Abstract
Nucleotide sequencing of the fusB-flanking regions in two fusidic acid-resistant Staphylococcus epidermidis isolates with the type IV aj1-leader peptide (LP)-fusB structure (lacking aj1) revealed that their fusB gene was located on novel phage-related islands inserted downstream of smpB and are here referred to as SeRIfusB-3692 and SePIfusB-857. The novel SePIfusB-857 structure was followed by SeCI857, forming a composite pathogenicity island which contained a putative virulence gene, vapE. The linkage of fusB and vapE may contribute to bacterial adaption.
TEXT
The horizontally acquired determinant fusB is the most frequent determinant responsible for fusidic acid resistance in Staphylococcus epidermidis and is often found associated with genomic resistance islands (RIs) (1, 2). We have previously found at least three types of structures of RIs, discriminated by their sequences flanking fusB (aj1-leader peptide [LP]-fusB), and we have identified different insertion sites, including sites downstream of groEL and rpsR (2). However, the genetic support of fusB in some isolates remains unknown. To analyze the unidentified structure and gain more understanding of the prevalence of various fusB-carrying elements, a total of 141 fusidic acid-resistant (MIC ≥ 2 μg/ml) S. epidermidis isolates were tested. The isolates were collected from a 3-year (2008 to 2010) collection in the Bacteriology Laboratory of the National Taiwan University Hospital, a 2,500-bed teaching hospital in northern Taiwan. The species of S. epidermidis was initially identified using the Phoenix Automated System and was then further confirmed by S. epidermidis-specific PCR (3).
Detection of fusB, fusC, and fusD (4), performed by PCR, revealed that the majority of isolates (136/141, 96.5%) possessed fusB. Only four isolates carried fusC, and one contained a fusA point mutation (resulting in P404L). Various types of aj1-LP-fusB fragments for 136 fusB-positive isolates were determined by PCR as previously described (2). Of them, 14 type I (full-length aj1), 58 type II (partial aj1 fragment, truncated from nucleotide position 93 to 421), 47 type III (a more truncated aj1 that retained only the last 37 bp), and 17 type IV (lacking aj1) isolates were identified. The fusidic acid MICs for 136 fusB-positive isolates ranged from 2 to 16 μg/ml. The type II isolates displayed significantly higher-level resistance to fusidic acid (the MIC for 41/58 [71%] isolates was 16 μg/ml) than type III isolates (the MIC for only 12/47 [26%] isolates was 16 μg/ml) (P < 0.05) (Table 1).
Table 1.
Distribution of fusidic acid resistance determinants and MICs among fusidic acid-resistant S. epidermidis isolates
| Resistance determinant | aj1-LP-fusB type | No. of isolates | No. of isolates for which the MIC (μg/ml) wasa: |
||||
|---|---|---|---|---|---|---|---|
| 2 | 4 | 8 | 16 | 32 | |||
| fusB | I | 14 | 0 | 1 | 8 | 5 | 0 |
| II | 58 | 0 | 1 | 16 | 41 | 0 | |
| III | 47 | 1 | 13 | 21 | 12 | 0 | |
| IV | 17 | 1 | 1 | 9 | 6 | 0 | |
| fusC | 4 | 0 | 0 | 0 | 1 | 3 | |
| fusA with point mutation | 1 | 0 | 0 | 0 | 1 | 0 | |
| Total | 141 | 2 | 16 | 54 | 66 | 3 | |
The MICs for isolates with a type II aj1-LP-fusB sequence were significantly (P < 0.05) higher than those for isolates with a type III aj1-LP-fusB sequence.
The isolates with type IV aj1-LP-fusB sequences differed from other types by the absence of aj1, but fusB's genetic environment was unknown. Two representative type IV isolates (NTUH-3692 and NTUH-857) were used for cloning and sequencing with a long accurate (LA)-PCR in vitro cloning kit (TaKaRa Shuzo Co. Ltd., Japan) (2) and by inverse PCR (see Table S1 in the supplemental material). The sequencing of amplification products was performed on an Applied Biosystems model 3100 DNA sequencer (Applied Biosystems, Foster City, CA) using the Taq BigDye-Deoxy Terminator cycle sequencing kit (Applied Biosystems).
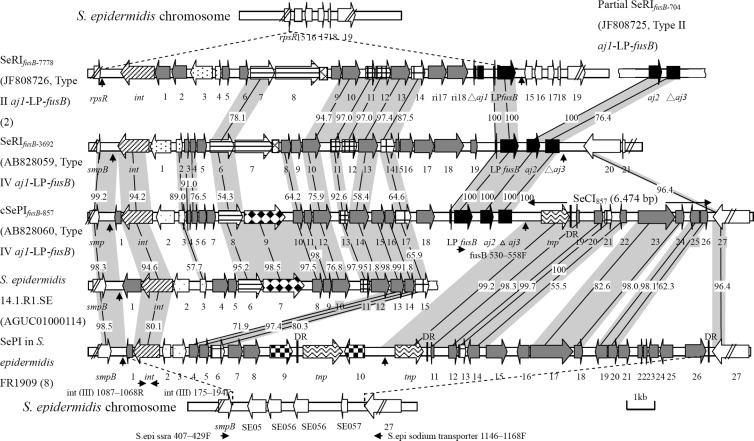
Sequencing results indicated that the fusB gene in NTUH-3692 was located on a 15,553-bp phage-related RI and is here referred to as SeRIfusB-3692, where “Se” signifies “S. epidermidis.” The fusB in NTUH-857 was located in a 21,003-bp composite island, here referred to as cSePIfusB-857 (where “PI” signifies “pathogenicity island”), which was composed by SePIfusB-857 (14,529 bp) and SeCI857 (where “CI” signifies “chromosomal insertion”) (6,474 bp) (Fig. 1). The sizes of SeRIfusB-3692 and SePIfusB-857 fit the criteria for a pathogenicity island (5). The GC contents of SeRIfusB-3692 and cSePIfusB-857 were 29.1% and 30.3% (SePIfusB-857, 29.8%, and SeCI857, 31.5%), respectively, slightly lower than that of the published whole-genome sequences of S. epidermidis (∼32%). Both SePIfusB-857 and SeRIfusB-3692 were flanked by direct repeats and contained conserved phage-related core genes (6). SeRIfusB-3692 and cSePIfusB-857 were located downstream of smpB, unlike with groEL and rpsR in previously found fusB RIs (2). The smpB insertion site has been found in SaPIm4 and SaPImw2 of Staphylococcus aureus (7) and in a composite PI in S. epidermidis (8).
Fig 1.
Genetic organization of SeRIfusB-3692 (GenBank accession no. AB828059) and SeRIfusB-857 (GenBank accession no. AB828060) compared with those of SeRIfusB-7778 (GenBank accession no. JF808726), partial SeRIfusB-704 (GenBank accession no. JF808725), S. epidermidis 14.1.R1.SE (GenBank accession no. AGUC01000114), and the PI in S. epidermidis FR1909 (GenBank accession no. AENR01000001 and AENR01000008). Genes are drawn according to their sequences and function. ▨, int and xis; ▩, transcription regulators; ▤, replication genes (including the primase gene[pri] and the replication initiator gene [rep]); ▩, the replication origin (ori);  , the terminase small-subunit encapsidation gene terS;
, the terminase small-subunit encapsidation gene terS;  , other encapsidation genes;
, other encapsidation genes;  , pif (phage interference); ■, aj1-LP-fusB regions;
, pif (phage interference); ■, aj1-LP-fusB regions;  , vapE;
, vapE;  , virulence factor genes;
, virulence factor genes;  , transposase genes; □, chromosome genes adjacent to SeRIfusB;
, transposase genes; □, chromosome genes adjacent to SeRIfusB;  , other genes coding hypothetical proteins. The predicted direct repeats are indicated by vertical arrows. The horizontal arrows represent the PCR primers used to determine the insertion sites (downstream of smpB). Homologous regions between resistance islands are shown with shaded connecting lines, and shaded numbers show the percentages of homology between the corresponding sequences. DR, direct repeat; S. epi, S. epidermidis; ssra, SsrA-binding protein encoded by smpB.
, other genes coding hypothetical proteins. The predicted direct repeats are indicated by vertical arrows. The horizontal arrows represent the PCR primers used to determine the insertion sites (downstream of smpB). Homologous regions between resistance islands are shown with shaded connecting lines, and shaded numbers show the percentages of homology between the corresponding sequences. DR, direct repeat; S. epi, S. epidermidis; ssra, SsrA-binding protein encoded by smpB.
Sequence data confirmed the lack of the aj1 gene in SeRIfusB-3692 or SePIfusB-857. Comparison of SeRIfusB-3692 and SePIfusB-857 to previously found SeRIfusB-7778 (2) revealed that the genetic organizations were similar and that the sequences of LP and fusB were identical (Fig. 1). All three islands carried conserved phage-related core genes (Fig. 1) (6). The open reading frames (ORFs) in regions upstream of fusB in SeRIfusB-3692 and SePIfusB-857 were in general similar. Sequences of ORFs in SePIfusB-857 showed high identity to those in an island in S. epidermidis 14.1.R1.SE (GenBank accession no. AGUC01000114) (Fig. 1).
Another important finding in this study is the presence of a putative virulence gene, vapE, in SePIfusB-857. The vapE gene was originally found in virulent Dichelobacter nodosus, a sheep pathogen that causes severe ovine foot rot (9). The vapE gene has also been found in PIs of S. aureus (10, 11) and in S. epidermidis 14.1.R1.SE but is here for the first time identified in a fusB element. Of 136 fusB-positive S. epidermidis isolates, vapE was detected in nine isolates (9/136, 6.6%), including three type I, one type II, and five type IV aj1-LP-fusB isolates. However, only in the five type IV isolates were vapE and fusB linked together and located downstream of smpB. In one type II isolate, vapE was located downstream of smpB, but fusB was located downstream of groEL. In the three type I isolates, the location of vapE was unknown. Bacterial PIs usually carry either virulence genes or antibiotic resistance genes; very few PIs carry both of them (12).
Unlike in other fusB RIs, SePIfusB-857 was followed by SeCI857. SeCI857 was similar to SeCI-1 in S. epidermidis FRI909 (8) (Fig. 1). Thus, cSePIfusB-857 may arise from two independent integration events.
To compare the characteristics of isolates carrying different structures in their fusB element, the antimicrobial resistance profile and the presence of virulence-related genes, including the biofilm-related icaAB locus (13), IS256 (14), and the resistance gene mecA (15), detected by PCR, were determined (Table 2). The above-named genes have been reported to be associated with nosocomial isolates but were detected in only a small subset of commensal isolates (14, 16). The rates of resistance to erythromycin and trimethoprim-sulfamethoxazole (SXT) were similar among the four types. The type IV isolates exhibited a lower occurrence of resistance to oxacillin, clindamycin, and gentamicin than type I, II, or III isolates. It has been previously reported that commensal isolates are less resistant to clindamycin, SXT, and oxacillin (16), and gentamicin resistance has been recognized as a marker of nosocomially acquired staphylococci (17). Resistance to more antibiotics and a high prevalence of the icaAB locus, IS256, and mecA in type II or III isolates suggests their clinical significance. For type IV isolates, although they less frequently carried the icaAB locus, IS256, and mecA genes than isolates of the other types, the presence of vapE may somehow confer an advantageous attribute.
Table 2.
Antimicrobial susceptibilities and distribution of the virulence-related icaAB locus and IS256 gene and mecA resistance gene among isolates with different aj1-LP-fusB types
| Antibiotic(s) to which isolates were resistant or virulence-related genotypea | No. of isolates (%) of aj1-LP-fusB type: |
|||
|---|---|---|---|---|
| I (n = 14) | II (n = 58) | III (n = 47) | IV (n = 17) | |
| Clindamycin | 9 (64) | 45 (78) | 35 (74) | 4 (24) |
| Erythromycin | 12 (86) | 45 (78) | 35 (74) | 12 (71) |
| Gentamicin | 13 (93) | 56 (97) | 42 (89) | 8 (47) |
| Oxacillin | 14 (100) | 57 (98) | 47 (100) | 15 (88) |
| Trimethoprim-sulfamethoxazole | 12 (86) | 54 (93) | 42 (89) | 14 (82) |
| icaAB (−) IS256 (−) mecA (−) | 0 (0) | 1 (2) | 0 (0) | 2 (12) |
| icaAB (−) IS256 (−) mecA (+) | 4 (29) | 1 (2) | 3 (6) | 10 (59) |
| icaAB (+) IS256 (−) mecA (+) | 1 (7) | 1 (2) | 0 (0) | 3 (18) |
| icaAB (−) IS256 (+) mecA (+) | 4 (29) | 0 (0) | 1 (2) | 2 (12) |
| icaAB (+) IS256 (+) mecA (+) | 5 (36) | 55 (95) | 43 (91) | 0 (0) |
(−), gene is absent; (+), gene is present.
Concluding remarks.
This study provides new data on the complexity and diversity of genetic elements associated with the fusB determinant in fusidic acid-resistant S. epidermidis strains. Novel SePIfusB-857 contained both an antibiotic resistance gene (fusB) and a putative virulence gene (vapE), which may provide an advantage for bacterial survival.
Nucleotide sequence accession number.
The nucleotide sequences of SeRIfusB-3692 and cSeRIfusB-857 have been deposited in GenBank under accession numbers AB828059 and AB828060, respectively.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grant NSC 100-2320-B-002-014-MY3 from the National Science Council of Taiwan.
Footnotes
Published ahead of print 26 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01433-13.
REFERENCES
- 1.McLaws F, Chopra I, O'Neill AJ. 2008. High prevalence of resistance to fusidic acid in clinical isolates of Staphylococcus epidermidis. J. Antimicrob. Chemother. 61:1040–1043 [DOI] [PubMed] [Google Scholar]
- 2.Chen HJ, Tsai JC, Hung WC, Tseng SP, Hsueh PR, Teng LJ. 2011. Identification of fusB-mediated fusidic acid resistance islands in Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 55:5842–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Swiatlo E, Austin FW, Lawrence ML. 2006. Use of a putative transcriptional regulator gene as target for specific identification of Staphylococcus epidermidis. Lett. Appl. Microbiol. 43:325–330 [DOI] [PubMed] [Google Scholar]
- 4.Chen HJ, Hung WC, Tseng SP, Tsai JC, Hsueh PR, Teng LJ. 2010. Fusidic acid resistance determinants in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 54:4985–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679 [DOI] [PubMed] [Google Scholar]
- 6.Novick RP, Christie GE, Penades JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 8:541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 8.Madhusoodanan J, Seo KS, Remortel B, Park JY, Hwang SY, Fox LK, Park YH, Deobald CF, Wang D, Liu S, Daugherty SC, Gill AL, Bohach GA, Gill SR. 2011. An enterotoxin-bearing pathogenicity island in Staphylococcus epidermidis. J. Bacteriol. 193:1854–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloomfield GA, Whittle G, McDonagh MB, Katz ME, Cheetham BF. 1997. Analysis of sequences flanking the vap regions of Dichelobacter nodosus: evidence for multiple integration events, a killer system, and a new genetic element. Microbiology 143(Part 2):553–562 [DOI] [PubMed] [Google Scholar]
- 10.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527–543 [DOI] [PubMed] [Google Scholar]
- 11.Yarwood JM, McCormick JK, Paustian ML, Orwin PM, Kapur V, Schlievert PM. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138–13147 [DOI] [PubMed] [Google Scholar]
- 12.Subedi A, Ubeda C, Adhikari RP, Penades JR, Novick RP. 2007. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology 153:3235–3245 [DOI] [PubMed] [Google Scholar]
- 13.Frebourg NB, Lefebvre S, Baert S, Lemeland JF. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohde H, Kalitzky M, Kroger N, Scherpe S, Horstkotte MA, Knobloch JK, Zander AR, Mack D. 2004. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42:5614–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YH, Tseng SP, Hu JM, Tsai JC, Hsueh PR, Teng LJ. 2007. Clonal spread of SCCmec type IV methicillin-resistant Staphylococcus aureus between community and hospital. Clin. Microbiol. Infect. 13:717–724 [DOI] [PubMed] [Google Scholar]
- 16.Conlan S, Mijares LA, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA. 2012. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13:R64. 10.1186/gb-2012-13-7-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer GL. 1991. Alteration of cutaneous staphylococcal flora as a consequence of antimicrobial prophylaxis. Rev. Infect. Dis. 13(Suppl. 10):S805–S809 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.