Abstract
Despite the emergence of non-O157 Shiga toxin-producing Escherichia coli (STEC) infections, E. coli serotype O157 is still the most commonly identified STEC in the world. It causes high morbidity and mortality and has been responsible for a number of outbreaks in many parts of the world. Various methods have been developed to detect this particular serotype, but standard bacteriological methods remain the gold standard. Here, we propose a new peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) method for the rapid detection of E. coli O157. Testing on 54 representative strains showed that the PNA probe is highly sensitive and specific to E. coli O157. The method then was optimized for detection in food samples. Ground beef and unpasteurized milk samples were artificially contaminated with E. coli O157 concentrations ranging from 1 × 10−2 to 1 × 102 CFU per 25 g or ml of food. Samples were then preenriched and analyzed by both the traditional bacteriological method (ISO 16654:2001) and PNA-FISH. The PNA-FISH method performed well in both types of food matrices with a detection limit of 1 CFU/25 g or ml of food samples. Tests on 60 food samples have shown a specificity value of 100% (95% confidence interval [CI], 82.83 to 100), a sensitivity of 97.22% (95% CI, 83.79 to 99.85%), and an accuracy of 98.33% (CI 95%, 83.41 to 99.91%). Results indicate that PNA-FISH performed as well as the traditional culture methods and can reduce the diagnosis time to 1 day.
INTRODUCTION
Escherichia coli strains include a genetically heterogeneous group of bacteria which are typically nonpathogenic (1, 2). However, a considerable number of strains are recognized as important pathogens. There are 6 classes of pathogenic E. coli: enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC), diffusely adherent E. coli (DAEC), and enterohemorrhagic E. coli (EHEC) (1). Among pathogenic E. coli strains, EHEC strains are perhaps the most important because of their virulence and association with life-threatening complications (1, 2). Within the EHEC group, E. coli serotype O157:H7 (the serotype is based on the O [Ohne] antigen, determined by cell wall lipopolysaccharide, and the H [Haunch] antigen due to the flagellum protein) is the most commonly isolated (2, 3).
The infectious dose of E. coli O157:H7 is reported to be as few as 10 cells, lower than that of most enteric pathogens (4, 5). Three major virulence factors have been identified in this bacterium, including the production of Shiga toxins, a pathogenicity island called the locus of enterocyte effacement, and an F-like plasmid, pO157 (4, 6). Among these virulence factors, the role of pO157 is the least understood (7). The most critical is the production of one or two phage-encoded Shiga toxins, called Stx1 and Stx2. These Shiga toxins are among the most potent cytotoxins currently known to affect eukaryotic cells (4, 6).
The effects of an EHEC infection range from asymptomatic to lethal. In severe cases the patients can develop serious diseases, such as hemolytic-uremic syndrome (HUS) and thrombocytopenic thrombotic purpura (TTP), or even die (4). For E. coli O157:H7 outbreaks reported in the United States, 25% of affected persons were hospitalized, 5 to 10% developed HUS or TTP, and 1% died (3).
Regarding the environmental reservoirs, cattle are considered the primary and natural reservoir, but other animals, such as goats, sheep, and pigs, may be carriers as well (8). Results from a study of 90 outbreaks occurring between 1982 and 2006 showed that in 42% of cases the source of transmission to humans was associated with food (such as ground beef, ready-to-eat products, and vegetables), 12.2% with dairy products (such as cheese and milk), 7.8% with animal contact, 6 to 7% with water, and 2.2% with the environment. The transmission source was unknown in 28.9% of the outbreaks (3).
Because of the importance of quality control in the food industry, the methods used to detect bacterial contaminants must be rapid, sensitive, and reliable, as well as versatile, in order to accommodate the dynamic needs of the food processing plant. Initially testing for sorbitol fermentation has been suggested as a simple means to screen for E. coli O157:H7, because it lacks the β-glucuronidase enzyme (9–11). Most of the existing culture methods were developed based on the above-mentioned feature, as well as the strain's inability to ferment rhamnose and its tolerance to tellurite (9, 10, 12). These plating techniques remain an integral aspect of quality control during food processing, because they are cost-effective and technically simple, with a high level of accuracy and sensitivity. However, they are very time-consuming and laborious, and they fail to detect E. coli O157:H7 bacteria that ferment sorbitol or are susceptible to tellurite. They also fail in the detection of samples with low numbers of pathogens (<200 CFU/g sample) (11, 13–15). Moreover, culture-based methods, such as ISO 16654:2001 (horizontal method for the detection of Escherichia coli O157), usually include an agglutination assay (detecting the O157 or H7 antigen) that is not specific, since the O157 and H7 antigens are present in other E. coli species. These antibodies can also cross-react with other E. coli serotypes, Escherichia species, and other members of the Enterobacteriaceae family (11, 16).
Fluorescence in situ hybridization (FISH) is a molecular assay that is widely applied for bacterial identification and localization within samples. This method is based on the specific binding of small oligonucleotides (probes) to particular rRNA regions due to its high cellular abundance, universal distribution, and use as a phylogenetic marker. More recently, peptide nucleic acid probes (PNA) have been developed for microbial detection (17, 18). These molecules mimic DNA and establish a stronger bond, since they have a neutrally charged repeated N-(2-aminoethil) glycine unit instead of the negatively charged sugar-phosphate backbone. The adequate use of this molecule in FISH technology has made the procedure more robust, quicker, and more efficient and allowed the development of several PNA-FISH methods for the detection of important pathogenic organisms (reviewed in reference 18).
Here, we have developed a new PNA-FISH-based method for the specific detection of E. coli O157 in food samples, and we have compared its performance to that of the traditional bacteriological method. To the best of our knowledge, this is the first PNA-FISH method developed to detect a specific serotype of E. coli.
MATERIALS AND METHODS
Bacterial growth and culture media.
The bacterial strains and species used in this study are listed in Table 1. All bacterial species were maintained on tryptic soy agar (TSA) (VWR, Portugal) at 37°C and streaked onto fresh plates every 48 h.
Table 1.
Results of the EcoPNA1169 probe specificity and sensitivity testc
| Straina | Serotype | Isolation origin | Verotoxin production | PNA FISH outcome |
|---|---|---|---|---|
| E. coli CECT 4267 | O157:H7 | Human stool from outbreak of hemorrhagic colitis | Stx1, Stx2 | + |
| E. coli CECT 4782 | O157:H7 | Human stool from outbreak of hemorrhagic colitis | Stx1, Stx2 | + |
| E. coli CECT 4783 | O157:H7 | Raw hamburger meat implicated in hemorrhagic colitis outbreak | Stx1, Stx2 | + |
| E. coli CECT 5947 | O157:H7 | stx2 gene has been replaced | + | |
| E. coli NCTC 12900 | O157:H7 | NT | + | |
| E. coli CCC-1-12 | O157:H7 | Fecal swab | Stx2 | + |
| E. coli CCC-5-12 | O157:H7 | Fecal swab | Stx2 | + |
| E. coli CCC-7-12 | O157:H7 | Fecal swab | Stx2 | + |
| E. coli CCC-10-12 | O157:H7 | Fecal swab | Stx2 | + |
| E. coli CCC-11-12 | O157:H7 | Milk filter | Stx2 | + |
| E. coli CCC-12-12 | O157:H7 | Milk filter | Stx2 | + |
| E. coli CCC-13-12 | O157:H7 | Milk filter | Stx2 | + |
| E. coli CCC-14-12 | O157:H7 | Bovine milk filter | Stx2 | + |
| E. coli CCC-15-12 | O157:H7 | Bovine milk filter | Stx2 | + |
| E. coli CCC-16-12 | O157:H7 | Caprine milk filter | Stx2 | + |
| E. coli CCC-18-12 | O157b | Bovine milk filter | Stx2 | + |
| E. coli CCC-23-12 | O157:H7 | Milk filter | NT | + |
| E. coli CCC-24-12 | O157:H7 | Milk filter | NT | + |
| E. coli CCC-25-12 | O157:H7 | Milk filter | NT | + |
| E. coli CCC-26-12 | O157b | Milk filter | NT | + |
| E. coli CECT 352 | O127a:K63(B8):H- | EPEC | − | |
| E. coli CECT 504 | O141:K85(B):H4 | Swine edema | ND | − |
| E. coli CECT 515T | O1:K1(L1):H7 | Human urine–cystitis | ND | − |
| E. coli CECT 533 | O103:K-:H- | ND | − | |
| E. coli CECT 727 | O111:K58(B4):H- | Infantile gastroenteritis | EPEC | − |
| E. coli CECT 730 | O55:K59(B5):H- | ND | − | |
| E. coli CECT 736 | O28a,28c:K73(B18):H- | Feces | ND | − |
| E. coli CECT 740 | O125a,125b:K70(B15):H19 | Gastroenteritis | ND | − |
| E. coli CECT 744 | O158:K-:h23 | Feces of infant with diarrhea | ND | − |
| E. coli CECT 832 | O111:K58(B4):H- | Infantile gastroenteritis | ND | − |
| E. coli CECT 4537 | O10:K5(L5):H4 | Human peritonitis | ND | − |
| E. coli CECT 4555 | O97:K-:H- | ND | − | |
| E. coli CCC-2-12 | O103 | Fecal swab | NT | − |
| E. coli CCC-3-12 | O26 | Fecal swab | Stx1, Stx2 | − |
| E. coli CCC-4-12 | O26 | Fecal swab | NT | − |
| E. coli CCC-8-12 | O26 | Fecal swab | NT | − |
| E. coli CCC-9-12 | O26 | Fecal swab | NT | − |
| E. coli CCC-19-12 | O26 | Caprine milk filter | Stx1 | − |
| E. coli CCC-20-12 | O26 | Caprine milk filter | Stx1 | − |
| E. coli CCC-21-12 | O26 | Bovine milk filter | Stx1 | − |
| E. coli CCC-22-12 | O26 | Bovine milk filter | Stx1 | − |
| E. coli CECT 434 | O6 | Clinical isolate | ND | − |
| E. coli N9 | ND | Porcine feces | ND | − |
| E. coli N5 | ND | Bovine feces | ND | − |
| E. coli ATCC 29425 (K12) | OR:H48:K- | ND | − | |
| Escherichia hermannii ATCC 33650 | NR | Human isolate | − | |
| Escherichia vulneris ATCC 29943 | Human wound | − | ||
| Shigella boydii ATCC 9207 | ND | − | ||
| Salmonella enterica serovar Typhimurium NCTC 12416 | − | |||
| Salmonella enterica serovar Typhi SGSC 3036 | − | |||
| Salmonella enteritidis SGSC 2476 | − | |||
| Cronobacter sakazakii CECT 858 | Child's throat | − | ||
| Cronobacter sakazakii | Milk | − | ||
| Klebsiella pneumoniae ATCC 11296 | − |
SGSC, Salmonella Genetic Stock Centre; ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures; CECT, Spanish Type Culture Collection.
E. coli O157 strains that tested negative for the presence of H7 antigen.
NT, nontoxigenic E. coli; EPEC, enteropathogenic E. coli (epidemiologically implicated as a pathogen, but the virulence mechanism is not related to the excretion of enterotoxins); ND, not determined; NR, nonrelevant information for the present study.
PNA probe design.
To identify potentially useful oligonucleotides to use as probes, 16S and 23S rRNA gene sequences available at the National Centre for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/) were chosen. This selection contained 6 Escherichia coli O157:H7 strains, 6 Escherichia coli non-O157:H7 strains, and 4 other strains from related species belonging to the Enterobacteriaceae family (see Fig. S1 in the supplemental material). The possible regions of interest were selected by sequence alignment using the ClustalW program, available from the European Bioinformatics Institute (EBI; http://www.ebi.ac.uk/Tools/msa/clustalw2/). A conserved region in the 23S rRNA of all E. coli O157:H7 isolates was identified. The criteria for the selection of the final probe sequence included Gibbs free energy, percentage of GC, lack of self-complementary structures, and melting temperature higher than 50°C. The selected sequence was synthesized (Panagene, Daejeon, South Korea), and the oligonucleotide N terminus was attached to Alexa Fluor 594 via a double 8-amino-3,6-dioxaoctanoic acid (AEEA) linker.
Theoretical evaluation of the PNA probe performance.
After the design of the probe, its performance was evaluated to determine the theoretical values for sensitivity and specificity. These parameters were evaluated with ProbeCheck software, available in the ARB Silva database (http://www.arb-silva.de/). For this theoretical estimation, only the good-quality sequences with at least 1,900 bp were considered along with E. coli strain sequences with the designated serotype. The probe was tested against the large-subunit ([LSU]; 23/28S) database and the small-subunit ([SSU]; 16/18S) database. Theoretical values were determined as previously reported by Almeida et al. (19). Briefly, specificity was calculated as (nECs/TnECs) × 100, where nECs is the number of non-Escherichia coli O157:H7 strains that did not react with the probe and TnECs is the total number of non-Escherichia coli O157:H7 strains examined. Sensitivity was calculated as (ECs/TECs) × 100, where ECs is the number of E. coli O157:H7 strains detected by the probe and TECs is the total number of E. coli O157:H7 strains present in the database. Accuracy was determined as the number of correct results divided by the number of all returned results (20).
Hybridization protocol optimization.
The PNA-FISH protocol was performed on glass slides as previously described (19), with some modifications. In order to understand the behavior of the probe and infer the best hybridization conditions, the hybridization temperature ranged between 53 and 61°C; the fixation step using ethanol was tested between 50 and 80%, and different hybridization times (30, 45, 60, and 90 min) were assessed.
After the optimization of all parameters described above, the procedure that was found to result in the strongest fluorescent signal was as follows. Smears of each strain were prepared by standard procedures and immersed in 4% (wt/vol) paraformaldehyde (Sigma), followed by 50% (vol/vol) ethanol for 10 min each, and allowed to air dry. The smears were then covered with 20 μl of hybridization solution containing 10% (wt/vol) dextran sulfate (Sigma), 10 mM NaCl (Sigma), 30% (vol/vol) formamide (Sigma), 0.1% (wt/vol) sodium pyrophosphate (Sigma), 0.2% (wt/vol) polyvinylpyrrolidone (Sigma), 0.2% (wt/vol) Ficoll (Sigma), 5 mM disodium EDTA (Sigma), 0.1% (vol/vol) Triton X-100 (Sigma), 50 mM Tris-HCl (pH 7.5; Sigma), and 200 nM EcoPNA1169 probe. Samples were covered with coverslips, placed in moist chambers, and incubated for 45 min at 59°C. Subsequently, the coverslips were removed and the slides were submerged in a prewarmed (59°C) washing solution containing 15 mM NaCl (Sigma), 1% (vol/vol) Triton X-100 (Sigma), and 5 mM Tris base (pH 10; Sigma). Washing was performed at 59°C for 30 min, and the slides were allowed to air dry. The slides were stored in the dark for a maximum of 24 h before microscopy. The experimental specificity and sensitivity of the probe were evaluated with the equations used for the theoretical calculation of specificity and sensitivity described above.
Characterization of the E. coli isolates.
The E. coli isolates were evaluated for the presence of the O157 and H7 antigens and also for the presence of verotoxin genes stx1 and stx2. For the serological evaluation, the latex agglutination test Wellcolex E. coli O157:H7 (Oxoid) was used according to the manufacturer's instructions. Briefly, the bacteria were grown in TSB until the exponential phase. Forty-μl drops then were placed in two circles on the reaction cards for each sample. For each test sample, one drop of the O157 or H7 test latex was added to one circle and one drop of the O157 or H7 test latex was added to the other circle. The contents of the samples were mixed and spread over the entire circle area. Samples were inspected for the presence of agglutination after 1 min.
The presence of the verotoxin genes was evaluated as previously described (21, 22). Briefly, 1-ml samples of TSB cultures, grown overnight, were pelleted, and genomic DNA was extracted using the DNeasy tissue kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions. DNA was screened for the presence of stx1 and/or stx2 genes by PCR (Peltier thermal cycler [PTC-200]) using the primers and conditions previously reported (21, 22). PCR products were separated by electrophoresis on a 1.5% (wt/vol) agarose gel and visualized under UV light (GelDoc 2000 system; Bio-Rad Laboratories, Hercules, CA) by ethidium bromide staining (10 mg/ml).
Cell inactivation treatment.
E. coli O157:H7 (CECT 4267) overnight culture aliquots (adjusted to an optical density at 600 nm [OD600] of ∼0.1) were used in the experiments of cell inactivation. The aliquots (1 ml) were heat treated in a thermoblock for 20 min at 72°C or autoclaved (20 min at 121°C) (23). Cells were immediately processed. For PNA-FISH analysis, 20 μl of both suspensions was fixed and hybridized according to the procedure described above. Loss of cell viability was evaluated by the inoculation of 100 μl of the treated cell suspensions into 10 ml of TSB (incubated at 37°C for 24 h, 120 rpm) and by agar plating of 100 μl of the same suspensions (TSA plates incubated at 37°C for 24 h).
Food sample inoculation and preenrichment.
For artificial food contamination, a loopful of E. coli O157:H7 CECT 4267 was transferred to 20 ml of tryptic soy broth (TSB) and incubated overnight (∼18 h) at 37°C and 120 rpm in an orbital incubator. Cells were then suspended in a phosphate-buffered saline (PBS) solution and adjusted to a cell density corresponding to approximately 1 × 108 cells/ml. Cells were further diluted in PBS to obtain the desired cell concentration for inoculation into food samples. Cell concentrations were confirmed by plating on TSA. For the testing on food samples, two matrices obtained from a local retailer (Pingo Doce, Braga, Portugal) were selected, ground beef and unpasteurized milk. Twenty-five g or ml from each type of food was mixed with 225 ml of prewarmed buffered peptone water (BPW; Liofilchem) or mTSB+N (modified tryptic soy broth supplemented with novobiocin; Oxoid) in sealed stomacher bags (with filters). The samples were then artificially contaminated with Escherichia coli O157:H7 at concentrations ranging from 0.01 to 100 CFU/25 g or ml of food. Samples were then homogenized with a stomacher (Seward 3500) for 1 min and transferred to 500-ml flasks. A noninoculated food sample was included for each experiment to check for any possible natural contamination with E. coli O157. This experiment was repeated with a different strain, the isolate E. coli O157:H7 CCC-05-12. Three independent assays were performed for each experiment and each strain.
Detection in food samples using conventional bacteriological methods.
The detection of E. coli O157:H7 by culture-based methods was performed according to ISO 16654:2001. Briefly, the artificially contaminated samples, prepared in mTSB+N as described above, were incubated overnight at 37 or 41.5°C with agitation at 120 rpm. After preenrichment, 1-ml samples were taken for the immunomagnetic separation step using a Dynabeads MAX E. coli O157 kit (Invitrogen). The samples were mixed with a 20-μl suspension of microspheres coated with anti-E. coli O157 antibodies, followed by a 3-min separation phase. After the separation phase, the enrichment medium is withdrawn and the coated microspheres washed and then resuspended in 100 μl of buffer (provided with the kit). This suspension was then inoculated in selective media, CT-SMAC (cefixime-tellurite sorbitol MacConkey agar; Oxoid) and CHROmagar O157, for 24 h at 37°C. Suspect colonies were inoculated in TSA and then tested by the Kovac reagent (Remel) and, in the case of a positive outcome, tested for immunoagglutination in latex (Oxoid).
Detection in food samples using PNA-FISH.
After an overnight preenrichment (18 to 24 h) at 37 or 41°C in BPW or mTSB+N, 20-μl samples were taken and placed directly in the microscope slide. Alternatively, 15-μl samples were mixed with 15 μl of a Triton X-100 solution (1%) directly on the microscope slides. A quick centrifugation step (10,000 × g for 5 min) of a 1-ml sample was also tested to try to remove some autofluorescence particles. Twenty μl then was also placed on the slide. All samples were dried (approximately 5 min at 59°C), and then hybridization was performed as described above.
Microscopy visualization.
The smears were mounted with one drop of nonfluorescent immersion oil (Merck) and analyzed using an Olympus BX51 (Olympus Portugal SA, Porto, Portugal) epifluorescence microscope equipped with one filter sensitive to the Alexa Fluor 594 molecule attached to the EcoPNA1169 probe (excitation, 530 to 550 nm; barrier, 570 nm; emission long-pass filter, 591 nm). Other filters present in the microscope that were not capable of detecting the EcoPNA1169 probe fluorescent signal were used in order to confirm that cells did not autofluoresce. For every experiment, a negative control was performed simultaneously for which all steps described above were carried out, but no probe was added during the hybridization procedure. All images were acquired using the Olympus CellB (Olympus Portugal) software with a magnification of ×1,000.
RESULTS AND DISCUSSION
Probe design and testing.
Based on the selection criteria described in Materials and Methods, the following probe sequence was selected: 5′-CAA CAC ACA GTG TC-3′. This sequence hybridizes between positions 1169 and 1183 of E. coli O157:H7 strain TW14359 (accession number CP_001368); hence, it was named EcoPNA1169.
The theoretical parameters of the probe were evaluated in silico using the probeCheck program coupled with the ARB SILVA rRNA database. The probe was aligned with a total of 180,344 sequences present in the large-subunit ([LSU]; 23/28S) database. EcoPNA1169 matched 80 E. coli O157:H7 sequences (all O157 sequences present in the database) and 11 nontarget sequences, with a total of 91 matches (last accession date, August 2012). The 11 non-E. coli O157 strains matched by EcoPNA1169 included 2 E. coli O55:H7 strains, 1 Escherichia hermannii strain, 7 Salmonella sp. strains, and 1 Cronobacter sakazakii strain. Despite the alignment of 7 Salmonella strains, this number represents less than 2% of the Salmonella sequences present in the database. Moreover, none of the Salmonella strains tested in this work have shown cross-hybridization with EcoPNA1169 (Table 1). The match of two E. coli O55:H7 isolates may represent a drawback, since this serotype, despite also being diarrheagenic, is an EPEC strain. This cross-hybridization with O55 strains has been reported for several other PCR and enzyme-linked immunosorbent assay (ELISA) methods developed to detect O157 (24, 25) and may happen because O55:H7 is the serotype most closely related to O157:H7 (14, 26). Both pathogens express the locus of enterocyte effacement (LEE) island that induces diarrhea via an attachment-effacement mechanism (27). E. coli (STEC) O157:H7 is believed to descend from E. coli O55:H7 that, during evolution, acquired bacteriophage encoding Stx2 and/or Stx1 toxins. Despite the importance of the O55:H7 serotype, EPEC strains are no longer an important cause of diarrhea in developed countries as they were a few decades ago (28).
Concerning the remaining nontarget sequences, despite the alignment with 1 E. hermannii and 1 Cronobacter sp. strain, no cross-hybridization was observed for any Escherichia or Cronobacter species included in the probe experimental test (Table 1).
Based on this in silico evaluation, the estimated theoretical specificity and sensitivity for the EcoPNA1169 probe were 99.95 and 100%, respectively.
To test the probe experimental specificity and sensitivity, the PNA-FISH procedure was applied to a total of 54 strains (Table 1). Twenty E. coli O157 and 25 E. coli non-O157 strains were included. Additionally, 9 strains belonging to the same genus and family (Escherichia, Salmonella, Enterobacter, Shigella, and Klebsiella) were also included. Results show that the hybridization only occurs with E. coli O157 (Table 1); therefore, specificity and sensitivity values for both were 100% (95% CI for specificity, 87.02 to 100%; 95% CI for sensitivity, 79.95 to 100%). As such, EcoPNA1169 has proven to be highly specific and sensitive for the detection of E. coli O157. On the other hand, 5 nontoxigenic O157 strains and two O157 non-H7 strains (both tested negative for H7 antigen) were detected. This indicates that the procedure is specific for O157 despite the H7 and toxin presence. Actually, this might be an important advantage for the identification of other E. coli (EHEC) O157 non-H7 strains. For instance, O157:H- strains, which are EH sorbitol-fermenting strains frequently isolated in Europe, are commonly associated with large outbreaks of HUS in Germany (15, 29, 30). Although they have not been included in this study, the detection of O157:H- strains is likely to occur.
Another issue in the development of molecular detection methods is the ability to distinguish dead from viable cells (31). This is not a crucial evaluation for this method, since a preenrichment step will be used to amplify the E. coli O157 population. Nonetheless, the heat-inactivated bacteria showed a weak fluorescence signal (see Fig. S2 in the supplemental material), indicating that misdetection is unlikely to occur.
Reduction of autofluorescence signal.
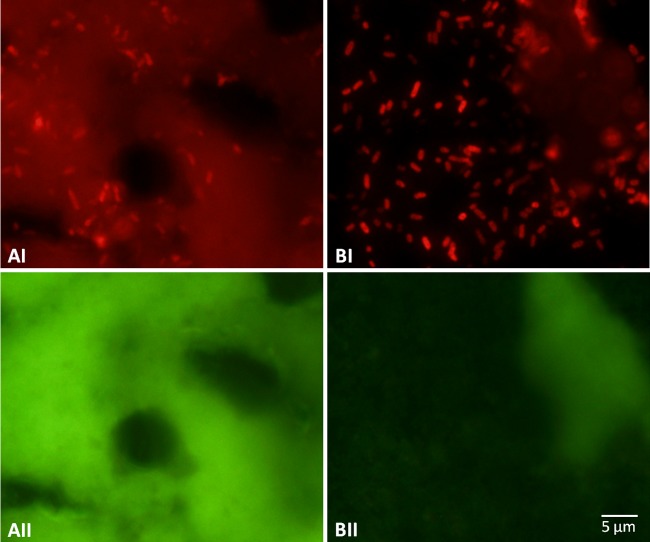
An important feature to bear in mind when optimizing FISH protocols is that some food components may present a strong autofluorescence signal, which can interfere with bacterial detection (32). To eliminate/reduce this phenomenon, an additional step can be added before the hybridization procedure. We have tested two different approaches: a centrifugation step (to remove some autofluorescent food particles) and the use of a detergent (1% [vol/vol] Triton X-100) to emulsify the fatty compounds. Both steps decreased the autofluorescence signal, but the detergent presented a stronger reduction and also seems to improve the fluorescence signal (Fig. 1). This may happen because detergents can also assist in cell permeabilization (33). Subsequent analyses included this additional step.
Fig 1.
PNA-FISH outcome for ground beef samples artificially inoculated with 10 CFU/25 g of E. coli O157:H7 CCC-05-12. Results were obtained using a direct hybridization protocol without any additional sample pretreatment (A) or using a pretreatment with 1% Triton X-100 (B). It is possible to observe a decrease in the autofluorescence intensity for panel B in both red (I) and green (II) channels.
Preenrichment optimization.
The preenrichment step is also recognized as a limiting step in several microbiological detection methods, mainly due to low numbers of the target bacteria, high levels of competing microflora, and technique detection limit (34, 35). A careful optimization of this step is of great important to achieve high values for sensitivity.
The food samples containing E. coli O157:H7 usually present low contamination levels. As the described PNA-FISH detection limit is approximately 105 cells per ml (27), an enrichment step is recommended. This enrichment step can be performed using several types of culture media, from complex rich media (such as TSB or BPW) to selective media, such as Gram-negative (GN) broth, R&F enrichment broth (R&F-EB), or E. coli (EC) broth (34).
TSB is reported as the most frequently used enrichment broth. Additionally, antibiotics, such as novobiocin (the most commonly used), cefixime, cefsulodin, and vancomycin, as well as other selective compounds (e.g., bile salts to inhibit the non-Enterobacteriaceae strains), are often added to enrichment broths. These media then are incubated for a period that usually ranges between 16 and 24 h (overnight growth) at 35 to 42°C. However, results relating to the enrichment protocol efficacy are rare and differ from one study to another.
Regarding the enrichment temperature, it appears that the incubation temperature is not related to the type of serogroup searched (34). Nevertheless, some authors have shown that O157:H7 strains usually present an optimal average temperature around 40°C, which means that temperature can be used to limit the background microflora and favor E. coli O157 growth. Actually, the ISO recommended for O157 detection in food samples (ISO 16654:2001) includes a preenrichment step in mTSB+N at 41.5°C.
In order to evaluate the influence of the enrichment medium and incubation temperature on the detection level of the PNA-FISH method, two different media (the selective mTSB+N and the complex BPW), at 37 and 41.5°C, were used. Additionally, the PNA-FISH method probe described in this work was tested in two different types of food samples: ground beef and unpasteurized milk (two matrices commonly associated with O157:H7 infections) artificially contaminated with low E. coli O157:H7 concentrations (Table 2).
Table 2.
PNA-FISH results obtained for the detection of E. coli O157:H7 on different food matrices inoculated with concentrations between 0.01 and 100 CFU per 25 g or mlc
| Concn (CFU/25 g or ml) | PNA-FISH result (PP/TPa) for: |
|||||
|---|---|---|---|---|---|---|
| Ground beef |
Unpasteurized milk | |||||
| 37°C |
41.5°C |
|||||
| mTSB+Nb | BPW | mTBS+N | BPW | 37°CmTBS+Nb | 41.5°CmTBS+N | |
| 100 | + (6/6) | + (6/6) | + (6/6) | + (6/6) | + (6/6) | + (6/6) |
| 10 | + (6/6) | + (4/6) | + (6/6) | + (2/6) | +(6/6) | + (6/6) |
| 1 | + (5/6) | − (0/6) | + (5/6) | − (0/6) | + (6/6) | + (6/6) |
| 0.1 | − (0/0) | − (0/0) | − (0/0) | − (0/0) | − (0/0) | − (0/0) |
| 0.01 | − (0/0) | − (0/0) | − (0/0) | − (0/0) | − (0/0) | − (0/0) |
PP/TP, number of samples that tested positive by PNA-FISH/total number of positive samples as determined by the culture method.
Results were considered for the determination of the method sensitivity, specificity, and accuracy.
Results for the three independent assays, performed for two E. coli O157:H7 strains, are provided.
As observed in Table 2, mTSB allowed the best detection limit, and the use of higher temperatures (41.5°C) did not seem to improve the detection rate of E. coli O157. The preenrichment in mTSB allowed a positive detection of 5 out of 6 samples for the lower concentration, while the BPW was not able to provide a concentration above the detection limit. The better performance of mTSB may be related to the selective nature of this medium, which includes novobiocin and bile salts that partially inhibit the growth of the competing microflora. However, after growth rate determination for both E. coli strains in BPW and mTSB, it was observed that the medium composition (not the selective factors) was the determinant factor. Growth rates between 1 and 0.8 h−1 were observed for the strains grown on mTSB, while for those grown on BPW the values ranged between 0.2 and 0.4 h−1 for both temperatures (data not shown).
Although the best performance was observed for mTSB+N, when a multiplex approach is desired, it should be possible to standardize the enrichment step to allow the simultaneous detection of different food-borne pathogens.
Performance in food samples.
After the selection of the best enrichment conditions, the PNA-FISH method performance was evaluated in two different matrices. For this, results obtained for 60 food samples evaluated by both techniques (see Table S1 in the supplemental material, samples enriched in mTSB+N at 37°C) were used to determine the sensitivity, specificity, and accuracy values for the PNA-FISH method. ISO 16654:2001 was considered the gold standard. Results were consistent with the inoculation levels for both techniques, with only one discrepant result observed (Table 2; see also Table S1 in the supplemental material). Based on these results, PNA-FISH specificity and sensitivity were 100% (95 CI%, 82.83 to 100%) and 97.22% (95 CI%, 83.79 to 99.85%), respectively. Based on these two values, an accuracy of 98.33% (95 CI%, 83.41 to 99.91%) was observed.
Regarding other molecular methods developed to detect E. coli O157, PCR protocols are probably the most widely used. It has recognized advantages over culture and other standard methods for the detection of microbial pathogens, such as specificity, rapidity, accuracy, and capacity to detect small amounts of target nucleic acid in a sample (36). However, a major drawback of PCR detection methods is the occurrence of false-negative results. Works comparing the performance of different PCR-based protocols for O157 detection have shown that some protocols failed to detect a number of samples that were positive by standard culture methods (24, 37, 38). This might be related to inefficient cell lysis (necessary for nucleic acid extraction), nucleic acid degradation, and, more commonly, PCR-inhibitory substances that might be present in food samples (24, 36, 37, 39). Usually PCR inhibition can be solved by diluting the sample 1:10. However, the majority of the published PCR-based approaches for detection of O157 lack an internal amplification control (IAC), which is required in order to monitor the presence of PCR inhibitors. Additionally, they usually are only relatively specific to O157, giving some cross-reactions with O26, O125, O126, O145, and especially with O55 (24, 37).
PNA-FISH has emerged more recently, but it is already established as a robust microbial identification/detection technique (17, 18). As it is not based on an amplification step, this technique is not susceptible to inhibitors. However, similar to what happens with PCR-based protocols, it can present some cross-hybridization with the O55 serotype. An important feature of the PCR protocol is its ability to evaluate the presence of Shiga toxin genes and to specifically identify EHEC. As PNA-FISH targets rRNA sequences, which are universal phylogenetic marks, EcoPNA1169 will have to be combined with a probe targeting a specific sequence of the EHEC group. Regarding the assay time and detection limit, the reported PCR detection limit varies between 104 CFU/ml and 1 CFU/35 g of food sample, but to achieve the desired detection limit of 1 CFU an enrichment step is also required (usually of 16 to 24 h) (24, 34, 40, 41). Sensitivity values are not present in some publications, but they seem to be diverse depending on the length of the enrichment step and PCR protocol. Arthur and coworkers have compared 3 PCR protocols and found sensitivity values between 53 and 98% (24). PCR procedures were not subjected to evaluation in this work; however, based on the reported results, it seems that the PNA-FISH method developed here performs at least as well as the existing PCR protocols.
Concluding remarks.
EcoPNA1169 is highly specific and sensitive to E. coli O157 strains; however, some cross-hybridization may occur with the closely related O55:H7 serotype. The use of selective compounds and higher temperature provided only a limited improvement on the method detection limit. Even so, the use of mTSB+N allowed the PNA-FISH method to reach the desired detection limit of 1 CFU/25 g or ml of food samples.
PNA-FISH performed well in the different matrices tested, and the inclusion of an additional step with Triton X-100 can significantly reduce the interference of autofluorescent food particles. The implementation of the PNA-FISH method can save at least 2 days in the detection of E. coli serotype O157 compared to the traditional bacteriological method; however, the effectiveness of the method detecting the O157:H7 serotype has not been proven. Finally, comparison of results to those of the standard culture method have shown high specificity and sensitivity, with an estimated accuracy of 98.33% (95 CI%, 83.41 to 99.91%).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Portuguese Institute Fundação para a Ciência e Tecnologia (FCT), project PIC/IC/82815/2007. C.A. acknowledges FCT for individual postdoctoral fellowship SFRH/BPD/74480/2010. We also acknowledge Biomode S.A. for providing some supplies for this project.
C.A., J.M.S., R.R., N.F.A., and M.J.V. have submitted a patent request for the PNA probes used in this study (INPI patent request no. 20131000032694, June 2013). L.C. and R.R. were employees of Biomode S.A. at the time of the study.
Footnotes
Published ahead of print 9 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01009-13.
REFERENCES
- 1.Fratamico PM, Smith JL. 2006 Escherichia coli infections, p 205–239 In Cliver DO, Riemann HP, Foodborne infections and intoxications, 3rd ed. Elsevier, New York, NY [Google Scholar]
- 2.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(13 Suppl.):E45–E62 [DOI] [PubMed] [Google Scholar]
- 3.Pennington H. 2010. Escherichia coli O157. Lancet 376:1428–1435 [DOI] [PubMed] [Google Scholar]
- 4.Robinson AL, McKillip J. 2010. Biology of Escherichia coli O157:H7 in human health and food safety with emphasis on sublethal injury and detection, p 1096–1105 In Mendez-Vilas A. (ed), Current research, technology and education topics in applied microbiology and microbial biotechnology. Formatex, Badajoz, Spain [Google Scholar]
- 5.Schmid-Hempel P, Frank SA. 2007. Pathogenesis, virulence, and infective dose. PLoS Pathog. 3:1372–1373. 10.1371/journal.ppat.0030147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect. Immun. 74:4685–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim JY, Yoon J, Hovde CJ. 2010. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 20:5–14 [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon Y, Sofos JN. 2008. Autoinducer-2 activity of gram-negative foodborne pathogenic bacteria and its influence on biofilm formation. J. Food Sci. 73:M140–M147 [DOI] [PubMed] [Google Scholar]
- 9.Zadik PM, Chapman PA, Siddons CA. 1993. Use of tellurite for the selection of verocytotoxigenic Escherichia coli O157. J. Med. Microbiol. 39:155–158 [DOI] [PubMed] [Google Scholar]
- 10.Feng P. 1995. Escherichia coli serotype O157:H7: novel vehicles of infection and emergence of phenotypic variants. Emerg. Infect. Dis. 1:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raji MA, Jiwa SF, Minga MU, Gwakisa PS. 2003. Escherichia coli O157:H7 reservoir, transmission, diagnosis and the African situation: a review. East Afr. Med. J. 80:271–276 [DOI] [PubMed] [Google Scholar]
- 12.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect. Immun. 76:5598–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng P, Lampel KA, Karch H, Whittam TS. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750–1753 [DOI] [PubMed] [Google Scholar]
- 15.Karch H, Bielaszewska M. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H(-) strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice EW, Sowers EG, Johnson CH, Dunnigan ME, Strockbine NA, Edberg SC. 1992. Serological cross-reactions between Escherichia coli O157 and other species of the genus Escherichia. J. Clin. Microbiol. 30:1315–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stender H. 2003. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 3:649–655 [DOI] [PubMed] [Google Scholar]
- 18.Cerqueira L, Azevedo NF, Almeida C, Jardim T, Keevil CW, Vieira MJ. 2008. DNA mimics for the rapid identification of microorganisms by fluorescence in situ hybridization (FISH). Int. J. Mol. Sci. 9:1944–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida C, Azevedo NF, Fernandes RM, Keevil CW, Vieira MJ. 2010. Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples. Appl. Environ. Microbiol. 76:4476–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida C, Cerqueira L, Azevedo NF, Vieira MJ.2013 Detection of Salmonella enterica serovar Enteritidis using real time PCR, immunocapture assay, PNA FISH and standard culture methods in different types of food samples. Int. J. Food Microbiol. 15;161:16–22 [DOI] [PubMed] [Google Scholar]
- 21.Monaghan A, Byrne B, Fanning S, Sweeney T, McDowell D, Bolton DJ. 2013. Serotypes and virulence profiles of atypical enteropathogenic Escherichia coli (EPEC) isolated from bovine farms and abattoirs. J. Appl. Microbiol. 114:595–603 [DOI] [PubMed] [Google Scholar]
- 22.Monaghan A, Byrne B, Fanning S, Sweeney T, McDowell D, Bolton DJ. 2012. Serotypes and virulotypes of non-O157 Shiga-toxin producing Escherichia coli (STEC) on bovine hides and carcasses. Food Microbiol. 32:223–229 [DOI] [PubMed] [Google Scholar]
- 23.Rudi K, Moen B, Drømtorp SM, Holck AL. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthur TM, Bosilevac JM, Nou X, Koohmaraie M. 2005. Evaluation of culture- and PCR-based detection methods for Escherichia coli O157:H7 in inoculated ground beef. J. Food Prot. 68:1566–1574 [DOI] [PubMed] [Google Scholar]
- 25.Sharma VK. 2002. Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J. Food Prot. 65:1371–1380 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, Feng L, Lan R, Reeves PR, Wang L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700. 10.1371/journal.pone.0008700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garmendia J, Frankel G, Crepin VF. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ammon A, Peterson LR, Karch H. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain E. coli O157:H-. J. Infect. Dis. 179:1274–1277 [DOI] [PubMed] [Google Scholar]
- 30.Alpers K, Werber D, Frank C, Koch J, Friedrich AW, Karch H, An Der Heiden M, Prager R, Fruth A, Bielaszewska M, Morlock G, Heissenhuber A, Diedler A, Gerber A, Ammon A. 2009. Sorbitol-fermenting enterohaemorrhagic Escherichia coli O157:H- causes another outbreak of haemolytic uraemic syndrome in children. Epidemiol. Infect. 137:389–395 [DOI] [PubMed] [Google Scholar]
- 31.Li B, Chen JQ. 2012. Real-time PCR methodology for selective detection of viable Escherichia coli O157:H7 cells by targeting Z3276 as a genetic marker. Appl. Environ. Microbiol. 78:5297–5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida C, Azevedo NF, Iversen C, Fanning S, Keevil CW, Vieira MJ. 2009. Development and application of a novel peptide nucleic acid probe for the specific detection of Cronobacter genomospecies (Enterobacter sakazakii) in powdered infant formula. Appl. Environ. Microbiol. 75:2925–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamur MC, Oliver C. 2010. Permeabilization of cell membranes. Methods Mol. Biol. 588:63–66 [DOI] [PubMed] [Google Scholar]
- 34.Vimont A, Vernozy-Rozand C, Delignette-Muller ML. 2006. Isolation of E. coli O157:H7 and non-O157 STEC in different matrices: review of the most commonly used enrichment protocols. Lett. Appl. Microbiol. 42:102–108 [DOI] [PubMed] [Google Scholar]
- 35.Vimont A, Vernozy-Rozand C, Montet MP, Lazizzera C, Bavai C, Delignette-Muller ML. 2006. Modeling and predicting the simultaneous growth of Escherichia coli O157:H7 and ground beef background microflora for various enrichment protocols. Appl. Environ. Microbiol. 72:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C. 2010. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol. Adv. 28:232–254 [DOI] [PubMed] [Google Scholar]
- 37.Fach P, Perelle S, Grout J, Dilasser F. 2003. Comparison of different PCR tests for detecting Shiga toxin-producing Escherichia coli O157 and development of an ELISA-PCR assay for specific identification of the bacteria. J. Microbiol. Methods 55:383–392 [DOI] [PubMed] [Google Scholar]
- 38.Holland JL, Louie L, Simor AE, Louie M. 2000. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 38:4108–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fode-Vaughan KA, Maki JS, Benson JA, Collins ML. 2003. Direct PCR detection of Escherichia coli O157:H7. Lett. Appl. Microbiol. 37:239–243 [DOI] [PubMed] [Google Scholar]
- 40.Burns F, Fleck L, Andaloro B, Davis E, Rohrbeck J, Tice G, Wallace M. 2011. DuPont Qualicon BAX system real-time PCR assay for Escherichia coli O157:H7. J. AOAC Int. 94:1117–1124 [PubMed] [Google Scholar]
- 41.Spano G, Beneduce L, Terzi V, Stanca AM, Massa S. 2005. Real-time PCR for the detection of Escherichia coli O157:H7 in dairy and cattle wastewater. Lett. Appl. Microbiol. 40:164–171 [DOI] [PubMed] [Google Scholar]
- 42.Fratamico PM, DebRoy C. 2010. Detection of Escherichia coli O157:H7 in food using real-time multiplex PCR assays targeting the stx(1), stx(2), wzy(O157), and the fliC(h7) or eae genes. Food Anal. Methods 3:330–337 [Google Scholar]
- 43.Gordillo R, Cordoba JJ, Andrade MJ, Luque MI, Rodriguez M. 2011. Development of PCR assays for detection of Escherichia coli O157:H7 in meat products. Meat Sci. 88:767–773 [DOI] [PubMed] [Google Scholar]
- 44.Ibekwe AM, Watt PM, Grieve CM, Sharma VK, Lyons SR. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauer WF, Tymciu S, Sidi CD, Sonigo P. 2009. Validation of iQ-Check E. coli O157:H7 real-time PCR test kit for detection of Escherichia coli O157:H7 in selected foods. J. AOAC Int. 92:1095–1104 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.