Abstract
Methylmercury (MeHg), a neurotoxic substance that accumulates in aquatic food chains and poses a risk to human health, is synthesized by anaerobic microorganisms in the environment. To date, mercury (Hg) methylation has been attributed to sulfate- and iron-reducing bacteria (SRB and IRB, respectively). Here we report that a methanogen, Methanospirillum hungatei JF-1, methylated Hg in a sulfide-free medium at comparable rates, but with higher yields, than those observed for some SRB and IRB. Phylogenetic analyses showed that the concatenated orthologs of the Hg methylation proteins HgcA and HgcB from M. hungatei are closely related to those from known SRB and IRB methylators and that they cluster together with proteins from eight other methanogens, suggesting that these methanogens may also methylate Hg. Because all nine methanogens with HgcA and HgcB orthologs belong to the class Methanomicrobia, constituting the late-evolving methanogenic lineage, methanogenic Hg methylation could not be considered an ancient metabolic trait. Our results identify methanogens as a new guild of Hg-methylating microbes with a potentially important role in mineral-poor (sulfate- and iron-limited) anoxic freshwater environments.
INTRODUCTION
Mercury (Hg) is a global environmental contaminant whose concentration in the biosphere is increasing as a result of industrial activity. Mercury enters the biosphere mostly in its inorganic form, Hg(II), but public health concerns are focused primarily on the neurotoxic substance monomethylmercury (MeHg). Since MeHg is environmentally persistent and is biomagnified in aquatic food webs (1), in situ methylation reactions critically affect the ecosystem and human health consequences of Hg contamination.
Methylation of Hg takes place in anoxic environments and is attributed largely to anaerobic microbes (2). To date, bacteria affiliated with the Deltaproteobacteria, including sulfate- and iron-reducing bacteria (SRB and IRB, respectively) have been identified as Hg methylators in environmental incubations and in pure cultures (3–5). Indeed, coexisting SRB and IRB may simultaneously contribute to MeHg production in river sediments (6). Evidence that other anaerobic microorganisms are capable of methylating Hg has remained elusive for decades. Methanogens were initially proposed to be Hg methylators as a conclusion of experiments showing MeHg synthesis in cell extracts of Methanobacterium bryantii (7–9). This idea was rejected after later studies identified SRB (3) and IRB (4–6) as the principal Hg methylators in salt marsh and freshwater sediments and failed to show methylation by methanogens in pure cultures (10). However, Hamelin et al. (11) recently showed that Hg methylation in periphyton collected from a freshwater lake could be attributed to methanogens, since methylation was abolished by the addition of 2-bromoethane sulfonic acid, a specific inhibitor of methanogenesis, and was stimulated 45-fold over methylation in unamended controls by the addition of molybdate. A corresponding change in the active-community structure, with a higher abundance of methanogens in molybdate-treated periphyton samples, was also noted (11). The authors, however, did not proceed to examine methylation by methanogens in pure cultures. This possibility was recently highlighted by Parks et al. (12), who identified putative Hg methylation genes in the genomes of several methanogens. These hgcA and hgcB gene homologs in the genomes of methanogens and other microbes could be used to explore the origin and evolution of Hg methylation by relating organisms and gene phylogenies to known events in the history of life on Earth (13). This approach is particularly powerful for examining the impact of Earth oxygenation on traits critical to the metabolism of redox-sensitive metals (14, 15). Here we clearly establish methanogens as a new guild of Hg methylators by showing that the methanogen Methanospirillum hungatei JF-1 (DSM 864) methylates Hg with methylation rates and yields similar to those of some known methylators from the SRB and IRB guilds. Furthermore, phylogenetic reconstruction of Hgc orthologs and their relationships to established archaeal phylogenies (16, 17) suggest that methylation is not an ancient metabolic trait.
MATERIALS AND METHODS
Microorganisms and culture conditions.
Strains M. hungatei JF-1 (DSM 864) and Desulfovibrio africanus subsp. africanus Benghazi (DSM 2603) were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Strain JF-1 was cultured as recommended by the DSMZ (medium 119, with H2 and Na-formate as electron donors and with Na-bicarbonate as the electron acceptor). D. africanus and Desulfovibrio desulfuricans ND132, a gift from C. Gilmour, were grown in the medium of Widdel and Bak (18) by using lactate as the electron donor and sulfate as the acceptor at 32°C. To compare the Hg methylation activities in sulfate-free medium, these two strains were also cultured in a modified SRB medium (19) containing 40 mM pyruvate (electron donor) and 40 mM fumarate (electron acceptor). Geobacter sulfurreducens PCA, a gift from D. Lovley, was cultured in modified ferric citrate-supplemented ATCC medium 1957 using acetate (60 mM) as an electron donor and a carbon source at 30°C. All cultures were grown in serum bottles (130 ml) (Wheaton, Millville, NJ) with a 50%-to-50% ratio of liquid to gas, by volume. The gas mixture was provided as recommended in the original medium formulations. Most media were reduced with 0.25 mM titanium(III) (in 100 μM nitrilotriacetic acid [NTA]) in order to minimize the influence of sulfide complexation with Hg on methylation (20). For example, the reducing agent Na2S in medium 119 for M. hungatei was replaced with Ti(III)-NTA. Prior to Hg methylation experiments, the purity of all strains was confirmed by sequencing of 16S rRNA genes. All manipulations were performed under strictly anaerobic conditions using O2-free gases obtained by passage through a reduced hot copper column for bench handling, or in an anaerobic glove box (Coy Laboratory Products Inc., Grass Lake, MI) with a gas mixture of 95% nitrogen and 5% hydrogen.
Mercury methylation.
Methylation assays were initiated with exponentially growing cultures. Because Desulfovibrio spp. were grown under sulfidogenic conditions for these assays, cultures of all strains were first washed with a sulfide-free medium under strictly anaerobic conditions to remove all traces of sulfide and other metabolic by-products. For each strain, the cells were diluted in a fresh growth medium to a final density of 2.5 × 104 to 2.8 × 104 cells ml−1 (for M. hungatei) or 4.0 × 105 to 4.6 × 105 cells ml−1 (for all other strains). Experiments were performed in triplicate and included heat-killed culture controls (80°C for 1 h) and medium blanks. All glassware used in methylation experiments was acid cleaned.
Radioactive 203HgCl2 (specific activity, 500.8 μCi μmol−1; kindly provided by Christy C. Bridges) was injected into 70-ml cultures in serum bottles to a final concentration of 9.7 to 10.3 nM, and cultures were then statically incubated at either 32 or 37°C in the dark. Ten-milliliter aliquots were withdrawn with a syringe at daily intervals for 5 days for MeHg extraction, and a similar volume of O2-free 100% N2 was added to the bottles to maintain constant pressure. CH3203Hg formed by cultures was extracted into a toluene phase as described previously (10). Levels of MeHg recovery ranged from 90 to 98%. Potential initial Hg methylation rates (fmol MeHg μg protein−1 day−1) were calculated from the linear range of lines describing MeHg concentrations versus time (0 to 32 h for M. hungatei at 32°C, D. desulfuricans, and D. africanus, and 0 to 12 h for G. sulfurreducens and M. hungatei at 37°C). Measured radioactivities were corrected for decay and were converted to the mass of MeHg by using the known specific activity of the isotope.
In order to confirm the production of MeHg, D. africanus and M. hungatei cultures and control treatments were spiked with nonradioactive HgCl2 (30 nM) as described above. MeHg was analyzed after 2 days of incubation by cold vapor atomic fluorescence spectrometry (CVAFS) with a Tekran model 2500 spectrophotometer (Tekran Instruments Corp., Toronto, ON, Canada) following aqueous ethylation with sodium tetraethylborate and separation by gas chromatography (GC) using a modification of EPA method 1630 (21). Percentages of MeHg recovery ranged from 80 to 97%. Differences among treatments or strains were analyzed by analysis of variance (ANOVA) followed by a post hoc test (Tukey's honest significant difference, one-way) using SAS software (SAS Institute, Cary, NC).
Protein assays.
Cell growth during Hg methylation assays was monitored by the change in protein concentrations. One-milliliter aliquots were removed at the same time points at which MeHg concentrations were determined and were centrifuged to pellet the cells. Cell pellets were stored at −20°C prior to analyses. Thawed cell pellets were resuspended in 0.3 ml of filtered (pore size, 0.22 μm) Milli-Q water and 0.1 ml of 0.5 N NaOH, and the suspensions were heated to 85°C for 30 min (22). Protein concentrations in processed samples, including similarly treated bovine serum albumin (BSA) standards, were then measured by the Bradford protein assay as described by the manufacturer (Bio-Rad Laboratories, Hercules, CA) in 96-well microtiter plates. The A595 of samples after 20 min of incubation was measured by a Tecan Sunrise microplate reader (Phenix Research Products) with advanced Magellan data reduction software.
Phylogenetic analysis.
The Mhun_0876 (HgcA ortholog) and Mhun_0875 (HgcB ortholog) loci in M. hungatei (12) were used as queries to identify homologous proteins in methanogens, known Hg methylators, and other microbes using BLASTp searches. A total of 33 HgcA and 10 HgcB orthologs (or paralogs thereof) from methanogens were included in the phylogenetic analysis. The classification of the orthologs and their similarities to the HgcA and HgcB proteins of strains D. desulfuricans ND132 and G. sulfurreducens PCA were analyzed by the Pfam database (23). Gene positions in genomes were checked by the Seed server (24).
HgcA and HgcB orthologs from methanogens, known Hg methylators, and other microbes, and their paralogs from Syntrophobacter fumaroxidans MPOB, a confirmed nonmethylator (25), used here as an outgroup, were aligned using Clustal W2 and Clustal X (26). The alignment blocks of HgcA and HgcB were concatenated (degenerated ends of concatenated HgcA and HgcB were trimmed from 444 to 422 amino acids) for multilocus sequence analysis (27–29). Phylogenetic reconstructions of HgcA, HgcB, and of concatenated HgcA and HgcB were performed by using PhyML (30) and PHYLIP, version 3.69 (31), with LG and JTT models for protein substitution, respectively. The phylogeny of concatenated HgcA and HgcB orthologs was also evaluated by MrBayes (version 3.2.1) (32) with default settings and the mixed models for amino acids with gamma-distributed rate variation across sites modified from the methods described by Wang et al. (33). Tree topologies were sampled every 1,000 generations over a total of 1,000,000 generations. A majority rule consensus tree is calculated from the set of trees that are left after discarding the initial 25% trees as the burn-in.
Phylogenetic trees were outgrouped by a paralog containing corrinoid and ferredoxin domains from S. fumaroxidans MPOB (accession no. YP_846677) for HgcA analyses (see Fig. S3 in the supplemental material), by a ferredoxin paralog from Methanoplanus petrolearius DSM 11571 (accession no. YP_003894799) for HgcB analyses (see Fig. S4 in the supplemental material), or by a group of paralogs from Pyrococcus furiosus DSM 3638, S. fumaroxidans MPOB, Methanococcus maripaludis C5, Methanobacterium formicicum DSM 3637, and/or Desulfotomaculum kuznetsovii DSM 6115 for the concatenated HgcA and HgcB analyses (see Fig. 3; see also Fig. S5 in the supplemental material). All maximum likelihood phylogenies were bootstrapped by 100 replicate analyses. Phylogenetic trees were depicted by TreeGraph 2 (34), Dendroscope 3 (35), or FigTree (version 1.4.0; http://tree.bio.ed.ac.uk/software/figtree/).
Fig 3.
Bayesian multilocus phylogeny of concatenated HgcA and HgcB proteins from known Hg-methylating species (shown in boldface) and orthologs identified by homology to the proteins in M. hungatei JF-1. The bar on the right identifies taxa at the phylum/class level; Chl stands for Chloroflexi, and the Deltaproteobacteria bracket distinguishes iron reducers, sulfate reducers, and two syntrophs. Note that the Chloroflexi sequences are embedded within the Deltaproteobacteria sequences. The tree is outgrouped by paralogs of Hgc proteins belonging to the CdhD family. The bar at the bottom indicates a branch length corresponding to 3 substitutions per 1,000 amino acids. Numbers at branching points represent the posterior probabilities of the Bayesian analyses.
RESULTS AND DISCUSSION
Mercury methylation by M. hungatei JF-1.
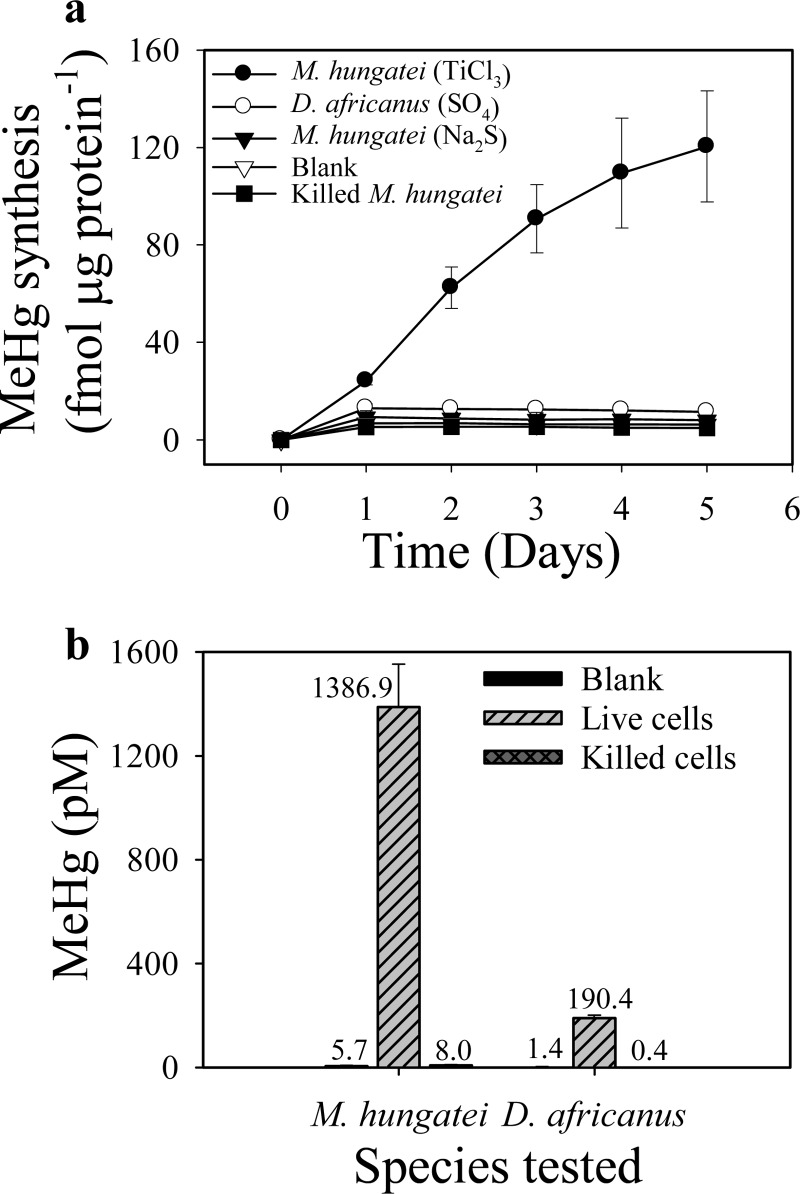
When grown in DSMZ medium 119, M. hungatei produced as much as 120 fmol of toluene-extractable Me203Hg per μg protein (approximately 1 nM Me203Hg) (see Fig. S1a in the supplemental material) over 5 days of incubation (Fig. 1a). In separate cultures, the production of MeHg by M. hungatei JF-1 was confirmed by sample distillation, GC separation of ethylated derivatives, and detection by cold vapor atomic fluorescence spectrometry (Fig. 1b). Here M. hungatei JF-1 produced 1.39 nM MeHg, an amount>170-fold greater than that in the killed controls and uninoculated media.
Fig 1.
Hg methylation by M. hungatei. (a) Hg methylation by M. hungatei (with TiCl3 or Na2S as the reducing agent) and D. africanus (with TiCl3 as the reducing agent and SO4 as an electron acceptor) at 32°C, measured by the conversion of 203Hg(II) to toluene-extractable Me203Hg. (b) Confirmation of MeHg synthesis by M. hungatei (with TiCl3) and D. africanus (with TiCl3 and SO4) after 2-day incubations. MeHg was analyzed by GC separation of ethylated derivatives, followed by CVAFS detection.
Interestingly, methylation was greatly enhanced when we substituted TiCl3 for the recommended reducing agent, Na2S (Fig. 1a). Methylation in the TiCl3-reduced medium was 15-fold higher than that in the Na2S-reduced medium, even though the 5-day growth yields of M. hungatei in the two media were similar (see Fig. S1b in the supplemental material). Decreased Hg methylation in the Na2S-reduced medium was likely due to the binding of Hg(II) by sulfide, which lowers Hg(II) bioavailability, as is well documented for methylating SRB in pure cultures and environmental incubations (36, 37). Indeed, we observed relatively low methylation activity in cultures of D. africanus (DSM 2603) grown under sulfate-reducing conditions (Fig. 1a and b). The inhibitory effect of sulfide on Hg(II) availability may explain why some prior studies failed to detect Hg methylation in methanogens.
Specific Hg methylation rates normalized to protein biomass levels and MeHg yields of M. hungatei were compared to those of model methylating bacteria: two SRB strains, D. desulfuricans ND132 (19) and D. africanus (38), grown without sulfate to avoid sulfide inhibition, and one IRB strain, G. sulfurreducens PCA (5), grown with ferric citrate and acetate (Fig. 2).
Fig 2.
Rates and yields of Hg methylation by M. hungatei and representative SRB and IRB strains. Shown are specific initial methylation rates (normalized to protein levels) obtained after 32 h of incubation (open bars) (left y axis) and maximum MeHg yields after 5 days of incubation (filled squares) (right y axis) of M. hungatei (M. h.), D. africanus (D. a.) (with TiCl3 as the reducing agent and fumarate as an electron acceptor), D. desulfuricans ND132 (D. d.) (with TiCl3 and fumarate), and G. sulfurreducens PCA (G. s.). Data were obtained by Me203Hg extraction from triplicate cultures.
The SRB strains were tested under sulfate-free conditions (with fumarate as the electron acceptor), which resulted in methylation yields higher than those obtained when the strains were grown with sulfate (19). This difference is apparent when the level of methylation by D. africanus grown with lactate and sulfate (Fig. 1) is compared to that in a medium with pyruvate and fumarate (Fig. 2). Obviously, the production of sulfide limited methylation under the former conditions. While M. hungatei had specific methylation rates lower than those of D. desulfuricans ND132 and D. africanus, and comparable to those of G. sulfurreducens, its 5-day maximum yield of MeHg at 32°C was significantly higher than those of D. africanus and G. sulfurreducens PCA (P, <0.01) (Fig. 2). At 37°C, this yield was still significantly higher than that of strain PCA but comparable to that of D. africanus. The relationships between specific rates and yields are greatly impacted by the higher cell biomass of strains PCA and JF-1, which grew actively during the 5 days of incubation, than of the two sulfate reducers (see Fig. S2b in the supplemental material).
To the best of our knowledge, this is the first report of Hg methylation by a pure methanogenic culture. The results reported here, together with those of Hamelin et al. (11) showing that methanogens played a role in Hg methylation in a lake periphyton community, clearly establish methanogens as a new microbial guild of Hg methylators in addition to previously identified methylating SRB and IRB.
The presence of the protein orthologs HgcA (Mhun_0876) and HgcB (Mhun_0875) in the genome of M. hungatei JF-1 may suggest that the Hg methylation mechanism of this methanogen is similar to that of bacterial methylation. Future knockout and complementation studies should clearly test the function of putative methylation genes in this and other methanogens with hgcA and hgcB homologs. Protein BLAST searches and genome comparison analyses showed that the putative HgcA (342 amino acids) of M. hungatei had similarities of 45% and 40% to the protein in G. sulfurreducens PCA and D. desulfuricans ND132, respectively. M. hungatei HgcB (102 amino acids) was 49% similar to the HgcB of strain PCA and 40% similar to that of strain ND132.
Phylogenetic analysis of HgcA and HgcB.
Previous phylogenetic analyses (based on 16S rRNA genes) failed to show evolutionary relationships among Hg-methylating bacteria (6, 25, 37). The identification of HgcA (a corrinoid iron-sulfur protein) and HgcB (a 2[4Fe-4S] ferredoxin) as two essential proteins for Hg methylation in strains ND132 (SRB) and PCA (IRB) (12) supports the hypothesis that corrinoid enzymes are involved in Hg methylation (39, 40) and enables a phylogenetic analysis of these putative functional proteins in microbial genomes among Hg methylators. This analysis may lead to an understanding of how Hg methylation has evolved and what ecological significance it might have (41).
We analyzed the similarities and possible evolutionary relationships of the putative Hg methylation proteins of M. hungatei and other methanogens with those of known methylators (see Fig. S3 and S4 in the supplemental material). Phylogenetic analyses showed that HgcA- and HgcB-like proteins from M. hungatei clustered tightly with orthologs from eight other species of methanogens and that this whole group of nine methanogens was strongly affiliated with confirmed Hg-methylating strains of the Geobacter and Desulfovibrio-Desulfobulbus groups (see Fig. S3 and S4). Paralogs of HgcA proteins from all other methanogens formed a large, distant group (see Fig. S3). The strongly supported clustering of HgcA with sequences of known proteobacterial methylators rather than with its CdhD paralogs from methanogens suggests that the eight untested methanogens may also methylate Hg. Multilocus sequence analyses of concatenated HgcA and HgcB orthologs from species that contain both HgcA- and HgcB-like proteins further confirmed the occurrence of M. hungatei within a cluster of nine methanogens that is closely associated with known SRB and IRB methylators (Fig. 3). Maximum likelihood analysis showed a similar clustering pattern of the concatenated HgcA and HgcB (see Fig. S5 in the supplemental material). Thus, the phylogenetic analysis is consistent with our finding that M. hungatei is a Hg-methylating microorganism and suggests that at least eight other methanogens, all belonging to the Methanomicrobia class in the Euryarchaeota, also methylate Hg.
The demonstration of methylation by M. hungatei, a methanogen with HgcA and HgcB orthologs, sheds new light on prior attempts to study methylation by methanogens. Pak and Bartha (42) failed to detect MeHg formation by Methanococcus maripaludis ATCC 43000, a result that could be now explained by the absence of HgcA and HgcB orthologs, although paralogous CdhD proteins are present in the genome of that strain (see Fig. S3 in the supplemental material). Surprisingly, the genome of M. bryantii, which was used by Wood et al. (7) in the first demonstration of microbial methylation in cell extracts, does not include either orthologs or paralogs of HgcA and HgcB. Hg methylation in M. bryantii may therefore follow a biochemical pathway different from that proposed for species with HgcA and HgcB. The possibility that one or more other methylation pathways may exist among methanogens that do not belong to the Methanomicrobia is supported by the findings of Hamelin et al. (11), who showed that methanogens affiliated with the Methanobacteriales and Methanococcales, both orders belonging to early evolving class I methanogens (17, 43), dominated the active community of lake periphyton, where methanogens were documented to be the principal methylators.
Evolutionary and ecological significance of mercury methylation by methanogens.
The phylogenetic analyses employed here do not clearly suggest an ancestry and evolutionary path for Hg methylation. While the two concatenated HgcA and HgcB phylogenies place the Hg-methylating methanogens basal to bacterial clades, the branching points separating the clades are poorly supported in both trees (Fig. 3; see also Fig. S5 in the supplemental material). Furthermore, the methanogenic HgcA cluster branches as a late lineage in the HgcA tree (see Fig. S3 in the supplemental material). Obviously, more-robust phylogenetic analyses are needed to support a hypothesis about how Hg methylation genes have evolved among anaerobic microbes.
Fossil evidence dates methanogenesis back to ∼2.8 billion years ago (43), and methanogens therefore are considered an ancient microbial guild (16, 17). Methanogens belong to two archaeal lineages: an earlier lineage consisting of Methanopyrales, Methanobacteriales, and Methanococcales (the so-called class I methanogens) and a later one (class II) consisting of Methanomicrobiales, Methanocellales, and Methonosarcinales (together, the class Methanomicrobia) (16, 43). All methanogens possessing hgcA and hgcB gene homologs are members of the Methanomicrobia (12). Because the Methanomicrobia likely share a common ancestor with the Halobacteria (16, 43), a lineage consisting of aerobic halophiles, they might have diversified after the oxygenation of Earth ∼2.4 billion years ago. Thus, methanogenic mercury methylation may not be an ancient physiological trait but rather a trait that arose during a period of redox transition in the biosphere. How this transition affected Hg speciation and gave rise to methylation may be one of the more interesting questions raised by our study.
Methanogens are widely distributed in natural habitats where Hg methylation is documented, such as wetland soils, peatlands, sediments of freshwater rivers and lakes, and near-shore and oceanic sea floor (44–46). Methanogenesis is a common terminal process of organic carbon mineralization in anoxic environments and a key process in the global carbon cycle that returns 1% of photosynthetically fixed CO2 to the atmosphere as methane (47), thus enhancing global warming. Many studies of Hg methylation in freshwater lakes and wetlands have focused on methanogenic environments (2, 48). Therefore, it is surprising that methanogens have been identified as methylators in environmental incubations only once (11) since 1985, while SRB have been identified multiple times as the principal methylators (3, 48). Thus, the discovery of Hg methylation by a methanogen raises questions regarding the environmental significance of this process. The discovery of proteins required for Hg methylation in SRB, IRB, and methanogens (12), including the Hg methylator M. hungatei, should assist in the delineation of processes and factors that control which microbial guilds methylate Hg in various environments.
Mercury methylation is a complicated biological process influenced by a variety of abiotic and biotic factors (19, 37). Our study suggests that methanogens could be important contributors to MeHg production in mineral-poor [sulfate- and Fe(III)-limited] anoxic environments. This discovery provides new insight into Hg biogeochemistry and should help elucidate patterns of environmental MeHg production and assist in MeHg remediation in contaminated environments.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Office of Science (BER), U.S. Department of Energy (grant DE503 FG02-08ER64544), and by the NSF Geobiology and Low-Temperature Geochemistry program (EAR-0952291).
We thank Christy C. Bridges for the supply of 203HgCl2, Cindy Gilmour and Derek Lovley for bacterial strains, Chengsheng Zhu for constructive suggestions regarding phylogenetic analyses, and Eric Boyd for stimulating discussions.
Footnotes
Published ahead of print 9 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01556-13.
REFERENCES
- 1.Selin NE. 2011. Science and strategies to reduce mercury risks: a critical review. J. Environ. Monit. 13:2389–2399 [DOI] [PubMed] [Google Scholar]
- 2.Lin C-C, Yee N, Barkay T. 2012. Microbial transformations in the mercury cycle, p 155–191 In Liu G, Cai YO', Driscoll N. (ed), Environmental chemistry and toxicology of mercury. John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 3.Compeau GC, Bartha R. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50:498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleming EJ, Mack EE, Green PG, Nelson DC. 2006. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl. Environ. Microbiol. 72:457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, Mason RP. 2006. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 72:7919–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu RQ, Flanders JR, Mack EE, Turner R, Mirza MB, Barkay T. 2012. Contribution of coexisting sulfate and iron reducing bacteria to methylmercury production in freshwater river sediments. Environ. Sci. Technol. 46:2684–2691 [DOI] [PubMed] [Google Scholar]
- 7.Wood JM, Kennedy FS, Rosen CG. 1968. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature 220:173–174 [DOI] [PubMed] [Google Scholar]
- 8.Bryant MP, Wolin EA, Wolin MJ, Wolfe RS. 1967. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 59:20–31 [DOI] [PubMed] [Google Scholar]
- 9.Boone DR. 1987. Request for an opinion: replacement of the type strain of Methanobacterium formicicum and reinstatement of Methanobacterium bryantii sp. nov. nom. rev. (ex Balch and Wolfe, 1981) with M.o.H. (DSM 863) as the type strain. Int. J. Syst. Bacteriol. 37:172–173 [Google Scholar]
- 10.Pak K, Bartha R. 1998. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 64:1987–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamelin S, Amyot M, Barkay T, Wang Y, Planas D. 2011. Methanogens: principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 45:7693–7700 [DOI] [PubMed] [Google Scholar]
- 12.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. 2013. The genetic basis for bacterial mercury methylation. Science 339:1332–1335 [DOI] [PubMed] [Google Scholar]
- 13.Glass JB, Wolfe-Simon F, Anbar AD. 2009. Coevolution of metal availability and nitrogen assimilation in cyanobacteria and algae. Geobiology 7:100–123 [DOI] [PubMed] [Google Scholar]
- 14.Saito MA, Sigman DM, Morel FMM. 2003. The bioinorganic chemistry of the ancient ocean: the co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean-Proterozoic boundary? Inorg. Chim. Acta 356:308–318 [Google Scholar]
- 15.Barkay T, Kritee K, Boyd E, Geesey G. 2010. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 12:2904–2917 [DOI] [PubMed] [Google Scholar]
- 16.Blank CE. 2009. Phylogenomic dating—the relative antiquity of archaeal metabolic and physiological traits. Astrobiology 9:193–219 [DOI] [PubMed] [Google Scholar]
- 17.Brochier-Armanet C, Forterre P, Gribaldo S. 2011. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr. Opin. Microbiol. 14:274–281 [DOI] [PubMed] [Google Scholar]
- 18.Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p 3353–3378 In Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH. (ed), The Prokaryotes. Springer-Verlag, New York, NY [Google Scholar]
- 19.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, Wall JD. 2011. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl. Environ. Microbiol. 77:3938–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jay JA, Murray KJ, Gilmour CC, Mason RP, Morel FM, Roberts AL, Hemond HF. 2002. Mercury methylation by Desulfovibrio desulfuricans ND132 in the presence of polysulfides. Appl. Environ. Microbiol. 68:5741–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. EPA January 2001. Method 1630: methyl mercury in water by distillation, aqueous ethylation, purge and trap, and CVAFS. EPA-821-R-01-020. Office of Water, Office of Science and Technology, Engineering and Analysis Division, U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 22.Hemme CL. 2004. Examination of metabolic and regulatory networks of Desulfovibrio species. Ph.D. dissertation. University of Missouri, Columbia, MO [Google Scholar]
- 23.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, Guyoneaud R. 2009. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol. J. 26:1–8 [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 27.Anderson I, Ulrich LE, Lupa B, Susanti D, Porat I, Hooper SD, Lykidis A, Sieprawska-Lupa M, Dharmarajan L, Goltsman E, Lapidus A, Saunders E, Han C, Land M, Lucas S, Mukhopadhyay B, Whitman WB, Woese C, Bristow J, Kyrpides N. 2009. Genomic characterization of methanomicrobiales reveals three classes of methanogens. PLoS One 4:e5797. 10.1371/journal.pone.0005797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cano-Gomez A, Hoj L, Owens L, Andreakis N. 2011. Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst. Appl. Microbiol. 34:561–565 [DOI] [PubMed] [Google Scholar]
- 29.Mo S, You M, Su YC, Lacap-Bugler DC, Huo YB, Smith GJ, Leung WK, Watt RM. 2013. Multilocus sequence analysis of Treponema denticola strains of diverse origin. BMC Microbiol. 13:24. 10.1186/1471-2180-13-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package), version 3.65 Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 32.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Boyd E, Crane S, Lu-Irving P, Krabbenhoft D, King S, Dighton J, Geesey G, Barkay T. 2011. Environmental conditions constrain the distribution and diversity of archaeal merA in Yellowstone National Park, Wyoming, U.S.A. Microb. Ecol. 62:739–752 [DOI] [PubMed] [Google Scholar]
- 34.Stover BC, Muller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. 10.1186/1471-2105-11-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61:1061–1067 [DOI] [PubMed] [Google Scholar]
- 36.Benoit JM, Gilmour CC, Mason RP. 2001. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl. Environ. Microbiol. 67:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benoit JM, Gilmour CC, Heyes A, Mason RP, Miller CL. 2003. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems, p 262–297 In Cai Y, Braids OC. (ed), Biogeochemistry of environmentally important trace elements, vol 835 ACS Symposium Series. American Chemical Society, Washington, DC [Google Scholar]
- 38.Ekstrom EB, Morel FM, Benoit JM. 2003. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl. Environ. Microbiol. 69:5414–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi SC, Chase T, Bartha R. 1994. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi SC, Chase T, Jr, Bartha R. 1994. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 60:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulain AJ, Barkay T. 2013. Cracking the mercury methylation code. Science 339:1280–1281 [DOI] [PubMed] [Google Scholar]
- 42.Pak KR, Bartha R. 1998. Mercury methylation and demethylation in anoxic lake sediments and by strictly anaerobic bacteria. Appl. Environ. Microbiol. 64:1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brocks JJ, Logan GA, Buick R, Summons RE. 1999. Archean molecular fossils and the early rise of eukaryotes. Science 285:1033–1036 [DOI] [PubMed] [Google Scholar]
- 44.Conrad R, Erkel C, Liesack W. 2006. Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil. Curr. Opin. Biotechnol. 17:262–267 [DOI] [PubMed] [Google Scholar]
- 45.Franklin MJ, Wiebe WJ, Whitman WB. 1988. Populations of methanogenic bacteria in a Georgia salt marsh. Appl. Environ. Microbiol. 54:1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ver Eecke HC, Butterfield DA, Huber JA, Lilley MD, Olson EJ, Roe KK, Evans LJ, Merkel AY, Cantin HV, Holden JF. 2012. Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc. Natl. Acad. Sci. U. S. A. 109:13674–13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thauer RK, Shima S. 2006. Biogeochemistry: methane and microbes. Nature 440:878–879 [DOI] [PubMed] [Google Scholar]
- 48.Gilmour CC, Henry EA, Mitchell R. 1992. Sulfate stimulation of mercury methylation in freshwater sediments. Environ. Sci. Technol. 26:2281–2287 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.