Abstract
One of the central regulators of oxidative stress in Saccharomyces cerevisiae is Yap1, a bZIP transcription factor of the AP-1 family. In unstressed cells, Yap1 is reduced and cytoplasmic, but in response to oxidative stress, it becomes oxidized and accumulates in the nucleus. To date, there have been no reports on the role of AP-1-like transcription factors in symbiotic fungi. An ortholog of Yap1, named YapA, was identified in the genome of the grass symbiont Epichloë festucae and shown to complement an S. cerevisiae Δyap1 mutant. Hyphae of the E. festucae ΔyapA strain were sensitive to menadione and diamide but resistant to H2O2, KO2, and tert-butyl hydroperoxide (t-BOOH). In contrast, conidia of the ΔyapA strain were very sensitive to H2O2 and failed to germinate. Using a PcatA-eGFP degron-tagged reporter, YapA was shown to be required for expression of a spore-specific catalase gene, catA. Although YapA-EGFP localized to the nucleus in response to host reactive oxygen species during seedling infection, there was no difference in whole-plant and cellular phenotypes of plants infected with the ΔyapA strain compared to the wild-type strain. Homologs of the S. cerevisiae and Schizosaccharomyces pombe redox-sensing proteins (Gpx3 and Tpx1, respectively) did not act as redox sensors for YapA in E. festucae. In response to oxidative stress, YapA-EGFP localized to the nuclei of E. festucae ΔgpxC, ΔtpxA, and ΔgpxC ΔtpxA cells to the same degree as that in wild-type cells. These results show that E. festucae has a robust system for countering oxidative stress in culture and in planta but that Gpx3- or Tpx1-like thiol peroxidases are dispensable for activation of YapA.
INTRODUCTION
Reactive oxygen species (ROS) play a very important biological role in fungus-plant interactions. Production of ROS by plasma membrane-localized NADPH oxidases at sites of pathogen invasion is one of the early defense responses of the plant host (1–3). This oxidative burst induces programmed cell death but also serves as a second messenger to activate the expression of various plant defense genes (4–6). Detoxification of this burst of ROS is vital for maintaining plant cellular integrity. Likewise, fungal plant pathogens must have efficient mechanisms for ROS detoxification if they are to successfully colonize the host (7, 8). Two strategies that are commonly used by plants and fungi to detoxify ROS are the use of antioxidants, such as ascorbate, glutathione, and carotenoids, and enzymatic scavenging of ROS by superoxide dismutase, catalase, ascorbate peroxidase, or glutathione peroxidase (9, 10). Production of “bursts” of ROS by specific NADPH oxidase isoforms are also important for polarized growth and multicellular development in both plants and fungi (11–16). As with the defense response, restoration of ROS homeostasis following differentiation signaling is vital for maintenance of cellular integrity. One of the key regulators mediating an oxidative stress response is the AP-1 class of basic leucine zipper (bZIP) transcription factors.
The Saccharomyces cerevisiae AP-1-like transcription factor Yap1 is the best-characterized member of the bZIP family of transcription factors. Yap1 contains two cysteine-rich domains (CRDs), at the N-distal (n-CRD) and C-proximal (c-CRD) termini, which are fundamental to its activation. In the absence of oxidative stress, the nuclear export sequence (NES) is able to interact with the Crm1 nuclear exportin, resulting in export of the protein from the nucleus to the cytoplasm. In cells exposed to H2O2 stress, two disulfide bonds are formed between the n- and c-CRDs, resulting in a conformational change that masks the interaction of the NES with Crm1 (17–19). As a result, oxidized Yap1 is retained in the nucleus and activates the expression of numerous genes involved in antioxidant defense (20).
Interdomain disulfide bond formation between the n- and c-CRDs after exposure to H2O2 is by far the most commonly recognized mechanism of Yap1 activation. However, sensing of the redox signal (H2O2) is mediated by the glutathione peroxidase Gpx3 (also known as Hyr1 [hydrogen peroxide resistance protein 1] or ORP1 [oxidant receptor peroxidase 1]) rather than by Yap1 itself, resulting in oxidation of a conserved Cys in Gpx3 to a sulfenic acid (21). The Cys-SOH then condenses with a Cys in the c-CRD of Yap1 to form an intermolecular disulfide bond, which is resolved by thiol disulfide exchange, resulting in release of Gpx3 and formation of an intramolecular disulfide bond between conserved cysteines in the c- and n-CRDs of Yap1 (21). The second disulfide bond, between two other conserved cysteines in the n- and c-CRDs, is formed by a second oxidation cycle with Gpx3. Transduction of the H2O2 signal requires formation of multiple interdomain disulfide bonds to generate a reduction-resistant Yap1 protein (22). In addition, Ybp1 (Yap1-binding protein) has an important role, together with Gpx3, in the cytoplasmic oxidation of Yap1 in response to H2O2 stress and in promotion of nuclear accumulation (23). In Schizosaccharomyces pombe, the thioredoxin peroxidase Tpx1, rather than a Gpx protein, is used as the redox sensor to regulate Pap1, the Yap1 homolog (24, 25). Activation of Pap1 occurs by a catalytic mechanism slightly different from that described for S. cerevisiae, as both catalytic cysteine residues are used in Tpx1, whereas just one is used in Gpx3. Whether homologs of Gpx3 or Tpx1 act as redox sensors for activation of AP-1-like proteins in filamentous fungi is not known. Yap1 and Pap1 can also be modified covalently by thiol-reactive electrophiles, such as diamide, N-ethylmaleimide, and diethylmaleate, independently of Gpx3 or Tpx1. Oxidation of conserved cysteines in the c-CRD, without the formation of disulfide bonds, is sufficient to disrupt interaction between Crm1 and either Yap1 or Pap1, promoting retention of the transcription factor in the nucleus (26–28).
Yap1 homologs have been identified in a number of other fungi, including Candida albicans (29), Aspergillus fumigatus (30), Cochliobolus heterostrophus (31), Alternaria alternata (32), Botrytis cinerea (33), Neurospora crassa (34), Magnaporthe oryzae (8), and Ustilago maydis (7). Disruption of these transcription factors commonly results in hyphal and conidial sensitivity to H2O2 and other oxidative stress-inducing compounds, such as tert-butyl hydroperoxide (t-BOOH), potassium superoxide (KO2), menadione, and diamide. Regulation through control of nuclear import and export is a shared feature of these AP-1-like proteins, a property that presumably reflects the conserved domain (bZIP, n-CRD, and c-CRD) and motif (NES and nuclear localization sequence [NLS]) structure of this family of proteins.
AP-1-like transcription factors have a key role in protecting phytopathogenic fungi from plant-generated ROS during host colonization, a function highlighted by retention of these proteins in the nucleus upon contact with the leaf surface and during appressorium-mediated penetration of the host (7, 31). While AP-1 deletion mutants of fungi are sensitive to H2O2 in culture, the plant colonization phenotype is very dependent on the growth lifestyle. AP-1 mutants of the biotrophic fungi U. maydis and M. oryzae are sensitive to a host oxidative burst and have reduced virulence (7, 8). In contrast, the AP-1 mutants of the necrotrophic fungi B. cinerea and C. heterostrophus are as virulent as the wild-type strains (31, 33). A. alternata is an exception to the generalization that lifestyle determines the host colonization outcome, as AP-1 mutants of this necrotrophic pathogen are defective in host colonization (32).
In M. oryzae, the homolog of GPX3/HYR1 also has a role in host virulence; the hyr1 mutant was shown to be less tolerant to ROS generated by a susceptible plant and formed significantly smaller lesions on both barley and rice (35). However, the study did not establish whether these effects were due solely to the antioxidant activity of this protein or because of a defect in redox signaling.
To date, there have been no reports on the role of AP-1-like transcription factors in fungus-plant symbiotic interactions, such as the mutualistic symbiotic interaction between the fungus Epichloë festucae (Ascomycota, Clavicipitaceae) and the plant Lolium perenne (36, 37). This biotrophic fungus systemically colonizes the vegetative and reproductive aerial tissues but not the roots of the plant. The growth of this fungus within the leaves is tightly regulated, with usually just a single hypha found between adjacent columns of plant cells. Hyphae that colonize the leaves are firmly attached to the plant cells and extend by intercalary division and extension, a mechanism of growth that synchronizes hyphal growth with plant leaf growth (38). Using both forward and reverse genetic strategies, we previously identified genes crucial for establishment and maintenance of this symbiosis, including genes encoding components of the NADPH oxidase and mitogen-activated protein kinase (MAPK) signaling complexes (15, 39–42).
The aim of this study was to test whether YapA-mediated signaling is required for maintenance of a mutualistic symbiotic interaction between E. festucae and L. perenne and to determine whether E. festucae GpxC and TpxA, which are homologs of Gpx3 and Tpx1, respectively, are redox sensors of H2O2 stress resulting in nuclear retention and activation of E. festucae YapA.
MATERIALS AND METHODS
Strains and growth conditions.
Escherichia coli cultures (see Table S1 in the supplemental material) were grown overnight in Luria-Bertani (LB) broth or on LB agar containing 100 μg/ml ampicillin as previously described (43).
E. festucae cultures (see Table S1 in the supplemental material) were grown on 2.4% (wt/vol) potato dextrose (PD) agar (44) under previously described conditions (45, 46). Liquid cultures were grown for 5 days at 22°C in 250-ml conical flasks containing 50 ml medium on an orbital shaker at 200 rpm. For collection of spores, E. festucae cultures were grown on PD agar for 10 days at 22°C. The spores were vigorously washed from the surface of the colony with PD broth, and the spore suspension was filtered through sterile glass wool.
S. cerevisiae cultures for complementation (see Table S1 in the supplemental material) were grown on synthetic complete (SC) medium (0.67% yeast nitrogen base without amino acids, 2% galactose, 1% raffinose, 0.01% [each] leucine, tryptophan, and uracil, 0.005% [each] histidine and methionine, and 2% agar) at 30°C for 3 days. For yeast recombinational cloning, strain FY834 was grown on YPD (1% yeast extract, 2% peptone, 2% d-glucose, 2% agar), and transformants were selected on synthetic dropout (SD) medium (1 M sorbitol, 0.67% yeast nitrogen base without amino acids, 0.08% uracil dropout supplement [Clontech], 2% glucose, and 2% agar).
DNA isolation, PCR, and sequencing.
Fungal genomic DNA was extracted from freeze-dried mycelium by the method of Byrd et al. (47). For small-scale experiments, rapid extracts of genomic DNA mycelia were grown in PD broth for 1 to 3 days, transferred to lysis buffer (150 mM EDTA, 50 mM Tris-HCl, 1% sodium lauroyl sarcosine), and incubated at 70°C for 30 min. DNA was isolated from the aqueous phase by sequential precipitations with 5 M potassium acetate, isopropanol, and 70% ethanol and then was resuspended in 20 μl H2O. Isolation of plasmid and cosmid DNAs was performed by alkaline lysis and extraction using a High Pure plasmid isolation kit (Roche) according to the manufacturer's instructions.
Standard PCR amplification was performed with Taq DNA polymerase (Roche). Where proofreading activity was required, the Expand High Fidelity PCR system (Roche) was used. Each reaction mixture (50 μl) contained 1× Taq reaction buffer or 1× Expand High Fidelity buffer, 0.2 μM (each) forward and reverse primers, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 1 ng template DNA, and 1 U of Taq polymerase or 1.75 U of Expand High Fidelity enzyme mix (Roche). The following cycle conditions were used: 1 cycle of 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 45 to 68°C for 30 s, and 72°C for 1 min per kb of sequence. The final extension consisted of one cycle at 72°C for 10 min, and then the tubes were stored at 4°C until analysis. The sequences of PCR primers are provided in Table S2 in the supplemental material.
DNA fragments were sequenced using the dideoxynucleotide chain termination method with a Big Dye Terminator ready reaction cycle sequencing kit, version 3.1 (Applied Biosystems), and were separated using an ABI3730 genetic analyzer (Applied Biosystems). Sequence data were assembled and analyzed using MacVector sequence assembly software, version 12.0.5.
DNA hybridizations.
E. festucae genomic digests were transferred to positively charged nylon/nitrocellulose membranes (Roche) by the method described by Southern (48). DNA was fixed by UV light cross-linking in a Cex-800 UV light cross-linker (Ultra-Lum) at 254 nm for 2 min. The DNA probes were synthesized by random priming with Klenow DNA polymerase and [α-32P]dCTP (3,000 Ci/mmol; Amersham Biosciences), using a High Prime DNA labeling kit (Roche), or by the incorporation of digoxigenin-11-dUTP (DIG-11-dUTP) into DNA by PCR, using a PCR DIG probe synthesis kit (Roche). Hybridizations were performed according to the manufacturer's instructions and visualized by either autoradiography (32P-labeled probes) or nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) color detection (DIG-labeled probes).
Cosmid clone 28E7 (yapA) was isolated from an Fl1 cosmid library (49) by colony hybridization as previously described (39), using a 1.7-kb fragment of yapA, amplified using the Expand High Fidelity PCR system and the yap5-yap6 primer pair, as a probe. The probe was labeled with [α-32P]dCTP as described above.
Preparation of complementation, deletion, and expression constructs.
The S. cerevisiae YAP1 (pGC8) and GPX3 (pGC7) genomic complementation constructs were prepared by PCR amplification of the 1.95-kb YAP1 gene and the 0.5-kb GPX3 gene, using the yap7-yap8 and gpx7-gpx8 primers, respectively, with the Expand High Fidelity PCR system. The E. festucae yapA (pGC6) and gpxC (pGC5) cDNA complementation constructs were prepared by PCR amplification of the 1.7-kb yapA cDNA and 0.5-kb gpxC cDNA from E. festucae total mRNA, using the yap5-yap6 and gpx5-gpx6 primer pairs, respectively. These fragments were then ligated into pCR4-TOPO (Invitrogen), sequenced, and subcloned into the pYES2 vector (Invitrogen).
The yapA replacement construct (pGC2) was prepared by sequentially ligating into pSF15.15 (a pII99-based vector containing the hph cassette) a 1.1-kb BglII/KpnI fragment 5′ of yapA, generated by PCR amplification using the yap1-yap2 primer pair, and a 1.2-kb HindIII/XhoI fragment 3′ of yapA, generated by PCR amplification using the yap3-yap4 primer pair, as shown in Fig. S1 in the supplemental material.
The gpxC replacement construct, plasmid pGC4, was prepared by sequentially ligating into pSF17.8 (a pII99-based vector containing the nptII cassette) a 1.1-kb BglII/KpnI fragment 5′ of gpxC, generated by PCR amplification using the gpx1-gpx2 primer pair, and a 1.4-kb SalI/SalI fragment 3′ of gpxC, generated by PCR amplification using the gpx3-gpx4 primer pair, as shown in Fig. S1 in the supplemental material.
The tpxA replacement construct, plasmid pGC12, was prepared by sequentially ligating into pSF15.15 a 2.3-kb BglII/KpnI fragment 5′ of tpxA, generated by PCR amplification using the tpx1-tpx2 primer pair, and a 2.45-kb fragment 3′ of tpxA, generated by PCR amplification using the tpx3-tpx4 primer pair, as shown in Fig. S1 in the supplemental material.
The 3.8-, 4.3-, and 6.2-kb linear products of pGC2 (yapA), pGC4 (gpxC), and pGC12 (tpxA) used for transformation were amplified with the yap1-yap4, gpx1-gpx4, and tpx1-tpx4 primer pairs, respectively, using the Expand High Fidelity PCR system (Roche) according to the manufacturer's instructions. A 3.7-kb EcoRV/EcoRI fragment extending 740 bp upstream from the transcription start site of yapA and 639 bp downstream from the transcription termination site was digested from cosmid 28E7 and subcloned into pSF17.8 to generate the pGC11 yapA complementation vector.
A YapA-EGFP fusion construct, pGC9, was prepared by cloning an EcoRI/ClaI fragment generated with the yap27-yap28 primer pair into the pPN94 vector. The translational stop codon of yapA was removed and replaced by the enhanced green fluorescent protein (EGFP) coding region, with its stop codon generated with the GCGFP1-GCGFP2 primer pair, creating a C-terminal in-frame fusion with the yapA gene.
A Yap1-EGFP fusion construct, pGC19, was prepared by amplifying overlapping Yap1 and EGFP fragments, using the scyap16-scyap18 and GCGFP3-GCGFP4 primer pairs, respectively, recombining them in yeast, and cloning the EcoRI/NotI Yap1-EGFP fragment into pPN94.
A PcatA-eGFP reporter construct, pGC13, was prepared by sequentially ligating into pSF17.8 an XbaI/EcoRI fragment containing the 1-kb region upstream of the catA start codon and a 0.7-kb XbaI/EcoRI fragment containing the EGFP coding sequence. The PcatA fragment was generated by PCR using the pcatA3-pcatA4 primer pair and ligation into pGC10, and the PcatA-eGFP XbaI/XhoI fragment was subcloned into pSF17.8. A destabilized version of the PcatA-eGFP reporter construct, PcatA-eGFP-CL1 (pGC14), was generated by adding a 16-amino-acid degron sequence (CL1 [ACKNWFSSLSHFVIHL]) to the C terminus of EGFP. Two-step PCR using the pCatA-CL1 R1 and pCatA-CL1 R2 primer pairs was used to generate the 1.2-kb SacII/NdeI fragment, which was then used to replace the corresponding region in the original PcatA-eGFP vector.
Fungal transformation.
S. cerevisiae transformation was carried out using the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (50). E. festucae protoplasts were prepared as previously described (51). Protoplasts were transformed with 3 to 5 μg of linear PCR-amplified or circular plasmid DNA by a previously described method (52). Transformants were selected on RG medium (regeneration medium, which is PD medium with 0.8 M sucrose) containing 150 μg/ml hygromycin or 200 μg/ml Geneticin and nuclear purified by three rounds of subculturing on hygromycin- or Geneticin- containing PD medium (51).
Putative yapA replacement mutants were screened by PCR using primers that flank the hph cassette (yap22 and yap23; 1 kb of wild-type sequence and 1.6 kb of replacement sequence) and the 5′ (yap9 and yap10; 2.6 kb)- and 3′ (yap11 and yap21; 2.4 kb)-flanking regions of the replacement. Putative gpxC replacement mutants were screened by PCR using primers that flank the nptII cassette (gpx15 and gpx16; 1 kb of wild-type sequence and 1.9 kb of replacement sequence) and the 5′ (gpx9 and gpx10; 2.3 kb)- and 3′ (gpx11 and gpx12; 2.9 kb)-flanking regions of the replacement. Putative tpxA replacement mutants were screened by PCR using primers that flank the hph cassette (tpx13 and tpx14; 1 kb of wild-type sequence and 1.6 kb of replacement sequence) and the 5′ (tpx15 and tpx16; 2.5 kb)- and 3′ (tpx17 and tpx18; 2.5 kb)-flanking regions of the replacement. For complementation of ΔyapA mutants, ΔyapA protoplasts were transformed with pGC11 and transformants selected on medium containing 200 μg/ml Geneticin. Reintroduction of the yapA gene was confirmed by PCR using the yap5-yap6 primer pair.
Transformants expressing EGFP or DsRed fusion proteins were selected using an Olympus BX51 stereomicroscope with Olympus U-MWIBA2 and U-MWIG2 filters to detect EGFP and DsRed, respectively.
Microscopy.
Cultures to be analyzed by microscopy were inoculated onto a thin layer of PD agarose (2% [wt/vol]) layered on top of the base layer of PD agar (1.5% [wt/vol]) and grown for 5 days. For examination of E. festucae spores, spore suspensions were spread onto the PD agarose and examined after 16 h. Square blocks were cut from the agarose and placed in an imaging chamber (CoverWell; 20-mm diameter by 0.5-mm depth) (Molecular Probes) filled with 500 μl of PD broth and sealed with a 22- by 60-mm glass coverslip.
Localization of YapA-EGFP and DsRed-StuA(NLS) in hyphae was analyzed by confocal laser scanning microscopy using a Leica SP5 DM6000B confocal microscope (488-nm argon laser, ×40 oil immersion objective, numerical aperture [NA] = 1.3). Differential interference contrast (DIC) imaging was used in conjunction with confocal microscopy, with the DIC image overlaid on the confocal fluorescence images.
Bioinformatic analysis.
E. festucae genes were identified by tBLASTn analysis of the E. festucae Fl1 (E894) genome (http://csbio-l.csr.uky.edu/ef894-2011) with S. cerevisiae and S. pombe protein sequences obtained from the SGC (http://www.yeastgenome.org/) and GeneDB (http://old.genedb.org/genedb/pombe/) databases, respectively. C. albicans and Yarrowia lipolytica protein sequences were obtained from the Candida (http://www.candidagenome.org/) and Genolevures (http://www.genolevures.org/yali.html) genome databases.
Other fungal sequences used in the phylogenetic analysis were retrieved from the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/) or the Broad Institute website (http://www.broad.mit.edu). Identity and similarity scores were calculated after ClustalW pairwise alignments of sequences by use of MacVector, version 12.0.5. Multiple-sequence alignments were edited using Jalview (http://www.jalview.org/). For phylogenetic relationships, Mega5.05 was used to generate maximum likelihood trees from ClustalW multiple-sequence alignments, using the default parameters in the program. Gene annotation and naming are given in accordance with the Broad Institute Aspergillus Comparative Database. The E. festucae genome sequence data, curated by C. L. Schardl at the University of Kentucky, are available at http://csbio-l.csr.uky.edu/ef894-2011/.
Nucleotide sequence accession numbers.
The yapA, gpxC, and tpxA gene sequences of E. festucae strain Fl1 (894) are available in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/index.html) under accession numbers KC121577, KC121578, and KC244374, respectively.
RESULTS
Identification and characterization of an AP-1-like transcription factor.
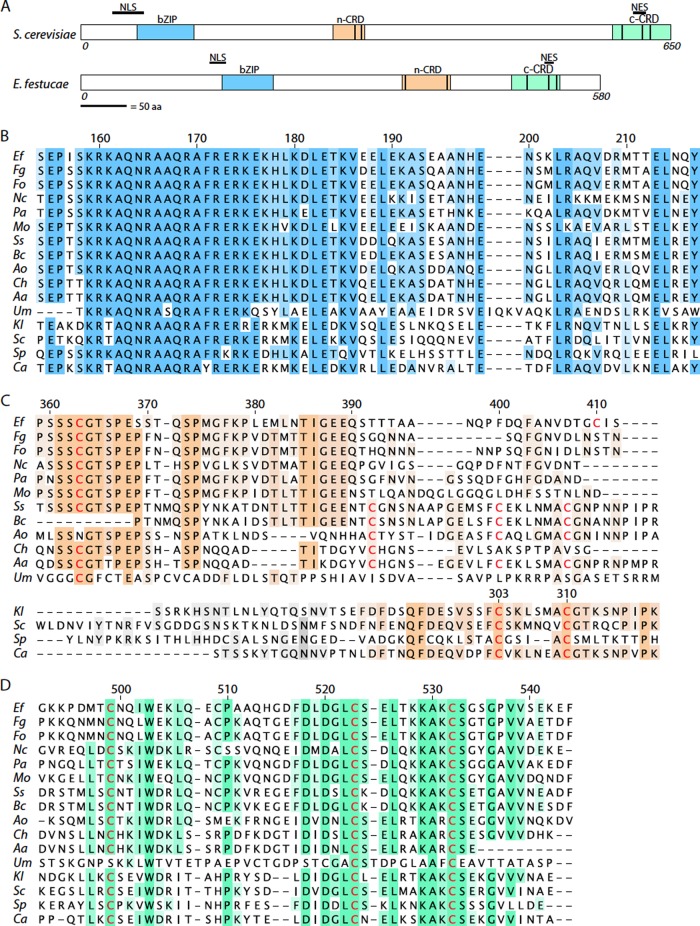
A tBLASTn analysis of the E. festucae genome with the S. cerevisiae Yap1 protein sequence identified a gene, designated yapA (accession number KC121577; E. festucae Fl1 gene model EfM2.092760), encoding a bZIP transcription factor that shares 57% identity with FgAP1 from Fusarium graminearum and 12 to 16% identity with AP-1 proteins from the more distantly related basidiomycete fungus Ustilago maydis and the yeast species S. cerevisiae, Kluyveromyces lactis, S. pombe, and C. albicans. The yapA coding sequence, comprising three exons separated by 72- and 64-bp introns, is predicted to encode a polypeptide of 580 amino acids that shares several conserved features with S. cerevisiae Yap1, including an NLS, an NES, a bZIP domain, and two cysteine-rich domains, n-CRD and c-CRD, that are distal and proximal, respectively, to the N and C termini of the protein (Fig. 1A). Both the bZIP and c-CRD motifs are highly conserved among all of the fungal AP-1 transcription factors analyzed (Fig. 1B and D). A multiple-sequence alignment of the n-CRDs from AP-1 homologs of various yeasts and filamentous fungi identified a major difference in the amino acid sequence of this domain between yeast and filamentous fungal species (Fig. 1C). While there are two conserved cysteine residues in the n-CRDs of yeast species, just one of these (corresponding to residue 310 in S. cerevisiae) is conserved in the filamentous fungi. The two cysteine residues in the c-CRD of the S. cerevisiae protein that form disulfide bonds with partners in the n-CRD upon oxidation are conserved in the filamentous fungi.
Fig 1.
Functional domains of E. festucae YapA. (A) Characteristic motifs of S. cerevisiae Yap1 and E. festucae YapA proteins. bZIP, basic leucine zipper DNA binding domain; n-CRD and c-CRD, N- and C-terminal cysteine-rich domains; NLS, nuclear localization signal; NES, nuclear export sequence. The length of each protein (in amino acids [aa]) is indicated. The vertical black bars within the proteins identify positions of conserved Cys residues. (B to D) Multiple-sequence alignments of bZIP domains, n-CRDs, and c-CRDs of fungal AP-1 proteins. Conserved cysteine residues are highlighted in red. Numbering indicates amino acid residue positions with respect to the E. festucae protein (bZIP [B], n-CRD [upper part of panel C], and c-CRD [D] sequences) or the S. cerevisiae protein (n-CRD [lower part of panel C] sequences). Ef, Epichloë festucae YapA (EfM2.092760; accession no. KC121577); Fg, Fusarium graminearum FGSG_08800.3 (accession no. XP_388976.1); Fo, Fusarium oxysporum FoAP1 (accession no. EGU84635.1); Nc, Neurospora crassa NcAp-1 (NCU03905.5; accession no. XP_957544.2); Pa, Podospora anserina PaAP1 (accession no. XP_001905945.1); Mo, Magnaporthe oryzae MoAP1 (MGG_12814.7; accession no. XP_001408783.1); Ss, Sclerotinia sclerotiorum SSAP1 (SS1G_09561.3; accession no. XP_001589839.1); Bc, Botrytis cinerea Bap1 (BC1G_14094.1; accession no. XP_001547321.1); Ao, Aspergillus oryzae AorAP1 (AO090001000627; accession no. XP_001819128.1); Ch, Cochliobolus heterostrophus CHAP1 (accession no. AAS64313); Aa, Alternaria alternata AaAP1 (accession no. ACM50933.1); Um, Ustilago maydis Yap1 (UM02191.1; accession no. XP_758338.1); Kl, Kluyveromyces lactis KlYap1 (accession no. XP_451077.1); Sc, Saccharomyces cerevisiae Yap1 (YML007W; accession no. CAA41536.1); Sp, Schizosaccharomyces pombe Pap1 (SPAC1783.07c; accession no. NP_593662.1); Ca, Candida albicans Cap1 (CaO19.1623; accession no. XP_721702.1).
Complementation of a yeast yap1 mutant.
To test whether YapA is a functional homolog of S. cerevisiae Yap1, a full-length cDNA encoding E. festucae YapA was cloned into the pYES2 yeast expression vector and transformed into the S. cerevisiae Δyap1 strain. In addition, the Δyap1 strain was transformed with pYES2 vector alone or pYES2 vector containing S. cerevisiae YAP1 (Fig. 2). Cells were grown on medium containing galactose and raffinose to activate the GAL1 promoter in pYES2, which was used to control expression of these genes in S. cerevisiae. All strains grew equally well on medium to which no H2O2 had been added. On medium containing 0.8 mM hydrogen peroxide, growth of the Δyap1 strain and the Δyap1/pYES2 strain was significantly inhibited in comparison to that of the wild-type strain. Expression of E. festucae YapA (Δyap1/EfyapA) or S. cerevisiae Yap1 (Δyap1/ScYAP1) in the S. cerevisiae Δyap1 background was able to restore growth of this mutant on H2O2 to levels comparable to those of the wild type (Fig. 2). These results demonstrate that E. festucae yapA is able to functionally complement the oxidative stress sensitivity defect of the S. cerevisiae Δyap1 mutant. However, it is evident from the number of colonies formed that complementation by yapA from E. festucae is less efficient than that achieved by S. cerevisiae YAP1 (Fig. 2).
Fig 2.
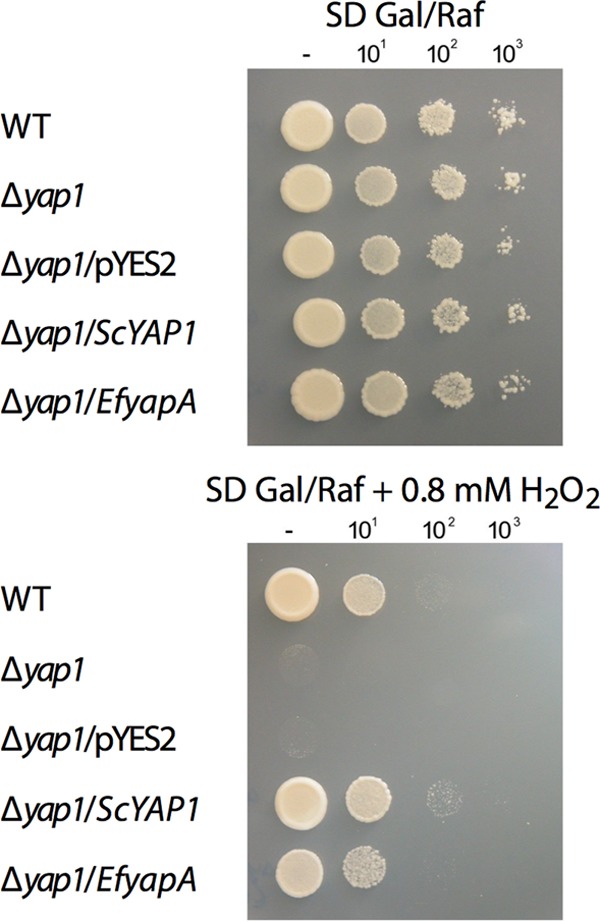
S. cerevisiae complementation by E. festucae yapA. The growth of S. cerevisiae BY4741 (PN2735) (wild type [WT]), S. cerevisiae BY4741-ΔYML007W (Δyap1; PN2736), and derivatives of this strain transformed with the empty vector pYES2 (PN2847), pYES2ScYAP1 (PN2845), or pYES2EfyapA (PN2846) was tested on SD plates containing galactose and raffinose (SD Gal/Raf) with and without 0.8 mM H2O2. Serial 10-fold dilutions of the cultures indicated on the left were spotted onto plates.
Oxidative stress phenotype of mycelia from E. festucae yapA deletion strains.
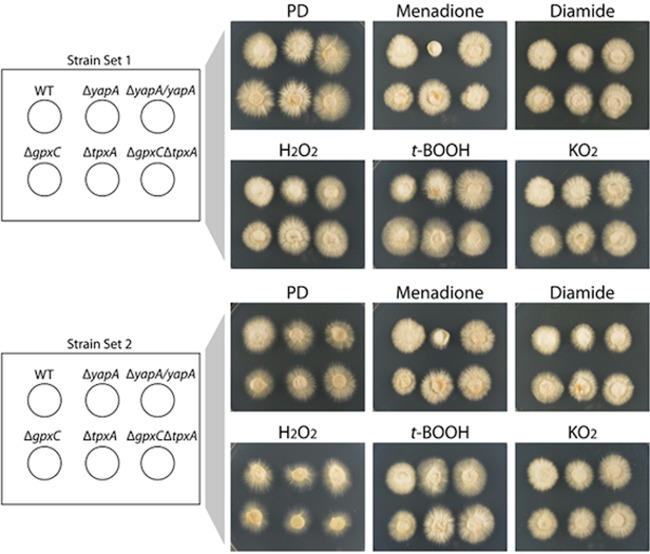
To investigate the role of the E. festucae YapA protein in conferring resistance to various oxidative stress-inducing compounds, yapA was deleted by targeted gene replacement (see Fig. S1 in the supplemental material). Two independent knockouts of yapA were generated, with each containing multiple copies of the deletion construct. Mycelial plugs of the wild-type and ΔyapA strains were inoculated onto PD agar supplemented with various stress-inducing compounds, including the thiol-oxidizing agents menadione and diamide as well as the peroxides H2O2 and t-BOOH and superoxide-generating KO2 (Fig. 3; see Fig. S2 in the supplemental material). Colony radial growth of two independently isolated ΔyapA mutants was severely reduced on 40 μM menadione and reduced to a lesser extent on 1 mM diamide in comparison to that of the wild type. Introduction of a wild-type allele of yapA into each of the ΔyapA strains restored growth on menadione and diamide to wild-type levels, confirming that loss of functional YapA confers sensitivity to these oxidizing agents. In contrast, addition of H2O2, t-BOOH, or KO2 had no effect on colony radial growth of ΔyapA strains (Fig. 3). At higher concentrations of these compounds, wild-type growth was reduced to the same extent as that of mutants (see Fig. S2 in the supplemental material). The lack of sensitivity of vegetative tissue to H2O2 was surprising given that, to date, all published studies of fungal AP-1 mutants report sensitivity to H2O2.
Fig 3.
Oxidative stress sensitivity of E. festucae deletion strains. Five-millimeter-diameter agar plugs of the indicated strains were inoculated onto PD medium and PD medium containing 40 μM menadione, 1 mM diamide, 4 mM H2O2, 0.25 mM t-BOOH, or 7 mM KO2 and cultured at 22°C for 7 days. (A) Strain set 1 included the wild-type (WT; PN2278), ΔyapA#145 (PN2740), ΔyapA#145/yapA (PN2788), ΔgpxC#10 (PN2741), ΔtpxA#105 (PN2821), and ΔgpxC ΔtpxA#22 (PN2831) strains. (B) Strain set 2 included the wild-type (WT), ΔyapA#243 (PN2739), ΔyapA#243/yapA (PN2787), ΔgpxC#34 (PN2742), ΔtpxA#157 (PN2822), and ΔgpxC ΔtpxA#168 (PN2830) strains (numbers in strain names indicate independent transformants).
Conidial germination sensitivity of E. festucae ΔyapA mutants to H2O2.
To investigate the possibility of a developmental stage-specific role for YapA in providing resistance against peroxides, E. festucae spores were spread onto PD medium containing H2O2. Spores derived from the wild-type strain were able to germinate on PD medium containing 0.5 mM H2O2, whereas spores of the ΔyapA strain were highly sensitive to H2O2 and were unable to germinate, as confirmed by light microscopy (Fig. 4). Reintroduction of yapA to the ΔyapA mutant restored H2O2 resistance of the spores back to wild-type levels. These results suggest that YapA is indispensable for conidial resistance to peroxide stress.
Fig 4.
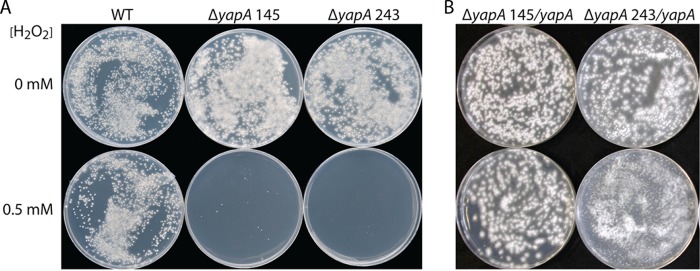
H2O2 sensitivity of E. festucae ΔyapA conidia. Spore suspensions (250 μl) of wild-type (WT), ΔyapA#145, and ΔyapA#243 strains (A) and complemented ΔyapA#145/yapA and ΔyapA#243/yapA strains (B) were spread over the surfaces of plates of PD medium and PD medium containing H2O2, and the spores were allowed to germinate at 22°C for 12 and 11 days, respectively.
Expression of catA in the ΔyapA mutant.
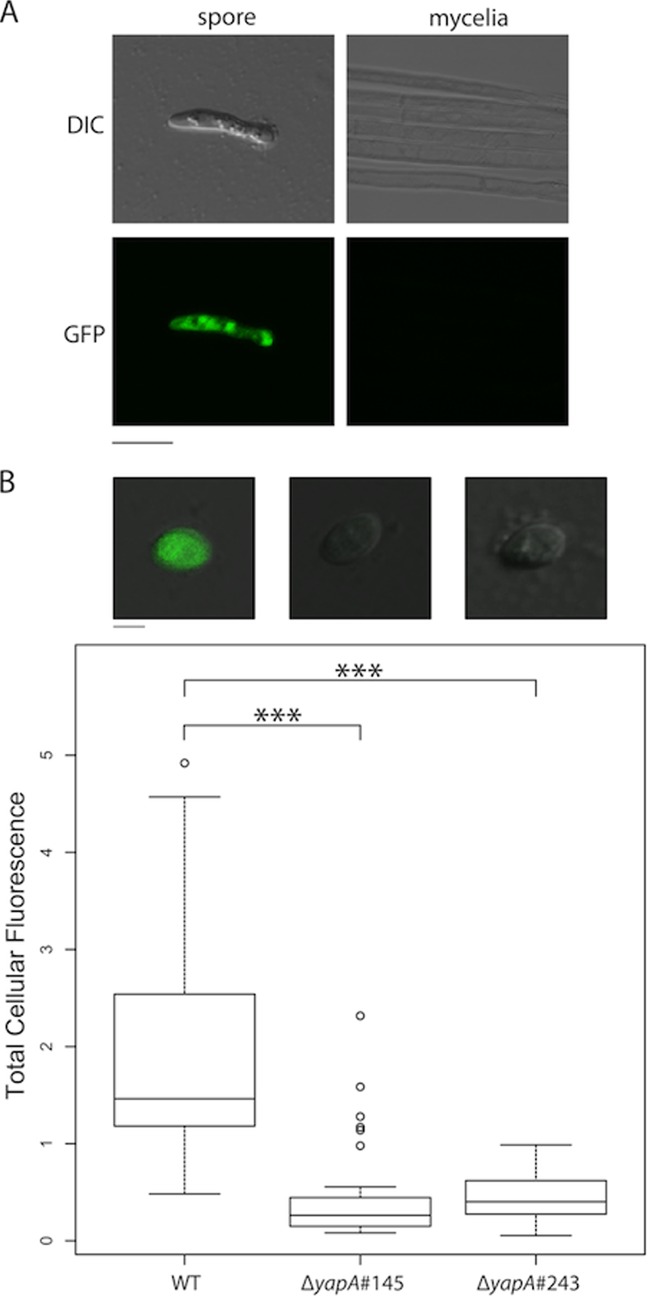
To explain the elevated sensitivity of ΔyapA spore germination to H2O2, we tested whether YapA was required for activation of a spore-specific catalase, an enzyme responsible for H2O2 detoxification. A homolog of the Aspergillus nidulans spore-specific catalase gene, catA (53), was identified in E. festucae (EfM2.069120). The predicted protein shares 74.1% similarity with A. nidulans CatA. The promoter of E. festucae catA was fused to the EGFP gene to monitor its expression. EGFP fluorescence was detected in wild-type spores but not vegetative mycelia, confirming that expression of E. festucae catA is spore specific (Fig. 5A). To enhance the sensitivity of the spore expression analysis, a second construct was prepared, in which the CL1 degron sequence was fused to the C terminus of the catA-EGFP construct. Addition of the CL1 degron sequence, specific for ubiquitination and degradation by the proteasome, is reported to confer a 20- to 30-min half-life on EGFP (54). Spores of the ΔyapA mutant strains showed a significant reduction (P ≪ 0.001) in EGFP fluorescence, indicating reduced catA promoter activity compared to that in wild-type spores (Fig. 5B). EGFP fluorescence was restored in the yapA-complemented ΔyapA strains, indicating that the reduction in EGFP reporter gene expression was due to a lack of yapA expression. This result suggests that YapA is required for activation of catA expression and that the sensitivity of ΔyapA conidial germination to H2O2 may be due to decreased expression of catA.
Fig 5.
Spore-specific expression of PcatA-eGFP. (A) Confocal and DIC microscopy images confirming the spore-specific expression of the PcatA-eGFP reporter, pGC13. Bar = 10 μm. (B) Spore-specific expression of the PcatA-eGFP-CL1 reporter, pGC14. The box plot shows the total amount of fluorescence per spore, indicating a significant decrease in EGFP fluorescence in ΔyapA spores relative to wild-type (WT) spores. Significance between samples was determined using Student's two-tailed t test: for the WT versus ΔyapA#145 strains, t100 = 9.6 and the P value was ≪0.0001 (***); for the WT versus ΔyapA#243 strains, t89 = 12.6 and the P value was ≪0.0001 (***); and for the ΔyapA#145 versus ΔyapA#243 strains, t34 = 0.3, and the difference was not significant. The total cellular fluorescence of each spore was quantified using ImageJ software. The image analysis was performed on maximum-intensity projection images that were generated from 5- by 1-μm confocal z stacks. Representative merged DIC and confocal fluorescence images showing EGFP expression in wild-type (WT), ΔyapA#145 (PN2740), and ΔyapA#243 (PN2739) spores are shown above the box plot. Bar = 2 μm. Multiple transformants, including Fl1::PcatA-eGFP-Cl1 (PN2838, PN2839, PN2840, and PN2841), ΔyapA#243::PcatA-eGFP-CL1 (PN2836, PN2837, and PN2844), and ΔyapA#145::PcatA-eGFP-CL1 (PN2842 and PN2843) transformants, were analyzed.
Activation of YapA by H2O2 in axenic culture.
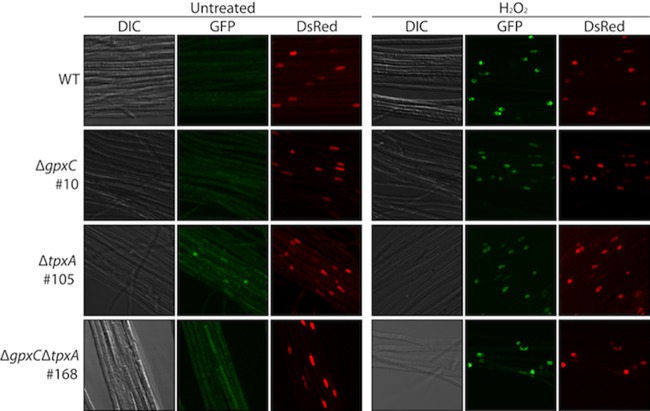
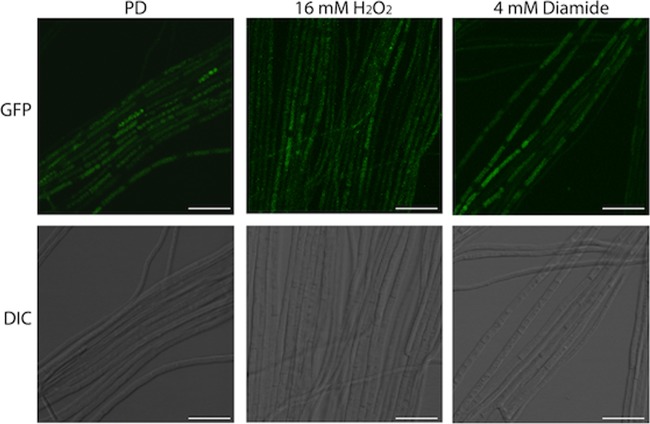
In S. cerevisiae, oxidation of the Yap1 protein by H2O2 correlates with its activation and nuclear accumulation. To monitor the cellular localization and inferred activation of H2O2-oxidized YapA, protoplasts of the wild type were transformed with a YapA-EGFP fusion construct, pGC9, together with a DsRed-StuA(NLS) construct, pJW19, to confirm nuclear localization. In the absence of oxidative stress, YapA-EGFP localized to the cytoplasm, but upon treatment with H2O2, YapA-EGFP was redistributed to the nucleus, where it colocalized with the nuclear marker DsRed-StuA(NLS) (Fig. 6).
Fig 6.
H2O2 activation of YapA-EGFP in axenic culture. E. festucae wild-type (WT; PN2790), ΔgpxC#10 (PN2789), ΔtpxA#105 (PN2823), and ΔgpxC ΔtpxA#168 (PN2851) cultures expressing YapA-EGFP and the nuclear marker DsRed-StuA(NLS) were examined using confocal laser scanning and DIC microscopy before (untreated) and 30 min after treatment with 16 mM H2O2. EGFP images were generated by maximum-intensity projection of confocal z stacks taken at 1-μm intervals from the top to the bottom of the section. Each box represents a 50-μm by 50-μm area.
Given that S. cerevisiae Gpx3 (glutathione peroxidase 3) and S. pombe Tpx1 (thioredoxin peroxidase 1) are responsible for the initial sensing of the H2O2 signal and activation of their respective AP-1 transcription factors (21, 24, 25), we tested whether homologs of these thiol peroxidases (peroxiredoxins) were required for activation of E. festucae YapA under conditions of oxidative stress. Homologs of S. cerevisiae GPX3 and S. pombe TPX1 in the E. festucae genome were identified by tBLASTn and reciprocal BLASTp analyses and designated gpxC (KC121578; EfM2.018640) and tpxA (KC244374; EfM2.113210), respectively. Multiple-sequence alignment and phylogenetic analyses confirmed that GpxC and TpxA are the most closely related proteins to S. cerevisiae Gpx3 and S. pombe Tpx1, respectively (see Fig. S3 and S4 in the supplemental material). However, in the case of the latter, filamentous fungal proteins are more closely related to S. cerevisiae Prx1, a 1-Cys peroxiredoxin, whereas S. pombe Tpx1 groups with S. cerevisiae Tsa1 and Tsa2, which are typical 2-Cys peroxiredoxins (see Fig. S4 in the supplemental material). Typical 2-Cys peroxiredoxins have both peroxidatic (C48 in S. pombe) and resolving (C169 in S. pombe) Cys residues and, in addition, contain the signature consensus sequence FTFVCPTEI for the first Cys (see Fig. S4 in the supplemental material). In contrast, the Tpxs of filamentous fungi have a single cysteine residue that forms part of the signature consensus sequence FTPVCTTEL. Two additional peroxiredoxins were identified in filamentous fungal genomes, but these appear to be homologs of S. pombe Pmp20 (EfM2.064230) and Bcp (EfM2.115510). These are atypical 2-Cys peroxiredoxins, indicating that typical 2-Cys peroxiredoxins appear to be absent from filamentous fungi.
Given that there are three different but closely related Gpxs in S. cerevisiae, i.e., Gpx1, Gpx2, and Gpx3, cDNA of E. festucae gpxC was expressed in an S. cerevisiae Δgpx3 strain to test if it could complement this mutant under conditions of oxidative stress. All strains grew equally well on medium to which no H2O2 had been added. On medium containing 1.25 mM hydrogen peroxide, growth of the Δgpx3 strain and the Δgpx3/pYES2 strain was significantly inhibited in comparison to that of the wild-type strain. Expression of E. festucae GpxC (Δgpx3/EfgpxC) or S. cerevisiae Gpx3 (Δgpx3/ScGPX3) in the S. cerevisiae Δgpx3 background was able to restore growth of this mutant on H2O2 to levels comparable to those of the wild type (Fig. 7). The slightly higher concentration of H2O2 used for these experiments (1.25 mM) than for the YAP1 complementation tests (0.8 mM) (Fig. 2) reflects the slightly greater tolerance of the Δgpx3 strain to oxidative stress, because of an alternative pathway for Yap1 activation (21). These results demonstrate that E. festucae gpxC is able to functionally complement the oxidative stress sensitivity defect of the S. cerevisiae GPX3 mutant.
Fig 7.
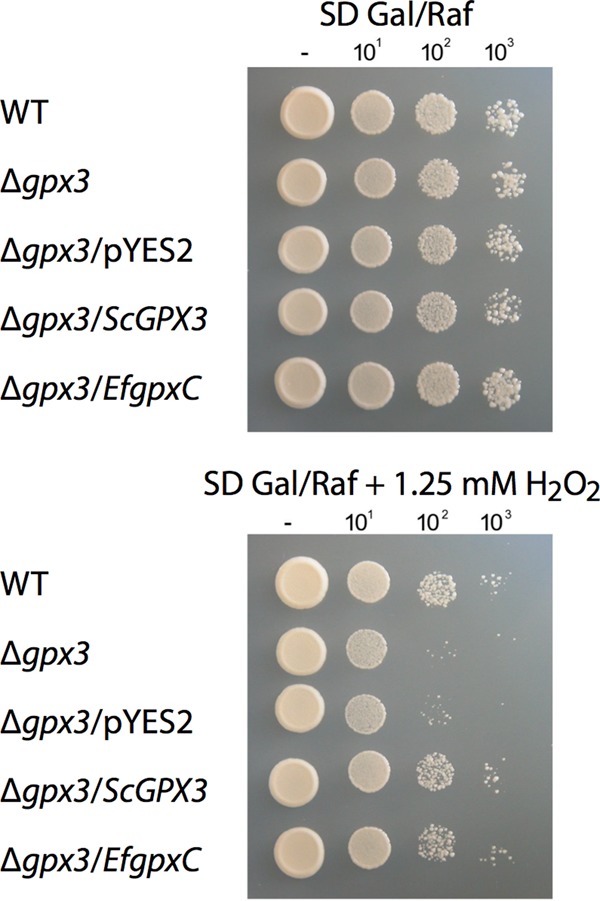
S. cerevisiae complementation by E. festucae gpxC. The growth of S. cerevisiae BY4741 (WT; PN2735), S. cerevisiae BY4741-ΔYIR037W (ΔgpxC; PN2737), and derivatives of this strain transformed with the empty vector pYES2 (PN2850), pYES2ScGPX3 (PN2848), or pYES2EfgpxC (PN2849) was tested on SD plates containing galactose and raffinose with and without 1.25 mM H2O2. Serial 10-fold dilutions of the cultures indicated on the left were spotted onto plates.
To test whether either GpxC or TpxA serves as a redox sensor for YapA activation in E. festucae, single (ΔgpxC or ΔtpxA) and double (ΔgpxC ΔtpxA) mutants of gpxC and tpxA were generated, and localization of YapA-EGFP was monitored in hyphae of these different mutant backgrounds (Fig. 6; see Fig. S1 in the supplemental material). In the ΔgpxC, ΔtpxA, and ΔgpxC ΔtpxA backgrounds, YapA-EGFP still translocated from the cytoplasm to the nucleus in response to H2O2 treatment, where it colocalized with the nuclear marker DsRed-StuA(NLS). These results demonstrate that neither GpxC nor TpxA is essential for YapA nuclear localization and that these proteins do not have overlapping functions (Fig. 6).
To assess whether E. festucae possesses a redox sensor capable of activating S. cerevisiae Yap1, a Yap1-EGFP fusion construct (pGC19) was expressed in E. festucae wild-type cells. However, this fusion failed to localize to the nucleus when cells were treated with H2O2 or diamide (Fig. 8). In a control experiment, the same Yap1-EGFP construct under the control of the S. cerevisiae GAL1 promoter localized to the nuclei of S. cerevisiae wild-type and Δyap1 cells, but not Δgpx3 cells, in response to treatment of the cells with H2O2 (21) (see Fig. S5 in the supplemental material), demonstrating the functional fidelity of the construct. These results suggest that either E. festucae lacks an H2O2 redox sensor or the sensor is unable to interact with the Yap1-EGFP fusion.
Fig 8.
H2O2 activation of Yap1-EGFP in axenic culture. An E. festucae wild-type strain (PN2874) expressing S. cerevisiae Yap1-EGFP was examined by confocal laser scanning and DIC microscopy in the absence of oxidative stress (PD) or 30 min after treatment with 16 mM H2O2 or 4 mM diamide. EGFP images were generated by maximum-intensity projection of confocal z stacks taken at 1-μm intervals from the top to the bottom of the section. Bars = 20 μm.
Plant phenotypes of E. festucae yapA, gpxC, and tpxA mutants.
To test whether disruption of yapA, gpxC, tpxA, or both gpxC and tpxA would affect the symbiotic interaction phenotype of E. festucae with L. perenne, seedlings were inoculated with each of these mutants, and whole-plant and in planta phenotypes were analyzed at 8 weeks postinfection. All mutants had a wild-type interaction phenotype at the whole-plant level (see Fig. S6 in the supplemental material) and subcellular level (see Fig. S7 in the supplemental material). Hyphae of all mutants were aligned parallel to the axes of the leaves and were similar to the wild type in morphology and growth habit. Given that inoculation of the plants required wounding of the seedlings by making a 2- to 3-mm longitudinal slit with a scalpel above the meristem of the hypocotyl, followed by placement of the inoculum on the wound site (55), we tested whether YapA was activated in the wild-type strain in response to the intense burst of ROS (H2O2) generated by wounding (56, 57). Consistent with the rapid generation of H2O2 in response to wounding, YapA-EGFP localized to the nuclei of wild-type hyphae within 30 min of inoculation (Fig. 9). After a period of about 6 h, YapA-EGFP was found predominantly in the cytoplasm, consistent with a return to homeostasis and recovery of the cells from the oxidative burst. This burst of ROS was insufficient to disrupt infection by the wild-type and ΔyapA mutant strains, despite the latter lacking a functional YapA oxidative stress signaling pathway. These results suggest that E. festucae has a highly redundant system for protection from oxidative stress.
Fig 9.
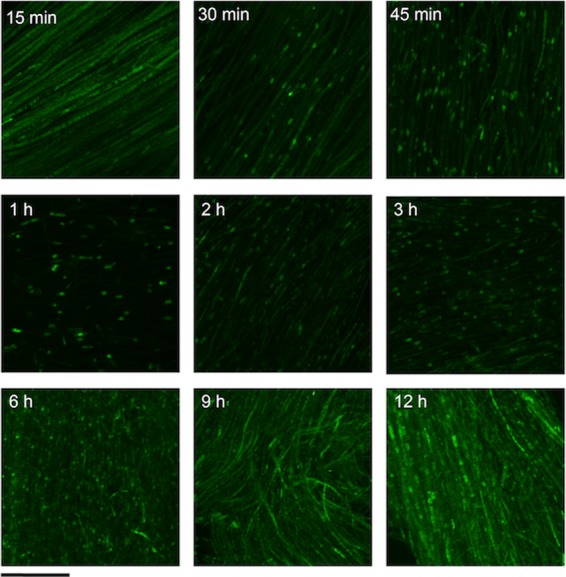
Time course of YapA-EGFP localization. An E. festucae wild-type strain expressing YapA-EGFP (PN2790) was inoculated into the meristematic region of grass seedlings, and the subcellular localization of YapA-EGFP was examined using confocal laser scanning microscopy at the indicated time points. EGFP images were generated by maximum-intensity projection of confocal z stacks taken at 1-μm intervals from the top to the bottom of the section. Bar = 10 μm.
DISCUSSION
The results described here demonstrate that E. festucae YapA is a bona fide AP-1-like bZIP transcription factor involved in the oxidative stress response. E. festucae YapA functionally complemented the H2O2 stress sensitivity defect of an S. cerevisiae Δyap1 mutant and translocated to the nuclei of E. festucae cells in response to H2O2, properties that demonstrate that YapA is capable of activating genes required for an H2O2-induced oxidative stress response in S. cerevisiae. However, unlike other filamentous fungi (8, 32, 33, 58), the E. festucae ΔyapA mutants did not show increased hyphal sensitivity to H2O2 compared to the wild type. Hyphae of the E. festucae ΔyapA mutants were also resistant to KO2 and t-BOOH but very sensitive to menadione and slightly sensitive to diamide. Thus, E. festucae is the exception among the filamentous fungi and yeast studied to date in that its AP-1-like transcription factor is not essential for hyphal resistance to oxidative stress induced by hydroperoxides. One possible explanation is the presence of functionally overlapping signaling pathways in E. festucae to protect against H2O2-induced oxidative stress.
The other well-studied pathway involved in fungal oxidative stress signaling is the Spc1/SakA (S. pombe/A. nidulans) MAPK pathway (59, 60). In this pathway, a multistep phosphorelay sensor-responder system transmits the H2O2 oxidative stress signal from the response regulator Mcs4/SskA to the Spc1/SakA MAPK, which phosphorylates and activates the bZIP transcription factor, Atf1/AtfA. Disruption of spc1 and atf1 in S. pombe (61, 62) or their homologs, sakA and atfA, in A. nidulans (58, 63, 64) leads to increased sensitivity to oxidative (H2O2) stress. Once again, E. festucae appears to be the exception among the filamentous fungi studied to date.
In contrast to the lack of sensitivity of vegetative mycelia to oxidative stress, conidia of the E. festucae ΔyapA mutant were very sensitive to H2O2 oxidative stress and failed to germinate. Introduction of the wild-type yapA gene into the mutant restored H2O2 tolerance of the germinating spores to wild-type levels, confirming a role for YapA signaling in spore adaptation to H2O2 oxidative stress. In A. nidulans, both SakA, through the transcription factor AtfA, and YapA pathways have been shown to be important for conferring spore resistance to H2O2 stress, as both atfA and napA (equivalent to yapA) mutants are sensitive to H2O2 stress (58, 64, 65). For the SakA pathway, tolerance to oxidative stress is mediated through a spore-specific catalase, CatA, whose expression is regulated by AtfA. Levels of catA mRNA are dramatically lower in the ΔatfA mutant than in the wild-type strain, suggesting that transcription of the catA gene is regulated in a spore-specific manner through AtfA (58, 64). The expression of E. festucae catA is also spore specific, but in this fungal species, as in Aspergillus ochraceus (66), catA appears to be regulated by YapA, as deletion of yapA confers a spore germination sensitivity phenotype under conditions of oxidative stress. How YapA regulates catA in E. festucae spores remains to be determined.
While infection of L. perenne with the E. festucae ΔsakA mutant results in a defective symbiotic interaction phenotype (42), yapA mutants are able to infect and colonize L. perenne seedlings as effectively as the wild type and give rise to mature plants with a phenotype similar to that with the wild type. These results contrast with the reduced virulence observed for AP-1 mutants of the biotrophic fungal pathogens U. maydis and M. oryzae, because of their sensitivity to the host oxidative burst (7, 8). However, an important lifestyle difference between these biotrophic fungal pathogens and E. festucae is the apparent absence of a leaf penetration mechanism for the latter. To infect endophyte-free seedlings of L. perenne with E. festucae in the laboratory, it is necessary to mechanically wound the seedlings to allow the endophyte to colonize the grass host (55), as the natural route of infection through the stigmata and styles of the flowers is a process that is difficult to reproduce under laboratory conditions (67). However, wounding of seedlings should generate a burst of ROS and a state of oxidative stress for hyphae inoculated at the wound site (56, 57). Wounding may also result in a release of phenolics that have been shown to induce relocalization of GFP-ChAP1 to the nucleus in Cochliobolus heterostrophus (68). Using YapA-EGFP as a redox sensor, we were able to show that YapA-EGFP hyphae in direct contact with the wound, as well as in cells more distant from the wound site, relocalized to the nucleus within 30 min of wounding. The localization of YapA-EGFP in the nucleus was transient, peaking at 1 h postinoculation, followed by a gradual redistribution to the cytoplasm over a 6- to 9-h period, indicating restoration of redox homeostasis. If this short period of oxidative stress does impair hyphal growth at the infection site, it is not sufficient to prevent subsequent host colonization and establishment of a mutualistic symbiotic interaction.
Although AP-1 transcription factors have been characterized in a range of filamentous fungi, the mechanism by which oxidative stress is sensed and transduced to the AP-1 protein is still not known (8, 30–34). The assumption has been that the immediate sensor of increased levels of H2O2 would be a thiol peroxidase (7, 33), as has been established for the well-characterized Yap1-Gpx3 and Pap1-Tpx1 redox relay systems that operate in the yeast fungi S. cerevisiae and S. pombe, respectively (21, 24, 25). The abundance of these proteins in the cell and their favorable kinetic properties compared to those of alternative thiol-reactive proteins support this hypothesis (69). In addition, signaling through a thiol peroxidase/peroxiredoxin (Prdx1) was recently demonstrated in a mammalian system (70). However, our results show that YapA fused to EGFP still localizes to the nucleus in mutants defective in either GpxC or TpxA, to the same extent as it does in the wild type, when mycelia are subjected to H2O2-induced oxidative stress. Furthermore, YapA-EGFP still localizes to the nucleus when both genes are deleted, ruling out the possibility of functional redundancy between GpxC and TpxA. These results were surprising given the high degree of primary amino acid sequence conservation between GpxC and Gpx3 and the ability of GpxC to functionally complement the Δgpx3 mutant of S. cerevisiae, as does the M. oryzae homolog Hyr1 (35).
While most fungi have a single Gpx protein, S. cerevisiae has three isoforms: Gpx1, Gpx2, and Gpx3 (71). All are classified as atypical 2-Cys peroxiredoxins because they form, as a consequence of the peroxidase reaction, an intramolecular (as opposed to intermolecular) disulfide bond (72) which is cleaved by glutathione or thioredoxin. GPX1 and GPX3 are paralogs that have arisen from whole-genome duplication (WGD) (http://wolfe.gen.tcd.ie/ygob/). Although Gpx1 and Gpx2 show a high degree of conservation with Gpx3 and have known peroxidase activity in vitro, Δgpx1 and Δgpx2 mutants have no oxidative stress phenotype in culture and no known involvement in Yap1 activation (71, 73). The preferential localization of Gpx3 to the cytoplasm (74), compared to peroxisomal matrix and mitochondrial localizations for Gpx1 and Gpx2, respectively, may be one explanation for why Gpx3 can promote oxidation of Yap1 (73, 75). Alternatively, subtle structural changes in Gpx3 may promote the interaction with Yap1 (76).
In S. pombe, Tpx1 rather than Gpx1 has been shown to be the crucial peroxiredoxin (Prx) required for redox sensing and signaling (24, 25). Phylogenetic analysis identified E. festucae TpxA as the closest homolog of Tpx1. However, Tpx1 is a typical 2-Cys Prx, whereas E. festucae TpxA and homologs from other filamentous fungi are all 1-Cys Prxs. Another important biochemical difference between the typical 2-Cys and 1-Cys Prxs is the ability of the former to undergo hyperoxidation, a property that was recently shown to be crucial for oxidative stress signaling (77). A sulfiredoxin (Srx1) has been identified in the yeast group of fungi that reduces the cysteine-sulfinic acid in Prx (Tpx1/Tsa) back to a cysteine-sulfenic acid (78), but interestingly, this protein appears to be absent from the filamentous fungi (79). The absence of typical 2-Cys peroxiredoxins in the filamentous fungi and the corresponding absence of a sulfiredoxin are particularly interesting given the recent hypothesis that oxidation-reduction cycles of 2-Cys peroxiredoxins constitute universal markers for circadian rhythms (80).
While several studies have established a key role for thiol peroxidases (peroxiredoxins) in protection of yeast to oxidative stress, very few studies have been carried out in filamentous fungi. Like the ΔyapA mutant, growth of the E. festucae ΔgpxC, ΔtpxA, and ΔgpxC ΔtpxA strains in culture was not impaired by addition of H2O2, once again emphasizing that this fungal symbiont has a robust oxidative stress protection system. In contrast, growth of the M. oryzae Δhyr1 mutant in culture was impaired by H2O2 (35). In S. cerevisiae, there are at least eight thiol peroxidases that appear to have overlapping functions for protection from a range of different types of oxidative stress: five of the atypical 2-Cys family, including Gpx1 to -3, Dot5 (Bcp in S. pombe), and AHP1 (Pmp20); two of the typical 2-Cys family, including Tsa1 and Tsa2 (Tpx1); and one from the 1-Cys family (Prx1). The numbers of thiol peroxidase family members in S. pombe and E. festucae are, by comparison, reduced. S. pombe has a single Gpx homolog, Gpx1, and three peroxiredoxin homologs, Tpx1, Bcp, and Pmp20 (81). Similarly, E. festucae has a single Gpx1 homolog, a Tpx1 protein which groups more closely with S. cerevisiae Prx1 than with S. pombe Tpx1, and Bcp and Pmp20 homologs. The E. festucae thiol peroxidases may also constitute a cellular signaling network, but this remains to be tested.
Like the ΔyapA mutant, the E. festucae ΔgpxC, ΔtpxA, and ΔgpxC ΔtpxA mutants were able to infect and colonize L. perenne seedlings as effectively as the wild type and gave rise to mature plants with the same phenotypes as those of the wild type, demonstrating that the GpxC and TpxA thiol reductases are not crucial for establishment and maintenance of this fungus-grass symbiosis. As discussed above, the need for mechanical wounding to introduce the wild type and these mutants into the host grass limits our screen to postinfection and host colonization phenotypes. The only other filamentous fungal thiol peroxidase that has been analyzed functionally is the Gpx3 homolog Hyr1 from M. oryzae. The Δhyr1 mutant had reduced virulence on both susceptible barley and rice leaves and was sensitive to ROS produced at the infection sites (35), but the host phenotype observed was not nearly as severe as that with the Δyap1 mutant, which is completely blocked in host leaf infection (8).
Given that neither GpxC nor TpxA is required for YapA localization to the nucleus, we conclude that H2O2 oxidative stress signaling in E. festucae occurs by a mechanism distinct from the classic Gpx3 and Tpx1 redox relay systems that occur in S. cerevisiae and S. pombe. One possibility is that YapA is directly oxidized by H2O2, as has been shown for OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella enterica serovar Typhimurium (82, 83). Interestingly, nearly all the Yap1 homologs found in filamentous fungi have just a single conserved Cys in the n-CRD, suggesting that formation of just a single disulfide bond may be sufficient to induce the necessary conformation change required for Yap to relocalize to the nucleus. Alternatively, oxidation of the conserved cysteines in the c-CRD alone may suffice. Whether there is an accessory protein similar to Ybp1 in S. cerevisiae that promotes Yap1 nuclear localization remains to be determined. A second possibility is that YapA is activated by a thiol peroxidase other than GpxC or TpxA. Although the genes for two additional atypical 2-Cys peroxiredoxins, homologs of yeast Dot5/Bcp and AHP1/Pmp20, were found in the genomes of E. festucae and other filamentous fungi, neither has been shown to function in redox signaling in yeast (81), and inactivation of Tpx1 alone is sufficient to completely abolish the transcriptional response to H2O2 stress in S. pombe (84). Given that peroxiredoxins have the most favorable kinetic and cell abundance properties (69), a mechanism that involves an alternative redox-active protein to activate YapA seems unlikely.
While our results do not support the hypothesis that a homolog of either S. cerevisiae Gpx3 or S. pombe Tpx1 is required for activation of E. festucae YapA in response to oxidative stress, we have shown that YapA readily relocalizes from the cytoplasm to the nucleus and is important in providing protection from oxidative stress in culture and in planta. Identifying the specific structural changes that occur upon oxidation of YapA to bring about the conformational change necessary to promote nuclear retention will be crucial for understanding how YapA is activated in filamentous fungi.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the Tertiary Education Commission (TEC) to the Bio-Protection Research Centre, Massey University, and by a Top Achiever doctoral scholarship to G.M.C. from TEC.
We thank Doug Hopcroft, Jianyu Chen, Jordan Taylor (all from Manawatu Microscopy and Imaging Centre, Massey University), and Ruth Wrenn for technical assistance, Yvonne Becker for technical advice, Murray Cox for statistical analysis, Evelyn Sattlegger (Massey University, Albany, NY) for provision of yeast strains, and Reinhard Fischer (Karlsruhe Institute of Technology) for providing PgpdA-DsRed-StuA(NLS). We also thank Rosie Bradshaw, Yvonne Becker, and Elizabeth Veal for comments on the manuscript.
The E. festucae DNA sequence was provided by Chris Schardl through grants EF-0523661 and EPS-0814194 from the U.S. National Science Foundation, NRI 2005-35319-16141 from the USDA, and 2 P20 RR-16481 from the NIH.
G.M.C. and B.S. designed the experiments, and G.M.C. performed the experiments. G.M.C. and B.S. jointly analyzed the results and wrote the paper.
Footnotes
Published ahead of print 26 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00129-13.
REFERENCES
- 1.Simon-Plas F, Elmayan T, Blein J-P. 2002. The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 31:137–147 [DOI] [PubMed] [Google Scholar]
- 2.Torres MA, Jones JDG, Dangl JL. 2005. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37:1130–1134 [DOI] [PubMed] [Google Scholar]
- 3.Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JDG, Doke N. 2003. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15:706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doke N. 1983. Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal wall components of Phytophthora infestans and specific inhibition of the reaction by suppressors of hypersensitivity. Physiol. Plant Pathol. 23:359–367 [Google Scholar]
- 5.Apostol I, Heinstein PF, Low PS. 1989. Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiol. 90:109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8:397–403 [DOI] [PubMed] [Google Scholar]
- 7.Molina L, Kahmann R. 2007. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19:2293–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo M, Chen Y, Du Y, Dong Y, Guo W, Zhai S, Zhang H, Dong S, Zhang Z, Wang Y, Wang P, Zheng X. 2011. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1001302. 10.1371/journal.ppat.1001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373–399 [DOI] [PubMed] [Google Scholar]
- 10.Foyer CH, Noctor G. 2005. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17:1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, Davies JM, Dolan L. 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422:442–446 [DOI] [PubMed] [Google Scholar]
- 12.Lara-Ortíz T, Riveros-Rosas H, Aguirre J. 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50:1241–1255 [DOI] [PubMed] [Google Scholar]
- 13.Malagnac F, Lalucque H, Lepère G, Silar P. 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41:982–997 [DOI] [PubMed] [Google Scholar]
- 14.Egan M, Wang Z-Y, Jones MA, Smirnoff N, Talbot NJ. 2007. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. U. S. A. 104:11772–11777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. 2006. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic association. Plant Cell 18:1052–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. 2008. Local positive feedback regulation determines cell shape in root hair cells. Science 319:1241–1244 [DOI] [PubMed] [Google Scholar]
- 17.Yan C, Lee LH, Davis LI. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood MJ, Andrade EC, Storz G. 2003. The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry 42:11982–11991 [DOI] [PubMed] [Google Scholar]
- 19.Wood MJ, Storz G, Tjandra N. 2004. Structural basis for redox regulation of Yap1 transcription factor localization. Nature 430:917–921 [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040–16046 [DOI] [PubMed] [Google Scholar]
- 21.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471–481 [DOI] [PubMed] [Google Scholar]
- 22.Okazaki S, Tachibana T, Naganuma A, Mano N, Kuge S. 2007. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol. Cell 27:675–688 [DOI] [PubMed] [Google Scholar]
- 23.Veal EA, Ross SJ, Malakasi P, Peacock E, Morgan BA. 2003. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 278:30896–30904 [DOI] [PubMed] [Google Scholar]
- 24.Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayte J, Toledano MB, Hidalgo E. 2005. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. U. S. A. 102:8875–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bozonet SM, Findlay VJ, Day AM, Cameron J, Veal EA, Morgan BA. 2005. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J. Biol. Chem. 280:23319–23327 [DOI] [PubMed] [Google Scholar]
- 26.Kuge S, Arita M, Murayama A, Maeta K, Izawa S, Inoue Y, Nomoto A. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castillo EA, Ayte J, Chiva C, Moldon A, Carrascal M, Abian J, Jones N, Hidalgo E. 2002. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 45:243–254 [DOI] [PubMed] [Google Scholar]
- 28.Azevedo D, Tacnet F, Delaunay A, Rodrigues-Pousada C, Toledano MB. 2003. Two redox centers within yap1 for H2O2 and thiol-reactive chemical signaling. Free Radic. Biol. Med. 35:889–900 [DOI] [PubMed] [Google Scholar]
- 29.Alarco AM, Raymond M. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao J, Kontoyiannis DP, Calderone R, Li D, Ma Y, Wan Z, Li R, Liu W. 2008. Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclophosphamide and triamcinolone. Med. Mycol. 46:773–782 [DOI] [PubMed] [Google Scholar]
- 31.Lev S, Hadar R, Amedeo P, Baker SE, Yoder OC, Horwitz BA. 2005. Activation of an AP1-like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell 4:443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin CH, Yang SL, Chung KR. 2009. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol. Plant Microbe Interact. 22:942–952 [DOI] [PubMed] [Google Scholar]
- 33.Temme N, Tudzynski P. 2009. Does Botrytis cinerea ignore H2O2-induced oxidative stress during infection? Characterization of botrytis activator protein 1. Mol. Plant Microbe Interact. 22:987–998 [DOI] [PubMed] [Google Scholar]
- 34.Tian C, Li J, Glass NL. 2011. Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157:747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM. 2011. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7:e1001335. 10.1371/journal.ppat.1001335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott B, Becker Y, Becker M, Cartwright G. 2012. Morphogenesis, growth and development of the grass symbiont Epichloë festucae, p 243–264 In Martin JP, Di PietroA. (ed), Morphogenesis and pathogenicity in fungi. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 37.Eaton C, Mitic M, Scott B. 2011. Signalling in the Epichloë festucae-perennial ryegrass mutualistic symbiotic interaction, p 143–181 In Perotto S, Baluska F. (ed), Signaling and communication in plant symbiosis, vol 11 Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 38.Christensen MJ, Bennett RJ, Ansari HA, Koga H, Johnson RD, Bryan GT, Simpson WR, Koolaard JP, Nickless EM, Voisey CR. 2008. Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genet. Biol. 45:84–93 [DOI] [PubMed] [Google Scholar]
- 39.Takemoto D, Tanaka A, Scott B. 2006. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 18:2807–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takemoto D, Kamakura S, Saikia S, Becker Y, Wrenn R, Tanaka A, Sumimoto H, Scott B. 2011. Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc. Natl. Acad. Sci. U. S. A. 108:2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka A, Takemoto D, Hyon GS, Park P, Scott B. 2008. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 68:1165–1178 [DOI] [PubMed] [Google Scholar]
- 42.Eaton CJ, Cox MP, Ambrose B, Becker M, Hesse U, Schardl CL, Scott B. 2010. Disruption of signaling in a fungal-grass symbiosis leads to pathogenesis. Plant Physiol. 153:1780–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44.Sanderson KE, Srb AM. 1965. Heterokaryosis and parasexuality in the fungus Ascochta imperfecta. Am. J. Bot. 52:72–81 [PubMed] [Google Scholar]
- 45.Moon CD, Tapper BA, Scott B. 1999. Identification of Epichloë endophytes in planta by a microsatellite-based PCR fingerprinting assay with automated analysis. Appl. Environ. Microbiol. 65:1268–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon CD, Scott B, Schardl CL, Christensen MJ. 2000. The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia 92:1103–1118 [Google Scholar]
- 47.Byrd AD, Schardl CL, Songlin PJ, Mogen KL, Siegel MR. 1990. The β-tubulin gene of Epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 18:347–354 [DOI] [PubMed] [Google Scholar]
- 48.Southern EM. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 49.Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. 2005. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57:1036–1050 [DOI] [PubMed] [Google Scholar]
- 50.Gietz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96 [DOI] [PubMed] [Google Scholar]
- 51.Young CA, Bryant MK, Christensen MJ, Tapper BA, Bryan GT, Scott B. 2005. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Gen. Genomics 274:13–29 [DOI] [PubMed] [Google Scholar]
- 52.Itoh Y, Johnson R, Scott B. 1994. Integrative transformation of the mycotoxin-producing fungus, Penicillium paxilli. Curr. Genet. 25:508–513 [DOI] [PubMed] [Google Scholar]
- 53.Navarro RE, Stringer MA, Hansberg W, Timberlake WE, Aguirre J. 1996. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr. Genet. 29:352–359 [PubMed] [Google Scholar]
- 54.Bence NF, Sampat RM, Kopito RR. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552–1555 [DOI] [PubMed] [Google Scholar]
- 55.Latch GCM, Christensen MJ. 1985. Artificial infection of grasses with endophytes. Ann. Appl. Biol. 107:17–24 [Google Scholar]
- 56.Orozco-Cardenas M, Ryan CA. 1999. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. U. S. A. 96:6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Deunff E, Davoine C, Le Dantec C, Billard JP, Huault C. 2004. Oxidative burst and expression of germin/oxo genes during wounding of ryegrass leaf blades: comparison with senescence of leaf sheaths. Plant J. 38:421–431 [DOI] [PubMed] [Google Scholar]
- 58.Hagiwara D, Asano Y, Yamashino T, Mizuno T. 2008. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 72:2756–2760 [DOI] [PubMed] [Google Scholar]
- 59.Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111–118 [DOI] [PubMed] [Google Scholar]
- 60.Quinn J, Malakasi P, Smith DA, Cheetham J, Buck V, Millar JB, Morgan BA. 2011. Two-component mediated peroxide sensing and signal transduction in fission yeast. Antioxid. Redox Signal. 15:153–165 [DOI] [PubMed] [Google Scholar]
- 61.Degols G, Shiozaki K, Russell P. 1996. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16:2870–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, Toda T, Millar JB, Jones N. 1996. The Atf1 transcription factor is a target for the StyI stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289–2301 [DOI] [PubMed] [Google Scholar]
- 63.Kawasaki L, Sánchez O, Shiozaki K, Aguirre J. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153–1163 [DOI] [PubMed] [Google Scholar]
- 64.Lara-Rojas F, Sánchez O, Kawasaki L, Aguirre J. 2011. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 80:436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asano Y, Hagiwara D, Yamashino T, Mizuno T. 2007. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:1800–1803 [DOI] [PubMed] [Google Scholar]
- 66.Reverberi M, Gazzetti K, Punelli F, Scarpari M, Zjalic S, Ricelli A, Fabbri AA, Fanelli C. 2012. Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl. Microbiol. Biotechnol. 95:1293–1304 [DOI] [PubMed] [Google Scholar]
- 67.Chung K-R, Schardl CL. 1997. Sexual cycle and horizontal transmission of the grass symbiont, Epichloë typina. Mycol. Res. 101:295–301 [Google Scholar]
- 68.Shanmugam V, Ronen M, Shalaby S, Larkov O, Rachamim Y, Hadar R, Rose MS, Carmeli S, Horwitz BA, Lev S. 2010. The fungal pathogen Cochliobolus heterostrophus responds to maize phenolics: novel small molecule signals in a plant-fungal interaction. Cell. Microbiol. 12:1421–1434 [DOI] [PubMed] [Google Scholar]
- 69.Winterbourn CC, Hampton MB. 2008. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 45:549–561 [DOI] [PubMed] [Google Scholar]
- 70.Jarvis RM, Hughes SM, Ledgerwood EC. 2012. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 53:1522–1530 [DOI] [PubMed] [Google Scholar]
- 71.Inoue Y, Matsuda T, Sugiyama K, Izawa S, Kimura A. 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274:27002–27009 [DOI] [PubMed] [Google Scholar]
- 72.Tanaka T, Izawa S, Inoue Y. 2005. GPX2, encoding a phospholipid hydroperoxide glutathione peroxidase homologue, codes for an atypical 2-Cys peroxiredoxin in Saccharomyces cerevisiae. J. Biol. Chem. 280:42078–42087 [DOI] [PubMed] [Google Scholar]
- 73.Ukai Y, Kishimoto T, Ohdate T, Izawa S, Inoue Y. 2011. Glutathione peroxidase 2 in Saccharomyces cerevisiae is distributed in mitochondria and involved in sporulation. Biochem. Biophys. Res. Commun. 411:580–585 [DOI] [PubMed] [Google Scholar]
- 74.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691 [DOI] [PubMed] [Google Scholar]
- 75.Ohdate T, Inoue Y. 2012. Involvement of glutathione peroxidase 1 in growth and peroxisome formation in Saccharomyces cerevisiae in oleic acid medium. Biochim. Biophys. Acta 1821:1295–1305 [DOI] [PubMed] [Google Scholar]
- 76.Zhang WJ, He YX, Yang Z, Yu J, Chen Y, Zhou CZ. 2008. Crystal structure of glutathione-dependent phospholipid peroxidase Hyr1 from the yeast Saccharomyces cerevisiae. Proteins 73:1058–1062 [DOI] [PubMed] [Google Scholar]
- 77.Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA. 2012. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol. Cell 45:398–408 [DOI] [PubMed] [Google Scholar]
- 78.Biteau B, Labarre J, Toledano MB. 2003. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425:980–984 [DOI] [PubMed] [Google Scholar]
- 79.Morel M, Kohler A, Martin F, Gelhaye E, Rouhier N. 2008. Comparison of the thiol-dependent antioxidant systems in the ectomycorrhizal Laccaria bicolor and the saprotrophic Phanerochaete chrysosporium. New Phytol. 180:391–407 [DOI] [PubMed] [Google Scholar]
- 80.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, Reddy AB. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JS, Bang MA, Lee S, Chae HZ, Kim K. 2010. Distinct functional roles of peroxiredoxin isozymes and glutathione peroxidase from fission yeast, Schizosaccharomyces pombe. BMB Rep. 43:170–175 [DOI] [PubMed] [Google Scholar]
- 82.Storz G, Tartaglia LA. 1992. OxyR: a regulator of antioxidant genes. J. Nutr. 122:627–630 [DOI] [PubMed] [Google Scholar]
- 83.Storz G, Tartaglia LA, Ames BN. 1990. The OxyR regulon. Antonie Van Leeuwenhoek 58:157–161 [DOI] [PubMed] [Google Scholar]
- 84.Veal EA, Findlay VJ, Day AM, Bozonet SM, Evans JM, Quinn J, Morgan BA. 2004. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol. Cell 15:129–139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.