Abstract
Salmonella enterica serovar Typhimurium is a Gram-negative bacterium able to invade and replicate inside eukaryotic cells. To cope with the host defense mechanisms, the bacterium has to rapidly remodel its transcriptional status. Regulatory RNAs and ribonucleases are the factors that ultimately control the fate of mRNAs and final protein levels in the cell. There is growing evidence of the direct involvement of these factors in bacterial pathogenicity. In this report, we validate the use of a Galleria mellonela model in S. Typhimurium pathogenicity studies through the parallel analysis of a mutant with a mutation in hfq, a well-established Salmonella virulence gene. The results obtained with this mutant are similar to the ones reported in a mouse model. Through the use of this insect model, we demonstrate a role for the main endoribonucleases RNase E and RNase III in Salmonella virulence. These ribonuclease mutants show an attenuated virulence phenotype, impairment in motility, and reduced proliferation inside the host. Interestingly, the two mutants trigger a distinct immune response in the host, and the two mutations seem to have an impact on distinct bacterial functions.
INTRODUCTION
Salmonella infections are a serious medical and veterinary problem worldwide. This pathogenic bacterium is able to invade and replicate within eukaryotic host cells. For infection, Salmonella relies upon a range of laterally acquired virulence regions, the so-called Salmonella pathogenicity islands (SPIs). Of these, SPI-1 and SPI-2 contain genes that encode type III secretion systems (TTSS), which deliver effector proteins into host cells to facilitate either cellular invasion or intracellular survival, respectively (for a review, see reference 1). Hundreds of genes are upregulated during infection and play important roles in adaptation, survival, and proliferation within mammalian cells (2). Transcriptome analysis of Salmonella enterica serovar Typhimurium within epithelial cells and macrophages revealed distinct patterns of expression linked to the different stages of infection (3, 4).
Both the evolutionarily close relationship with Escherichia coli and the pathogen-specific aspects make Salmonella a very good model for studying the influence of RNA determinants in bacterial pathogenicity. In addition to transcriptional control, regulation of RNA decay has emerged as a major pathway in the fast adaptive process of bacteria to changes in the environment. RNAs may also act as regulatory molecules that can directly sense environmental clues and modulate the expression of target RNAs (for a review, see reference 5). The fate of RNA transcripts can be also controlled by proteins, including ribonucleases (RNases) and RNA chaperones.
RNases are enzymes that govern the maturation and degradation of RNA molecules. RNA decay in Gram-negative bacteria usually begins with an endonucleolytic cleavage at one or more internal sites on the RNA molecule. This cleavage is normally performed by RNase E and/or RNase III (6, 7). The single-stranded specific endoribonuclease RNase E is an essential enzyme involved in many aspects of RNA metabolism, including mRNA decay, tRNA processing, rRNA maturation, and small noncoding RNA (sRNA) processing and decay (6–9). The C terminus of the enzyme forms a scaffold for interactions with other proteins, which together form the degradosome, an important RNA degradation complex. RNase III is member of a highly conserved family of double-stranded RNA-specific enzymes with essential roles in RNA processing and decay (7). Bacterial RNase III is primarily known for its roles in rRNA maturation, mRNA degradation, and sRNA processing, turnover, and sRNA-dependent mRNA degradation (6, 10–13). Several genome-wide analyses reported that the absence of RNases E and III in E. coli (14) and RNase III in Bacillus subtilis (15) or Staphylococcus aureus (16, 17) affects the abundance of a high number of mRNAs and sRNAs. Namely, in Salmonella and in other bacteria, RNases E and III have roles in the control of a number of sRNAs implicated in the regulation of outer membrane proteins (18–20) and important virulence factors (11, 21, 22).
In the present work, it was our aim to determine the influence of these two main endoribonucleases on the virulence capacity of S. Typhimurium. For this, we have used the greater wax moth, Galleria mellonella, as a host model. The possibility of addressing many aspects of mammalian innate immunity in invertebrates has expanded their use as models to study human infections (23, 24). The human and insect immune systems demonstrate many similarities (24), with most insect species containing specialized cells, known as hemocytes, that phagocytose bacterial pathogens and form aggregates, to encapsulate and neutralize foreign microorganisms (25). The hemocyte-mediated response involves the trigger of a phenoloxidase melanization cascade and the synthesis of antimicrobial peptides by the insect's fat bodies (26). These molecules are rapidly released into the hemolymph, where they act synergistically against the microorganisms. A very good correlation between bacterial pathogenicity in G. mellonela and mammalian models of infection has been established (27–30), favoring the emergence of larvae of the greater wax moth G. mellonella as a reliable insect model host to study pathogenesis of a wide range of Gram-positive/negative bacteria (23, 27, 29, 30) and fungi (31, 32). Galleria mellonella combines the advantages of invertebrate host models with several other unique benefits, such as larger size, enabling easy manipulation and injection, low maintenance and breeding costs, status as an ethically acceptable animal model, and growth at 37°C. This is the temperature at which human pathogens are adapted and which is essential for synthesis and release of many pathogenicity factors (32, 33).
There is growing evidence of an effect of ribonucleases on bacterial virulence. We were interested in evaluating the role of two of those enzymes, RNases E and III, in the virulence of S. Typhimurium. In parallel we wanted to establish G. mellonella as a host model for S. Typhimurium pathogenicity studies, as already reported for other bacteria.
MATERIALS AND METHODS
Bacterial strains, insects, and growth conditions.
All bacterial strains used in this study are listed in Table 1. All Salmonella strains used are isogenic derivatives of the wild-type Salmonella enterica serovar Typhimurium strain SL1344. Larvae were reared on their natural food, beeswax, and pollen grains at 25°C in darkness prior to use. Larvae weighing 250 ± 25 mg were used.
Table 1.
List of strains used in this work
| Strain | Relevant marker(s) or genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-hsdR17 supE44 relA1 Δ(lacZYA-argF)U169 ϕ80dlacZΔM15 | New England Biolabs |
| MG1693 | thyA715 | 79 |
| S. Typhimurium | ||
| SL1344 | Strr hisG rpsL xyl | 54 |
| CMA-537 | SL1344 rne-537 (Δrne::Cmr) | 19 |
| CMA-551 | SL1344 rnc-14::ΔTn10 (Tcr) | 20 |
| JVS-0255 | SL1344 hfq (Δhfq::Cmr) | 45 |
| CMA-700 | CMA-537 carrying pSVA-8 plasmid | This study |
| CMA-701 | CMA-551 carrying pSVA-7 plasmid | This study |
| CMA-702 | JVO-0255 carrying pSVA-6 plasmid | This study |
Bacteria were grown in Luria-Bertani (LB) broth at 37°C and 220 rpm, unless stated otherwise. SOC medium (Super Optimal Broth with catabolite repression) was used to recover E. coli and Salmonella transformants before plating. Growth medium was supplemented with the following antibiotics when appropriate: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), streptomycin (90 μg/ml), and tetracycline (25 μg/ml). For growth of the E. coli MG1693 strain, LB medium was supplemented with thymine (50 μg/ml).
For the Galleria mellonella infection experiments, cultures grown overnight in LB medium were diluted 1/100 (to an optical density at 600 nm [OD600] of ∼0.05) in 5 ml of high-salt LB medium (0.3 M NaCl) and further grown in 15-ml Falcon tubes with a tightly closed lid for 5 h at 37°C with shaking. To compare the growth behaviors of the Salmonella wild type and mutants, each diluted culture (OD600 of ∼0.05) was split into 12 aliquots and further grown, under the aforementioned conditions, for 12 h. Each aliquot was opened only once to measure the OD600 every 60 min.
Plasmids.
All plasmids used in this study are listed in Table 2. For construction of the pWSK29hfq plasmid (pSVA-6) expressing hfq, a PCR fragment containing the entire hfq sequence was amplified from the SL1344 chromosome, digested with the enzymes HindIII and XbaI, and ligated into plasmid pWSK29 digested with the same enzymes. For the construction of plasmid pWSK29rnc (pSVA-7), the pSVDA-01 plasmid expressing His-tagged RNase III (20) was digested with BamHI and XbaI to obtain the rnc fragment, which was ligated with pWSK29 digested with the same enzymes. In the case of plasmid pSVA-8 expressing RNase E, a PCR fragment containing the entire rne sequence was amplified from SL1344 chromosome and was cloned into the XbaI and EcoRI sites of the pSE420 vector (Invitrogen). Competent E. coli DH5α cells (New England BioLabs) were used for cloning procedures during plasmid construction. The selected clones were sequenced (at STAB Vida, Portugal) to confirm the presence of the correct gene sequence and transferred to the respective SL1344 derivative strain. Primers used for gene amplifications are presented in Table S1 in the supplemental material.
Table 2.
List of plasmids used in this work
| Plasmid | Description | Origin/marker | Reference |
|---|---|---|---|
| pWSK29 | Low-copy-no. plasmid | pSC101/Ampr | 80 |
| pSE420 | IPTG-inducible plasmid | Ampr | Invitrogen |
| pSVDA-01 | pET-15b encoding His-RNase III | Ampr | 20 |
| pSVA-6 | pWSK29 expressing Hfq | pWSK29/Ampr | This study |
| pSVA-7 | pWSK29 expressing RNase III | pWSK29/Ampr | This study |
| pSVA-8 | pSE-420 expressing RNase E | pSE420/Ampr | This study |
Galleria mellonella killing assay.
Cultures of S. Typhimurium and E. coli were grown under the aforementioned conditions for 5 h. The optical density of the cultures was measured, and the appropriate volume was collected to contain all of the strains with the same OD value. Cells were then harvested by centrifugation and resuspended in 10 mM MgSO4 in a series of 10-fold serial dilutions corresponding to the number of CFU per volume of injection. A micrometer was adapted to control the volume of a microsyringe and inject 3.5-μl aliquots of each dilution into G. mellonella, via the hindmost left proleg, which had been previously surface sterilized with 70% (vol/vol) ethanol. Control larvae were injected with the same volume of 10 mM MgSO4 to monitor any problem associated with the injection process. Following injection, larvae were placed in glass petri dishes and stored in the dark at 37°C for 4 days. For each condition, we have used 10 larvae and followed its survival and appearance at 24-h intervals. Caterpillars were considered dead when they displayed no movement in response to touch.
CFU count of S. Typhimurium.
Each bacterial suspension used for worm infection was serially diluted in 10 mM MgSO4 and plated. This was done to verify that all samples injected had similar numbers of cells. To determine intracellular bacterial load, hemolymph was collected from three living larvae at 1, 5, and 16 h after injection by puncturing the larval abdomen with a sterile needle. The outflowing plasma was immediately transferred into a sterile microtube containing a few crystals of phenylthiourea to prevent melanization. The hemolymph collected was serially diluted in 10 mM MgSO4 and plated. In both cases, dilutions were plated in duplicate in LB-agar plates with the respective antibiotics, and CFU were determined after incubation at 37°C for 24 h.
G. mellonella RNA extraction.
Briefly, sets of 20 larvae were infected with S. Typhimurium at 105 CFU/larva as described above. For each time point (1, 5, and 16 h after injection), three living larvae per set were cryopreserved, sliced, and homogenized in 1 ml of TRIzol reagent (Sigma-Aldrich). Whole-animal RNA was extracted according to the manufacturer's protocol. After extraction, RNA was treated with Turbo DNase (Ambion, Applied Biosystems). The purified RNA was quantified spectrophotometrically (NanoDrop ND-1000).
Quantitative RT-PCR.
The transcriptional levels of genes encoding the G. mellonella antimicrobial peptides gallerimycin, galliomycin, inducible metalloproteinase inhibitor (IMPI), and lysozyme were determined with the 7500 real-time PCR (RT-PCR) system (Applied Biosystems), using Power SYBR green master mix (Applied Biosystems), cDNA synthesized from 200 ng of purified RNA with the TaqMan kit (Roche, Applied Biosystems), and specific primers (see Table S1 in the supplemental material) as described previously (34). All samples were analyzed in triplicate, and the amount of mRNA detected was normalized to control actin mRNA values. Relative quantification of gene expression was calculated by using the threshold cycle (ΔΔCT) method (35).
In vitro cultivation of hemocytes of G. mellonella.
To isolate G. mellonella hemocytes, hemolymph was collected from larvae previously anesthetized on ice and surface sterilized with ethanol by puncturing the larval abdomen with a sterile needle (36). The outflowing hemolymph was immediately transferred into a sterile microtube containing anticoagulant buffer (98 mM NaOH, 145 mM NaCl, 17 mM EDTA, 41 mM citric acid [pH 4.5]) in a 1:1 proportion. The hemolymph was centrifuged at 250 × g for 10 min at 4°C to pellet hemocytes. The supernatant was taken off, and the pellet was washed twice with 0.9% NaCl and centrifuged at 250 × g for 5 min at 4°C. The hemocyte pellet was then suspended gently in 1 ml of Grace insect medium (GIM) (Sigma) supplemented with 10% fetal bovine serum, 1% glutamine, and 1% antibiotic or antimycotic solution (10,000 U penicillin G, 10 mg streptomycin, 25 mg/liter amphotericin B). Suspended hemocytes were counted with a hemocytometer and incubated at 26°C in 24-well plates at a concentration of 2 × 105 cells/ml. Monolayers of primary Galleria hemocytes were used for experiments the next day. All preparations and assays were carried out under sterile conditions.
Determination of in vitro bacterial load of hemocytes.
Cultures of S. Typhimurium cells were grown under the above-mentioned conditions for 5 h. The optical density of the cultures was measured, and the appropriate volume was collected to have 4 × 103 bacteria/ml in 0.9% NaCl. Galleria hemocyte monolayer medium was replaced with GIM without antibiotics, and then cells were infected with the bacterial suspensions. After 1 h of infection at 37°C, the hemocytes were carefully washed twice with cell culture medium, followed by the addition of GIM containing 100 mg/liter of gentamicin to kill the extracellular bacteria. After 1 h, supernatants were plated to confirm the effectiveness of antibiotic treatment and the medium was replaced with GIM containing 10 mg/liter of gentamicin. The quantification of viable intracellular bacteria was achieved 2, 4, and 20 h after infection. Cell monolayers were lysed with 0.5% Triton X-100, and CFU were determined by plating dilutions of cell lysates on LB-agar plates supplemented with the respective antibiotics followed by incubation at 37°C for 24 h.
Motility assays.
Bacterial strains were grown for 5 h under the aforementioned conditions, the optical density of the cultures was measured, and the appropriate volume collected to contain all of the strains with the same OD value. The different strains were inoculated (3 μl) in motility agar plates, incubated at 37°C for the time specified on the respective figure, and photographed. The experiment was repeated more than 3 times with independent cultures. The swimming medium was composed of 10 g/liter tryptone, 5 g/liter NaCl, and 0.3% (wt/vol) agar. The swarming medium was composed of LB supplemented with 0.5% (wt/vol) agar and 0.5% glucose.
RESULTS
Comparison of the growth properties of S. Typhimurium wild-type and mutant strains.
It was our main aim to evaluate the virulence potential of Salmonella mutant strains deficient in the endoribonucleases RNase E and RNase III. RNase E is encoded by the essential gene rne, and mutations in the N-terminal catalytic domain are lethal. An RNase E mutant with a deletion of the C-terminal scaffold of the enzyme (rne-537 mutation) was used (19). A similar mutation in E. coli (rne-131) was reported to cause a significant mRNA stabilization (37), and the mutant is defective in both the assembly of a functional degradosome and the interaction with the RNA chaperone Hfq (38, 39). This mutation also affects processing and decay of several sRNAs in E. coli and Salmonella (19). In the case of RNase III, strain CMA-551 (20), which is an SL1344 equivalent of the rnc-14 mutant of E. coli (40), was used. Loss of Salmonella RNase III function results in a defect in rRNA processing (41) and has an effect on sRNA and sRNA-dependent mRNA degradation (20).
The growth characteristics of the S. Typhimurium wild-type and mutant strains were analyzed under the experimental conditions used in this study. Invasive Salmonella strains control the expression of the proteins that stimulate entry into mammalian cells based upon environmental clues, which include osmolarity, oxygen levels in the growth medium, and the bacterial growth state. To mimic these conditions in the lab, cells were grown under SPI-1-inducing conditions—i.e., high osmolarity and low oxygen (42). The growth of the strains was monitored for a particular period; the results presented in Fig. 1 show that the growth profiles of the strains were similar. The growth of the wild-type strain is faster at the early stage, but after 5 h of growth (the time at which the bacteria were collected for injection), the optical densities of all strains were very similar. In addition, the viable counts of the bacterial samples were determined prior to injection for each survival assay. These numbers were always comparable between the strains.
Fig 1.
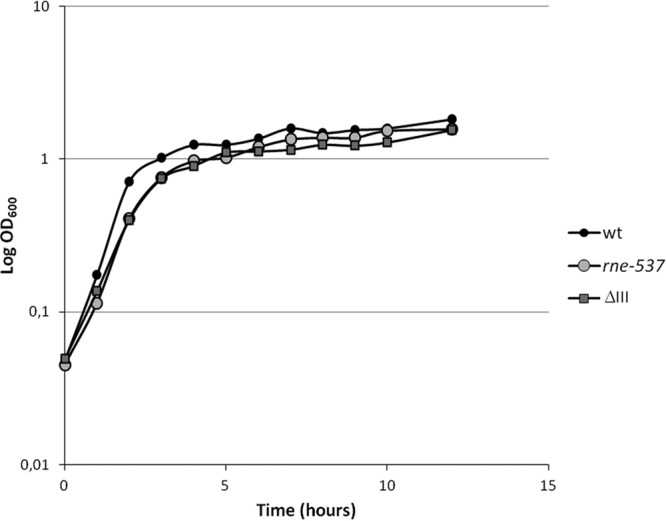
Growth characteristics of S. Typhimurium wild-type and mutant strains. To compare the growth behaviors of the Salmonella wild-type (wt) and rne (rne-537) and rnc (ΔIII) mutant strains under SPI-1-inducing conditions, the OD600 of the cultures was registered every 60 min until 12 h of growth.
Mortality of Salmonella-infected G. mellonella is dose dependent.
An aim of this work was to evaluate the pathogenicity level of S. Typhimurium to G. mellonella larvae and to determine the optimal dosage for virulence studies with this insect model. For this, larvae were injected with 108, 106, 105, 104, 103, and 102 CFU of bacteria/larva and incubated at 37°C, and mortality was monitored daily. As a control, larvae were also injected with the MgSO4 buffer alone. Infection of insects is accompanied by the generation of melanin, which becomes deposited around pathogens (43), and as a consequence, infected G. mellonella larvae change from their normal cream color to a dark brown. As represented in Fig. 2, infection with S. Typhimurium causes a strong melanization of the larvae in comparison with the noninfected ones. At doses of S. Typhimurium higher than 105 CFU/larva, all wax moth larvae were killed after 24 h. At doses lower than 105, the mortality was null or near zero. Since the dose of 105 CFU/larva induces an intermediate level of virulence, it should allow us to discern the differences in the virulence potentials of the different strains and was chosen as the inoculation dose for the subsequent assays in this study. No deaths were recorded when larvae were injected with 105 CFU/larva of the nonpathogenic E. coli K-12 derivative strain MG1693.
Fig 2.
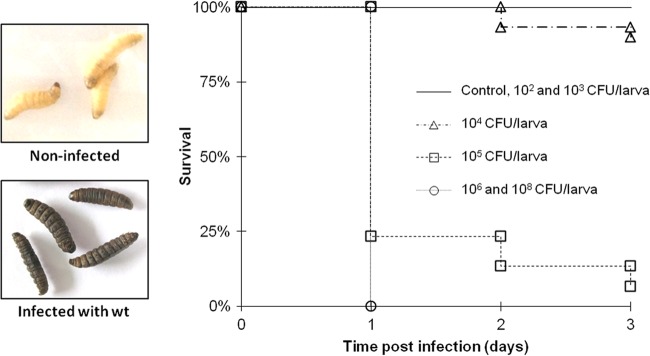
Dose-dependent survival of Galleria mellonella after Salmonella infection. Larvae were injected with 108, 106, 105, 104, 103, and 102 CFU/larva (CFU contained in 3.5 μl) of the wild-type S. Typhimurium strain and incubated at 37°C, and mortality was monitored daily. Kaplan-Meier survival curves were determined from three independent experiments. The control corresponds to the injection of MgSO4 alone. “Non-infected” corresponds to the larvae injected with the MgSO4 alone (control), and “Infected with wt” corresponds to the inoculation with 105 CFU/larva.
Correlation between Galleria mellonella and mouse models of Salmonella infection.
To establish G. mellonella as a host model for Salmonella infection, the effect of the deletion of a gene known to have a marked influence on Salmonella virulence was tested. Hfq acts as a pleiotropic regulator of Salmonella gene expression, controlling the expression of nearly one-fifth of its genes (44). Deletion of the hfq gene attenuates the ability of S. Typhymurium to infect mice, to invade epithelial cells, to secrete virulence factors, and to survive inside cultured macrophages (45). Therefore, the hfq mutant strain was included in the present study to confirm the reduced virulence phenotype of this mutant in our model.
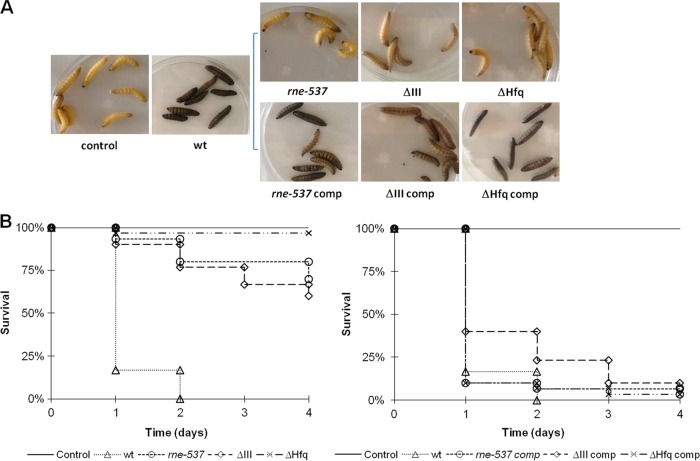
The larvae of the wax great moth G. mellonella were inoculated with 105 CFU/larva of wild-type and hfq mutant strains and incubated at 37°C, and mortality was monitored daily. The effects of infection by the wild-type Salmonella strain were rapidly seen after the first 24 h of infection, with strong melanization of the caterpillars (Fig. 3A) and a reduction of survival to about 20% of the initial number of larvae (Fig. 3B). After 48 h of infection, all of the wild-type-infected larvae were dead. In contrast, the larvae injected with the hfq mutant show very weak signs of melanization, and after 4 days of infection, the survival of G. mellonella injected with this mutant was over 95% (Fig. 3B). Complementation of the mutant with the hfq gene in trans restores the wild-type phenotype (Fig. 3). These results confirm the prominent virulence attenuation phenotype seen before with this mutant in a murine model (45). The agreement of these observations with the previous data validates G. mellonella as a good host model of infection for S. Typhimurium pathogenicity studies. The hfq mutant was used henceforth as a positive control in our assays.
Fig 3.
Effects of Galleria mellonella infection by different Salmonella strains. Survival of larvae injected with 105 CFU/larvae of S. Typhimurium wild-type, RNase E (rne-537), RNase III (ΔIII), and Hfq (ΔHfq) mutant strains. The control corresponds to the injection of MgSO4 alone. (A) Larvae after 24 h of infection. (B) Kaplan-Meier survival curves of larvae after Salmonella infection. The results represent three independent experiments.
Mutations in endoribonucleases RNase E and RNase III reduce Salmonella virulence capacity.
In order to analyze the influence of endoribonucleases RNase E and RNase III on S. Typhimurium virulence in the host model Galleria mellonella, larvae were injected with 105 CFU/larva (Fig. 2) of the wild-type, RNase E (rne-537) and RNase III (ΔIII) mutant strains, incubated at 37°C, and their mortality was monitored daily. The larvae infected with the mutant strains show modest signs of melanization, and more than 90% remained alive after 24 h of injection (Fig. 3). The survival of the larvae was monitored for 4 days after inoculation, and in the end, there were survival rates of about 60 and 70% for larvae injected with the RNase III and RNase E mutants, respectively. To corroborate that the endoribonuclease mutations were the main cause of the attenuated virulence phenotype of the mutants, it was tested whether it could be complemented by the expression of the respective genes in trans. A strong melanization of both complemented strains is evident after 24 h of infection (Fig. 3A), and there was a marked reduction of host survival to levels comparable to those observed with the wild-type strain (Fig. 3B).
The bacterial load of endoribonuclease mutants remains constant during G. mellonella colonization.
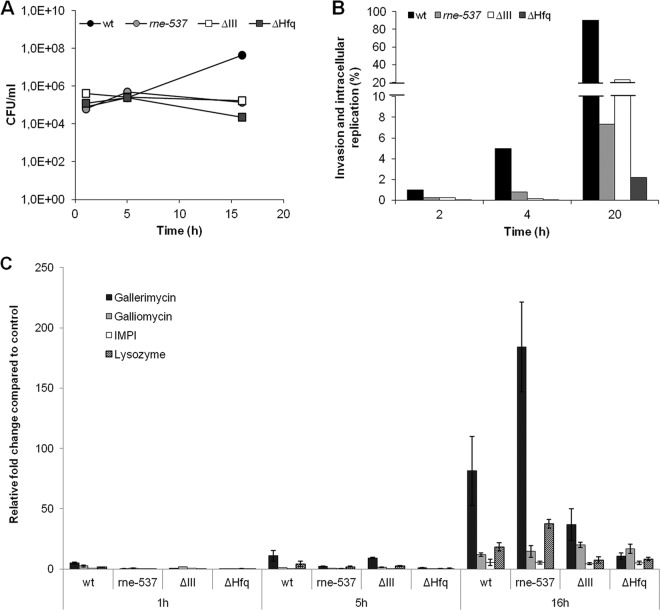
In order to evaluate the proliferation of S. Typhimurium within the insect hemocoel, the viable bacterial load within the hemolymph of larvae was determined at three distinct time points in the course of infection. A total of 105 CFU/larva of the S. Typhimurium wild-type and RNase E, RNase III, and Hfq mutant strains were used to promote infection, and at the time points indicated in Fig. 4A, hemolymph from three living larvae was collected and pooled, and the number of CFU was determined. All three mutants were able to persist in larval hemolymph to the point of 16 h postinfection; however, they did not show signs of proliferation. On the contrary, infection of larvae with the wild-type strain resulted in a 100-fold increase of CFU for the same period of time, indicating that S. Typhimurium is able to replicate inside G. mellonella. These results are in accordance with the results of the survival of infected larvae (Fig. 3B), since at 48 h postinfection all infected insects were killed by the wild-type strain but infection with the mutants led to the insect's death in only 10 to 25%.
Fig 4.
Repercussion of Salmonella infection inside the G. mellonella model. (A) The viable bacterial load was determined in the hemolymph of larvae infected with 105 CFU/larva of the S. Typhimurium wild-type strain or RNase E (rne-537), RNase III, and Hfq mutant strains, during a period within the time course of infection. (B) In vitro infection of hemocytes with S. Typhimurium wild-type and mutant strains (MOI, 50:1). The results are shown as percentages of the bacterial input. Experiments were repeated at least three times, and one representative experiment is shown. (C) Transcriptional activation of immune-responsive genes of G. mellonella at 1, 5, and 16 h postinfection with 105 CFU/larva of S. Typhimurium wild-type, RNase E (rne-537), RNase III (ΔIII), and Hfq (ΔHfq) mutant strains. The transcriptional levels of gallerimycin, galliomycin, IMPI, and lysozyme were determined by quantitative RT-PCR analysis and are shown relative to the expression levels in noninfected larvae injected with 10 mM MgSO4. Results were normalized to the expression of the housekeeping actin gene.
In vitro infection of hemocytes.
To further analyze the host-pathogen interaction in this insect model, hemocytes were extracted from G. mellonella hemolymph and used for in vitro cultures. The cultured hemocytes were infected with the S. Typhimurium wild type and the endonuclease mutant strains at a multiplicity of infection (MOI) of 50:1 (bacteria to hemocytes). The bacterial load of hemocytes was evaluated after 2, 4, and 20 h. The Hfq mutant strain was analyzed under the same conditions and functioned as a control. To evaluate the effectiveness of gentamicin treatment, we plated the supernatant before lysis, and no significant CFU were obtained.
At 2 h postinfection, there were ∼5-fold fewer intracellular bacteria in hemocytes infected with the mutant strains compared with the wild type, likely reflecting the reduced invasion rate of these strains (Fig. 4B).
Within the next 2 h of infection, the number of wild-type bacteria increased 5-fold compared to the previous time point, whereas the number of mutant bacteria remained almost unchanged. This suggests an intracellular growth defect in addition to an invasion defect. At 20 h postinfection, the number of wild-type bacteria reached nearly 100% of the input of infection. Despite the initial reduction of cellular uptake of the mutants compared to the wild type, the increase in intracellular CFU of the RNase III mutant was comparable. In contrast, slower intracellular replication was noticed with the RNase E mutant (Fig. 4B). The defect in invasion and intracellular replication of the Hfq mutant, previously observed for epithelial and macrophage cell lines (45), was confirmed in the present study. The same assays were performed with mutants defective in SPI-1 and SPI-2 expression (see Fig. S2 in the supplemental material). The SPI mutant showed a strong invasion defect. In the SPI-2-defective strain, the cellular uptake was lower than that for the wild type and the intracellular replication was also defective. However, analysis of survival of the larvae with both strains showed only a modest attenuation of virulence for the SPI-1-defective mutant (see Fig. S1 in the supplemental material).
Mutations in endoribonucleases E and III trigger a different immune response by G. mellonella.
Antimicrobial peptides play a crucial role in insect innate immunity against invading pathogens. In fact, G. mellonella comprises a remarkable antimicrobial peptide arsenal that is released into the hemolymph, where it attacks elements of the bacterial or fungal cell wall (46). Therefore, it is expected that antimicrobial peptides are induced in Galleria upon S. Typhimurium infection. To check this, larvae were infected with this bacterium and the gene expression of four selected antimicrobial peptides was monitored 1, 5, and 16 h postinfection. The selected genes coded for the cysteine-rich antifungal peptide gallerimycin (47), a defensin called galliomycin (48), lysozyme, and an inducible metalloproteinase inhibitor (IMPI) (47). Gene expression was determined by quantitative RT-PCR analysis of the total RNA extracted for each postinfection time point. The results showed that infection with the wild-type S. Typhimurium strain lead to upregulation of the immune-related peptides tested compared to the case in control larvae injected with 10 mM MgSO4 (Fig. 4C), and the induction is more pronounced at 16 h after injection. This result is in agreement with the 100-fold increase in bacterial load observed in the hemolymph of insects infected with wild-type Salmonella for the same length of infection.
To analyze the possible effect of the endoribonucleases RNase E and RNase III in the immune response of the larvae against S. Typhimurium infection, the same experiment was carried out using the RNase E, RNase III, and Hfq (as a control) mutant strains. At 16 h postinfection, all of the mutants exhibited increased expression levels of the four immune-related peptides analyzed, compared to the uninfected larvae (Fig. 4C). Galliomycin and IMPI levels are identical for the wild-type and mutant strains. In contrast, the expression levels of gallerimycin and lysozyme in the infections with RNase III and Hfq mutants were lower than with the wild-type strain. Surprisingly, infection with the RNase E mutant lead to a stronger increase in the expression levels of these two peptides (more of gallerimycin) than infection with the wild-type strain.
RNase mutants are less motile.
The ability to move is a main advantage of bacteria, namely, to obtain nutrients, disperse more effectively and avoid unfavorable environments. Not surprisingly, motility constitutes a virulence factor for many pathogenic bacteria, namely, Salmonella (49–51). Two types of bacterial movement were evaluated in the Salmonella mutants studied. Swimming represents the individual movement of cells in aqueous or semisolid environments and is dependent on flagella (52). Swarming consists of the mechanism of propagation of a group of cells in a coordinated form on semisolid surfaces, which is dependent both on the flagella and on the production of specific extracellular compounds that reduce the superficial tension and allow the organism's motility (52). The medium's requirements for swarming motility depend on the organism being considered. Salmonella enterica specifically requires the presence of glucose in an energy-rich semisolid medium to swarm, and swarmer cells become longer and hyperflagellated (53).
The results obtained show that both rne and rnc genes are important for both types of motility. The respective mutant strains present a reduced swimming and swarming motility (Fig. 5). The wild-type cells are motile in swimming plates, forming concentric motility rings around the point of inoculation that can be observed soon after 8 h of incubation at 37°C (Fig. 5A, upper panel). After 24 h of incubation, the wild-type cells have already covered all the plate (Fig. 5A, lower panel). Both RNase E and RNase III mutants displayed an impaired motility, by forming a barely detectable motility ring after 8 h of incubation that remained much smaller than the one formed by the wild type after 24 h of incubation. In the RNase III mutant, swarming is less affected than in the RNase E mutant. Complementation of the strains with the respective gene expressed from a plasmid restored the motility capabilities and characteristics of the wild-type strain.
Fig 5.
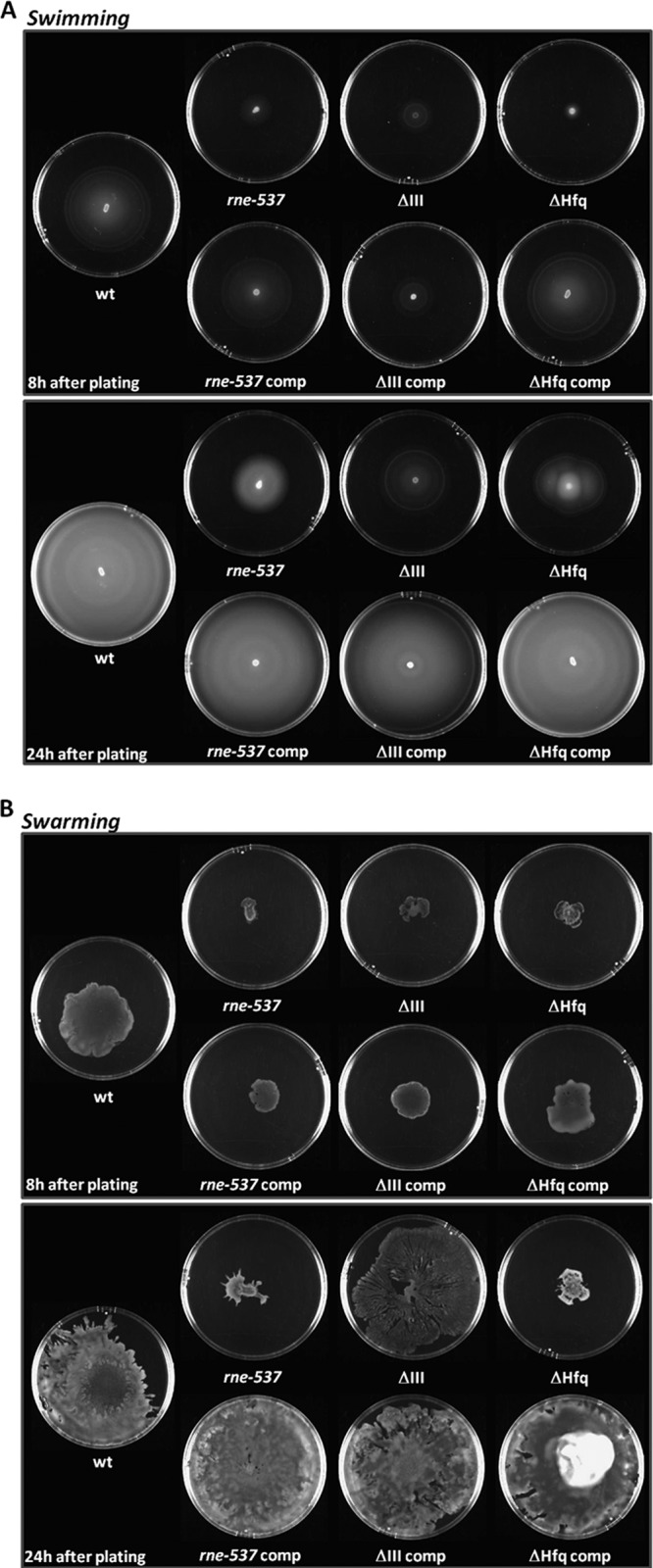
RNase E and RNase III mutant strains are impaired in motility. To measure motility, equal numbers of bacteria from each strain were inoculated onto swimming (A) and swarming (B) agar plates. The plates were incubated at 37°C for the time indicated on each figure and photographed.
It had been seen before that Salmonella Δhfq cells are nonmotile in motility agar plates (45). Under the experimental conditions used, the Δhfq cells showed a very limited motility on swimming and swarming plates that was fully restored in the complemented strain.
DISCUSSION
Salmonella is a facultative intracellular pathogen that can survive the host immune response and replicate inside host cells, most prominently in phagocytic cells, such as macrophages. The ease of genetic manipulation coupled with a detailed understanding of core metabolism has made S. Typhimurium an excellent model for studying host-pathogen interactions and intracellular survival.
Adequate infection models that approximate human disease are the key for the analysis of the molecular basis of bacterial pathogenesis. The possibility of addressing many aspects of mammalian innate immunity in invertebrates has expanded their use as models to study human infections (23, 24). One of the aims of this study was to examine the effectiveness of the larvae of the wax moth G. mellonella as a model of infection for the S. Typhimurium strain SL1344 (isolate virulent in mice) (54). There is only one very old report of the use of Galleria in a Salmonella study performed with the LT2 strain (not virulent in rats) (55). Therefore, we found it important to validate the use of this system with Salmonella model strain SL1344.
For this, a gene well known as being important for Salmonella virulence (hfq) was also included in our studies. The ubiquitous RNA-binding protein Hfq has been shown to be required for the fitness and virulence of an increasing number of bacterial pathogens (56–59). Transcriptome analysis suggests that Hfq regulates the expression of nearly a fifth of all Salmonella genes (44), and deletion of the hfq gene was shown to attenuate the ability of S. Typhimurium to infect mice, to invade epithelial cells, to secrete virulence factors, and to survive inside cultured macrophages. The present work confirms the results obtained before with the Hfq mutant, in a murine model, namely, the attenuated virulence phenotype and reduced proliferation inside the host cells. These results validate G. mellonella as a model for the identification of virulence genes.
These results enabled us to use this insect model to analyze the impact of two main endoribonuclease activities on S. Typhimurium SL1344 pathogenesis. There are already reports about the influence of ribonucleases, the key players in the RNA decay process, in the virulence of different pathogens (see reference 60 for a review). The influence of endoribonucleases RNase E and RNase III in Salmonella infection was investigated for the first time in this work, using G. mellonella as a host infection model. The level of infection of G. mellonella by the SL1344 wild-type strain is shown to be dose dependent and causes a high percentage of mortality, soon after 24 h of infection. The infection of insect larvae with RNase E and RNase III mutants is highly attenuated. Estimation of the proliferation of Salmonella strains inside the host hemolymph showed that wild-type bacteria persisted and replicated within the larvae, while the mutants did not replicate, despite remaining viable up to 16 h of infection. The assays with hemocyte cell lines also revealed that the endoribonuclease mutants are defective for invasion of the phagocytic cells compared to the wild-type bacteria.
Analysis of antimicrobial host immune response revealed that Salmonella infection of G. mellonella resulted in upregulation of the expression of the antimicrobial peptides tested compared to the uninfected control larvae. Interestingly, in studies of the pathogenic activity of Listeria (61), Legionella pneumophila (62), and Burkholderia cenocepacia (34) in Galleria mellonella, gallerimycin was also highly expressed in response to the infection by each of these bacteria, indicating a prominent role in antibacterial defense. These results show that G. mellonella develops an immune response to Salmonella that nonetheless is not effective in clearing wild-type bacteria, as shown by the proliferation of the bacteria inside the host and the high mortality rates. In comparison, the response of the host in terms of gallerimycin and lysozyme expression is much higher for the RNase E mutant (Fig. 4C). It could be observed that in the larva hemolymph, there is no significant replication of this mutant despite its persistence (Fig. 4A), and the reduced bacterial counts inside the hemocytes reveal defects in invasion and intracellular replication (Fig. 4B). All of these factors together seem to result in a more effective neutralization and clearance of the mutant reflected by a weaker effect on mortality in Galleria larvae (Fig. 3). In fact, in the case of RNase III mutant, for which the expression of antimicrobial peptides is much lower (Fig. 4C) and the replication inside the hemocytes is higher than for RNase E mutant (Fig. 4B), the virulence attenuation effect is lower (Fig. 3). RNase E was shown to regulate the expression of several outer membrane proteins through the action of sRNAs. Mutations in the enzyme may alter bacterial envelope composition, triggering a higher immune response from the host.
To enable pathogenesis, Salmonella has an array of specific virulence genes that are upregulated during infection and play important roles in adaptation, survival, and proliferation within mammalian cells. The analysis of the Salmonella transcriptome inside macrophages and epithelial cells revealed the differential regulation of at least one of the type three secretion systems (TTSS), SPI-1, SPI-2 and the flagellar systems (3, 4). The expression of a few randomly chosen representative genes of the systems known to be relevant for Salmonella pathogenicity was evaluated (see Fig. S3 in the supplemental material). Two growth conditions showing maximal Salmonella invasiveness have been defined: growth in LB with aeration to the early stationary phase (ESP) and growth in low-oxygen, high-salt medium (SPI-1 inducing). In fact, the expression of the genes analyzed was much lower at the late stationary phase (LSP) than under the other two conditions analyzed. Of note, it was observed that the expression of these genes changes according to the growth conditions and is affected by mutations in the RNase E and RNase III genes in different ways.
Among these were the SPI-1 genes hilA and prgH. HilA is the SPI-1 major transcriptional activator, which is responsible for most of the SPI-1 TTSS and effector gene expression (63); PrgH is required for the secretion of invasion effector proteins and is regulated by HilA (64, 65). The expression of both genes was differentially affected by RNase III mutation, depending on the growth condition (see Fig. S3 in the supplemental material). The expression of ssav, an SPI-2 gene needed for the secretion of SPI-2 effectors, and gmm (or wcaH), which is necessary for the production of the colanic acid capsule (66), was also affected by the RNase mutations. Overproduction of the capsule was reported to cause attenuation of Salmonella virulence in mice (67, 68); the higher expression of gmm (or wcaH) in the RNase mutants is in agreement with this observation.
Flagella are responsible for a number of bacterial behaviors, including motility, biofilm formation, and chemotaxis. On one hand, flagella provide a great advantage at the early stages of infection by increasing Salmonella invasiveness (69); on the other hand, flagellin monomers induce the host immune response (70, 71) to induce bacterial clearance from the host. Consequently, Salmonella cells tightly control their flagellum expression because flagella can mediate virulence. Expression of flagella is induced in response to a number of stimuli and is normally downregulated inside the host (72). There are reports showing that both the overexpression and downregulation of flagella can significantly attenuate Salmonella virulence (73–75). In S. Typhimurium, expression of flagella is controlled by phase variation, a mechanism by which the organism alternately expresses two different types of flagellin subunit proteins, FljB and FliC (76). The expression of these two genes was differentially affected by the endoribonuclease mutations under the different growth conditions tested (see Fig. S3 in the supplemental material): so was the expression of csgD gene, the transcriptional regulator which controls expression of biofilm-associated matrix compounds (77), mostly in the RNase III mutant. Altogether, these data show that both endoribonucleases have an effect on the expression of these virulence-related genes, as revealed by their differential expression in the mutants analyzed in comparison to the wild type. However, this effect is divergent and seems to indicate that these enzymes affect distinct bacterial functions.
Bacterial motility behavior has been reported to contribute to virulence in Salmonella and many other bacteria (49–51). This type of community motility seems to enable migration and expansion through sites of infection, contributing to facing host defenses. Therefore, the motility of the mutant strains was also evaluated. The nonmotile phenotype of the Salmonella Hfq mutant, previously shown to be associated with reduced expression of FliC flagellin (44, 45), was confirmed. In E. coli, it was already reported that RNase III was involved in motility (78). The present results show that RNase E and RNase III mutants have significantly reduced motility compared to wild-type Salmonella. The attenuated virulence phenotype of the endoribonuclease mutant strains studied was coincident with the reduced motility of these strains.
This study gives evidence that the function of endoribonucleases E and III is necessary for Salmonella virulence and sheds light on some of the functions that may underlie this fact. These RNases can act directly on virulence-related messages or impact their expression through sRNA regulation. Future genome-wide analysis of these mutants should reveal the main S. Typhimurium genes affected, on the basis of the observed attenuated virulence phenotype.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andreia Aires and Nuno Bernardes for technical assistance. We are also grateful to Joerg Vogel (IMIB, Wurzburg, Germany) for Δhfq strain JVS-0255, Nelson Simões (Centro de Investigação de Recursos Naturais of Universidade dos Açores, Ponta Delgada, Portugal) for the Galleria mellonella larvae, and Francisco García-del Portillo (CNB, Madrid, Spain) for strains MD0706 and MD1111 (SL1344 ΔssaV::aphT).
This work was supported by Fundação para a Ciência e Tecnologia (FCT) (Portugal). During the realization of this work, S. C. Viegas was the recipient first of an FCT Postdoctoral Fellowship (SFRH/BPD/30766/2006) and later of a grant from European Commission (FP7-KBBE-2011-1-289326); D. Mil-Homens was the recipient of an FCT Postdoctoral Fellowship (SFRH/BPD/43390/2008). The work at ITQB was supported by grants from FCT, Portugal (including grant PEst-OE/EQB/LA0004/2011), and by grant FP7-KBBE-2011-1-289326 from the European Commission. The work at IBB/IST was supported by FCT, Portugal (grants PTDC/EBB-BIO/100326/2008 and PTDC/BIA-MIC/118386/2010).
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02044-13.
REFERENCES
- 1.Hansen-Wester I, Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549–559 [DOI] [PubMed] [Google Scholar]
- 2.Hebrard M, Kroger C, Sivasankaran SK, Handler K, Hinton JC. 2011. The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium. Curr. Opin. Biotechnol. 22:200–210 [DOI] [PubMed] [Google Scholar]
- 3.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 4.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10:958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34:883–923 [DOI] [PubMed] [Google Scholar]
- 7.Arraiano CM, Mauxion F, Viegas SC, Matos RG, Seraphin B. 2013. Intracellular ribonucleases involved in transcript processing and decay: precision tools for RNA. Biochim. Biophys. Acta 1829:491–513 [DOI] [PubMed] [Google Scholar]
- 8.Masse E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva IJ, Saramago M, Dressaire C, Domingues S, Viegas SC, Arraiano CM. 2011. Importance and key events of prokaryotic RNA decay: the ultimate fate of an RNA molecule. Wiley Interdiscip. Rev. RNA 2:818–836 [DOI] [PubMed] [Google Scholar]
- 10.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. 2005. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33:1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, Jacquier A, Vandenesch F, Romby P. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Blasi U, Kaberdin VR. 2008. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 14:454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel J, Argaman L, Wagner EG, Altuvia S. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14:2271–2276 [DOI] [PubMed] [Google Scholar]
- 14.Stead MB, Marshburn S, Mohanty BK, Mitra J, Castillo LP, Ray D, van Bakel H, Hughes TR, Kushner SR. 2011. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 39:3188–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durand S, Gilet L, Bessieres P, Nicolas P, Condon C. 2012. Three essential ribonucleases—RNase Y, J1, and III—control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 8:e1002520. 10.1371/journal.pgen.1002520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, Solano C, Gingeras TR. 2011. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U. S. A. 108:20172–20177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lioliou E, Sharma CM, Caldelari I, Helfer AC, Fechter P, Vandenesch F, Vogel J, Romby P. 2012. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 8:e1002782. 10.1371/journal.pgen.1002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viegas SC, Arraiano CM. 2008. Regulating the regulators: How ribonucleases dictate the rules in the control of small non-coding RNAs. RNA Biol. 5:230–243 [DOI] [PubMed] [Google Scholar]
- 19.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35:7651–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. 2011. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 39:2918–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee EJ, Groisman EA. 2010. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 76:1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champion OL, Cooper IA, James SL, Ford D, Karlyshev A, Wren BW, Duffield M, Oyston PC, Titball RW. 2009. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155:1516–1522 [DOI] [PubMed] [Google Scholar]
- 24.Kavanagh K, Reeves EP. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28:101–112 [DOI] [PubMed] [Google Scholar]
- 25.Marmaras VJ, Lampropoulou M. 2009. Regulators and signalling in insect haemocyte immunity. Cell. Signal. 21:186–195 [DOI] [PubMed] [Google Scholar]
- 26.Cerenius L, Soderhall K. 2004. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 198:116–126 [DOI] [PubMed] [Google Scholar]
- 27.Champion OL, Karlyshev AV, Senior NJ, Woodward M, La Ragione R, Howard SL, Wren BW, Titball RW. 2010. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201:776–782 [DOI] [PubMed] [Google Scholar]
- 28.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joyce SA, Gahan CG. 2010. Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology 156:3456–3468 [DOI] [PubMed] [Google Scholar]
- 30.Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. 2011. Virulence of serotype M3 group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2:111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs BB, O'Brien E, Khoury JB, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482 [DOI] [PubMed] [Google Scholar]
- 32.Mowlds P, Barron A, Kavanagh K. 2008. Physical stress primes the immune response of Galleria mellonella larvae to infection by Candida albicans. Microbes Infect. 10:628–634 [DOI] [PubMed] [Google Scholar]
- 33.Cheng LW, Portnoy DA. 2003. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell. Microbiol. 5:875–885 [DOI] [PubMed] [Google Scholar]
- 34.Mil-Homens D, Bernardes N, Fialho AM. 2012. The antibacterial properties of docosahexaenoic omega-3 fatty acid against the cystic fibrosis multiresistant pathogen Burkholderia cenocepacia. FEMS Microbiol. Lett. 328:61–69 [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 36.Brivio MF, Mastore M, Nappi AJ. 2010. A pathogenic parasite interferes with phagocytosis of insect immunocompetent cells. Dev. Comp. Immunol. 34:991–998 [DOI] [PubMed] [Google Scholar]
- 37.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33:188–199 [DOI] [PubMed] [Google Scholar]
- 38.Morita T, Maki K, Aiba H. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19:2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ow MC, Liu Q, Kushner SR. 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 38:854–866 [DOI] [PubMed] [Google Scholar]
- 40.Takiff HE, Chen SM, Court DL. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171:2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattatall NR, Sanderson KE. 1998. RNase III deficient Salmonella typhimurium LT2 contains intervening sequences (IVSs) in its 23S rRNA. FEMS Microbiol. Lett. 159:179–185 [DOI] [PubMed] [Google Scholar]
- 42.Eichelberg K, Galan JE. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect. Immun. 67:4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nappi AJ, Christensen BM. 2005. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem. Mol. Biol. 35:443–459 [DOI] [PubMed] [Google Scholar]
- 44.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63:193–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown SE, Howard A, Kasprzak AB, Gordon KH, East PD. 2009. A peptidomics study reveals the impressive antimicrobial peptide arsenal of the wax moth Galleria mellonella. Insect Biochem. Mol. Biol. 39:792–800 [DOI] [PubMed] [Google Scholar]
- 47.Altincicek B, Vilcinskas A. 2006. Metamorphosis and collagen-IV-fragments stimulate innate immune response in the greater wax moth, Galleria mellonella. Dev. Comp. Immunol. 30:1108–1118 [DOI] [PubMed] [Google Scholar]
- 48.Wojda I, Kowalski P, Jakubowicz T. 2009. Humoral immune response of Galleria mellonella larvae after infection by Beauveria bassiana under optimal and heat-shock conditions. J. Insect Physiol. 55:525–531 [DOI] [PubMed] [Google Scholar]
- 49.Allen-Vercoe E, Sayers AR, Woodward MJ. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605–614 [DOI] [PubMed] [Google Scholar]
- 51.Ottemann KM, Miller JF. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109–1117 [DOI] [PubMed] [Google Scholar]
- 52.Inoue T, Shingaki R, Fukui K. 2008. Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. 281:81–86 [DOI] [PubMed] [Google Scholar]
- 53.Kim W, Surette MG. 2005. Prevalence of surface swarming behavior in Salmonella. J. Bacteriol. 187:6580–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 55.Kurstak E, Vega CE. 1968. Bacterial infection due to Salmonella typhimurium in an invertebrate, Galleria mellonella L. Can. J. Microbiol. 14:233–237 [DOI] [PubMed] [Google Scholar]
- 56.Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345–354 [DOI] [PubMed] [Google Scholar]
- 57.Geng J, Song Y, Yang L, Feng Y, Qiu Y, Li G, Guo J, Bi Y, Qu Y, Wang W, Wang X, Guo Z, Yang R, Han Y. 2009. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS One 4:e6213. 10.1371/journal.pone.0006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson GT, Roop RM., Jr 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690–700 [DOI] [PubMed] [Google Scholar]
- 59.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217–228 [DOI] [PubMed] [Google Scholar]
- 60.Lawal A, Jejelowo O, Chopra AK, Rosenzweig JA. 2011. Ribonucleases and bacterial virulence. Microb. Biotechnol. 4:558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. 2010. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 76:310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harding CR, Schroeder GN, Reynolds S, Kosta A, Collins JW, Mousnier A, Frankel G. 2012. Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect. Immun. 80:2780–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lostroh CP, Bajaj V, Lee CA. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300–315 [DOI] [PubMed] [Google Scholar]
- 64.Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703–714 [DOI] [PubMed] [Google Scholar]
- 65.Pegues DA, Hantman MJ, Behlau I, Miller SI. 1995. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol. Microbiol. 17:169–181 [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Calderon CB, Casadesus J, Ramos-Morales F. 2007. Rcs and PhoPQ regulatory overlap in the control of Salmonella enterica virulence. J. Bacteriol. 189:6635–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Calderon CB, Garcia-Quintanilla M, Casadesus J, Ramos-Morales F. 2005. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151:579–588 [DOI] [PubMed] [Google Scholar]
- 68.Mouslim C, Delgado M, Groisman EA. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386–395 [DOI] [PubMed] [Google Scholar]
- 69.Schmitt CK, Ikeda JS, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, O'Brien AD. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Nunez G. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7:576–582 [DOI] [PubMed] [Google Scholar]
- 71.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575 [DOI] [PubMed] [Google Scholar]
- 72.Sano G, Takada Y, Goto S, Maruyama K, Shindo Y, Oka K, Matsui H, Matsuo K. 2007. Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 189:8224–8232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robertson JM, McKenzie NH, Duncan M, Allen-Vercoe E, Woodward MJ, Flint HJ, Grant G. 2003. Lack of flagella disadvantages Salmonella enterica serovar Enteritidis during the early stages of infection in the rat. J. Med. Microbiol. 52:91–99 [DOI] [PubMed] [Google Scholar]
- 74.Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, Russell JM, Henry T, Tran QT, Lawhon SD, Gomez G, Bevins CL, Russmann H, Monack DM, Adams LG, Baumler AJ. 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect. Immun. 77:1904–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang X, Thornburg T, Suo Z, Jun S, Robison A, Li J, Lim T, Cao L, Hoyt T, Avci R, Pascual DW. 2012. Flagella overexpression attenuates Salmonella pathogenesis. PLoS One 7:e46828. 10.1371/journal.pone.0046828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, Adams P, O'Connor CD, O'Brien AD. 2001. Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect. Immun. 69:3021–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gerstel U, Romling U. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154:659–667 [DOI] [PubMed] [Google Scholar]
- 78.Apirion D, Watson N. 1978. Ribonuclease III is involved in motility of Escherichia coli. J. Bacteriol. 133:1543–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arraiano CM, Yancey SD, Kushner SR. 1988. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 170:4625–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.