Abstract
Because of the significant morbidity and lethality caused by alcoholic liver disease (ALD), there remains a need to elucidate the regulatory mechanisms that can be targeted to prevent and treat ALD. Towards this goal, minimally invasive biomarker discovery represents an outstanding approach for these purposes. The mechanisms underlying ALD include hepatic lipid accumulation. As the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) has been shown to inhibit steatosis, the present study examined the role of PPARβ/δ in ALD coupling metabolomic, biochemical and molecular biological analyses. Wild-type and Pparβ/δ-null mice were fed either a control or 4% ethanol diet and examined after 4–7 months of treatment. Ethanol fed Pparβ/δ-null mice exhibited steatosis after short-term treatment compared to controls, the latter effect appeared to be due to increased activity of sterol regulatory element binding protein 1c (SREBP1c). The wild-type and Pparβ/δ-null mice fed the control diet showed clear differences in their urinary metabolomic profiles. In particular, metabolites associated with arginine and proline metabolism, and glycerolipid metabolism, were markedly different between genotypes suggesting a constitutive role for PPARβ/δ in the metabolism of these amino acids. Interestingly, urinary excretion of taurine was present in ethanol-fed wild-type mice but markedly lower in similarly treated Pparβ/δ-null mice. Evidence suggests that PPARβ/δ modulates pyridoxal kinase activity by altering Km, consistent with the observed decreased in urinary taurine excretion. These data collectively suggest that PPARβ/δ prevents ethanol-induced hepatic effects by inhibiting hepatic lipogenesis, modulation of amino acid metabolism, and altering pyridoxal kinase activity.
Keywords: metabolomics, alcoholic liver disease, peroxisome proliferator-activated receptor-β/δ (PPARβ/δ), pyridoxal kinase
1. Introduction
Ethanol is consumed by 75% of the United States population, 7% of who become alcoholics. Each year there are approximately one hundred thousand ethanol-related deaths in the United States out of which 21% are non-accident-related. From all of the non-accident ethanol related deaths, 15,183 deaths were directly due to alcoholic liver disease (ALD) in 2010 (Stubbs and Morgan 2011). This is a 22% increase as compared to statistics from 2003 (Stubbs and Morgan 2011). Chronic ethanol consumption induces oxidative stress, lipid peroxidation, and acetaldehyde toxicity. Initially, ethanol abuse leads to steatosis (fatty liver) and if left untreated to alcoholic hepatitis, fibrosis, and cirrhosis. The early stages of ALD are reversible as shown from studies showing that abstinence from ethanol leads to amelioration of lesions, whereas liver damage observed with later stages of ALD are irreversible (Bruha et al. 2012; Woo and O’Brien 2012). Thus, reliable early ALD-specific diagnostic tools are the first line of defense against progression toward irreversible stages of ALD. However, the current diagnostic biochemical assays are not specific enough to detect ALD at its early stages and the ongoing biomarker discovery efforts although promising, have not produced new clinical assays. Therefore, there remains a need to elucidate the mechanisms by which ethanol alters liver function, in particular during the early stages of ALD when the disease may be reversible.
Examination of null mouse models of transcription factors that regulate lipid metabolism have been instrumental in identifying new molecular targets for the treatment and prevention of ALD. For example, Pparα-null mice exhibit hepatotoxicity in response to dietary exposure to ethanol and the phenotype is remarkably similar to that observed in humans (Nakajima et al. 2004). This is of interest to note because it is well known that PPARα is a key transcription factor that increases fatty acid catabolism suggesting that reduced capacity to catabolize lipids may enhance ALD. In contrast, sterol response element binding protein 1 (Srebp-1)-null mice exhibit reduced lipid accumulation in liver after ethanol consumption as compared to controls (Ji et al. 2006). As SREBP-1 is a key transcription factor that increases hepatic lipogenesis, this suggests that endogenous lipid synthesis may be important in the etiology of ALD. Interestingly, recent evidence has also shown that PPARβ/δ can also regulate hepatic function. Activation of PPARβ/δ inhibits the lipogenic actions of SREBP-1 by directly increasing expression of Insig-1 (Qin et al. 2008). Since steatosis is an important early event in the etiology of ALD, this suggests that PPARβ/δ could have an important function in the liver and for the prevention of ALD. This hypothesis is consistent with recent studies showing that Pparβ/δ-null mice exhibit enhanced sensitivity to hepatotoxicants (Shan et al. 2008a; Shan et al. 2008b). Thus, in this study, the hypothesis that PPARβ/δ is important in the etiology of early events associated with ALD was examined.
2. Materials and methods
2.1. Chemicals
Debrisoquine sulfate, 4-nitrobenzoic acid (4-NBA), xanthurenic acid, hippuric acid, urocanic acid, L-pipecolate, 2-methylglutaric acid, glycerophosphocholine, pyroglutamic acid, oxoglutaric acid, sodium pyruvate, N-acetylglutamine, and taurine were purchased from Sigma Chemical Company (St. Louis, MO). L-glyceric acid, hexanoylglycine, propionylcarnitine, glutaconic acid, ethyl-β-d-glucuronide and 2,3-dimethyl-3-hydroxyglutaric acid were obtained from Santa Cruz Biotechnology (Dallas, TX). Isobutyrylglycine, hydroxyphenylacetylglycine, and 2-methylbutyrylglycine were obtained from Molport (Riga, Latvia). The UPLC-grade solvents were purchased from Fisher Scientific (Hampton, NH). Stable isotopes of taurine and creatinine were obtained from Cambridge Isotope Laboratories, Inc. (Tewksbury, MA) and CDN Isotopes (Quebec, Canada) respectively.
2.2. Animals and treatments
Male wild-type or Pparβ/δ-null mice on a C57BL/6 genetic background (Peters et al. 2000), aged 6–8 weeks were housed in a temperature controlled environment (25 °C) with a 12-h light/dark cycle. Mice were fed either a control ethanol-containing liquid diet (Dyets, Inc. Bethlehem, PA) as previously described (Lieber and DeCarli 1994). Mice were fed the control diet for one week to acclimate to the liquid diet and then fed either the control or ethanol-containing liquid diet (4% v/v) for either four or seven months. Five to six animal per group were used. A separate cohort of mice from both genotypes were pair-fed with the control liquid diet based on the average intake of diet in the ethanol diet fed mice.
Twenty-four hour urine samples were obtained from mice individually housed in metabolic cages. Samples of urine were collected after the one week acclimation period and every month after the initiation of feeding the ethanol-containing diet. Urine samples were frozen at −80 °C until further use. After 4–6 months of treatment, mice were euthanized by over exposure to carbon dioxide. Blood was collected for isolation of serum and frozen until further analysis. Livers were carefully dissected, weighed, and then either snap frozen or fixed in 10% neutral buffered formalin (PBF).
2.3. Histopathology
Fixed sections of liver were prepared and stained with hematoxylin and eosin and examined under a light microscope. The entire representative liver section from the same lobe from at least five mice per group were examined and scored by a pathologist.
2.4. Serum markers of hepatic function
Serum was obtained from mice and the concentration of alkaline phosphatase (ALP), alanine amino transferase (ALT), bile acids and blood urea nitrogen were measured using the mammalian liver profile wheel for the Vetscan analyzer (Abaxis, Inc., Union City, CA).
2.5. Liver triglycerides
Liver triglycerides were quantified using a triglyceride colormetric assay (Cayman Chemical Company, Ann Arbor, MI).
2.6. Western blot analysis
For analysis of SREBP1c and pyridoxal kinase (PDXK) expression, liver samples were homogenized with RIPA buffer (50 mM Tris, 150 mM sodium chloride, 1% Nonidet P40 (v/v), and 0.1% sodium dodecyl sulfate (SDS)) including protease inhibitor cocktail (Complete Mini, Rosh, IN). For analysis of cytochrome P450 2E1 (CYP2E1), liver samples were homogenized in MENG buffer (25 mM MOPS, 2 mM EDTA, 0.02% sodium azide, and 10% glycerol, pH 7.5) containing a protease inhibitor cocktail. Samples were centrifuged at 1000 x g for 15–30 minutes and the supernatants removed. The supernatant was used for analysis of SREBP1 or PDXK, or for analysis for CYP2E1, the supernatant was removed and centrifuged at 100,000 X g for one hour. The resulting pellet containing the microsomes was resuspended in buffer (50 mM Tris HCl, pH 7.5, 1 mM EDTA, 20% glycerol, and 1 mM dithiothreitol). The protein concentration was quantified (BCA kit; Pierce, Rockford, IL), and between 10 and 50 μg of protein was separated on a 10%-12% gels by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After blocking in Tris-buffered saline with Tween 20 (TBST)–5% milk at room temperature, the membrane was incubated overnight at 4°C with an either an anti-PDXK (Lifespan Biosciences, Seattle, WA), anti-SREBP-1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-CYP2E1 (Abcam, Cambridge, MA) or anti- lactate dehydrogenase (LDH; Rockland, Gilbertsville, PA) antibody. Membranes were then washed with TBST and then incubated with biotinylated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Immunoreactive proteins on the membranes were detected after incubation with 125I-labeled streptavidin (Amersham Biosciences, Piscataway, NJ). Hybridization signals for specific proteins were normalized to the hybridization signal of the housekeeping protein LDH. A minimum of triplicate, independent samples were analyzed for each treatment group.
2.7. Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was isolated from liver samples using TRIzol reagent and the manufacturer’s recommended protocol. The mRNAs encoding insulin-induced gene 1 (Insig1), NM_153526.5, Cyp2e1, NM_021282.2, uridine diphosphate glucuronyltransferase (UDP)-glucuronosyltransferase 1a1(Ugt1a1), NM_201645.2, Ugt1a2, NM_013701.3, Ugt1a5, NM_201643.2, Ugt1a6b, NM_201410.3, Ugt1a7c, NM_201642.4, Ugt1a9, NM_201644.2, Ugt1a10, NM_201641.2, Pdxk, NM_172134.2, and glyceraldehyde 3-phosphate dehydrogenase (Gapdh, NM_008084.2) were measured by qPCR analysis. cDNA was generated from 2.5 μg total RNA using MultiScribe Reverse Transcriptase kit (Applied Biosystems, Foster City, CA). Real-time PCR primers for the above genes were designed using Integrated DNA Technologies (Coralville, IA) SciTools. The qPCR analysis was carried out using SYBR Green PCR master mix (Finnzymes, Espoo, Finland) in the iCycler and detected using the MyiQ Realtime PCR Detection System (Bio-Rad Laboratories, Hercules, CA). The following PCR reaction was used for all genes: 95°C for 10 s, 60°C for 30 s, 72°C for 30 s, and repeated for 45 cycles. Each PCR reaction included a no template control reaction to control for contamination, and all real-time PCR reactions had greater than 85% efficiency. The relative mRNA value for each gene was normalized to the relative mRNA value for the housekeeping gene Gapdh.
2.8. Preparation of urine samples for UPLC-ESI-QTOF-MS analysis
Urine was diluted 1:4 in a 50% acetonitrile solution containing 30 μM of 4-nitrosobenzoic acid and 2 μM of debrisoquine. The protein content was precipitated out by centrifugation at maximum speed. A 5 μL aliquot of the recovered supernatant was then injected into a reverse-phase 50 × 2.1 mm Acquity 1.7-μm C18 column (Waters Corp, Milford, MA) coupled to a time of flight mass spectrometry (TOF-MS). The binary UPLC system consisted of 2% acetonitrile in water containing 0.1% formic acid (aqueous solvent) and 2% water in acetonitrile containing 0.1% formic acid (organic solvent). The 10-minute long mobile phase gradient switched from 100% aqueous solvent to 100% organic at a flow rate of 0.5 mL/min. The QTOF Premier mass spectrometer was operated in electrospray ionization positive (ESI+) and negative (ESI−) modes. Capillary voltage was set at 3200 V while the cone voltage was kept at 20 V in the ESI−mode and 35 V in the ESI+ mode. Source temperature was set at 120 °C and the desolvation temperature was at 350 °C at a flow rate of 800 L/h. Sulfadimethoxine was used as the lock mass (m/z 311.0814+) for accurate mass calibration in real time. The MS data were acquired in centroid mode and processed using MassLynx software (Waters Corp, Milford, MA).
2.9. MS/MS validation of select metabolites
Metabolites were initially putatively identified by comparing their m/z to those published in online databases such as Human Metabolome Database (HMDB) and Madison Metabolomics Consortium Database (MMCD) using a predefined monoisotopic mass error window of 7 ppm. The retention time of each metabolite was considered to narrow down the possibilities of the putative identifications. Several of the putatively identified metabolites (P < 0.05, Mann-Whitney U test) were then chosen based on their biological relevance for further MS/MS validation against commercially available pure standards. The pure standards were diluted in 50% acetonitrile to a final concentration of 25 μM and fragmented using Q-TOFMS Premier with ramping collision energy of 5 to 50 eV. Validation was confirmed for metabolites that produced fragmentation patterns of the parent ions matching those of pure standards using UPLC-TOF-MS/MS.
2.10. Quantification of validated metabolites
The QTOF Premier was used for quantification of metabolites using pure chemical standards to establish a standard curve. Stable isotopes of taurine and 4-NBA were used as standards in the ESI− mode and the stable isotopes of creatinine and debrisoquine in the ESI+ mode. These chemical standards were made into a 100 μM stock mixture in 50% acetonitrile and diluted serially resulting in dilutions of 1:2, 1:4, 1:8, and 1:16. The standard mixture was spiked into a urine sample at the above concentration to ensure stability, reproducibility of signal, and optimization of the concentrations. A standard curve was obtained by plotting the peak areas of internal standards against their known concentration in each mode using QuanLynx (Waters Corporation, Milford, MA). A linear regression with a forced axis intercept of zero was obtained. The urine samples for each study group were then pooled and spiked with the optimized concentrations of internal standards and analyzed by QTOF. The observed peak area of the desired metabolites after creatinine normalization and the slope of the linear regression from the standard curve were used to determine the concentration of the metabolites normalized to the concentration of creatinine.
2.11. Data analysis
MarkerLynx software (Waters Corp, Milford, MA) was used to extract information such as retention time, m/z, and the abundance value (via the normalized peak area) for each ion from raw MS data. The abundance value of each ion was normalized with respect to the total ion count (TIC). Statistical analysis was conducted via in-house developed software, written in Python and the R statistical computing language. When comparing data from control versus ethanol-treated animals, ions with non-zero abundance values in at least 70% of samples in both groups were first identified (dubbed full-presence ions). Data from these ions were then log-transformed, and analyzed for statistical significance via the nonparametric Mann-Whitney U statistical hypothesis test (p-value < 0.05). Statistical significance for ions with non-zero abundance values in at least 70% of the samples in only one group (partial-presence ions) were analyzed categorically for presence status (i.e. non-zero abundance) via Fisher’s exact test (p-value < 0.05). The log-transformed data for statistically significant full-presence ions was then utilized for principal component analysis via singular value decomposition after zero centering and unit variance scaling. In addition, kernel principal component analysis utilizing a 3rd degree polynomial kernel was also conducted via the ‘kernlab’ package for R (Karatzoglou et al. 2004).
2.12. Metabolic pathway analysis
Both statistically significant full-presence and partial-presence ions were putatively identified via in-house software written in Python utilizing the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Ogata et al. 1999). Ions were identified by elucidating their neutral mass from the list of possible adducts, which for positive mode data included adducts formed by the addition of H+, Na+, and/or NH4+. These neutral masses were then compared to the exact mass of small molecules in the KEGG COMPOUND database (Kanehisa et al. 2006), from which putative metabolites were identified (with a mass error of 20 ppm or less). KEGG annotated pathways associated with these putative metabolites were also identified, from which a histogram of top pathways by number of putative metabolite hits was generated. Selected metabolites from these pathways were later validated via MS/MS as described above.
2.13. PDXK enzyme assay
Liver or cell samples were homogenized with phosphate-buffered saline containing 0.1% NP40 and then incubated for 20 minutes on ice followed by centrifugation at 14,000 rpm 15 min. Stable Huh7 cells containing the control Migr1-eGFP vector or the bi-cistronic Migr1-eGFP-hPPARβ/δ vector were obtained as previously described (Borland et al. 2011; Foreman et al. 2011). The supernatants were used as the enzyme extract. This enzyme preparation procedure was carried out at 4°C. The protein concentration was quantified as described above. Measurement of PDXK activity was performed as described in (Kwok and Churchich 1979; Sussmane and Koontz 1995) with some modifications. Briefly, the enzyme extract was added to a reaction mixture containing 0.1 M potassium phosphate, 1 mM ATP and 0.1 mM zinc chloride, pH 6.4, and then pyridoxal was added at a final concentrations from 1.6 μM to 2000 μM. The mixture was incubated at 37°C for 1 hour with shaking, then quickly put on ice, and hydroxylamine was added to a final concentration of 1 mM. The fluorescence was recorded using an excitation wavelength of 365 nm and an emission wavelength of 450 nm. Fluorescence was measured with GloMax-Multi+ detection system (Promega, Inc. Madison, WI).
2.14. Statistical analyses
Statistical differences were determined by ANOVA with a post hoc test using Tukey-Kramer procedures for the comparison of each group (Prism 5.0, GraphPad Software Inc., La Jolla, CA). Enzyme kinetics, Vmax and Km were calculated using Prism 5.0 software (GraphPad Software Inc., La Jolla, CA).
3. RESULTS
3.1. Animal monitoring and liver histology
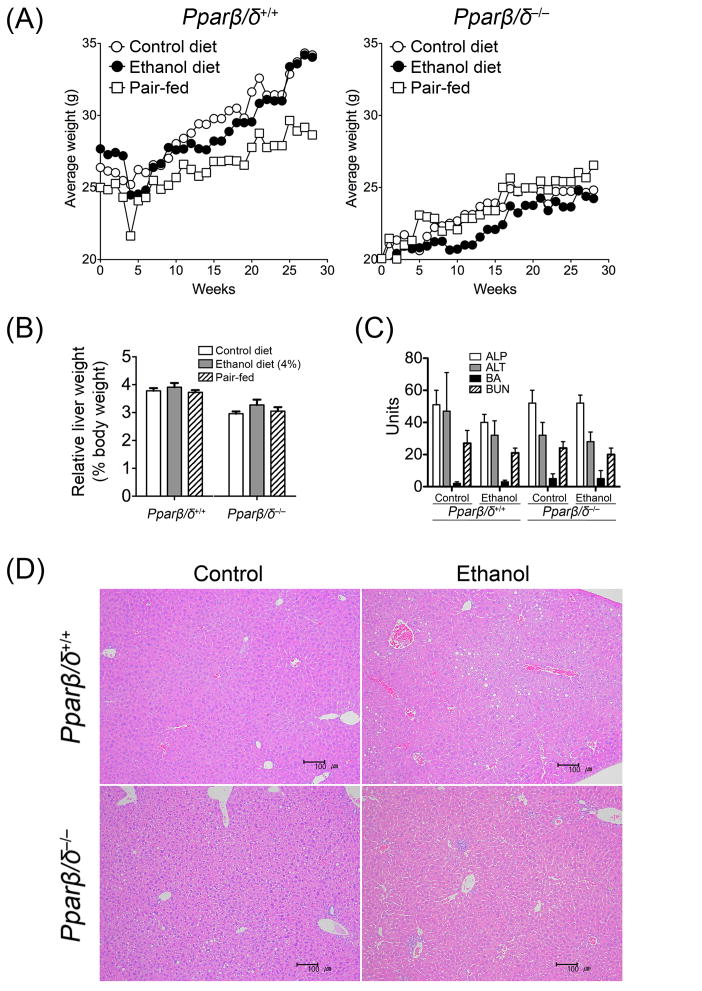
While mice fed ethanol showed a tendency of gaining less weight compared to respective control, this difference was not statistically significant between either genotype (Fig. 1A). Similarly, there were no statistically significant differences between the pair-fed mice and controls (Fig. 1A) due in large part to significant variation between all groups. Mice fed ethanol also exhibited a tendency of higher average liver weight compared to respective control, but this was not statistically significant between either genotype (Fig. 1B). Additionally, there were no statistically significant differences in average liver weight between the pair-fed mice and controls (Fig. 1B). Average serum concentrations of ALP, ALT and blood urea nitrogen were not different between control and mice fed ethanol in either genotype (Fig. 1C). While there was a trend for higher bile acids in Pparβ/δ-null mice fed ethanol compared to controls and both groups of wild-type mice, this trend was not statistically significant (Fig. 1C). Histopathological analysis revealed an increase in the incidence of lipid accumulation in wild-type mice after 7 months of consuming the ethanol diet as compared to control, although not remarkable in severity (Fig. 1D). In contrast, lipid accumulation was observed in the liver of Pparβ/δ-null mice fed the control diet (Fig. 1D). Interestingly, there was a tendency of immune cell infiltrate observed in the liver of Pparβ/δ-null mice fed the ethanol diet and this effect was not found in similarly treated wild-type mice (Fig. 1D). Results from this analysis indicate that other than an increase in hepatic lipid accumulation was observed in wild-type C57BL/6 mice fed ethanol compared to controls, this effect was only observed in control Pparβ/δ-null mice but not Pparβ/δ-null mice fed ethanol. In contrast, an enhanced inflammatory response was observed at the histopathological level in Pparβ/δ-null mice fed ethanol compared to controls and all groups of wild-type mice. This suggests that PPARβ/δ modulates the effects of ethanol differentially.
Fig. 1.
Effect of ethanol feeding on body weight, relative liver weight, serum markers of liver toxicity and liver histology in control, ethanol-fed, or pair-fed wild-type (Pparβ/δ+/+) and Pparβ/δ-null (Pparβ/δ−/−) mice after seven months of treatment. (A) Average body weight and (B) average relative liver weight in control, ethanol-fed, or pair-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. (C) Average serum concentrations of ALP (U/L), ALT (U/L), BA (μM) and BUN (mg/dL) in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. Values represent the mean (body weight) or mean ± S.E.M. (relative liver weight and serum markers of liver toxicity). (D) Liver histology (HE staining) of Pparβ/δ+/+ and Pparβ/δ−/− mice after seven months of control (left panel) and 4% ethanol containing liquid diet (right panel) showing significant increase in lipid deposition in the ethanol-treated wild-type and control Pparβ/δ-null mice. Note immune cell infiltrate in the liver of Pparβ/δ-null mice fed the ethanol diet, an effect not found in similarly treated wild-type mice. Bar = 100 μm.
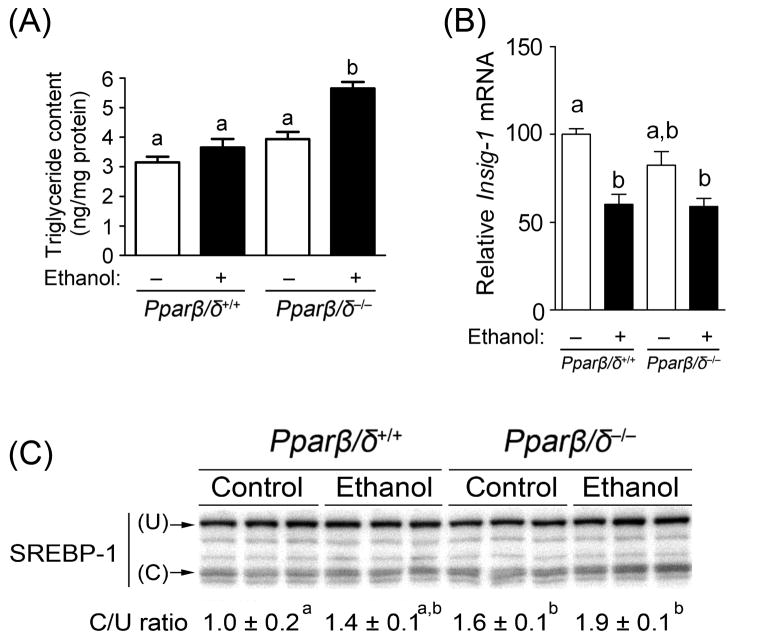
To begin to examine how PPARβ/δ influences the etiology of ethanol-induced hepatic effects, a separate cohort of mice were fed ethanol for only four months and examined (Fig. 2). Whereas heightened lipid accumulation was observed at the histopathological level in wild-type mice fed ethanol after 7 months of treatment but not in Pparβ/δ-null mice, this effect was reversed after only 4 months of ethanol feeding (Fig. 2A). As compared to control and both groups of wild-type mice, ethanol feeding resulted in increased accumulation of hepatic triglycerides in Pparβ/δ-null mice fed ethanol but not in similarly treated wild-type mice (Fig. 2A). Since a previous study showed that activation of PPARβ/δ can prevent lipogenesis by increasing expression of Insig1 causing decreased activity of SREBP1 activity, expression of these lipogenic markers was examined. Expression of Insig1 mRNA was not different between wild-type and Pparβ/δ-null mice, and ethanol feeding caused a decrease in expression of Insig1 mRNA in both genotypes (Fig. 2B). Interestingly, there was an increase in the active form of SREBP1 in control and ethanol-fed Pparβ/δ-null mice as compared to wild-type mice (Fig. 2C). This is consistent with the higher accumulation of triglycerides observed in ethanol-fed Pparβ/δ-null mice compared to controls (Fig. 2B).
Fig. 2.
PPARβ/δ-dependent modulation of hepatic triglyceride accumulation and active SREBP1 after four months of treatment. (A) Average concentration of hepatic triglycerides in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. (B) Average expression of hepatic mRNA encoding Insig1 in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. Average triglyceride concentrations and normalized hybridization values represent the mean ± S.E.M.. (C) Western blot analysis of inactive (uncleaved-U) and active (cleaved-C) SREBP1 in the liver of control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. Values with different superscripts are statistically different at P ≤ 0.05.
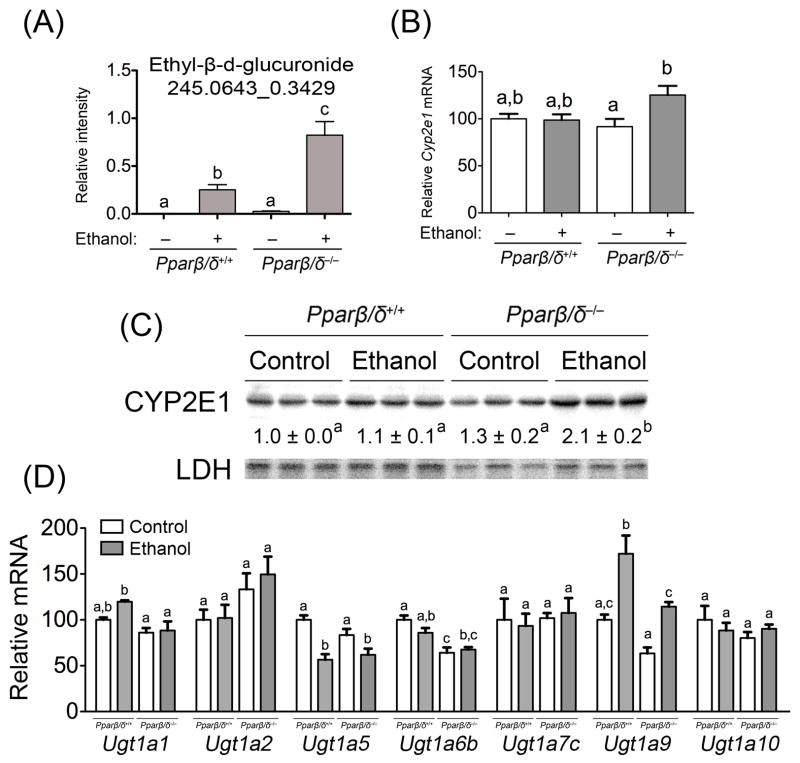
There was an increase in the relative intensity of urinary ethyl-β-d-glucuronide (MS/MS validated) in both ethanol-fed wild-type and Pparβ/δ-null mice compared to controls (Fig. 3A). This demonstrates that both wild-type and Pparβ/δ-null mice were exposed to ethanol, but suggests that PPARβ/δ differentially regulates the metabolic fate of ethanol. To begin to examine potential mechanisms for this observed difference, the expression of Cyp2e1 mRNA and protein was examined. Surprisingly, expression of Cyp2e1 mRNA and CYP2E1 protein in the liver was higher in Pparβ/δ-null mice fed ethanol but not in similarly treated wild-type mice (Figs. 3B, 3C). To determine if the difference in urinary ethyl-β-d-glucuronide was due to altered expression of UGTs, the mRNA expression of the most commonly expressed UGTs in the liver was examined. While some differences in expression of mRNAs encoding Ugts were noted in response to ethanol feeding, increased expression in ethanol-fed Pparβ/δ-null mice was not found as compared to ethanol-fed wild-type mice (Fig. 3D).
Fig. 3.
PPARβ/δ-dependent modulation of a urinary ethanol metabolite and ethanol metabolizing enzymes after four months of feeding. (A) Relative urinary abundance of MS/MS validated ethyl-β-d-glucuronide in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. (B) Average expression of hepatic mRNA encoding Cyp2e1 in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. Average urinary concentration of ethyl-β-d-glucuronide and normalized Cyp2e1 mRNA values represent the mean ± S.E.M.. (C) Western blot analysis of CYP2E1 in the liver of control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. (D) Average expression of hepatic mRNA encoding different Ugt mRNAs in control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice. Normalized values represent the mean ± S.E.M.. Values with different superscripts are statistically different at P ≤ 0.05.
3.2 Metabolomic data and pathway analysis
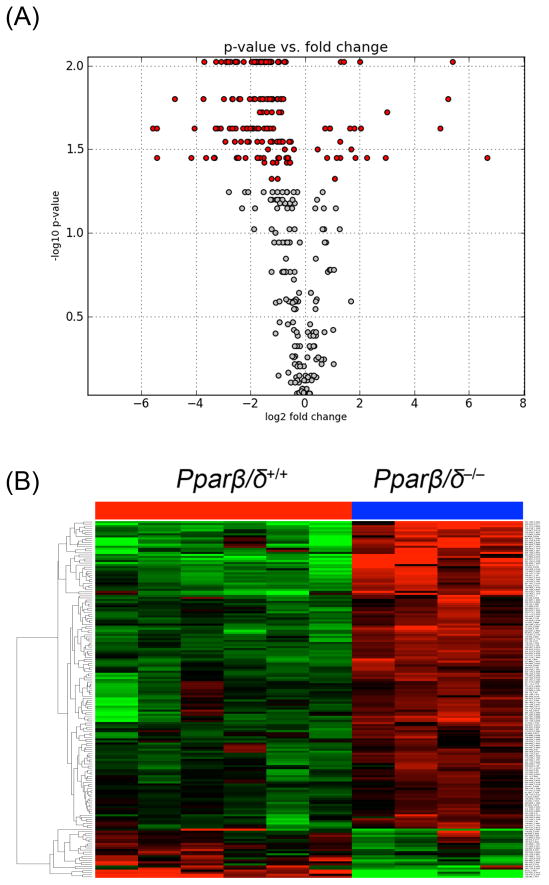
412 ions were selected in positive ESI mode (ESI+) and 520 ions in negative ESI mode (ESI−) for a battery of statistical tests from all of the urinary metabolites detected in control wild-type and Pparβ/δ-null mice. The volcano plot depicts the statistically significant ions in red from ESI– data (Fig. 4A). The non-statistically significant ions are shown in grey (Fig. 4A). The heatmap shows clustering of significantly different urinary ions between genotypes including those that were lower in wild-type mice compared to Pparβ/δ-null mice and vice versa (Fig. 4B). Further pathway analysis revealed that tryptophan, arginine, proline, sugar and lipid metabolism were the top metabolic processes affected by the absence of PPARβ/δ expression (data not shown).
Fig. 4.
Differential presence of urinary ions between control Pparβ/δ+/+ and Pparβ/δ−/− mice after four months of feeding. (A) Volcano plot (ESI–) and (B) heatmap (ESI–) showing up- and down-regulation of several ions in Pparβ/δ−/− mice compared to Pparβ/δ+/+ mice fed the control liquid diet. The volcano plot depicts the statistically significant ions in red and the non-statistically significant ions are shown in grey.
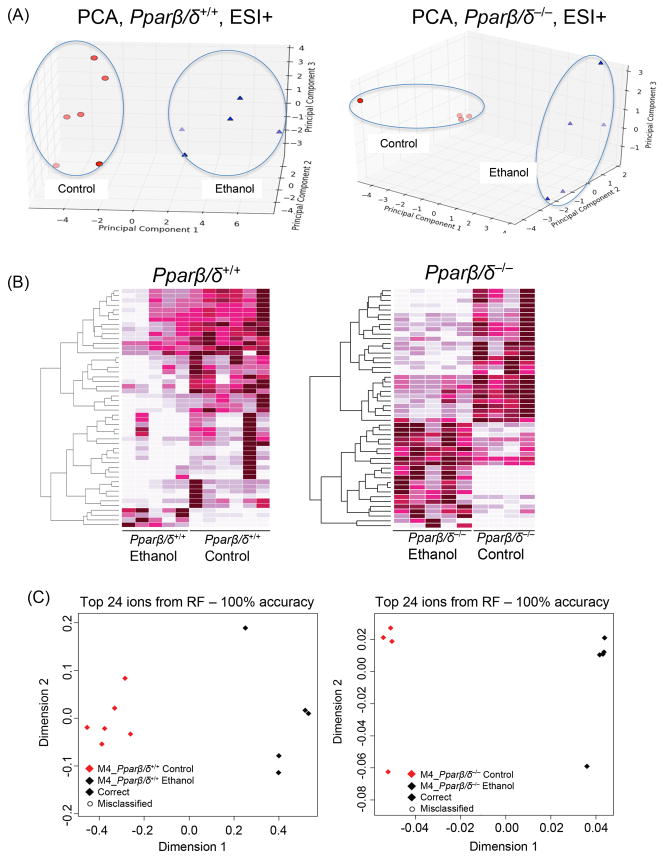
The principal component analyses (PCA) show clear separation between control and ethanol-fed mice (Fig. 5A). This result was confirmed by the machine-learning algorithm Random Forests as depicted in heatmaps (Figure 5B). The Pparβ/δ-null mice show a cleared separation between the two diets as compared to wild-type mice (Fig. 5C). Both genotypes gave a 100% accuracy of classification as shown in the multi-dimensional scaling (MDS) plots with the top 24 significant ions (Fig. 5C). Our in-house software compiled and sorted the ions that contributed the most to the separation between genotypes and treatments (Table 1). Significantly different metabolites were mapped to organic acid and amino acid metabolism (particularly lysine, tyrosine, and tryptophan) pathways in both genotypes (data not shown). The role of PPARβ/δ in ethanol-induced hepatic effects was furthered studied by analyzing metabolites whose levels changed as a result of ethanol feeding in both genotypes (Fig. 6). The 50 ions displayed on the heatmap (Fig. 6) are the most significantly different ions between the two genotypes and/or ethanol treatment.
Fig. 5.
Differential presence of urinary ions between control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice after four months of feeding. (A) Score scatter plot from PCA of the urinary metabolomic signature showing separation of metabolic profiles of mice on control and ethanol diets after 4 months of feeding in ESI+ mode. (B) Random Forest generated heatmaps showing ions contributing the most to the separation of metabolomic profiles of normal and ethanol-fed mice for both genotypes. (C) The MDS plot below each heatmap indicates the separation accuracy for the top 24 contributing ions. We further focused our analysis on the metabolites that contributed to the dietary separation observed in Pparβ/δ+/+ mice whose levels did not change in Pparβ/δ−/− mice.
Table 1.
List of urinary ions exhibiting differential presence between genotypes and/or ethanol feeding.
| Comparison | Marker | m/z | RT (min)* | P value** | Fold change*** | Confirmed ID**** |
|---|---|---|---|---|---|---|
| Wild-type vs. Ppar!/! -null controls | N2 | 105.0187 | 0.3197 | 0.0357 | 61.6 | L-Glyceric acid |
| Wild-type vs. Ppar!/! -null controls | N4 | 145.049 | 1.4471 | 0.0095 | 40.01 | 2-Methylglutaric acid, Adipic acid |
| Wild-type vs. Ppar!/! -null controls | N1 | 431.183 | 6.3109 | 0.024 | 3.6 | |
| Wild-type vs. Ppar!/! -null controls | N3 | 553.3382 | 5.5091 | 0.0095 | 3.2 | |
| Wild-type vs. Ppar!/! -null controls | N9 | 161.0438 | 0.5371 | 0.0095 | 2.72 | |
| Wild-type vs. Ppar!/! -null controls | P2 | 281.0999 | 2.1687 | 0.016 | 0.57 | |
| Wild-type vs. Ppar!/! -null controls | N7 | 144.0654 | 0.8706 | 0.016 | 0.36 | Isobutyrylglycine |
| Wild-type vs. Ppar!/! -null controls | N8 | 128.034 | 1.0348 | 0.016 | 0.34 | Pyroglutamic acid |
| Wild-type vs. Ppar!/! -null controls | P1 | 172.0968 | 2.4299 | 0.036 | 0.28 | |
| Wild-type vs. Ppar!/! -null controls | N5 | 172.0963 | 3.1093 | 0.024 | 0.16 | Hexanoylglycine |
| Wild-type vs. Ppar!/! -null controls | N6 | 216.1231 | 3.7551 | 0.0095 | 0.13 | Propionylcarnitine |
| Wild-type controls vs. ethanol-fed wild-type | N11 | 129.0182 | 0.4204 | 0.03 | 2.22 | Glutaconic acid |
| Wild-type controls vs. ethanol-fed wild-type | P4 | 160.096 | 1.6379 | 0.016 | 1.6 | |
| Wild-type controls vs. ethanol-fed wild-type | P5 | 206.0452 | 1.6903 | 0.015 | 0.47 | Xanthurenic acid |
| Wild-type controls vs. ethanol-fed wild-type | N12 | 145.0129 | 0.3431 | 0.0043 | 0.36 | |
| Wild-type controls vs. ethanol-fed wild-type | N10 | 186.0758 | 1.5359 | 0.0043 | 0.25 | |
| Ppar!/! -null controls vs. ethanol-fed Ppar!/! -null | P6 | 211.0708 | 0.3344 | 0.036 | 2.96 | N-Acetylglutamine |
| Ppar!/! -null controls vs. ethanol-fed Ppar!/! -null | N14 | 87.0084 | 0.3171 | 0.036 | 0.6 | Pyruvic acid |
| Ppar!/! -null controls vs. ethanol-fed Ppar!/! -null | N13 | 175.0597 | 1.92 | 0.1016 | 0.52 | |
| Ppar!/! -null controls vs. ethanol-fed Ppar!/! -null | N15 | 161.0462 | 0.9361 | 0.036 | 0.31 | |
| Ppar!/! -null controls vs. ethanol-fed Ppar!/! -null | N16 | 124.0061 | 0.2852 | 0.36 | 0.3 | Taurine |
RT = retention time in minutes (min)l.
P-values as compared to respective control.
Fold change = (Average normalized intensity of second listed group)/(Average normalized intensity of the first listed group). If the fold change for a particular metabolite is less than unity it means that the average intensity value of that metabolites in first group was higher than in second group. These metabolites are specific to the comparison between each genotype/treatment comparison, and do not depict the same changes in their urinary excretion levels made in other comparisons.
Confirmed by MS/MS as described in Materials and methods.
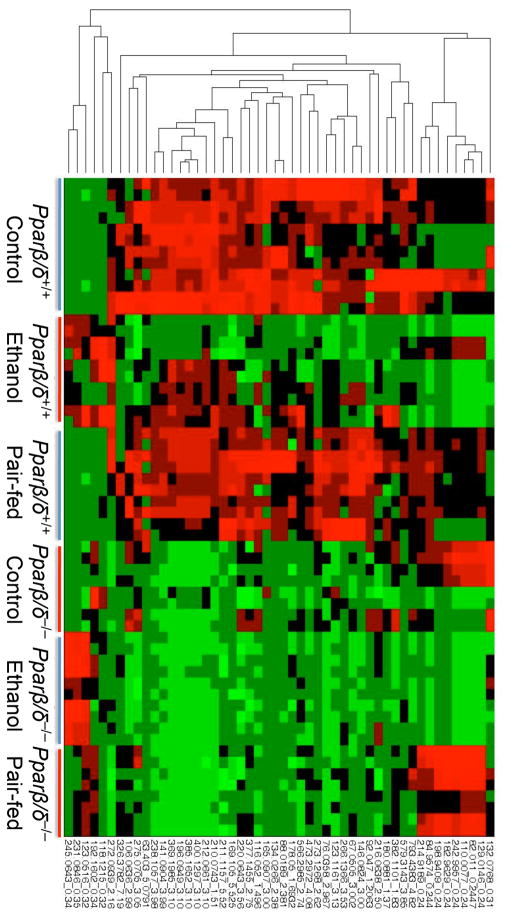
Fig. 6.
Heatmap representation of ions with altered urinary excretion levels as a result of four months of ethanol consumption in control, ethanol-fed, or pair-fed wild-type (Pparβ/δ+/+) and Pparβ/δ-null (Pparβ/δ−/−) mice.
3.3 PPARβ/δ-dependent modulation of hepatic PDXK activity partially underlies the differences in metabolic signature of urine following ethanol feeding
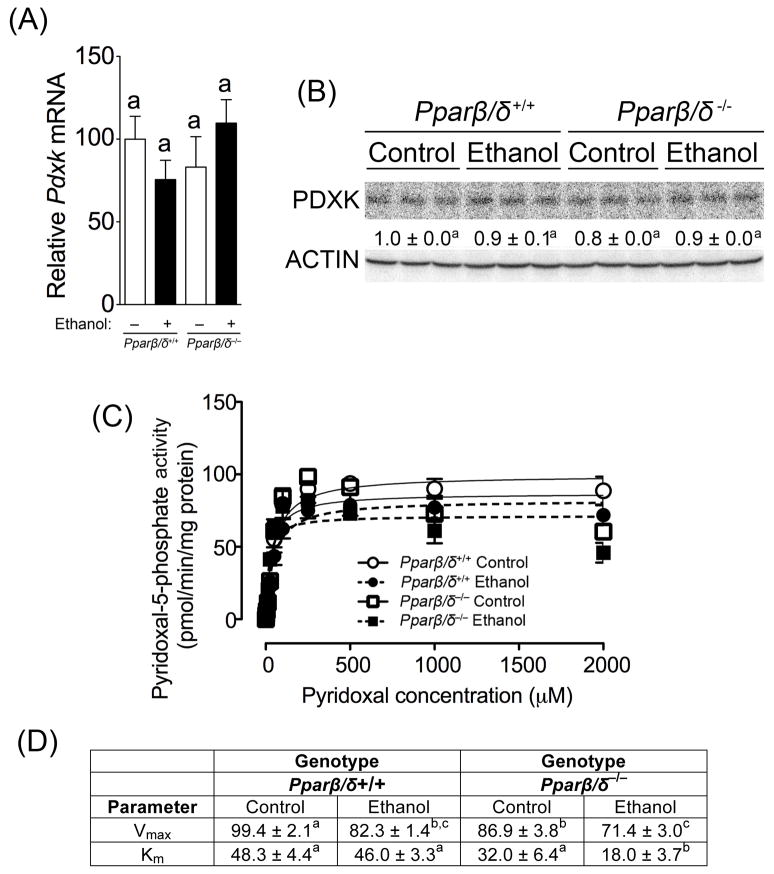
To begin to examine the mechanisms by which PPARβ/δ modulates the urinary metabolome in response to ethanol exposure, the metabolic signature of urine was evaluated for candidates that are typically coordinately regulated. Of particular interest was the finding that urinary taurine was decreased in ethanol-fed Pparβ/δ-null mice as compared to similarly treated wild-type mice (Table 1). Since a previous study showed that vitamin B6 deficiency can cause a decrease in urinary taurine excretion in rats (Koyanagi and Obori 1965), the expression and activity of the rate limiting enzyme PDXK, responsible for generating the active form vitamin B6, pyridoxal-5-phosphate, was examined. While expression of Pdxk mRNA and PDXK protein was unchanged by ethanol in either genotype (Figs. 7A, 7B), ethanol feeding caused a decrease in PDXK activity as compared to controls in both genotypes (Figs. 7C, 7D). This is consistent with studies showing reduced vitamin B6 status and PDXK activity in human alcoholics (Fonda et al. 1989; Halsted and Medici 2011). Interestingly, the Km of PDXK in liver was markedly lower in ethanol-fed Pparβ/δ-null mice as compared to ethanol-fed wild-type mice and controls of both genotypes (Fig. 7D). This is of interest because the decrease in Km indicates that PDXK is capable of producing more active pyridoxal-5-phosphate (active vitamin B6), thus potentially increasing activity of many enzymes that utilize vitamin B6 as a co-factor, since many of these enzymes are involved in amino acid metabolism (Toney 2011), and differences in amino acid ions were detected in the urinary metabolome of ethanol-fed mice. For example, taurine-2-oxoglutarate transaminase is pyridoxal-dependent enzyme that catalyzes the formation of sulfoacetaldehyde and L-glutamate from taurine and 2-oxoglutarate. The potential increase in taurine-2-oxoglutarate transaminase is consistent with the decrease in urinary taurine observed in ethanol-fed Pparβ/δ-null mice as compared to control-fed Pparβ/δ-null mice (Table 1). The decrease in urinary taurine concentration in ethanol-fed Pparβ/δ-null mice as compared to control-fed Pparβ/δ-null mice was quantified and confirmed to be 0.87 μM/mM creatinine versus 0.31 μM/mM creatinine, respectively. Moreover, this is also consistent with the observed decrease in urinary 2,3-dimethyl-3-hydroxyglutaric acid derivative and increase in urinary N-acetylglutamine, which could reflect further metabolism of the taurine-2-oxoglutarate transaminase substrate 2-oxoglutarate and the taurine-2-oxoglutarate transaminase product L-glutamate. The increase in urinary N-acetylglutamine concentration in ethanol-fed Pparβ/δ-null mice as compared to control-fed Pparβ/δ-null mice was quantified and confirmed to be 0.07 μM/mM creatinine versus 0.12 μM/mM creatinine, respectively.
Fig. 7.
Ethanol decreases PDXK Vmax and causes a PPARβ/δ-dependent maintenance of PDXK Km. (A) Relative hepatic mRNA expression of Pdxk in liver of control or ethanol-fed wild-type (Pparβ/δ+/+) and Pparβ/δ-null (Pparβ/δ−/−) mice after four months. (B) Western blot analysis of PDXK in the liver of control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice after 4 months. (C) Michaelis-Menten kinetic parameters were derived from the enzyme assay for PDXK activity using liver enzyme extracts from control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice after 4 months. (D) Vmax and Km values calculated from the enzyme assay for PDXK activity using liver enzyme extracts from control and ethanol-fed Pparβ/δ+/+ and Pparβ/δ−/− mice after 4 months. Values represent the mean ± S.E.M.. Values with different letters or superscripts are significantly different at P ≤0.05.
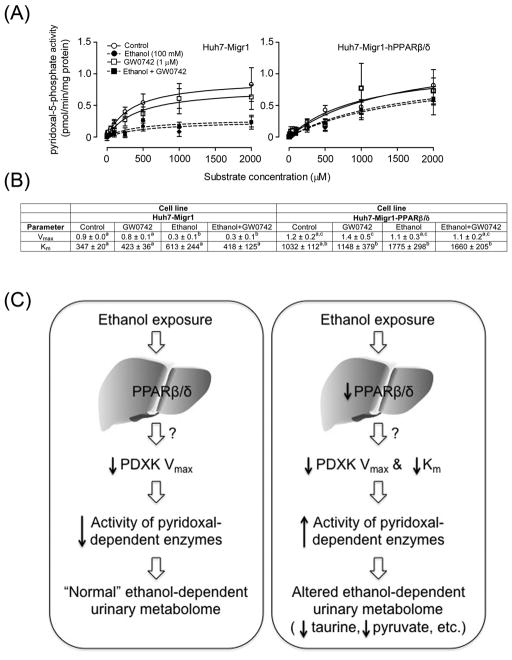
To determine whether PPARβ/δ can directly influence PDXK activity in hepatic cells, PDXK activity was examined in control Huh7 cells or Huh7 cells that over-express PPARβ/δ, in the presence or absence of ethanol. Similar to the effects observed in vivo, ethanol treatment caused a decrease in Vmax of PDXK in control Huh7 cells (Figs. 8A, 8B). The observed decrease in PDXK Vmax was not found in Huh7 cells over-expressing PPARβ/δ, with and without the PPARβ/δ ligand GW0742 (Figs. 8A, 8B). Moreover, over-expression of PPARβ/δ in the presence or absence of GW0742 also caused an increase in the Km of PDXK (Figs. 8A, 8B). These results indicate that PPARβ/δ is directly involved in the modulation of PDXK resulting from ethanol exposure.
Fig. 8.
Over-expression and ligand activation of PPARβ/δ prevents the ethanol-induced decrease in PDXK Vmax and maintains Km. (A) Michaelis-Menten kinetic parameters were derived from the enzyme assay for PDXK activity using extracts control Huh7 cells (Huh7-Migr1) or Huh7 cells over-expressing PPARβ/δ (Huh-Migr1-hPPARβ/δ) in the presence or absence of ethanol, the highly specific PPARβ/δ ligand GW0742, or both ethanol and GW0742. (B) Vmax and Km values calculated from the enzyme assay for PDXK activity using control Huh7 cells (Huh7-Migr1) or Huh7 cells over-expressing PPARβ/δ (Huh-Migr1-hPPARβ/δ) in the presence or absence of ethanol, the highly specific PPARβ/δ ligand GW0742, or both ethanol and GW0742. (C) Hypothetical model showing how PPARβ/δ alters ethanol metabolism by altering PDXK activity. In normal conditions (left panel), ethanol exposure causes a decrease in hepatic PDXK activity by decreasing Vmax, likely causing decreased activity of a number of pyridoxal- dependent enzymes leading to alterations in the urinary metabolome. In the absence of PPARβ/δ expression and/or the presence of a loss-of-function polymorphism (right panel), ethanol exposure causes an increase in hepatic PDXK activity by markedly decreasing PDXK Km, likely causing increased activity of a number of pyridoxal-dependent enzymes leading to alterations in the urinary metabolome including decreased taurine and pyruvate. These data collectively indicate that PPARβ/δ decreases PDXK activity by maintaining Km in response to ethanol exposure.
4. Discussion
Results from the present studies are the first to demonstrate a role for PPARβ/δ in modulating the hepatic effects induced by exposure to ethanol. Chronic exposure to ethanol caused hepatic lipid accumulation in both wild-type and Pparβ/δ-null mice after 7 months of treatment, but this effect was not as striking in Pparβ/δ-null mice. This could be due to the fact that the observed increase in hepatic CYP2E1 activity induced by ethanol in Pparβ/δ-null mice can cause increased generation of reactive oxygen species and inflammation (Halsted and Medici 2011). This is consistent with the histopathology observed in ethanol-fed Pparβ/δ-null mice compared to similarly treated wild-type mice after 7 months of treatment. The enhanced expression of CYP2E1 found in ethanol-fed Pparβ/δ-null mice compared to ethanol-fed wild-type mice could also be related to the increased presence of urinary ethyl-β-d-glucuronide, but does not appear to be due to differences in expression of UGTs. The mechanism by which PPARβ/δ represses CYP2E1 expression requires further investigation but repression by the presence of PPARβ/δ near the promoters of target gene chromatin has been observed previously (Adhikary et al. 2011; Khozoie et al. 2012).
Examination of the liver also revealed that at an earlier time point (4 month of ethanol exposure), accumulation of triglycerides was higher in ethanol-fed Pparβ/δ-null mice compared to ethanol-fed wild-type mice. A previous study indicated that PPARβ/δ represses hepatic lipogenesis by increasing expression of INSIG1 causing decreased cleavage and activity of the lipogenic transcription factor SREBP1 (Qin et al. 2008). In the present study, ethanol feeding caused a decrease in Insig1 mRNA in both genotypes but no change in constitutive expression of Insig1 mRNA between wild-type and Pparβ/δ-null mice. However, it is worth noting that ethanol feeding did result in enhanced presence of active SREBP1 in Pparβ/δ-null mice, consistent with a previous study indicating that PPARβ/δ inhibits hepatic lipogenesis and the observed increase in hepatic triglycerides. Further studies are needed to determine whether ligand activation of PPARβ/δ regulates Insig1 expression and/or SREBP1 activity that could in theory prevent hepatic lipid accumulation induced by ethanol feeding. Why higher lipid accumulation was found in wild-type mice but not Pparβ/δ-null mice after seven months of feeding cannot be determined from these studies. No changes in active SREBP1 was detected after seven month of feeding (data not shown). However, higher lipid accumulation found in wild-type mice but not Pparβ/δ-null mice after seven months of feeding could be due to increased presence of pro-inflammatory lipids that promoted the observed enhanced inflammatory infiltrates causing reduced triglycerides and relatively lower total lipids detected using the methods utilized in this study. Alternatively, the observed difference in lipid accumulation and inflammation could be due in part to increased CYP2E1-mediated metabolism of ethanol observed in the Pparβ/δ-null mice. Further studies are needed to examine this hypothesis.
Metabolomic analysis of urinary ions in wild-type and Pparβ/δ-null mice revealed significant differences between genotypes in both control and ethanol-fed mice. An increase in the glycerolipid pathway ions in Pparβ/δ-null mice fed the control diet compared to wild-type mice was observed. For example, an ion confirmed as glyceric acid, was present in the urine of Pparβ/δ-null control mice at significantly higher levels than in control wild-type mice, and closer to the levels observed in ethanol-fed wild-type. This increase in urinary excretion of lipid-related ions in Pparβ/δ-null mice is consistent with the results of liver histopathology suggesting a protective role for PPARβ/δ against lipid deposition. This is consistent with the mechanism describe above. In the absence of PPARβ/δ expression, mice can accumulate more lipids in their liver and model the phenotype of ALD patients. Thus, Pparβ/δ-null mice may provide a new model to study the mechanisms underlying the onset, etiology and prevention of ALD. In addition to lipid-related ions, the metabolism of amino acids was also altered in Pparβ/δ-null mice. This is of interest to note because amino acid metabolism is closely linked to vitamin B6-dependent enzymes.
The role of ethanol diet on the urinary metabolomics profile of both genotypes was determined by examining and comparing ions contributing the most to the profile differences in wild-type and Pparβ/δ-null mice control diet compared to those on ethanol diet. Pathway analysis revealed that ions involved in lysine, tryptophan, cysteine and methionine metabolism were among the top metabolic pathways affected by the ethanol diet, suggesting a direct or indirect link between ethanol metabolism/detoxification and metabolism of these amino acids. Interestingly, the early inflammation marker xanthurenic acid, a product of tryptophan catabolism, was lower in the urine of wild-type mice fed the ethanol diet compared to control wild-type mice on control diet. Urinary excretion of xanthurenic acid was unchanged in Pparβ/δ-null mice compared to controls. In contrast, Pparα-null mice on a C57BL/6 genetic background (Akiyama et al. 2001) fed ethanol exhibit a ~2-fold increase in urinary xanthurenic acid compared to control wild-type and Pparα-null mice (Manna et al. 2010). In the same study, mice on an Sv/129 genetic background (Akiyama et al. 2001) showed minimal elevation of the xanthurenic acid after ethanol treatment. In another study examining the urinary metabolome of interleukin 10 (Il10)-null mice that exhibit chronic inflammation, urinary xanthurenic acid was decreased in Il10-null mice as compared to wild-type mice (Otter et al. 2011), similar to results observed in ethanol-fed wild-type mice in the present study. Thus, decreased urinary xanthurenic may serve as a marker of ethanol-induced hepatic effects. Combined, these observations suggest that urinary excretion of xanthurenic acid may be strain dependent and/or PPARα specific marker of ethanol exposure.
The importance of identifying early non-invasive biomarkers for ethanol-induced liver damage is due to the large number of patients admitted to hospitals due to ethanol related liver disease and the reported death rate associated with this disease (Stubbs and Morgan 2011). Thus, the findings that PPARβ/δ can modulate PDXK enzyme activity that is related to the urinary metabolome is of striking importance (Fig. 8C). It is also important to note that the urinary metabolome can be influenced by both mammalian enzymes in tissues as well as microbial enzymes present in the gut (Wikoff et al. 2009). This may account for some of the changes observed in the present study. The decrease in urinary taurine and the increase in urinary N-acetylglutamine observed in ethanol-fed Pparβ/δ-null mice compared to controls, could reflect alterations in the metabolism catalyzed by the pyridoxal-dependent taurine-2-oxoglutarate transaminase (Fig. 8C). Issues with sensitivity and/or the approach used for this analysis may have prevented us from detecting all of the substrates and products of pyridoxal-dependent taurine-2-oxoglutarate transaminase. Moreover, PDXK regulates the production of pyridoxal-5-phosphate, the active form of vitamin B6 required as a co-factor for many enzymes (Toney 2011). Enzyme activity is the collective result of both Vmax and Km. Thus, it is important to note, that paradoxically, ethanol consumption caused decreased activity of PDXK (Vmax), which would presumably increase the presence of urinary taurine through pyridoxal-dependent taurine-2-oxoglutarate transaminase due to decreased catabolism of taurine as an enzyme substrate. However, since Km was reduced in ethanol-fed Pparβ/δ-null mice, this would explain the apparent decrease in urinary taurine and increase in N-acteylglutamine due to increased activity of pyridoxal-dependent taurine-2-oxoglutarate transaminase mediated by the lower Km of PDXK. This also suggests that other metabolites of pyridoxal-dependent enzymes could serve as early biomarkers of ethanol-induced hepatic effects that could predict later events including steatosis, fibrosis, and cirrhosis (Fig. 8C). This idea deserves further investigation. Additionally, whether activation and/or approaches that could lead to increased expression of PPARβ/δ could be targeted for the prevention or treatment of ALD should be examined. For example, since Huh7 cells express low levels of ethanol metabolizing enzymes (i.e. ALDH, CYP2E1) (Plumlee et al. 2005), this suggests that the inhibition of PDXK and alteration of PDXK Km are due to secondary changes induced by ethanol metabolites. Thus, results from this in vitro model suggest that over-expression and/or activation of PPARβ/δ would be effective for preventing ethanol metabolite-induced changes in PDXK activity and/or ALD. Lastly, given that polymorphisms exist in the PPARβ/δ gene (Burch et al. 2010; Burch et al. 2009; Holzapfel et al. 2006; Jguirim-Souissi et al. 2010; Stefan et al. 2007), it remains possible that some humans could be at more or less risk for developing ALD, depending on the functionality of the polymorphic PPARβ/δ they express. Further studies of this possibility, including whether activating PPARβ/δ can prevent or inhibit progression of ALD, also deserve further evaluation.
Acknowledgments
Funding information
This research was supported by R01 AA018863 from the National Institutes of Health (Fornace and Peters) and U01 ES016013 (Fornace).
The authors gratefully acknowledge the Proteomic and Metabolomic Shared Resources at Georgetown University and Yue Luo for UPLC-TOFMS operation and sample analysis and Susan Mitchel for technical support for these studies.
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adhikary T, Kaddatz K, Finkernagel F, Schonbauer A, Meissner W, Scharfe M, Jarek M, Blocker H, Muller-Brusselbach S, Muller R. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) PLoS ONE. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama TE, Nicol CJ, Fievet C, Staels B, Ward JM, Auwerx J, Lee SS, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-α regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J Biol Chem. 2001;276:39088–39093. doi: 10.1074/jbc.M107073200. [DOI] [PubMed] [Google Scholar]
- Borland MG, Khozoie C, Albrecht PP, Zhu B, Lee C, Lahoti TS, Gonzalez FJ, Peters JM. Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes. Cell Signal. 2011;23:2039–2050. doi: 10.1016/j.cellsig.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruha R, Dvorak K, Petrtyl J. Alcoholic liver disease. World journal of hepatology. 2012;4:81–90. doi: 10.4254/wjh.v4.i3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch LR, Donnelly LA, Doney AS, Brady J, Tommasi AM, Whitley AL, Goddard C, Morris AD, Hansen MK, Palmer CN. Peroxisome proliferator-activated receptor-delta genotype influences metabolic phenotype and may influence lipid response to statin therapy in humans: a genetics of diabetes audit and research Tayside study. J Clin Endocrinol Metab. 2010;95:1830–1837. doi: 10.1210/jc.2009-1201. [DOI] [PubMed] [Google Scholar]
- Burch LR, Zhou K, Donnelly LA, Doney AS, Brady J, Goddard C, Morris AD, Hansen MK, Palmer CN. A single nucleotide polymorphism on exon-4 of the gene encoding PPARδ is associated with reduced height in adults and children. J Clin Endocrinol Metab. 2009;94:2587–2593. doi: 10.1210/jc.2009-0392. [DOI] [PubMed] [Google Scholar]
- Fonda ML, Brown SG, Pendleton MW. Concentration of vitamin B6 and activities of enzymes of B6 metabolism in the blood of alcoholic and nonalcoholic men. Alcohol Clin Exp Res. 1989;13:804–809. doi: 10.1111/j.1530-0277.1989.tb00426.x. [DOI] [PubMed] [Google Scholar]
- Foreman JE, Chang WC, Palkar PS, Zhu B, Borland MG, Williams JL, Kramer LR, Clapper ML, Gonzalez FJ, Peters JM. Functional characterization of peroxisome proliferator-activated receptor-β/δ expression in colon cancer. Mol Carcinog. 2011;50:884–900. doi: 10.1002/mc.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsted CH, Medici V. Vitamin-dependent methionine metabolism and alcoholic liver disease. Adv Nutr. 2011;2:421–427. doi: 10.3945/an.111.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel J, Heun R, Lutjohann D, Jessen F, Maier W, Kolsch H. PPARD haplotype influences cholesterol metabolism but is no risk factor of Alzheimer’s disease. Neuroscience letters. 2006;408:57–61. doi: 10.1016/j.neulet.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Jguirim-Souissi I, Jelassi A, Hrira Y, Najah M, Slimani A, Addad F, Hassine M, Hamda KB, Maatouk F, Rouis M, Slimane MN. +294T/C polymorphism in the PPAR-δ gene is associated with risk of coronary artery disease in normolipidemic Tunisians. Genetics and molecular research: GMR. 2010;9:1326–1333. doi: 10.4238/vol9-3gmr831. [DOI] [PubMed] [Google Scholar]
- Ji C, Chan C, Kaplowitz N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. Journal of hepatology. 2006;45:717–724. doi: 10.1016/j.jhep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatzoglou A, Smola A, Hornik K, Zeileis A. Kernlab – An S4 package for kernel methods in R. Journal of Statistical Software. 2004;11:1–20. [Google Scholar]
- Khozoie C, Borland MG, Zhu B, Baek S, John S, Hager GL, Shah YM, Gonzalez FJ, Peters JM. Analysis of the peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) cistrome reveals novel co-regulatory role of ATF4. BMC genomics. 2012;13:665. doi: 10.1186/1471-2164-13-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi T, Obori H. Effect of pantothenic acid and vitamin B6 on the excretion of taurine in the urine of rats. The Tohoku journal of experimental medicine. 1965;86:394–396. doi: 10.1620/tjem.86.394. [DOI] [PubMed] [Google Scholar]
- Kwok F, Churchich JE. Brain pyridoxal kinase. Purification, substrate specificities, and sensitized photodestruction of an essential histidine. J Biol Chem. 1979;254:6489–6495. [PubMed] [Google Scholar]
- Lieber CS, DeCarli LM. Animal models of chronic ethanol toxicity. Methods Enzymol. 1994;233:585–594. doi: 10.1016/s0076-6879(94)33061-1. [DOI] [PubMed] [Google Scholar]
- Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, Fornace AJ, Jr, Gonzalez FJ. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. Journal of proteome research. 2010;9:4176–4188. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Kamijo Y, Tanaka N, Sugiyama E, Tanaka E, Kiyosawa K, Fukushima Y, Peters JM, Gonzalez FJ, Aoyama T. Peroxisome proliferator-activated receptor α protects against alcohol-induced liver damage. Hepatology. 2004;40:972–980. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter D, Cao M, Lin HM, Fraser K, Edmunds S, Lane G, Rowan D. Identification of urinary biomarkers of colon inflammation in IL10−/− mice using Short-Column LCMS metabolomics. Journal of biomedicine & biotechnology. 2011;2011:974701. doi: 10.1155/2011/974701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Lee SST, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Growth, adipose, brain and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β/δ. Molecular and Cellular Biology. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumlee CR, Lazaro CA, Fausto N, Polyak SJ. Effect of ethanol on innate antiviral pathways and HCV replication in human liver cells. Virology journal. 2005;2:89. doi: 10.1186/1743-422X-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Xie X, Fan Y, Tian J, Guan Y, Wang X, Zhu Y, Wang N. Peroxisome proliferator-activated receptor-δ induces insulin-induced gene-1 and suppresses hepatic lipogenesis in obese diabetic mice. Hepatology. 2008;48:432–441. doi: 10.1002/hep.22334. [DOI] [PubMed] [Google Scholar]
- Shan W, Nicol CJ, Ito S, Bility MT, Kennett MJ, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-β/δ protects against chemically induced liver toxicity in mice. Hepatology. 2008a;47:225–235. doi: 10.1002/hep.21925. [DOI] [PubMed] [Google Scholar]
- Shan W, Palkar PS, Murray IA, McDevitt EI, Kennett MJ, Kang BH, Isom HC, Perdew GH, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor β/δ (PPARβ/δ) attenuates carbon tetrachloride hepatotoxicity by downregulating proinflammatory gene expression. Toxicol Sci. 2008b;105:418–428. doi: 10.1093/toxsci/kfn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan N, Thamer C, Staiger H, Machicao F, Machann J, Schick F, Venter C, Niess A, Laakso M, Fritsche A, Haring HU. Genetic variations in PPARD and PPARGC1A determine mitochondrial function and change in aerobic physical fitness and insulin sensitivity during lifestyle intervention. J Clin Endocrinol Metab. 2007;92:1827–1833. doi: 10.1210/jc.2006-1785. [DOI] [PubMed] [Google Scholar]
- Stubbs MA, Morgan MY. Managing alcohol dependence and alcohol-related liver disease: a problem for the hepatologist, psychiatrist or economist? Clin Med. 2011;11:189–193. doi: 10.7861/clinmedicine.11-2-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussmane S, Koontz J. A fluorometric assay for pyridoxal kinase applicable to crude cell extracts. Anal Biochem. 1995;225:109–112. doi: 10.1006/abio.1995.1115. [DOI] [PubMed] [Google Scholar]
- Toney MD. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim Biophys Acta. 2011;1814:1407–1418. doi: 10.1016/j.bbapap.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo GA, O’Brien C. Long-term management of alcoholic liver disease. Clinics in liver disease. 2012;16:763–781. doi: 10.1016/j.cld.2012.08.007. [DOI] [PubMed] [Google Scholar]