Abstract
In this surveillance study, we identified the genotypes, carbapenem resistance determinants, and structural variations of AbaR-type resistance islands among carbapenem-resistant Acinetobacter baumannii (CRAB) isolates from nine Asian locales. Clonal complex 92 (CC92), corresponding to global clone 2 (GC2), was the most prevalent in most Asian locales (83/108 isolates; 76.9%). CC108, or GC1, was a predominant clone in India. OXA-23 oxacillinase was detected in CRAB isolates from most Asian locales except Taiwan. blaOXA-24 was found in CRAB isolates from Taiwan. AbaR4-type resistance islands, which were divided into six subtypes, were identified in most CRAB isolates investigated. Five isolates from India, Malaysia, Singapore, and Hong Kong contained AbaR3-type resistance islands. Of these, three isolates harbored both AbaR3- and AbaR4-type resistance islands simultaneously. In this study, GC2 was revealed as a prevalent clone in most Asian locales, with the AbaR4-type resistance island predominant, with diverse variants. The significance of this study lies in identifying the spread of global clones of carbapenem-resistant A. baumannii in Asia.
INTRODUCTION
Although Acinetobacter baumannii had been viewed as a colonizer and was often ignored in the clinical setting until the 1980s, the pathogen has emerged as one of the major causal agents of health care-associated infections, particularly in immunocompromised patients and patients in intensive care units (ICUs) (1). Although A. baumannii is responsible for urinary tract infections, skin and soft tissue infections, and bloodstream infections, it mainly causes pneumonia. In the United States, the proportion of Acinetobacter spp. causing ICU pneumonia increased from 1.4% in 1975 to 6.9% in 2003 (2). A recent study showed that Acinetobacter sp. is a major pathogen causing hospital-acquired pneumonia (HAP) in several Asian locales (3). Acinetobacter sp. was the third most frequently isolated among 2,554 cases of HAP in 10 Asian countries or locales, and it was the most frequently isolated in ventilator-associated pneumonia (VAP). In particular, it was the most prevalent HAP-causing pathogen in Thailand and mainland China (3). With respect to treatment of Acinetobacter infections, resistance to carbapenems is of great concern. Although carbapenems, such as imipenem and meropenem, are usually recommended as potent antimicrobial agents against Acinetobacter infections (4), carbapenem resistance in A. baumannii has been emerging in many parts of the world, and the resistance rate has increased to about 30% (5). In Asia, the prevalence of carbapenem-resistant A. baumannii has been gradually increasing (6).
Since the 1990s, three European clones of A. baumannii (EC I, II, and III) have been reported (7). Of these, EC I and EC II have disseminated worldwide (8) and have been dubbed global clone 1 (GC1) and GC2. GC2, which includes clonal complex 92 (CC92) in the multilocus sequence typing (MLST) scheme of Bartual et al. and Woodford et al. (9, 10), has been identified among carbapenem-resistant A. baumannii isolates in Asian countries, including South Korea and China (11, 12), as well as in European countries and Australia (7, 13). Carbapenem resistance in A. baumannii has also been reported in limited regions, such as South Korea, China, and Taiwan. While the class D oxacillinase OXA-23 is most prevalent in South Korea and China, the most frequently identified oxacillinases in Taiwan are OXA-58 and OXA-24 (6). In A. baumannii isolates from South Korea and Taiwan, blaOXA-23 is accompanied by an AbaR4-type resistance island (14, 15). However, little is known of the clonality and resistance determinants of carbapenem-resistant A. baumannii (CRAB) isolates from many Asian countries. In addition, although AbaR-type resistance islands have been reported in A. baumannii isolates from many regions, little is known about the compositions of different AbaRs in isolates from Asian countries.
In the present study, genotypes, carbapenem resistance determinants, and structural variations of AbaR-type resistance islands among A. baumannii isolates from nine Asian locales were investigated to understand the spread of global clones of carbapenem-resistant A. baumannii in Asia.
MATERIALS AND METHODS
A. baumannii isolates.
All isolates used in this study were collected during the Asian Network for Surveillance of Resistant Pathogens (ANSORP) surveillance study of HAP (3). Two hundred seventy Acinetobacter sp. isolates were available. Initial identification was done with the Vitek2 system (bioMérieux, Hazelwood, MO), and final identification was performed using partial rpoB gene sequences (16, 17). As a result of species identification, 253 isolates (93.7%) were identified as A. baumannii. Ten Acinetobacter nosocomialis (formerly Acinetobacter genomic species 13TU) isolates, five Acinetobacter pittii (formerly Acinetobacter genomic species 3) isolates, one Acinetobacter bereziniae isolate, and one Acinetobacter genomic species “close to 13TU” (Aba-like species or Aba-B group in our previous studies) (18) isolate were identified as non-baumannii Acinetobacter.
In vitro susceptibility testing.
In vitro antimicrobial susceptibility testing was performed with all A. baumannii isolates by measuring the MIC using the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (19). Eleven antimicrobial agents were tested: imipenem, meropenem, polymyxin B, colistin, gentamicin, ceftazidime, cefotaxime, cefepime, ciprofloxacin, trimethoprim-sulfamethoxazole, and piperacillin-tazobactam. CSLI susceptibility interpretive criteria were used (19). Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as control strains.
Genotyping.
For 108 CRAB isolates, genes encoding oxacillinase (OXA) carbapenemases, such as blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like, were identified as previously described (20). The presence of ISAba1 upstream of the bla genes was investigated by PCR, as described previously (21, 22). In addition, blaIMP and blaVIM were also tested by PCR as previously described (22). MLST was performed for 108 CRAB isolates as previously described (9), with some modifications (18). Among 127 CRAB isolates from Thailand, oxacillinase gene assays and MLST were performed for 30 isolates selected at random. On the other hand, all CRAB isolates from locales other than Thailand were subjected to oxacillinase gene assay and MLST analysis. Clonal complexes were determined using the eBURST program (23).
Identification of the resistance island.
To identify the presence of the AbaR-type resistance island and to determine its structure, PCR amplification was carried out for 68 CRAB isolates using previously published primers (14, 24–27). Integration of AbaR-type resistance islands in other sites was investigated based on the method of Šeputienė et al. (28).
RESULTS
Antimicrobial resistance.
Among 253 A. baumannii isolates, 208 (82.5%) isolates were carbapenem resistant, that is, imipenem resistant (202 isolates; 80.2%) or meropenem resistant (206 isolates; 81.8%). The resistance rates of the other antimicrobial agents, except polymyxins, were also very high: 86.5% (ceftazidime), 86.5% (cefotaxime), 82.6% (cefepime), 76.2% (gentamicin), 89.7% (ciprofloxacin), 83.0% (trimethoprim-sulfamethoxazole), and 86.1% (piperacillin-tazobactam). Among 17 non-baumannii Acinetobacter sp. isolates, however, only one to three isolates were resistant to antimicrobial agents except polymyxins (Table 1). All Acinetobacter sp. isolates were susceptible to polymyxin B and colistin. CRAB isolates showed very high resistance rates, except to polymyxins: 85.1% (gentamicin), 96.6% (ceftazidime), 96.6% (cefotaxime), 94.2% (cefepime), 97.6% (ciprofloxacin), 90.9% (trimethoprim/sulfamethoxazole), and 98.1% (piperacillin-tazobactam). One hundred fifty-four A. baumannii isolates (61.1%) were resistant to all antimicrobial agents except polymyxins.
Table 1.
Antimicrobial resistance among Acinetobacter sp. isolates from Asian locales
| Antimicrobial agent | Resistance rate (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
A. baumannii (n = 253)a |
Non-A. baumannii (n = 17) | |||||||||
| Total | HK (n = 16) | IN (n = 14) | KR (n = 18) | MY (n = 42) | PH (n = 5) | SG (n = 2) | TW (n = 14) | TH (n = 141) | ||
| Imipenem | 80.2 | 25.0 | 85.7 | 44.4 | 92.9 | 40.0 | 100 | 64.3 | 90.1 | 11.8 |
| Meropenem | 81.8 | 31.3 | 85.7 | 50.0 | 92.9 | 40.0 | 100 | 71.4 | 90.8 | 11.8 |
| Ceftazidime | 86.5 | 50.0 | 100 | 72.2 | 88.1 | 40.0 | 100 | 92.9 | 91.5 | 5.9 |
| Cefotaxime | 86.5 | 50.0 | 100 | 72.2 | 88.1 | 40.0 | 100 | 92.9 | 91.5 | 5.9 |
| Cefepime | 82.6 | 43.8 | 78.6 | 66.7 | 88.1 | 40.0 | 50.0 | 78.6 | 90.1 | 5.9 |
| Gentamicin | 76.2 | 37.5 | 100 | 55.6 | 76.2 | 20.0 | 50.0 | 92.9 | 81.6 | 17.6 |
| Ciprofloxacin | 89.7 | 75.0 | 100 | 72.2 | 88.1 | 80.0 | 100 | 92.9 | 92.9 | 17.6 |
| Trimethoprim-sulfamethoxazole | 83.0 | 68.8 | 100 | 66.7 | 83.3 | 80.0 | 100 | 85.7 | 84.4 | 11.8 |
| Piperacillin-tazobactam | 86.1 | 56.3 | 92.9 | 72.2 | 92.9 | 40.0 | 100 | 92.9 | 89.4 | 5.9 |
| Polymyxin B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Colistin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| All antibiotics except polymyxins | 60.9 | 6.3 | 71.4 | 33.3 | 71.4 | 20.0 | 50.0 | 57.1 | 68.8 | 0 |
| None of the antibiotics | 7.9 | 18.8 | 0 | 27.8 | 7.1 | 20.0 | 0 | 7.1 | 5.0 | 58.8 |
HK, Hong Kong; IN, India; KR, South Korea; MY, Malaysia; PH, Philippines; SG, Singapore; TW, Taiwan; TH, Thailand. One A. baumannii isolate from India was resistant to ceftazidime, cefotaxime, cefepime, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, and piperacillin-tazobactam, but not to imipenem and meropenem.
OXA carbapenemase genes.
The blaOXA-51-like gene was identified in all 108 CRAB isolates tested in this study. Among these, only seven isolates (6.5%) had the ISAba1-activated blaOXA-51-like gene. These isolates were from Hong Kong (one isolate), India (three isolates), South Korea (one isolate), and Taiwan (two isolates). While the blaOXA-23-like gene was identified in most CRAB isolates (102 isolates; 94.4%), the blaOXA-24-like gene was identified in only seven isolates (6.5%). Most isolates harboring the blaOXA-23-like gene were positive when the ISAba1-blaOXA-23-like gene was amplified (98/102 isolates; 96.1%). Six isolates without the blaOXA-23 gene were collected from Hong Kong (one isolate), India (one isolate), South Korea (one isolate), and Taiwan (three isolates). All seven CRAB isolates harboring the blaOXA-24-like gene, which were negative for the ISAba1-blaOXA-24-like gene, were from Taiwan. Among these, four CRAB isolates from Taiwan harbored three oxacillinase genes (blaOXA-51-like, blaOXA-23-like, and blaOXA-24-like) simultaneously.
Genotype.
For 108 CRAB isolates subjected to the Oxford MLST scheme, 36 sequence types (STs) were identified (Table 2). Although the Oxford scheme (9) has been shown to have limitations of recombination involving the gpi locus (28), we used it in order to compare our data to other reports. Most CRAB isolates (83 isolates; 76.9%) belonged to CC92, including 18 STs in this study. CC92 included the CRAB isolates from all eight Asian locales in the surveillance study. Among the STs of CC92, ST92 and ST195 were the most frequently identified. However, their prevalence patterns were different from each other: while ST92 isolates were found in five Asian locales, namely, India, Malaysia, the Philippines, Taiwan, and Thailand, ST195 isolates were found in only two locales, Malaysia and Thailand. All STs of CC92 except ST208 shared the same alleles in five gene loci and differed only in the gyrB and gpi gene loci. Eight CRAB isolates from five STs belonged to CC108. CC108 included the isolates from three Asian locales, namely, India, Malaysia, and Taiwan. In particular, five CRAB isolates from India belonged to CC108 (ST108, ST419, and ST422) (Table 2).
Table 2.
Results of the Oxford MLST scheme and oxacillinases of 108 CRAB isolates from Asian locales
| CC | ST | Allelic profile | No. of isolatesb |
No. of isolates with bla gene: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 108) | HK (n = 5) | IN (n = 12) | KR (n = 9) | MY (n = 38) | PH (n = 2) | SG (n = 2) | TW (n = 9) | TH (n = 31) | Only OXA-51 | OXA-23 OXA-51 | OXA-24 OXA-51 | OXA-23, OXA-24, OXA-51 | |||
| 92 (n = 83) | 92 | 1-3-3-2-2-7-3 | 19 | 2 | 12 | 1 | 2 | 2 | 17 | 2 | |||||
| 195 | 1-3-3-2-2-96-3 | 18 | 7 | 11 | 18 | ||||||||||
| 346 | 1-3-3-2-2-56-3 | 7 | 4 | 3 | 5 | 2 | |||||||||
| 426 | 1-64-3-2-2-12-3 | 7 | 7 | 7 | |||||||||||
| 75 | 1-3-3-2-2-11-3 | 5 | 5 | 5 | |||||||||||
| 118 | 1-3-3-2-2-3-3 | 4 | 1 | 3 | 1 | 1 | 2 | ||||||||
| 88 | 1-3-3-2-2-10-3 | 3 | 3 | 3 | |||||||||||
| 138 | 1-3-3-2-2-50-3 | 3 | 3 | 3 | |||||||||||
| 365 | 1-15-3-2-2-59-3 | 3 | 3 | 3 | |||||||||||
| 393 | 1-3-3-2-2-80-3 | 3 | 3 | 3 | |||||||||||
| 395 | 1-3-3-2-2-58-3 | 3 | 3 | 3 | |||||||||||
| 208 | 1-3-61-2-2-97-3 | 2 | 1 | 1 | 1 | 1 | |||||||||
| 76 | 1-12-3-2-2-10-3 | 1 | 1 | 1 | |||||||||||
| 136 | 1-3-3-2-2-16-3 | 1 | 1 | 1 | |||||||||||
| 392 | 1-43-3-2-2-10-3 | 1 | 1 | 1 | |||||||||||
| 425 | 1-3-3-2-2-100-3 | 1 | 1 | 1 | |||||||||||
| 421 | 1-17-3-2-2-7-3 | 1 | 1 | 1 | |||||||||||
| 423 | 1-46-3-2-2-142-3 | 1 | 1 | 1 | |||||||||||
| 108 (n = 8) | 108 | 10-12-4-6-4-9-5 | 2 | 2 | 2 | ||||||||||
| 419 | 10-12-4-6-4-95-5 | 2 | 2 | 1 | 1 | ||||||||||
| 397 | 10-53-4-6-4-80-5 | 2 | 1 | 1 | 2 | ||||||||||
| 396 | 10-53-74-6-4-80-5 | 1 | 1 | 1 | |||||||||||
| 422 | 10-53-4-6-4-58-5 | 1 | 1 | 1 | |||||||||||
| 119 (n = 3) | 119 | 1-15-12-6-28-59-40 | 2 | 2 | 2 | ||||||||||
| 399 | 1-15-12-6-28-14-40 | 1 | 1 | 1 | |||||||||||
| 254 (n = 3) | 254 | 21-15-3-2-35-111-4 | 2 | 2 | 2 | ||||||||||
| 398 | 21-15-3-2-35-76-4 | 1 | 1 | 1 | |||||||||||
| Singletons (n = 11) | 96 | 1-43-50-31-1-50-26 | 2 | 2 | 2 | ||||||||||
| 194 | 1-15-4-6-4-58-4 | 2 | 2 | 2 | |||||||||||
| 20 | 1-15-13-12-4-12-2 | 1 | 1 | 1 | |||||||||||
| 110 | 1-15-2-28-1-52-32 | 1 | 1 | 1 | |||||||||||
| 297 | 1-64-109-1-23-12-26 | 1 | 1 | 1 | |||||||||||
| 424 | 1-17-8-10-28-117-32 | 1 | 1 | 1 | |||||||||||
| 420 | 12-17-12-1-29-95-2 | 1 | 1 | 1 | |||||||||||
| 400 | 1-81-11-48-18-115-43 | 1 | 1 | 1 | |||||||||||
| nSTa | 1-12-C1-D1-16-F1-50 | 1 | 1 | 1 | |||||||||||
| Total | 108 | 5 | 12 | 9 | 38 | 2 | 2 | 9 | 31 | 3 | 98 | 3 | 4 | ||
nST, unassigned ST. C1, D1, and F1 indicate the novel unassigned alleles of the gdhB, recA, and gpi genes, respectively.
HK, Hong Kong; IN, India; KR, South Korea; MY, Malaysia; PH, Philippines; SG, Singapore; TW, Taiwan; TH, Thailand.
blaOXA-24 genes were identified in three STs of CC92, ST92 (two isolates), ST118 (three isolates), and ST346 (two isolates). Of these, ST118 and ST346 each included two CRAB isolates with blaOXA-51, blaOXA-23, and blaOXA-54 genes.
Resistance islands identified.
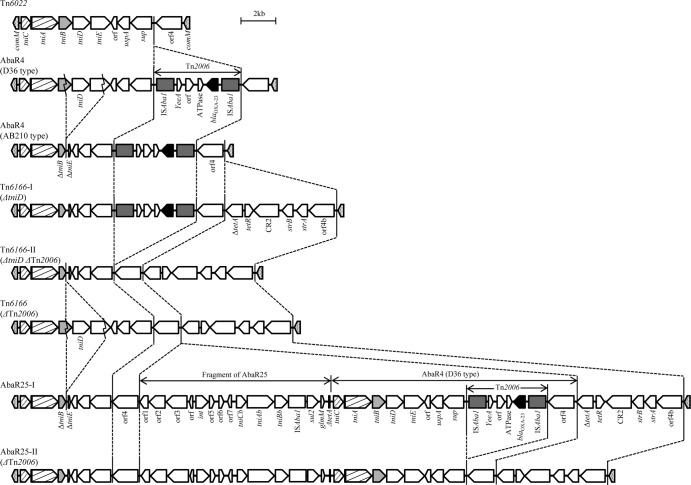
The distribution and structures of AbaR-type resistance islands identified in this study are presented in Table 3, Fig. 1, Fig. 2, and Fig. 3. Among 68 CRAB isolates, AbaR-type resistance islands were identified in 66 isolates. AbaR4-like resistance islands were subdivided into eight subtypes (Fig. 1). The simplest resistance island was Tn6022, which may be a backbone of AbaR4-like resistance islands and includes only nine genes. In two AbaR4-type resistance islands (D36 and AB210 types), Tn2006 included the blaOXA-23-like gene incorporated into the Tn6022 backbone. The AbaR4-D36 type differs from the AbaR4-AB210 type in that it produces 3,238-bp amplicons spanning tniB to tniE rather than 388-bp amplicons lacking tniD and some of tniB and tniE (29). In this study, three subtypes of Tn6166 were included in AbaR4-like resistance islands. A typical Tn6166 (Tn6166-I type) harbors a region including six genes (ΔtetA, tetR, CR2, strB, strA, and orf4b) in addition to an AbaR4-AB210-type resistance island. Unlike the typical Tn6166 harboring Tn2006 with blaOXA-23-like, Tn2006 was absent in other Tn6166 subtypes (Tn6166-II and Tn6166-III types). The Tn6166-II type contains intact tniB, tniD, and tniE, unlike the Tn6166-I and -III types (Fig. 1). AbaR25-type resistance islands were also subdivided into two subtypes, AbaR25-I and AbaR25-II, based on the presence or absence of Tn2006. While the AbaR25-I type included Tn2006, the AbaR25-II type did not.
Table 3.
Distribution of AbaR-type resistance islands
| Resistance island | Subtype | Total (n = 66) (%) | No. of clones |
Countrya (no. of isolates) | ||
|---|---|---|---|---|---|---|
| CC92 (n = 47) | CC108 (n = 6) | Other (n = 13) | ||||
| AbaR4-like | Tn6022 | 5 (7.5) | 4 | 1 | KR (4), TW (1) | |
| AbaR4, D36 type | 9 (13.6) | 3 | 5 | MY (3), IN (5), TH (1) | ||
| AbaR4, AB210 type (ΔtniD) | 12 (18.2) | 10 | 2 | MY (5), IN (3), TH (4) | ||
| Tn6166-I (ΔtniD) | 2 (3.0) | 2 | IN (1), TH (1) | |||
| Tn6166-II (ΔtniD ΔTn2006) | 5 (7.5) | 5 | HK (3), TW (2) | |||
| Tn6166-III (ΔTn2006) | 2 (3.0) | 2 | PH(1), TW(1) | |||
| AbaR25-I | 23 (34.8) | 21 | 2 | TH (11), MY (7), KR (2), SG (1), TW (1), HK (1) | ||
| AbaR25-II (ΔTn2006) | 3 (4.5) | 3 | TW (3) | |||
| AbaR3-like | AbaR3 + AbaR4-D36 type | 1 (1.5) | 1 | IN (1) | ||
| AbaR3ΔMARR + AbaR4-D36 type | 2 (3.0) | 2 | MY (1), SG (1) | |||
| AbaR3ΔMARR | 2 (3.0) | 2 | HK (2) | |||
HK, Hong Kong; IN, India; KR, South Korea; MY, Malaysia; PH, Philippines; SG, Singapore; TW, Taiwan; TH, Thailand.
Fig 1.
Structures of resistance islands identified in this study. Eight subtypes of AbaR4-like resistance islands are shown. AbaR4-D36, AbaR4-AB210, Tn6166, and AbaR25 include Tn2006 with a blaOXA-23-like gene. In AbaR4-AB210, Tn6166ΔtniD, AbaR25, and AbaRΔTn2006, a region including tniD and its upstream and downstream sequence was partially deleted.
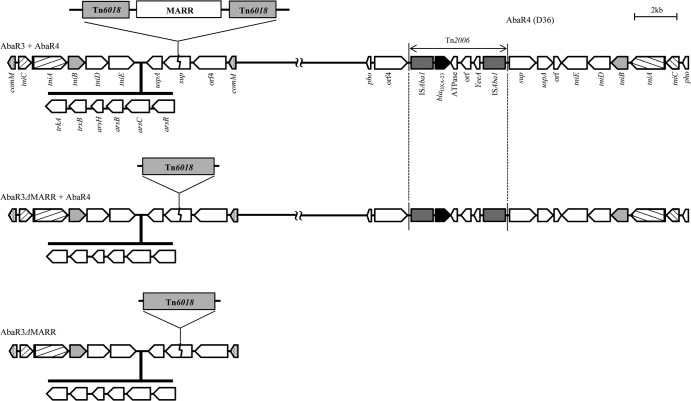
Fig 2.
Structures of resistance islands identified in this study. Three subtypes of AbaR3-like resistance islands are represented. In three isolates, AbaR4-D36 was also found in another region. While the AbaR3-like resistance island of one isolate includes a MARR, a MARR was not found in those of the other four isolates.
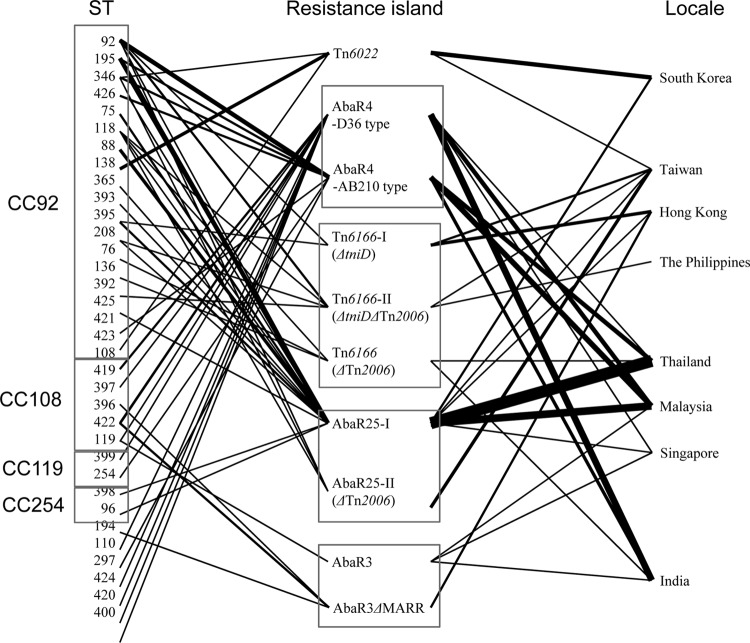
Fig 3.
Relationships among STs, resistance islands, and locales of isolates used in the analysis of resistance islands. The thickness of the lines is proportional to the number of isolates.
Two subtypes were identified in AbaR3-like resistance islands; one includes a MARR (multiple antibiotic resistance region) interrupting Tn6018, and another possesses an intact Tn6018 (Fig. 2). Interestingly, both AbaR3-like and AbaR4-like resistance islands were identified simultaneously in three isolates (Table 3 and Fig. 2). While the AbaR3-type resistance island interrupted the comM gene, as is typical in those isolates, the AbaR4-type resistance island (D36 type) inserted into the pho target.
Distribution of resistance islands.
While most isolates (64/66; 97.0%), which were from all locales investigated in this study, possessed the AbaR4-like resistance islands, AbaR3-like resistance islands were found in only five isolates from India, Malaysia, Singapore, and Hong Kong. All 47 CC92 CRAB isolates were shown to harbor AbaR4-like resistance islands. The most prevalent AbaR-type resistance island was the AbaR25-I type, which was found in 23 CRAB isolates (34.8%) from six Asian locales, including Thailand, Malaysia, South Korea, Singapore, Taiwan, and Hong Kong (Table 3 and Fig. 3). Following the AbaR25-I type, the AbaR4-AB210 type was identified in 12 isolates from Malaysia, India, and Thailand. While most isolates with the AbaR4-AB210 type belonged to CC92, no AbaR4-D36 type was identified in any CC92 isolate. All six CC108 isolates possessed AbaR4-D36-type resistance islands, and an AbaR3-type resistance island was found simultaneously in three isolates (Table 3, Fig. 2, and Fig. 3).
Figure 3 shows the relationships among the genotypes of isolates, resistance islands, and isolation locales. It shows no distinct relationships among them, except association between CC108 and the AbaR4-D36 type.
DISCUSSION
In this study, we have shown that >60% of A. baumannii isolates causing HAP were resistant to all antimicrobial agents except the polymyxins, which indicates the continuing increase of highly multidrug-resistant (MDR) A. baumannii in Asia (3).
Two international lineages (GC1 and GC2) of CRAB isolates have become disseminated worldwide (30). In the Oxford MLST scheme (1), GC1 and GC2 might correspond to CC109 and CC92, respectively (23, 27). While the AbaR3-type resistance island and its variants are found frequently in GC1 isolates, AbaR4-type resistance islands are common in GC2 isolates (29, 31). AbaR resistance islands have evolved into diverse types through multiple events of insertion, deletion, and recombination (24, 32). Only a few studies on the distribution of clones and their characteristics have been reported in Asian countries. For example, clonal dissemination and diversification of CC92 and the predominance of AbaR4-type resistance islands among CRAB isolates have been reported in South Korea (14, 33). CC92 was also the predominant clone in CRAB isolates from China (34), and the widespread occurrence of the AbaR4-type resistance island among CRAB isolates was also shown in Taiwan (15). However, the clones and AbaR resistance islands have not been compared among A. baumannii isolates from several Asian countries.
The present study revealed the predominance of CC92 (i.e., GC2) among CRAB isolates in Asian locales. As CC92 isolates were identified in all Asian locales included in this study, they might be disseminated widely in Asia, including China. Resistance to multiple antimicrobials may confer a selective advantage and drive their expansion (30). However, diverse STs within CC92 may indicate rapid diversification or evolution for a long period after introduction into Asia. CC108 corresponds to the GC1 lineage, as ST108 differs at one locus (recA) from ST109, an ST of a representative strain (D13) of GC1 (31). Although CC108 was the minor clone in most Asian locales, it was predominant in India. Thus, the origin of CRAB isolates in India may be different from that in other Asian locales.
OXA-23-like oxacillinase might be most responsible for carbapenem resistance in A. baumannii isolates from most Asian locales, except Taiwan. Among isolates in which resistance islands were found, all blaOXA-23-like genes were located within Tn2006. Seven Taiwanese CRAB isolates contained blaOXA-24-like genes. blaOXA-58-like and blaOXA-24-like have been frequently identified in Taiwan (6). Most isolates with the blaOXA-24-like gene displayed very high MICs (>64 mg/liter) of both imipenem and meropenem. Although all belonged to CC92, they represented three STs: ST92, ST118, and ST346. Thus, CRAB isolates with the blaOXA-24-like gene might not be disseminated clonally in Taiwan.
As in a previous study (28), AbaR4-type resistance islands were common among CC92 CRAB isolates, and several variants were also identified among them. The predominant subtypes, AbaR25-I type and AbaR4-AB210 type, were identified in CRAB isolates from diverse Asian locales and diverse STs (Table 3 and Fig. 3). The most predominant ST, ST92, represented five subtypes of AbaR4-like resistance islands, and ST195 represented two subtypes, AbaR4-AB210 type and AbaR25-I type. Thus, it might be that AbaR4-like resistance islands have been incorporated into A. baumannii isolates frequently. AbaR4-like resistance islands might have diversified by insertion or deletion of Tn2006; a region including tniC, tinD, and tniA; a region unique to Tn6166 (ΔtetA to orf4b); and a region unique to AbaR25 (orf1 to ΔtetA), which has been accompanied by the diversification of a clone. Actually, Tn2006 can be transferred independently and can be mobile, even in a given genome (35). The AbaR25-type resistance island, which was the most prevalent in this study, was identified in A. baumannii isolates belonging to GC2 from Lithuania (28). A. baumannii isolates with AbaR25-type resistance islands may now disseminate worldwide, along with isolates with AbaR4-type resistance islands. In addition, it can be inferred that more diverse AbaR-type resistance islands will emerge via insertion or deletion of mobile fragments. Furthermore, the lack of correlation between the genotype and the type of resistance island indicates that the insertion and deletion of mobile elements may occur frequently. Because the regions transferred horizontally in resistance islands carry many antimicrobial resistance determinants, including blaOXA-23-like and strA, strB, and tetR, their transfer is worrisome.
One of interesting results of this study was the identification of the simultaneous existence of AbaR3- and AbaR4-type resistance islands in three CC108 CRAB isolates from three different countries, India, Malaysia, and Singapore. The genotypes of three isolates harboring both AbaR3- and AbaR4-type resistance islands were different from one another: ST396, ST397, and ST422. The simultaneous existence of two AbaR-type resistance islands was seen in one previous study (36). The integration of an AbaR4-type resistance island into the pho gene was also reported in a previous study (28). In that study, a pho-integrated AbaR4-type resistance island was identified in A. baumannii isolates belonging to European clone I. It may confirm the mobility of the AbaR-type resistance island and may indicate that integration of a resistance island into the pho gene might not be rare.
In summary, CRAB isolates belonging to GC2, that is, CC92, are prevalent in most Asian locales. AbaR4-like resistance islands were predominant, and several variants of them were also identified. The simultaneous existence of AbaR3- and AbaR4-type resistance islands was also identified in different chromosomal regions in three isolates.
ACKNOWLEDGMENTS
All Acinetobacter isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID) (Seoul, South Korea).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A2A2A0101413).
Footnotes
Published ahead of print 12 August 2013
REFERENCES
- 1.Munoz-Price LS, Weinstein RA. 2008. Acinetobacter infection. N. Engl. J. Med. 358:1271–1281 [DOI] [PubMed] [Google Scholar]
- 2.Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41:848–854 [DOI] [PubMed] [Google Scholar]
- 3.Chung DR, Song JH, Kim SH, Huang SG, Wang H, So TM, Yasin RM, Hsueh PR, Carlos CC, Hsu LY, Buntaran L, Lalitha MK, Kim MJ, Choi JY, Kim SI, Ko KS, Kang CI, Peck KR, ANSORP Study Group 2011. High prevalence of multidrug-resistant non-fermenters in hospital-acquired pneumonia in Asia. Am. J. Respir. Crit. Care Med. 184:1409–1417 [DOI] [PubMed] [Google Scholar]
- 4.Gilad J, Carmeli Y. 2008. Treatment options for multidrug-resistant Acinetobacter species. Drugs 68:165–189 [DOI] [PubMed] [Google Scholar]
- 5.Giske CG, Monnet DL, Cars O, Carmeli Y, ReAct-Action on Antibiotic Resistance 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jean SS, Hsueh PR. 2011. High burden of antimicrobial resistance in Asia. Int. J. Antimicrob. Agents 37:291–295 [DOI] [PubMed] [Google Scholar]
- 7.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, Verhoef J, Brisse S. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105–112 [DOI] [PubMed] [Google Scholar]
- 8.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238 [DOI] [PubMed] [Google Scholar]
- 9.Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]
- 11.He C, Xie Y, Fan H, Kang M, Tao C, Zhang R, Hu Y, Chen Z, Wang L. 2011. Spread of imipenem-resistant Acinetobacter baumannii of European clone II in Western China. Int. J. Antimicrob. Agents 38:257–260 [DOI] [PubMed] [Google Scholar]
- 12.Park YK, Lee GH, Baek JY, Chung DR, Peck KR, Song JH, Ko KS. 2010. A single clone of Acinetobacter baumannii, ST22, is responsible for high antimicrobial resistance rates of Acinetobacter spp. isolates that cause bacteremia and urinary tract infections in Korea. Microb. Drug Resist. 16:143–149 [DOI] [PubMed] [Google Scholar]
- 13.Hamidian M, Hall RM. 2011. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 66:2484–2491 [DOI] [PubMed] [Google Scholar]
- 14.Kim DH, Park YK, Ko KS. 2012. Variations of AbaR4-type resistance island in Acinetobacter baumannii isolates from South Korea. Antimicrob. Agents Chemother. 56:4544–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HY, Chang RC, Su LH, Liu SY, Wu SR, Chuang CH, Chen CL, Chiu CH. 2012. Wide spread of Tn2006 in an AbaR4-type resistance island among carbapenem-resistant Acinetobacter baumannii clinical isolates in Taiwan. Int. J. Antimicrob. Agents 40:163–167 [DOI] [PubMed] [Google Scholar]
- 16.La Scola B, Gundi VAKB, Khamis A, Raoutl D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J. Clin. Microbiol. 44:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J. Antimicrob. Chemother. 60:1163–1167 [DOI] [PubMed] [Google Scholar]
- 18.Park YK, Choi JY, Jung SI, Park KH, Lee H, Jung DS, Heo ST, Kim SW, Chang HH, Cheong HS, Chung DR, Peck KR, Song JH, Ko KS. 2009. Two distinct clones of carbapenems-resistant Acinetobacter baumannii isolates from Korean hospitals. Diagn. Microbiol. Infect. Dis. 64:389–395 [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement, M100–S21 CLSI, Wayne, PA: [PubMed] [Google Scholar]
- 20.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]
- 21.Kim YJ, Kim SI, Kim YR, Hong KW, Wie SH, Park YJ, Jeong H, Kang MW. 2012. Carbapenem-resistant Acinetobacter baumannii: diversity of resistant mechanisms and risk factors for infection. Epidemiol. Infect. 140:137–145 [DOI] [PubMed] [Google Scholar]
- 22.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Post V, White PA, Hall RM. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1162–1170 [DOI] [PubMed] [Google Scholar]
- 25.Turton JF, Baddal B, Perry C. 2011. Use of accessory genome for characterization and typing of Acinetobacter baumannii. J. Clin. Microbiol. 49:1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigro SJ, Hall RM. 2012. Tn6167, an antibiotic resistance island in an Australian carbapenem-resistant Acinetobacter baumannii GC2, ST92 isolate. J. Antimicrob. Chemother. 67:1342–1346 [DOI] [PubMed] [Google Scholar]
- 27.Saule M, Samuelsen Ø, Dumpis U, Sundsfjord A, Karlsone A, Balode A, Miklasevics E, Karah N. 2013. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob. Agents Chemother. 57:1069–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Šeputienė V, Povilonis J, Suźiedėlienė E. 2012. Novel variants of AbaR resistance islands with a common backbone in Acinetobacter baumannii isolates of European clone II. Antimicrob. Agents Chemother. 56:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nigro SJ, Hall RM. 2012. Antibiotic resistance islands in A320 (RUH134), the reference strain for Acinetobacter baumannii global clone 2. J. Antimicrob. Chemother. 67:335–338 [DOI] [PubMed] [Google Scholar]
- 30.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Post V, Hamidian M, Hall RM. 2012. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J. Antimicrob. Chemother. 67:1039–1049 [DOI] [PubMed] [Google Scholar]
- 32.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, NISC Comparative Sequence Program, Murray PR, Segre JA. 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. U. S. A. 108:13758–13763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park YK, Jung SI, Park KH, Kim DH, Choi JY, Kim SH, Ko KS. 2012. Changes in antimicrobial susceptibility and major clones of Acinetobacter calcoaceticus-baumannii complex isolates from a single hospital in Korea over 7 years. J. Med. Microbiol. 61:71–79 [DOI] [PubMed] [Google Scholar]
- 34.Fu Y, Zhou J, Zhou H, Yang Q, Wei Z, Yu Y, Li L. 2010. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65:644–650 [DOI] [PubMed] [Google Scholar]
- 35.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]