transcription | |||
| Rn.33229 | STAT1α | 76 | GG |
| Translation | |||
| Rn.144660 | |||
| Rn.965 | EF1α | 7 | NP/GG |
| Rn.55145 | EF2 | 3 | NP |
| Chaperones | |||
| IPI00358052 | putative, Hsp40/DnaJ familyc | 5 | GG |
| Rn.120392 | |||
| Rn.201298 | Hspa8, Hsp70 family | 5 | GG |
| Rn.11088 | Hspa5, Hsp70 family | 2 | NP |
| Rn. 1950 | Hspa1, Hsp70 family | 2 | GG |
| Cytoskelton and Myofibrillar Proteins | |||
| Rn.82732 | actin, alpha 1 | 59 | NP/GG |
| Rn.94978 | actin, beta | 5 | GG |
| Rn.101923 | dynactin 2 | 4 | NP |
| Rn.84920 | myosin, light chain 1 | 17 | GG |
| Rn.87540 | tropomyosin 1, alpha | 7 | GG |
| Rn.17580 | tropomyosin 2 | 9 | GG |
| Rn.43529 | troponin C2 | 2 | GG |
| Rn.15488 | troponin T3 | 10 | GG |
| Rn.2710 | vimentin | 2 | GG |
| Metabolism | |||
| Rn.129558 | glyceraldehyde 3-phosphate dehydrogenase | 13 | NP/GG |
| Rn.122663 | aldolase A-like | 8 | GG |
| Rn.1774 | aldolase | 7 | GG |
| Rn.1556 | pyruvate kinase | 4 | NP |
| IPI00388191 | enolasec | 2 | NP |
| Rn.144555 | |||
| Rn.37838 | triosephosphate isomerase | 2 | GG |
| Rn.10756 | creatine kinase | 2 | GG |
| Rn.3001 | catalase | 8 | NP |
| Rn.9857 | arginase 1 | 4 | NP |
| Rn.10039 | transglutaminase 1 | 2 | NP |
| Rn.53317 | transglutaminase 3 | 2 | NP |
| Rn.9486 | fatty acid synthase | 2 | NP |
| Miscellaneous | |||
| Rn.22682 | similar to thioredoxin family Trp26 | 27 | GG |
| Rn.1792 | annexin A1 | 8 | NP |
| Rn.90546 | annexin A2 | 16 | NP/GG |
| Rn.141102 | premature ovarian failure 1B | 5 | NP/GG |
| Rn.203179 | glioblastoma amplified sequence | 7 | GG |
| Rn.92965 | ATP synthase | 2 | NP |
| Rn.218029 | ATPase, Ca2+ transporting | 2 | GG |
| Rn.91512 | glial fibrillary acidic protein | 6 | NP/GG |
| IPI00231340 | histone foldc | 3 | NP |
| Rn.11207 | thrombospondin 4 | 2 | NP |
| Rn.2845 | peroxiredoxin 1 | 2 | NP |
| Rn.2511 | peroxiredoxin 2 | 2 | NP |
| Rn.68633 | tubulin tyrosine ligase-like family | 2 | NP |
| Rn.24735 | nuclear RNA export factor 7 | 2 | NP |
| Rn.172813 | Mab-21-like 2 | 2 | GG |
| Rn.161841 | |||
| Rn.2202 | cytochrome c | 2 | GG |
| IPI00367223 | similar to CG1193-PAc | 2 | GG |
Sum of unique peptides identified by indicated methods: NP (proteins identified in bands of doubly and singly capped proteasomes after native PAGE); GG (proteins identified in 26S peak of glycerol gradient). Databases: IPI database (http://www.ebi.ac.uk/IPI/IPIhelp.html); UniGene (http://www.ncbi.nlm.nih.gov/UniGene). Although cytoskeleton proteins are often found as contaminants or unspecifically interacting proteins in the study of protein complexes using mass spectrometry, identified candidates from this protein family are listed as hypothetical proteasome interacting proteins.
Not annotated in UniGene, termed HUWE in IPI database, but has no sequence similarity with HUWE1/Rn.12130.
Not annotated in UniGene.
In total, we detected 62 proteasome-associated proteins, 19 of which have previously been reported, namely, the DUBs Uch37 and USP14, the HEAT-repeat protein Ecm29, the proteasomal activator PA200, γ-interferon-induced activators PA28α and PA28γ, the proteasome inhibitor PI31, the p62/sequestosome, which has been reported to play a role in autophagy and ubiquitin-mediated degradation, the translation elongation factor EF1α, which was proposed to mediate degradation of cotranslationally damaged proteins (for references see ref 3), and the E3 ligase E6-AP (10), as well as a member of the thioredoxin family TRP26 (11). The translation elongation factor EF2, proteins of the Hsp70 family, and four enzymes of the glycolytic pathway, fructose-1,6-bisphosphate (FBP) aldolase, glyceraldehyde phosphate (GAP) dehydroxygenase (DH), pyruvate kinase, and enolase, have previously been reported on the yeast proteasome by a cross-linking approach (8).
Other proteins that were detected with the 26S proteasome after native gel electrophoresis or glycerol gradient centrifugation have not been reported before to interact with the 26S (Table 1). Some of them like cytoskeletal and myofibrillar components are very abundant proteins in the cell and possibly present contaminants or residual ubiquitinated substrates not removed by the UIM-elution step. Of greater interest are the deubiquitinating enzyme USP7/HAUSP, the ubiquitin ligases p600/UBR4 and Mule/ARF-BP, and the transcription factor STAT1α, which were present in significant (but substoichiometric) amounts relative to standard subunits of the 26S proteasome. To assess stoichiometry, the proteins eluted from the UBL column were separated on SDS gel and silver stained. Single bands were cut out of the gel and analyzed by mass spectrometry. However, many other of the novel proteasome-associated proteins overlap in size with proteasome subunits; thus their stoichiometric ratios could not be determined by this method (data not shown). Many of the proteasome-associated proteins in muscle were also identified in proteasomes isolated from rat liver and Hela cells and thus are not tissue-specific (data not shown). Their potential function at the proteasome will be discussed below.
Some USP5 and p97 Complexes Transiently Interact with the Proteasome
Although the isolation of p97 complexes and USP5 by this affinity approach was primarily due to direct binding to the UBL domain, our mass spectrometric analysis after glycerol gradient centrifugation and native PAGE revealed that some USP5 and p97–adaptor complexes were colocalized with the proteasome (Table 1). The p97 adaptors UFD1 and SAKS1 were detected with 26S proteasomes in native gels, and p97 as well as Ufd1-Npl4, p47, SAKS1, and FAF1 were present in the 26S fraction after glycerol gradient separation (Table 1). We confirmed this result by Western blot analysis of the respective TCA-precipitated fractions and indeed found traces of p97 and USP5 in fractions that contained 26S proteasomes (Figure 6C). We conclude that USP5, p97 complexes, and 26S proteasomes exist largely as separate entities and were isolated together due to their direct interactions with the UBL domain of hHR23B, but a fraction seemed to interact transiently with the 26S proteasome.
DISCUSSION
UBL-Affinity Purification and Its Applications
The method described here makes it possible to characterize the activity and composition of 26S proteasomes from different tissues or cells and different cellular compartments. In related studies, we have used this method successfully on muscle, brain, spleen, liver, Hela cells, Xenopus oocytes, and even yeast.
It has generally been assumed that the conjugation of ubiquitin is rate limiting in proteasome-mediated degradation, but methods have not been available to determine whether changes in degradation rates in normal or pathological states may also be due to changes in proteasomal activity. Unlike approaches involving genetic modification of proteasome subunits with affinity tags (6–8), the UBL method avoids the assumption that such tags do not alter 26S function. It allows the analysis of 26S proteasomes from tissues in which the capacity of the UPS may be altered (e.g., during drug treatments, aging, neurodegeneration, or muscle atrophy) and organisms where genetic manipulations are not feasible (e.g., human tissues).
Recently, after these studies were completed, an analogous method for isolation of 26S proteasomes was reported that used the UBL domain from hHR23A (45). However, that article did not describe the isolation of p97/adaptor complexes or ubiquitin conjugates and reported the isolation of a DNA-repair complex that was not isolated by our approach.
By combining the UBL method with additional isolation steps for 26S proteasomes, we present the first comprehensive study of proteasome-associated proteins from a differentiated cell (skeletal muscle). We identified 62 proteins, 43 of which have not been reported to interact with it previously. Surprisingly, the proteasome-associated E3 ligase Hul5/E3C (6, 10) and the proteasome subunit gankyrin (Nas6 in yeast) (46), though readily detectable in UBL eluates, were not detected on the 26S proteasomes after native gel electrophoresis or glycerol gradient centrifugation. Thus, there probably are even more proteasome-associated proteins in the UBL interactome, but their association with the proteasome is too weak to survive this additional separation step or may require specific conditions or cofactors. Also, some of the 26S proteasomes seemed to fall apart during the lengthy glycerol gradient ultracentrifugation, since some 19S and 20S subunits were detected by mass spectrometry in p97-containing fractions that showed no proteasome activity. Further investigations of proteasome-associated proteins using cross-linking reagents might reveal even more proteasome-as well as p97-associated proteins.
The Proteasome-Associated DUBS Include USP5/Isopeptidase T and USP7/HAUSP
One unexpected DUB, USP5/isopeptidase T, was isolated from muscle extracts by its association with the UBL domain of Rad23. This isopeptidase in vivo appears to disassemble free ubiquitin chains released by the proteasome (47). An interaction of USP5 with Rad23 or another UBL-containing protein has not been described before. In addition, we found USP5 associated with the proteasome. These observations suggest at least two independent functions of USP5, one on the proteasome and another through a complex with Rad23 or a similar UBL protein. In vitro, USP5 disassembles K48-linked unanchored ubiquitin chains (48, 49). Deletion of Ubp14 (the homologue of USP5 in yeast) in vivo leads to accumulation of unanchored ubiquitin chains and inhibition of the ubiquitin-dependent proteolysis (47), probably because the free ubiquitin chains compete with ubiquitinated substrates for binding to the proteasome. The association of this DUB with the 26S probably may be important to prevent such competition and to ensure efficient proteolysis.
Surprisingly, overexpression of Ubp14, as well as of a catalytically inactive form of Ubp14, inhibits ubiquitin-mediated degradation as well (50). These observations resemble results obtained in yeast with the proteasome-associated DUB Ubp6. Its association with the 19S particle reduces the rate of degradation of ubiquitinated proteins, independently from its catalytic activity (51). Another proteasome-associated DUB is Uch37 (43). RNAi of either Uch37 or USP14 (the mammalian homologue of yeast Ubp6) accelerates protein degradation. Thus both DUBs might restrict protein breakdown at the proteasome in mammalian cells (52). The deubiquitinating activity of both USP14/Ubp6 (6, 39) and Uch37 (6, 9, 39, 53) is activated upon association with the proteasome. However, in Hela cell extracts, a large pool of cellular Uch37 and USP14 is found outside the proteasome and presumably serves other functions in the cell (52).
A failure to maintain a high level of free ubiquitin in cells can limit the cell’s capacity for ubiquitin-dependent protein degradation. Ubiquitin levels are regulated at least in part by the deubiquitinating enzymes residing at the proteasome, although the biochemical mechanisms regulating these processes remain unclear. The functions of the other novel DUB we identified on the proteasome, USP7/HAUSP, are even less clear. USP7 is localized to the nucleus and counteracts ubiquitination of p53 by Mdm2 (54), as well as negatively regulates the transcriptional activity of FOXO4 upon oxidative stress (55). Thus, unlike USP5, USP14, and Uch37, USP7 might be serving a rather specific role at nuclear 26S proteasomes in context of transcriptional regulation.
Ubiquitin Ligases Associated with the Proteasome
Even more surprising is the presence of the E3 ligases ARF-BP1/HUWE1 and p600/UBR4 on the proteasome in significant, but substoichiometric, amounts. Mule/ARF-BP1 is a HECT domain E3 ligase also called HUWE1 for “HECT, UBA, and WWE domain containing 1” and reported to be critical for the suppression of p53 in unstressed cells (56). p600 is a huge (574 kDa) protein that interacts with retinoblastoma (57) and the E7 protein of human papillomavirus 16 (58, 59) and is associated with microtubules and the ER in neurons (60). Another study identified p600 as a member of the UBR family and found p600/UBR4 to be the major N-end rule E3 ligase in UBR1−/− and UBR2−/− fibroblasts (61). Intriguingly, UBR1, the major N-end rule ligase in yeast has previously been found to interact with the 26S proteasome (62).
Several other ubiquitin ligases have been reported to associate with the proteasome, among them the HECT E3 Hul5, the RING E3 parkin, which contains a UBL motif, and the multisubunit E3 ligases SCFCdc4, APC, and VHL, plus several E2s (reviewed in ref 3). In addition, the HECT E3 ligase E3A/E6-AP was recently detected on affinity-tagged proteasomes from 293 cells (10). The number of E3s detected at the proteasome is increasing, and while some like the SCF seem to interact with the proteasome weakly and transiently (63), others like E6-AP, ARF-BP1/HUWE1, and p600/UBR4 seem to be associated rather stably and in significant substoichiometric amounts. Some ubiquitin ligases may bind to the proteasome indirectly by remaining associated with their ubiquitinated substrates or due to their tendency to autoubiquitinate. Another possibility is that some E3s permanently reside at the proteasome and ubiquitinate their substrates. How these ligases bind to the proteasome remains to be studied. Both mechanisms might help to protect substrates from deubiquitination and ensure rapid degradation upon ubiquitination. The proteasome by degrading the substrate would release the substrate from its E3 ligase.
There is already some evidence for regulation of the proteasome function by E3 ligases. Skowyra and co-workers have argued that the SCF complex can directly deliver a substrate to the 26S proteasome in vitro, which subsequently leads to disassembly of the 19S complex (63). In yeast, the ubiquitin ligase Hul5 (E3C or KIAA0010 in mammals) resides on the proteasome and extends ubiquitin chains on already ubiquitinated substrates (64). This activity can be counteracted by Ubp6, and it has been proposed that Hul5 and Ubp6 might serve as a mechanism that prevents the release of potentially toxic partially degraded proteins from the proteasome (65). The tight association with the 26S of p600/URB4, an E3 with “N-end rule” specificity, could also serve such a function since it should ubiquitinate specifically protein fragments with unusual N-terminals.
Roles of p97–Adaptor Complexes in Ubiquitin-Mediated Degradation
To isolate as many proteasome-associated proteins as possible, we removed salt from our purification buffer during these experiments. Under these conditions, the UBL domain of hHR23B also bound the AAA ATPase p97 together with its adaptor proteins Ufd1-Npl4, p47, SAKS1, and FAF1 (all of which contain ubiquitin-interacting motifs) as well as ubiquitinated proteins. The direct interaction of these complexes with the UBL domain of Rad23 has not been described before and is most likely mediated through their ubiquitin-binding motifs.
In addition, we detected a small amount of p97 and adaptor proteins associated with the 26S proteasome after glycerol gradient purification or native gel electrophoresis. The yeast homologue of p97, CDC48, has previously been isolated with affinity-purified 26S in yeast (7), and in human B cell lines, p97 was reported to be immunoprecipitated with the proteasome (66). In both cases the interactions might have been indirect and mediated by binding to residual ubiquitin conjugates not removed by the UIM domain. However, no ubiquitin conjugates were detectable in the TCA-precipitated fractions after glycerol gradient centrifugation or on native gels by Western blot (data not shown).
In ER-associated degradation (ERAD), p97 in complex with Ufd1-Npl4 extracts from the ER membrane ubiquitinated substrates which are then degraded by the proteasome (24). p97 also plays a role in degradation of ubiquitin fusion (UFD) and N-end rule substrates (67), and in yeast the p97 homologue CDC48 limits the length of ubiquitin chains generated on UFD substrates by the CDC48-associated ubiquitin ligase UFD2 (68). In addition, we recently found that CDC48 together with UFD1 and NPL4 is required to degrade newly synthesized proteins damaged by heat or oxygen radicals (40).
How ubiquitinated substrates are transferred from p97 to the proteasome is not clear. Our finding that some p97 complexes can associate with the 26S proteasome suggests that some p97–adpator complexes might directly deliver ubiquitin conjugates to the 26S, as previously proposed to occur during degradation of IκB (66). Another possibility is that Rad23 (or another UBL-UBA protein) might shuttle proteins from CDC48/p97 to the proteasome (24). Interestingly, p97 complexes were prevented from binding to the UBL domain of Rad23 with low salt concentrations and thus seemed to have a lower affinity for the UBL domain of Rad23 than the proteasome. In contrast, p97 complexes displayed a higher affinity for ubiquitin conjugates, since, unlike the proteasomes, p97 was enriched on the UIM domain in the last step of the purification. In a very recent study it was found that those p97 complexes interacting with ubiquitin conjugates also interacted with a large number of E3 ligases, suggesting that p97 might be involved in the turnover of various proteins (69). It is noteworthy that in our hands binding to the UBL domain of hHR23B did not seem to interfere with binding of ubiquitin conjugates to the p97 complexes. These findings support a model in which Rad23 can bind to p97, interact with ubiquitin conjugates, and subsequently deliver them to the proteasome. However, it remains to be proven that the ubiquitinated proteins on p97 are precursors to those associated with the 26S, and this affinity approach should greatly facilitate such studies.
Interestingly, the transcription of the various proteins that we isolated as a group with the UBL method, including proteasome subunits, USP5, p97, p47, SAKS1, NPL4, UFD1, and several others, is coordinately upregulated together with Rad23 upon proteasome inhibition in Drosophila (70). Therefore, it seems likely that the proteins identified here as the UBL interactome comprise a dynamic network of proteins functionally linked to ubiquitin-mediated degradation. The coisolation of p97 complexes and 26S proteasomes with the UBL method provides a highly valuable tool to study the properties and roles of these molecular machines in proteolysis in cultured cells and differentiated tissues in diverse physiological and pathological conditions.
Recently, after these studies were completed, an analogous method was reported that used the UBL domain from hHR23A to isolate proteasomes from HEK-293 cells (45). In comparison, the UBL of hHR23B used in our study was 20-fold more efficient in isolating proteasome from cell extracts (on average 40% of total proteasomal activity compared to about 2%). Also, that article did not describe the isolation of p97/adaptor complexes or ubiquitin conjugates and reported the isolation of a DNA–repair complex that was not isolated by our approach. These differences might be due to different buffer conditions used in the two studies or reflect different functions of hHR23A and -B in mammalian cells or different tissues.
Supplementary Material
ACKNOWLEDGMENT
We thank Björn Mückschel for valuable support in optimizing the UBL method during an internship in our laboratory, supported by the German Academic Exchange Program (DAAD).
Footnotes
The work of H.C.B. was supported by a postdoctoral fellowship of the German Academic Exchange Program (DAAD). W.H. was funded by Charles A. King Trust, Bank of America, Co-Trustee.
Abbreviations: AAA, ATPases associated with diverse cellular activities; DTT, 1,4-dithio-dl-threitol; GST, glutathione S-transferase; GSH, glutathione; LC-MS/MS, microcapillary liquid chromatography mass spectrometry; LLVY, N-succinyl-Leu-Leu-Val-Tyr-AMC (7-amino-4-methylcoumarin); Ni-NTA, nickel nitrilotriacetic acid; UBA, ubiquitin-associated domain; UBL, ubiquitin-like domain; UBX, ubiquitin-regulatory X domain; UIM, ubiquitin-interacting motif; TCA, trichloroacetic acid.
SUPPORTING INFORMATION AVAILABLE
Further information on purification efficiency and purity as well as the complete mass spectrometric data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glickman MH, Raveh D. Proteasome plasticity. FEBS Lett. 2005;579:3214–3223. doi: 10.1016/j.febslet.2005.04.048. [DOI] [PubMed] [Google Scholar]
- 6.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 7.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: Quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (qtax) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Vicente M, Sovak G, Cuervo AM. Protein degradation and aging. Exp. Gerontol. 2005;40:622–633. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 15.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 16.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 17.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 18.Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Lowe T, Hoppe T. Protein quality control gets muscle into shape. Trends Cell Biol. 2008;18:264–272. doi: 10.1016/j.tcb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- 21.Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 22.Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 2008 doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin. Cell Dev. Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J. Biol. Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 26.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- 27.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 28.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 30.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 33.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J. Biol. Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 34.Kisselev AF, Kaganovich D, Goldberg AL. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. J. Biol. Chem. 2002;277:22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 36.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 38.Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J. Biol. Chem. 2004;279:54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- 39.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y, Zhang N, Koepp DM, Walters KJ. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J. Mol. Biol. 2007;365:1093–1101. doi: 10.1016/j.jmb.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol. Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, Baumeister W. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000;150:119–130. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 45.Scanlon TC, Gottlieb B, Durcan TM, Fon EA, Beitel LK, Trifiro MA. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp. Cell Res. 2009;315:176–189. doi: 10.1016/j.yexcr.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 46.Hori T, Kato S, Saeki M, DeMartino GN, Slaughter CA, Takeuchi J, Toh-e A, Tanaka K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- 47.Hadari T, Warms JV, Rose IA, Hershko A. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem. 1992;267:719–727. [PubMed] [Google Scholar]
- 48.Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 49.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 2008 doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions non-catalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 52.Koulich E, Li X, Demartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 55.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 56.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakatani Y, Konishi H, Vassilev A, Kurooka H, Ishiguro K, Sawada J, Ikura T, Korsmeyer SJ, Qin J, Herlitz AM. p600, a unique protein required for membrane morphogenesis and cell survival. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Munger K. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeMasi J, Huh KW, Nakatani Y, Munger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shim SY, Wang J, Asada N, Neumayer G, Tran HC, Ishiguro K, Sanada K, Nakatani Y, Nguyen MD. Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J. Neurosci. 2008;28:3604–3614. doi: 10.1523/JNEUROSCI.5278-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst P, Buranda T, Sklar LA, Baumler J, Gogol E, Skowyra D. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 64.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 65.Kraut DA, Prakash S, Matouschek A. To degrade or release: ubiquitin-chain remodeling. Trends Cell Biol. 2007;17:419–421. doi: 10.1016/j.tcb.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J. Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 67.Wojcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, Kujawa M, DeMartino GN. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell. 2006;17:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lundgren J, Masson P, Realini CA, Young P. Use of RNA interference and complementation to study the function of the Drosophila and human 26S proteasome subunit S13. Mol. Cell. Biol. 2003;23:5320–5330. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.