Abstract
Here, we isolated and characterized a new ginsenoside-transforming β-glucosidase (BglQM) from Mucilaginibacter sp. strain QM49 that shows biotransformation activity for various major ginsenosides. The gene responsible for this activity, bglQM, consists of 2,346 bp and is predicted to encode 781 amino acid residues. This enzyme has a molecular mass of 85.6 kDa. Sequence analysis of BglQM revealed that it could be classified into glycoside hydrolase family 3. The enzyme was overexpressed in Escherichia coli BL21(DE3) using a maltose binding protein (MBP)-fused pMAL-c2x vector system containing the tobacco etch virus (TEV) proteolytic cleavage site. Overexpressed recombinant BglQM could efficiently transform the protopanaxatriol-type ginsenosides Re and Rg1 into (S)-Rg2 and (S)-Rh1, respectively, by hydrolyzing one glucose moiety attached to the C-20 position at pH 8.0 and 30°C. The Km values for p-nitrophenyl-β-d-glucopyranoside, Re, and Rg1 were 37.0 ± 0.4 μM and 3.22 ± 0.15 and 1.48 ± 0.09 mM, respectively, and the Vmax values were 33.4 ± 0.6 μmol min−1 mg−1 of protein and 19.2 ± 0.2 and 28.8 ± 0.27 nmol min−1 mg−1 of protein, respectively. A crude protopanaxatriol-type ginsenoside mixture (PPTGM) was treated with BglQM, followed by silica column purification, to produce (S)-Rh1 and (S)-Rg2 at chromatographic purities of 98% ± 0.5% and 97% ± 1.2%, respectively. This is the first report of gram-scale production of (S)-Rh1 and (S)-Rg2 from PPTGM using a novel ginsenoside-transforming β-glucosidase of glycoside hydrolase family 3.
INTRODUCTION
Ginseng has been used as an herbal medicine in eastern Asian countries for more than 2,000 years, and it has become more popular in the Western Hemisphere over the past few decades (1, 2). The therapeutic effects of ginseng, which include antineoplastic, antistress, and antioxidant activities, are mainly attributed to a group of biologically active components called ginsenosides (3–7). More than 180 different ginsenosides have been identified, and they can be categorized as protopanaxadiol (PPD-), protopanaxatriol (PPT-), and oleanane saponin-type ginsenosides based on the structure of their aglycon, which has a dammarane skeleton (4, 5). They can also be classified as major or minor ginsenosides based on the amount found in cultivated ginseng (8).
Among the PPT-type ginsenosides, Rh1 and Rg2 have shown significant pharmaceutical effects. Rh1 displays antiallergic, anti-inflammatory, and antimetastatic activities; appears to increase memory and hippocampal excitability; acts as a phytoestrogen and activates the estrogen receptor signaling pathway; and has the potential to be developed into novel chemotherapeutic agents against malignant cancers (9–12). Rg2 can protect human erythrocytes against hemin-induced hemolysis (13); act as an antioxidant; prolong the lag time of hemolysis (14); exert effects on the immune response to ovalbumin (OVA) in mice (15); act as a prooxidant (16); and protect against memory impairment (17). Finally, both Rh1 and Rg2 have been shown to protect the rheological functions of erythrocytes by inhibiting the oxidation of SH groups in band-3 protein (12). These pharmacological activities give Rh1 and Rg2 therapeutic potential against serious diseases, such as malignant cancers, neuroinflammatory diseases, and type II diabetes.
Unfortunately, however, Rh1 and Rg2 are rare minor ginsenosides that comprise less than 0.034% of dried ginseng (18, 19). Thus, producing large quantities of pure Rh1 and Rg2 from ginseng is extremely difficult and costly, forming a bottleneck for research on the pharmacological activities of these ginsenosides. Some physiochemical methods, such as heating (20), acid treatment (21), and alkali treatment (22) of major ginsenosides, could be used to generate larger quantities of these minor ginsenosides. However, these approaches produce nonspecific stereoisomeric mixtures of rare ginsenosides that have low yields and are very difficult to separate. As an alternative, enzymatic methods have been explored as a way to more specifically convert major ginsenosides into pharmacologically active rare ginsenosides (23–33, 52).
Furthermore, methods have been developed to efficiently separate a PPT-type ginsenoside mixture (PPTGM) and a PPD-type ginsenoside mixture (PPDGM) from ginseng extracts (34, 35). The total contents of ginsenosides Rg1 and Re in PPTGM can reach over 82%, and they can be transformed into Rh1 and Rg2, respectively, by cleavage of the glucose moiety at the C-20 position of the aglycon, often by heating or enzymatic hydrolysis (19, 32, 36, 37). To date, however, low product yield and purity, the need for expensive substrates, and inefficient enzyme productivity have limited the mass production of Rh1 and Rg2.
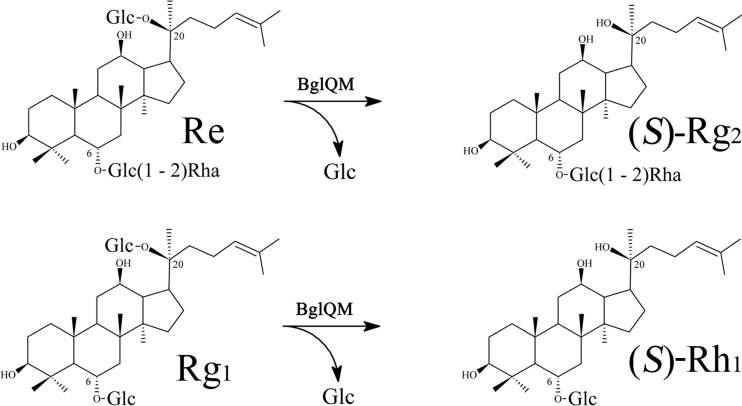
Here, we sought to overcome these disadvantages. From a dozen bacterial strains capable of transforming ginsenosides, we selected Mucilaginibacter sp. strain QM49 as having a high activity for transforming Re and Rg1 into (S)-Rg2 and (S)-Rh1, respectively. A genome fosmid library was constructed, the key ginsenoside-transforming β-glucosidase (BglQM) was identified, its novel gene was cloned for production as a recombinant enzyme, and the enzymatic properties and substrate specificities of the recombinant enzyme were thoroughly investigated. Our results revealed that BglQM belongs to glycoside hydrolase family 3, which shows efficient ginsenoside-transforming activities against various PPD- and PPT-type ginsenosides, especially for the transformations of Re and Rg1 into (S)-Rg2 and (S)-Rh1, respectively (Fig. 1). Treatment of PPTGM with BglQM followed by purification yielded gram-scale (S)-Rg2 and (S)-Rh1 of high purity. This is the first report of the gram-scale production of high-purity (S)-Rg2 and (S)-Rh1 by the application of a recombinant glycoside hydrolase to a crude substrate.
Fig 1.
Biotransformation pathway of ginsenosides Re and Rg1 into (S)-Rg2 and (S)-Rh1, respectively.
MATERIALS AND METHODS
Chemicals.
Standard-grade ginsenosides, including Rb1, Rc, Rb2, Rd, (S)-Rg3, (S)-Rh2, (R)-Rh2, F2, compound K (C-K), protopanaxadiol (PPD), Rg1, Re, (S)-Rg2, (S)-Rh1, F1, and protopanaxatriol (PPT), were purchased from Nanjing Zelang Medical Technology Co., Ltd. (China). The PPT-type ginsenoside mixture (PPTGM) from the root of Panax quinquefolius together with Panax ginseng C. A. Meyer, containing Re (32.4%; based on molar ratio), Rg1 (31.9%), Rb1 (2.8%), Rb3 (4.8%), Rd, (16.7%), (S)-Rg3 (1.7%), and other ginsenosides (9.7%), was kindly donated by F. X. Jin (Dalian Polytech University, China) and was dissolved in 50 mM phosphate buffer, which could solubilize PPTGM at up to 100 mg/ml. 5-Bromo-4-chloro-3-indolyl β-d-glucopyranoside (X-Glc) was obtained from Wako Co. Ltd. (Japan). High-performance liquid chromatography (HPLC)-grade methanol and acetonitrile were obtained from Duksan Co., Ltd. (South Korea). The other chemicals used in this study were of analytical grade or better.
Isolation and preparation of strain QM49 for ginsenoside bioconversion.
Soil samples taken from KAIST Hill, Daejeon, South Korea, were thoroughly suspended in 50 mM phosphate buffer (pH 7.0), serially diluted with 50 mM phosphate buffer (pH 7.0), and spread on half-strength R2A agar plates. The plates were incubated at 30°C for 2 weeks. Bacterial strains were obtained by transferring single colonies to one-half-strength R2A agar plates. Isolates were routinely cultured on R2A agar (Difco, Detroit, MI) at 30°C and stored as glycerol suspensions (20%, wt/vol) at −70°C. To screen for strains with high β-glucosidase activity, isolated colonies were transferred to esculin-R2A plates (R2A agar containing 1.0 g esculin and 0.5 g ferric citrate per liter). Black colonies growing on the esculin-R2A agar (i.e., those that exhibited β-glucosidase activity) were tested for their ability to transform the ginsenosides Rb1, Re, and Rg1. Liquid R2A broth cultures of these β-glucosidase-positive bacterial strains in the logarithmic phase of growth were mixed with 200 μl of autoclaved R2A broth containing 1 mg/ml of Rb1, Re, or Rg1 in a 1.5-ml Eppendorf tube and then incubated at 30°C. At 24-h intervals, 50-μl aliquots were taken, extracted with the same volume of water-saturated n-butanol, and then analyzed by thin-layer chromatography (TLC). Strain QM49, which was positive for the desired ginsenoside-hydrolyzing β-glucosidase activity, was selected for further experiments.
Taxonomic characterization of strain QM49.
Strain QM49 was cultivated on R2A agar (Difco) at 30°C unless noted otherwise. Cell morphology was observed under a Nikon light microscope at ×1,000 magnification using cells grown for 3 days. The Gram reaction was performed using the nonstaining method of Buck (38). Physiological and biochemical characterizations, including Gram staining, anaerobic growth, and catalase activity assays, were carried out as described by Smibert and Krieg (39). Enzymatic activities, the assimilation of substrates as the sole carbon source, acid production from the substrates, and other physiological and biochemical characteristics were determined using the API 20NE, API 32GN, and API ZYM kits (bioMérieux, France). Growth at different temperatures (4, 15, 20, 25, 30, 37, 42, and 45°C) and various pH levels (pH 4.5 to 10.0 at intervals of 0.5 pH unit) was assessed after a 5-day incubation period. Salt tolerance was tested on R2A medium supplemented with 1 to 10% (wt/vol) NaCl after incubation for 5 days. Growth on nutrient agar and Trypticase soy agar (Difco) was evaluated at 30°C. Isoprenoid quinones were extracted with chloroform-methanol (2:1, vol/vol), evaporated under a vacuum, and then reextracted in n-hexane–water (1:1, vol/vol). The crude n-hexane–quinone solution was purified using silica Sep-Pak Vac cartridges (Waters) and subsequently analyzed by high-performance liquid chromatography (HPLC) as previously described by Collins et al. (40). Cellular fatty acid profiles were determined for strain QM49T grown on Trypticase soy agar at 30°C for 3 days. Cellular fatty acids were saponified, methylated, and then extracted according to the protocol of the Sherlock microbial identification system (MIDI). The fatty acids were then analyzed by gas chromatography (model 6890; Hewlett Packard) using the Microbial Identification software package (41). The phylogenetic relatedness of strain QM49 to other bacteria was determined by analyzing a portion of the 16S rRNA gene, which was PCR amplified using the universal primers 27F (5′AGAGTTTGATCMTGGCTCAG3′) and 1492R (5′TACGGYTACCTTG TTACGACTT3′) and then sequenced (SolGent, Daejeon, South Korea) as described by Im et al. (42). A near-complete 16S rRNA gene sequence was compiled using the SeqMan program in the DNASTAR package (DNASTAR, Madison, WI). Sequences of the 16S rRNA genes of related taxa were obtained from the GenBank database, and multiple alignments were performed using the CLUSTAL_X program (43). Gaps were edited in the BioEdit program (44), and evolutionary distances were calculated using the Kimura two-parameter model (45). A phylogenetic tree was constructed using the neighbor-joining method (46) in the MEGA4 program (47), with bootstrap values based on 1,000 replicates (48). To determine the GC content of strain QM49, genomic DNA was enzymatically degraded into nucleotides and analyzed as described by Mesbah et al. (49). The nucleotide mixture was separated by reverse-phase HPLC using a Nova-pak C18 column (Waters) and 0.2 M (NH4)H2PO4 containing 7.5% acetonitrile as a solvent. E. coli DNA (Sigma) was used as a calibration reference.
Fosmid library construction and fosmid sequencing.
A CopyControl fosmid production kit (Epicentre Technologies, WI) was used to construct a fosmid library from Mucilaginibacter sp. strain QM49, and this library was used to clone the ginsenoside-hydrolyzing glycosidase gene. Infected E. coli cells were transferred to LB plates supplemented with 40 μg/ml X-Glc and 12.5 μg/ml chloramphenicol and then incubated at 37°C for 16 h. Blue colonies were selected as containing putative ginsenoside-hydrolyzing clones. After confirmation of the ginsenoside-hydrolyzing activity by a TLC assay, one clone was selected for fosmid sequencing. Fosmid DNA was purified using the Fosmid MAX DNA purification kit (Epicentre, WI) and sequenced by Macrogen Co., Ltd. (Korea). The full sequence of the positive fosmid clone was determined by insertion of a transposon into the fosmid clone using a HyperMu KAN-1 insertion kit (Epicentre, WI), followed by bidirectional sequencing of the fosmid using primers homologous to the ends of the inserted transposon. The DNA sequencing reactions were performed using ABI PRISM BigDye Terminator chemistry and ABI 3730XL capillary DNA sequencers (both from Applied Biosystems, Foster City, CA). The final sequences were assembled using the SeqMan program in the DNASTAR package, which yielded a 36.7-kb contig.
Molecular cloning, expression, and purification of recombinant BglQM.
The assembled DNA sequence was analyzed using the ORF Finder program on the NCBI website (www.ncbi.nlm.nih.gov/gorf). Predicted open reading frames (ORFs) were subjected to similarity searches using BLASTP, which identified two putative ORFs for β-glucosidases, BglQM and BglQM1, which belonged to glycoside hydrolase family 3 and consisted of 781 and 795 predicted amino acids, respectively. To clone the genes encoding these proteins (bglQM and bglQM1), PCR amplification was conducted using primers containing SalI (bglQM) or BamHI and EcoRI (bglQM1) restriction sites, respectively (in boldface). The primer sequences were the following: bglQMF, 5′-CGGAATTCAGAATGAGCTACAAATTACACGTTG-3′; bglQMR, 5′-CGGAATTCTTACTTTAGTTTTAGTTCGTTTATGG-3′; bglQM1F, 5′-CGGAATTCATCATGGCCAAACCAATTAAATTG-3′; and bglQM1R, 5′-ACGCGTCGACTTATGGCTTCACCACGCTCTTCAC-3′. The amplified fragment was digested with SalI (bglQM) or BamHI and EcoRI (bglQM1) and inserted into the pMAL-c2x vector containing the tobacco etch virus (TEV) proteolytic cleavage site to generate maltose binding protein (MBP)-bglQM and -bglQM1 fusion genes.
E. coli BL21(DE3) was transformed with pMAL-c2x-TEV-BglQM and -BglQM1 and grown in LB-ampicillin medium at 37°C until the culture reached an optical density at 600 nm (OD600) of 0.6, and then protein expression was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The bacterial cells were incubated further for 24 h at 18°C and then harvested by centrifugation at 10,000 × rpm for 15 min at 4°C. The cells were washed twice with 50 mM sodium phosphate, 5 mM EDTA, and 1% Triton X-100 (pH 7.0) and then resuspended in 50 mM sodium phosphate (pH 7.0). The cells were disrupted by ultrasonication (Vibra-cell; Sonics & Materials, CT). Intact cells and debris were removed by centrifugation at 13,000 × rpm for 30 min at 4°C to obtain a crude cell extract. The MBP-tagged fusion protein was purified by affinity chromatography with an amylose column (GE Healthcare). The MBP tag was removed by incubation with TEV protease. BglQM was further purified by DEAE-cellulose DE-52 chromatography (Whatman). Protein homogeneity was assessed by 10% SDS-PAGE and Coomassie blue staining.
Characterization of recombinant BglQM.
The specific activity of purified BglQM was determined using p-nitrophenyl-β-d-glucopyranoside (PNPGlc) as a surrogate substrate in 50 mM sodium phosphate buffer (pH 8.0) at 30°C. Reactions were stopped after 5 min by the addition of Na2CO3 (final concentration, 0.5 M), and the release of p-nitrophenol was measured immediately using a microplate reader at 405 nm (model 680; Bio-Rad, Hercules, CA). One unit of activity was defined as the amount of protein required to generate 1 μmol of p-nitrophenol per min. Specific activity was expressed as units per milligram of protein. Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce, Rockford, IL), with bovine serum albumin (Sigma) as the standard. All assays were performed in triplicate.
The effect of pH on enzymatic activity was determined using 2.0 mM PNPGlc as a substrate in the following buffers (each at 50 mM): KCl-HCl (pH 2.0), glycine-HCl (pH 3.0), sodium acetate (pH 4.0 and 5.0), sodium phosphate (pH 6.0 and 7.0), Tris-HCl (pH 8.0 and 9.0), and glycine-sodium hydroxide (pH 10.0). The pH stability of recombinant BglQM was determined by measuring enzymatic activity after incubation in each buffer (containing 2.0 mM PNPGlc in 50 mM potassium buffer as a substrate) for 24 h at 4°C. The results are expressed as a percentage of the activity obtained at the optimum pH. The effect of temperature on enzymatic activity was tested by incubating the enzyme at various temperatures, ranging from 4 to 90°C at the optimum pH for 5 min in 50 mM potassium phosphate buffer containing 2.0 mM PNPGlc. The thermostability of the enzyme was examined by incubating the enzyme in 50 mM potassium phosphate buffer for 2 h at different temperatures. After cooling the sample on ice for 10 min, activity was determined using PNPGlc as the substrate.
The effects of metals and other chemicals on BglQM activity were also determined. BglQM activity was tested in the presence of 1 or 10 mM (final concentration) of HgCl2, MnCl2, CaCl2, CoCl2, MgCl2, EDTA, NaCl, KCl, CuCl2, SDS, dithiothreitol (DTT), or β-mercaptoethanol for 30 min at 25°C. The remaining activity was determined using PNPGlc as a substrate, and activities are expressed as a percentage of the activity obtained in the absence of the compound.
Substrate preference was examined using 2.0 mM chromogenic o-nitrophenyl (ONP) and PNP as substrates at 37°C for 5 min, with one activity unit being defined as the release of 1 μmol o-nitrophenol or p-nitrophenol per min. The following substrates were tested: PNP-β-d-glucopyranoside, PNP-β-d-galactopyranoside, PNP-β-d-fucopyranoside, PNP-N-acetyl-β-d-glucosaminide, PNP-β-l-arabinopyranoside, PNP-β-d-mannopyranoside, PNP-β-d-xylopyranoside, PNP-α-d-glucopyranoside, PNP-α-l-arabinofuranoside, PNP-α-l-arabinopyranoside, PNP-α-l-rhamnopyranoside, PNP-α-d-mannopyranoside, PNP-α-d-xylopyranoside, ONP-β-d-glucopyranoside, ONP-β-d-galactopyranoside, ONP-β-d-fucopyranoside, and ONP-α-d-galactopyranoside (all from Sigma).
Kinetic studies were performed with freshly purified enzymes using PNPGlc at 0.1 to 10.0 mM and Re and Rg1 at concentrations from 0.5 to 25.0 mM. One unit of activity was defined as the amount of protein required to generate 1 μmol of p-nitrophenol or to convert 1 μmol of Re or Rg1 per min. All assays were performed in triplicate. The parameters Km and Vmax were determined using the enzyme kinetics program described by Cleland (50).
Biotransformation of major ginsenosides by BglQM.
The initial biotransformation experiments using Re and Rg1 as substrates revealed that the fusion of MBP to the enzyme did not affect the activity of BglQM. Therefore, the fusion protein (MBP-BglQM) was used to determine the specificity and selectivity of this enzyme for hydrolyzing the glucose moieties attached at the C-3 and C-20 sites of four PPD-type ginsenosides (Rb1, Rb2, Rc, and Rd) and two PPT-type ginsenosides (Re and Rg1). The enzyme solution (0.1 mg/ml in 50 mM sodium phosphate buffer, pH 7.0) was reacted with an equal volume of Rb1, Rb2, Rc, Rd, Re, or Rg1 (0.1%, wt/vol, in 50 mM sodium phosphate buffer, pH 7.0) at 25°C. Samples were withdrawn at regular intervals. For TLC analysis, an equal volume of water-saturated n-butanol was added to stop the reaction, and the reactant present in the n-butanol fraction was analyzed. For HPLC analysis, an equal volume of methanol was added to stop the reaction, the mixture was centrifuged at 15,000 × g for 10 min, and the supernatant was used for analysis. See below for details on the TLC and HPLC assays.
Optimization of substrate concentration.
To determine the optimal conditions for the biotransformation of PPTGM to (S)-Rg2 and (S)-Rh1, the substrate concentration of PPTGM in the reaction mixture was optimized. The final crude BglQM concentration was fixed to 39.6 mg/ml (corresponding to 0.5 U/ml for PNPGlc) and reacted with an equal volume of 10, 20, 40, 60, or 100 mg/ml PPTGM (final substrate concentration, 5, 10, 20, 30, and 50 mg/ml, respectively). Samples were taken at regular intervals and analyzed by TLC and HPLC.
Preparation of recombinant BglQM using high-cell-density culture.
For the production of recombinant BglQM, LB medium supplemented with ampicillin (100 μg/ml) was used to cultivate E. coli harboring pMAL-bglQM in a 10-liter stirred-tank reactor (5-liter working volume; Biotron Co., Seoul, South Korea) at 500 rpm, according to the previously described fed-batch mode (51). The pH of the medium was adjusted to 7.0 using 100 mM sodium phosphate. The culture was incubated at 37°C until it reached an OD600 of 3.0, and protein expression was induced with IPTG (final concentration, 0.1 mM). The bacterial cells were incubated for a further 24 h at 18°C, harvested by centrifugation at 5,000 × g for 20 min at 4°C, suspended in 100 mM phosphate buffer (pH 7.0), and then disrupted by sonication. Intact cells and debris were removed by centrifugation at 5,000 rpm (Component R; Hanil Science Co. Ltd., South Korea) for 20 min at 4°C. The crude recombinant BglQM (supernatant) was diluted to the desired concentration with 100 mM sodium phosphate buffer (pH 7.0) and used to biotransform the PPTGM.
Gram-scale PPTGM transformation by crude BglQM.
The scaled-up biotransformation was performed in a 10-liter stirred-tank reactor (3.0-liter working volume; Biotron Co., Seoul, South Korea) under optimal conditions (200 rpm for 50 h at pH 8.0 and 37°C). The initial reaction mixture contained 20 mg/ml of substrate ginsenosides (PPTGM; total of 60 g) and 1.5 liter of recombinant crude BglQM in 100 mM phosphate buffer (pH 8.0). Samples were collected at regular intervals, and HPLC was used to monitor the biotransformation of Re and Rg1 to (S)-Rg2 and (S)-Rh1, respectively.
Purification of Rg1 and Re using silica cartridges.
Following the scaled-up biotransformation of (S)-Rg2 and (S)-Rh1, an equal volume of water-saturated n-butanol was added to stop the reaction, and the tubes were laid down to separate the n-butanol from the mixture. This n-butanol extraction was carried out twice, and the butanol extracts were pooled and evaporated in vacuo. The resulting powder was purified using a Biotage SNAP flash chromatography cartridge (180 by 70 mm; Biotage KP-Sil) packed with 340 g silica resin (230 to 400 mesh). The cartridge was equilibrated with chloroform, and 15.0 g of powdered metabolites was dry loaded into a self-packed sample cartridge. Elution was performed with 6 bed volumes (BVs) of chloroform, 6 BVs of chloroform-methanol (9:1, vol/vol), and 6 BVs of chloroform-methanol (8:2, vol/vol). Fractions were taken for every 340-ml elution (0.5 BV), and the results were analyzed by TLC.
TLC analysis.
TLC was performed using 60F254 silica gel plates (Merck, Germany) with CHCl3-CH3OH-H2O (65:35:10, vol/vol/vol, lower phase) as the solvent. The spots on the TLC plates were visualized by spraying with 10% (vol/vol) H2SO4 followed by heating at 110°C for 5 min, and the results were determined by comparison to ginsenoside standards.
HPLC analysis.
HPLC was performed using an HPLC system with a quaternary pump, automatic injector, single-wavelength UV detector (model 730D), and Younglin's AutoChro 3000 software for peak identification and integration (all from Younglin Co., Ltd., South Korea). Separation was carried out on a Prodigy ODS(2) C18 column (5 μm; inner diameter, 150 by 4.6 mm; Phenomenex) with a guard column (Eclipse XDB C18; 5 μm; inner diameter, 12.5 by 4.6 mm; Agilent). The mobile phases were A (acetonitrile) and B (water). Gradient elution was performed as the following: 17% solvent A and 83% solvent B for 12 min, 25% solvent A and 75% solvent B from 12 to 20 min, 32% A and 68% B from 20 to 30 min, 55% A and 45% B from 30 to 35 min, 60% A and 40% B from 35 to 40 min, 80% A and 20% B from 40 to 45 min, 100% solvent A from 45 to 50 min, holding at 100% solvent A from 50 to 54 min, changing to 17% A and 83% B from 54.0 to 54.1 min, and holding at 17% and 83%, respectively, from 54.1 to 65 min. The flow rate was 1.0 ml/min, and detection was performed by monitoring absorbance at 203 nm with an injection volume of 25 μl.
MS analysis.
Electrospray ionization mass spectra (ESI-MS) were measured for the purified biotransformed ginsenosides [(S)-Rg2 and (S)-Rh1] using a triple-quadrupole tandem mass spectrometer (API-2000; Applied Biosystems, Foster City, CA) in the negative-ion mode. The ESI parameters were the following: ion spray voltage, −4,200 V; ion source gas 1 (GS1), 20; curtain gas (CUR), 20; and collision gas (CAD), 2. The declustering potential (DP), focusing potential (FP), entrance potential (EP), collision cell exit potential (CXP), and collision energy (CE) varied with regard to the measured ginsenosides. For full-scan MS analysis, the spectra were recorded in the m/z range of 400 to 1,000.
Nucleotide sequence accession numbers.
The sequences for the 16S rRNA and bglQM and bglQM1 genes from Mucilaginibacter sp. strain QM49 were deposited in GenBank/EMBL/DDBJ under accession numbers HM204922, JX403802, and JX403803, respectively.
RESULTS
Isolation and characterization of strain QM49.
After three rounds of screening, 15 bacterial strains were isolated from the grass soil of KAIST Hill in Daejeon, South Korea. We used a three-step screening process, consisting of (i) isolating the bacteria from soil, (ii) screening the bacteria for β-glucosidase activity based on esculin hydrolysis, and (iii) TLC-based selection of strains with biotransformation activity for the ginsenosides Rb1, Re, and Rg1 (see Table S1 in the supplemental material). Among the isolated bacteria, strain QM49 was confirmed to hydrolyze Rg1 and Re to (S)-Rh1 and (S)-Rg2, respectively (Fig. 2), and was selected for further study.
Fig 2.
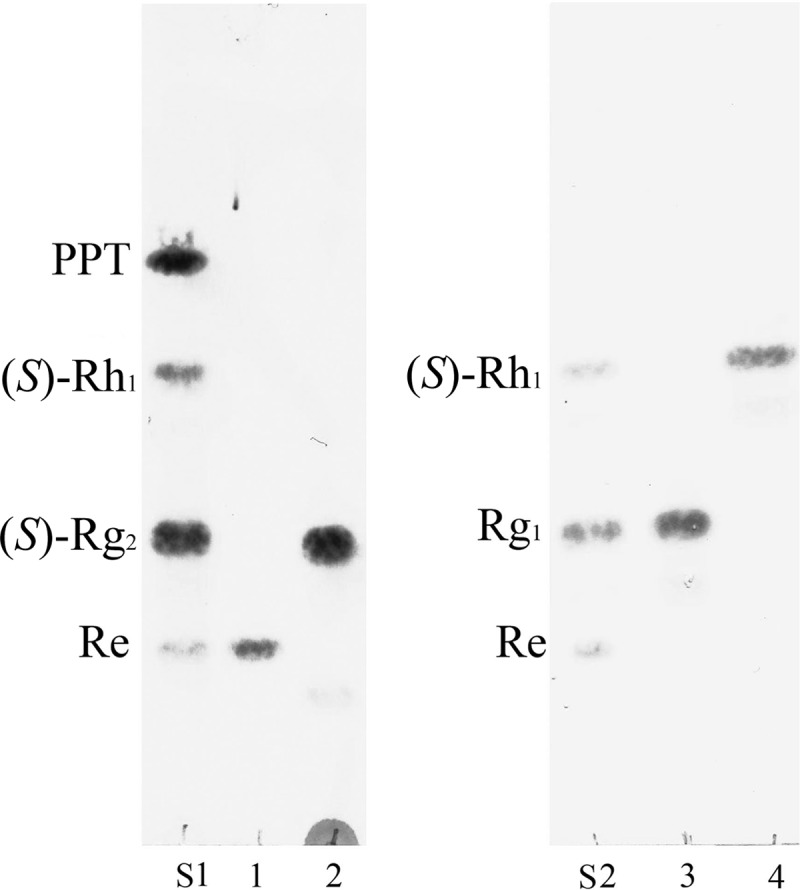
Thin-layer chromatography (TLC)-based analyses of the biotransformation of Re and Rg1 by strain QM49. The reaction time was 48 h, and the developing solvent was CHCl3-CH3OH-H2O (65:35:10, vol/vol/vol, lower phase). Lanes: S1 and S2, ginsenoside standard; 1, substrate Re; 2, reaction mixture of Re; 3, substrate Rg1; 4, reaction mixture of Rg1.
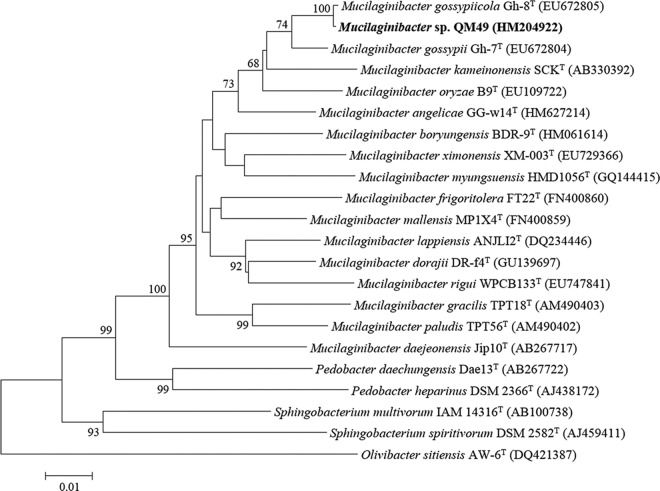
Cells of strain QM49 were Gram negative, aerobic, non-spore forming, nonmotile, rod shaped, oxidase positive, and catalase positive. Colonies grown on R2A agar plates for 3 days were smooth, transparent, circular, pale pink in color, convex, and 1 to 1.5 mm in diameter. On R2A agar, QM49 was able to grow at 4 to 37°C but not at 42°C. The isolate grew on nutrient agar and TSA but did not grow on MacConkey agar. Other biochemical characteristics are presented in the supplemental material (available online). Phylogenetic analysis based on 16S rRNA gene sequences indicated that the isolate belongs to the genus Mucilaginibacter in the phylum Bacteroidetes and is most closely related to Mucilaginibacter gossypiicola Gh-48T (99.86% similarity), followed by M. gossypii GH-67T (98.20%), M. kameinonensis SCKT (97.53%), M. oryzae B9T (97.23%), and M. polysacchareus DRP28T (97.07%). The relationships between strain QM49 and other members of the genus Mucilaginibacter were also evident in the phylogenetic tree, which used over 1,380 bp of sequence information (Fig. 3). The predominant menaquinone was MK-7. The fatty acids (>5% total fatty acids) contained C16:1 ω7c and/or C16:1 ω6c (23.1%), iso-C15:0 (18.3%), C16:1 ω6c and/or C16:1 ω7c (17.2%), C16:0 (12.1%), iso-C17:0 3OH(10.0%), and C16:1 ω5c (5.1%). The GC content of the genomic DNA was 45.2 mol%. Collectively, the physiological and chemotaxonomic characteristics of strain QM49 are consistent with the suggestion that it is a member of the genus Mucilaginibacter.
Fig 3.
Rooted phylogenetic tree based on the 16S rRNA gene sequences of strain QM49 and related bacteria in genus Mucilaginibacter. The tree was constructed by the neighbor-joining method. A bar represents 0.01 substitutions per nucleotide position. Bootstrap values (expressed as a percentage of 1,000 replications) are shown at the branch points.
Cloning of bglQM and bglQM1 from strain QM49.
To clone the ginsenoside-transforming β-glucosidase, genomic DNA from Mucilaginibacter sp. strain QM49 was subjected to phenol-chloroform extraction and used to generate a fosmid library. The average insert size of the fosmid clones was approximately 40 kb. Out of approximately 1,000 clones in the fosmid library, 24 conferred a blue color to E. coli transformants grown on LB agar plates containing 12.5 μg/ml of chloramphenicol and 27 μg/ml of X-Glc. One of these fosmid clones was positive for the ability to transform the ginsenosides Re and Rg1, and this clone was subjected to further analysis.
The positive clone contained a 36.7-kb genomic fragment. Nucleotide sequencing and analysis revealed that this fragment contained a total of 33 putative ORFs (data not shown), two of which were homologous to glycoside hydrolase genes. The first ORF encoded a putative β-glucosidase gene that we designated bglQM. Analysis of the amino acid sequences of BglQM indicated that it was 75.8% identical to the glycoside hydrolase of Spirosoma linguale DSM 74T (GenBank accession number YP_003386107), which belongs to glycoside hydrolase family 3. The enzymatic activity of the glycoside hydrolase from Spirosoma linguale DSM 74T has not yet been characterized. The closest characterized glycoside hydrolase found in the CAZy database (carbohydrate active enzymes; http://www.cazy.org) was BglY from Paenibacillus sp. strain C7 (53), which was 64.9% identical to BglQM based on amino acid sequences. The second ORF was a putative β-glucosidase gene that we designated bglQM1. Analysis of the amino acid sequences of BglQM1 indicated that it was 57.8% identical to the glycoside hydrolase of Niastella koreensis GR20-10T (GenBank accession number AEV97206).
Glycosyl hydrolases are classified according to their amino acid sequence similarities, which reflect the structural features and substrate specificities of the enzymes (http://www.cazy.org/fam/acc_GH.html). Glycoside hydrolase family 3 was subdivided into six subfamilies (54). In order to determine the evolutionary position of bglQM and bglQM1 within the characterized enzymes in glycoside hydrolase family 3, we carried out a phylogenetic analysis using the neighbor-joining method (46) in the MEGA4 program (47) with bootstrap values based on 1,000 replications (48). The resulting consensus tree is presented in Fig. S1 in the supplemental material. BglQM clustered within subfamily 6 and formed a separate, well-supported clade (bootstrap of 100) with Paenibacillus sp. strain C7 β-glucosidase (53) and Bacillus sp. strain GL1 β-glucosidase (55), which were characterized previously. Several ginsenoside-hydrolyzing β-glucosidases in glycoside hydrolase family 3 have previously been cloned, including a β-glucosidase (rApy-H11) from Bifidobacterium longum H-1 (8), a β-glucosidase (Bgp1) from Microbacterium esteraromaticum (31), a β-glucosidase (BgpA) from Terrabacter ginsenosidimutans (23), a β-glucosidase (BGL1) from Aspergillus niger (56), a β-glucosidase (BglAm) from Actinosynnema mirum (24), and a β-glucosidase (BglSk) from Sanguibacter keddieii (26). The relationship between BglQM and these ginsenoside-hydrolyzing β-glucosidases is presented in Fig. S1 in the supplemental material. Multiple sequence alignments of BglQM with family 3 glycoside hydrolases allowed identification of the active sites (see Table S2 in the supplemental material). BglQM shared many conserved amino acid residues with other β-glucosidases, which confirmed that BglQM belonged to the glycoside hydrolase family 3. Furthermore, Asp241 and Glu457 were thought to be the active sites of BglQM.
Expression of and ginsenoside biotransformation by BglQM.
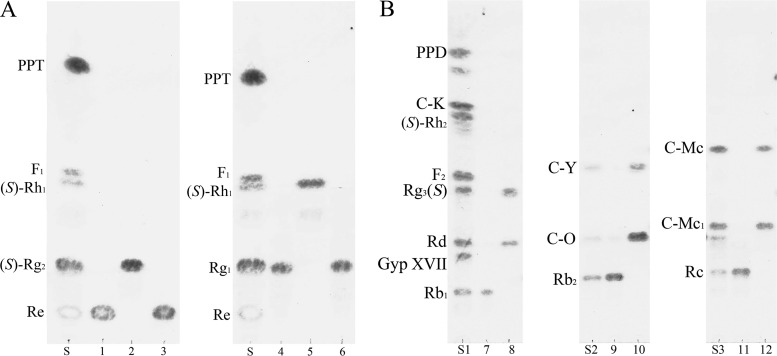
The two putative β-glucosidase genes, bglQM and bglQM1, were PCR amplified and subcloned into pMAL-c2x (TEV) to generate MBP-bglQM and MBP-bglQM1 fusion genes that could be expressed in E. coli C41(DE3) under the control of the IPTG-inducible promoter Ptac. We tested different induction conditions and found that induction with 0.1 mM IPTG at 18°C for 12 h produced the maximum level of soluble active fusion enzyme (data not shown). To verify the bioconversion abilities of MBP-BglQM and MBP-BglQM1 against the PPT-type major ginsenosides Re and Rg1, TLC analysis was performed on enzyme hydrolysates. As shown in Fig. 4A, MBP-BglQM could transform Re and Rg1 (see the Rf values). In contrast, MBP-BglQM1 showed no activity for transforming these ginsenosides; thus, it was excluded from subsequent experiments.
Fig 4.
TLC-based analyses of biotransformation by BglQM and BglQM1. S, S1, S2, and S3, ginsenoside standards; 1, substrate Re; 2, biotransformation of Re by BglQM; 3, biotransformation of Re by BglQM1; 4, substrate Rg1; 5, biotransformation of Rg1 by BglQM; 6, biotransformation of Rg1 by BglQM1; 7, substrate Rb1; 8, biotransformation of Rb1 by BglQM; 9, substrate Rb2; 10, biotransformation of Rb2 by BglQM; 11, substrate Rc; 12, biotransformation of Rc by BglQM.
MBP-BglQM could clearly transform six major ginsenosides (Re, Rg1, Rb1, Rb2, Rc, and Rd), as shown by the Rf values of our TLC analysis (Fig. 4) and the retention times in HPLC (data not shown). BglQM could efficiently hydrolyze the glucose moiety at the C-20 position of Re and Rg1, transforming them into (S)-Rg2 and (S)-Rh1, respectively (Fig. 4A). In addition to these PPT-type ginsenosides, BglQM could also transform the PPD-type ginsenosides Rb1, Rb2, Rc, and Rd. The proposed biotransformation pathways for the actions of MBP-BglQM on the PPD-type ginsenosides are Rb1→Rd→(S)-Rg3 via stepwise hydrolysis of the outer and inner glucoses at the C-20 position. Furthermore, BglQM transformed Rb2 and Rc to C-Y and C-Mc via C-O and C-Mc1, respectively, through consecutive cleavages of the outer and inner glucose moieties at the C-3 position (Fig. 4B). However, BglQM could not further hydrolyze C-Y or C-Mc due to the l-arabinopyranoside and l-arabinofuranoside moieties attached at the C-20 site. Although BglQM has arabinofuranoside-hydrolyzing activity, as detected with PNP substrates, it could not hydrolyze the arabinofuranoside of Rc. This may be due to structural differences between Rc and PNP-arabinofuranoside. Overall, BglQM hydrolyzed Rb1, Rb2, Rc, Rd, Re, and Rg1 as the following: Re→(S)-Rg2, Rg1→(S)-Rh1, Rb1→Rd→(S)-Rg3, Rb2→C-O→C-Y, and Rc→C-Mc1→C-Mc (see Fig. S2 in the supplemental material).
The metabolic pathways of ginsenoside hydrolysis by BglQM are similar to those of a β-glucosidase (Bgpl) from Microbacterium esteraromaticum (32, 57), except that Bgpl directly hydrolyzes Rb1 to (S)-Rg3, whereas BglQM hydrolyzed Rb1 into (S)-Rg3 via Rd. Re and Rg1 were deglucosylated when exposed to a culture of Mucilaginibacter sp. strain QM49, and recombinant BglQM has the same effect, presumably via the hydrolyzing pathways. This suggests that the ginsenoside-hydrolyzing activity of Mucilaginibacter sp. strain QM49 arises from the expression of BglQM.
When Rb1, Rd, Re, and Rg1 (1.0 mg/ml) were used as substrates, they were biotransformed within 10 min by 10 mg/ml of crude recombinant MBP-BglQM. Several other ginsenoside-hydrolyzing recombinant enzymes have been reported, but most were only able to hydrolyze the outer or inner glucose moiety at the C-3 position of the aglycon (23, 24, 26). One previously described enzyme from Microbacterium esteraromaticum KCTC 12040BP (32) had the same ability as BglQM, but the authors conducted only a simple enzymatic characterization without further scale up or process engineering. Thus, we sought to engineer our BglQM reaction for the industrially relevant gram-scale production of (S)-Rg2 and (S)-Rh1.
Purification and characterization of recombinant BglQM.
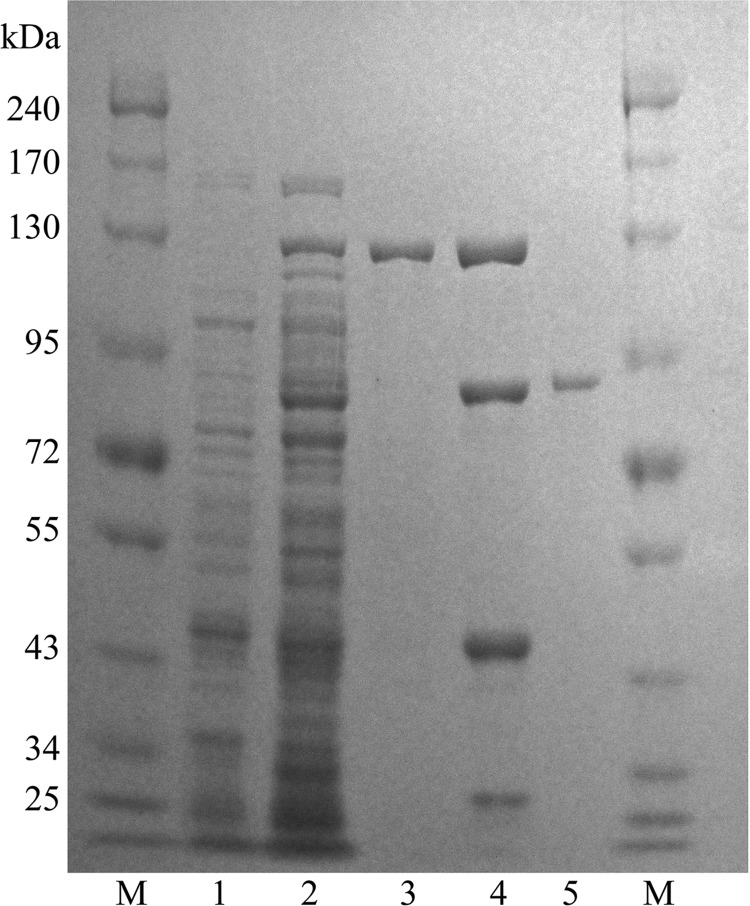
To maximize the yield of the fusion protein, we tested different induction conditions. Induction with 0.1 mM IPTG at 18°C for 18 h produced the maximum level of soluble active fusion enzyme (data not shown). BglQM fusion proteins purified by amylose-affinity column chromatography were digested by TEV protease at room temperature for 4 h to remove the MBP tag, and the resulting BglQM was further purified by chromatography on DEAE-cellulose. This procedure resulted in a 285-fold purification and a recovery of 29% from the crude extract (Table 1). The predictive molecular mass (85.6 kDa) of the native β-glucosidase was determined via SDS-PAGE (Fig. 5).
Table 1.
Purification scheme
| Purification step | Vol (ml) | Total protein (mg) | Sp act (U/mg) | Total act (Ua) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude extract | 30 | 1,188 | 0.01 | 14.9 | 100 | 1 |
| Amylose affinity | 18 | 99 | 0.82 | 8.1 | 54 | 65 |
| DEAE-cellulose | 4 | 1.23 | 3.57 | 4.4 | 29 | 285 |
One unit of BgpA was defined as the amount of enzyme liberating 1 μmol/min of p-nitrophenol.
Fig 5.
SDS-PAGE analysis of recombinant BglQM. Lanes: M, molecular mass standard; 1, crude extract of BL21 carrying pMAL-BglQM prior to induction with IPTG; 2, after induction with IPTG; 3, the MAL-BglQM fraction after amylose column purification; 4, MAL-BglQM after treatment with TEV; 5, the BglQM fraction after DEAE cellulose column and amylose column purification.
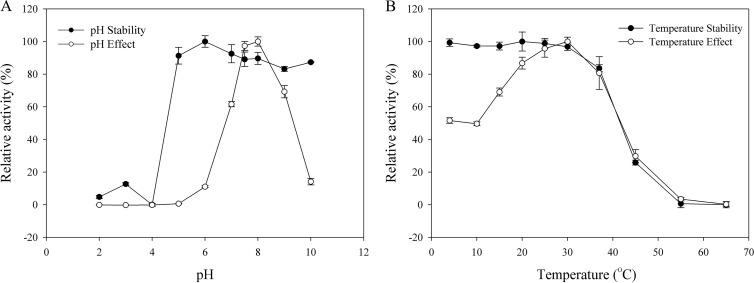
The optimum pH and temperature were determined using purified BglQM. The enzyme was active over a range of pH 7.0 to 9.0 and was stable at pH 5.0 to 10.0. The optimum pH was pH 8.0 in sodium phosphate buffer (Fig. 6A). The optimal temperature for BglQM activity was 30°C. The enzyme was stable at temperatures lower than 30°C, and about 67% of the activity was lost after incubation at 45°C for 120 min (Fig. 6B). The enzyme is probably mesophilic and stable at a wide pH range (Fig. 6A). These optimal conditions are consistent with the soil environment from which Mucilaginibacter sp. strain QM49 was isolated. In addition, the near-neutral optimal pH and mild optimal temperature of BglQM are similar to those of other ginsenoside-hydrolyzing family 3 β-glucosidases from bacteria (23, 32, 56).
Fig 6.
Effects of pH (A) and temperature (B) on the stability and activity of recombinant BglQM.
The effects of metal ions, EDTA, β-mercaptoethanol, and SDS on BglQM activity were also investigated (Table 2). The enzyme did not require Mg2+ for activity, lost its activity in the presence of 10 mM Cu2+, and was significantly inhibited by 1 mM Cu2+, 1 mM EDTA, 10 mM Co2+, or 10 mM SDS. The activity of BglQM was not affected by the presence of β-mercaptoethanol, which is a well-known thiol group inhibitor, suggesting that the sulfhydryl groups are not involved in the catalytic center of the enzyme but rather are essential for maintaining the three-dimensional structure of the active protein. The chelating agent, EDTA, inhibited the activity of BglQM, indicating that divalent cations are required for its enzymatic activity.
Table 2.
Effects of metal ions and chemical agents on the activity of purified recombinant BglQM
| Metal ion and reagent | Relative activity ± SD (%) at: |
|
|---|---|---|
| 1 mM | 10 mM | |
| NaCl | 91.1 ± 1.27 | 93.6 ± 3.14 |
| KCl | 92.0 ± 5.78 | 90.9 ± 1.61 |
| MgCl2 | 93.8 ± 3.18 | 85.0 ± 2.52 |
| CaCl2 | 99.6 ± 2.55 | 102.5 ± 5.21 |
| CoCl2 | 92.9 ± 1.68 | 76.1 ± 1.35 |
| CuCl2 | 46.4 ± 0.95 | NDa |
| SDS | 84.9 ± 2.26 | 16.5 ± 0.62 |
| EDTA | 64.3 ± 1.36 | 59.5 ± 0.87 |
| Beta-mercaptoethanol | 94.6 ± 2.22 | 95.4 ± 3.58 |
| Control | 100.0 ± 2.36 | 100.0 ± 2.36 |
ND, not determined.
The substrate specificity of BglQM was tested using 2.0 mM PNP- and ONP-glycosides with α and β configurations. The results, which are summarized in Table 3, showed that BglQM was maximally active against PNPGlc followed by ONPGlc but had little effect on various other PNP- and ONP-glycosides, with the exception of PNP-α-l-arabinofuranoside and PNP-β-d-fucopyranoside. The specific activity of BglQM against PNP-α-l-arabinofuranoside is not seen in ginsenoside-transforming glycoside hydrolase family 3 members from Terrabacter ginsenosidimutans (23) and Microbacterium esteraromaticum (32). However, BglQM cannot hydrolyze the arabinofuranoside at the outer position of the C-20 of the ginsenoside Rc, perhaps due to structural differences between the molecules.
Table 3.
Relative activity of purified recombinant BglQM toward various chromogenic substrates as measured by ONP or PNP release at 30°C
| Substrate | Relative activity ± SD (%) |
|---|---|
| PNP-α-d-glucopyranoside | 0 |
| PNP-α-d-mannopyranoside | 0 |
| PNP-α-d-xylopyranoside | 0 |
| PNP-α-l-arabinofuranoside | 36.67 ± 1.05 |
| PNP-α-l-arabinopyranoside | 0 |
| PNP-α-l-rhamnopyranoside | 0 |
| PNP-β-d-fucopyranoside | 12.17 ± 0.52 |
| PNP-β-d-glactopyranoside | 0 |
| PNP-β-d-glucopyranoside | 0 |
| PNP-β-d-glucosaminide | 0 |
| PNP-β-d-mannopyranoside | 0 |
| PNP-β-d-xylopyranoside | 0 |
| PNP-β-l-arabinopyranoside | 0 |
| ONP-α-d-galactopyranoside | 0 |
| ONP-β-d-fucopyranoside | 49.43 ± 0.71 |
| ONP-β-d-glucopyranoside | 100.00 ± 2.42 |
The Km values of BglQM for p-nitrophenyl-β-d-glucopyranoside, Re, and Rg1 were 37.0 ± 0.4 μM, 3.22 ± 0.15, and 1.48 ± 0.09 mM, respectively, and the Vmax values were 33.4 ± 0.6 μmol min−1 mg−1 of protein and 19.2 ± 0.2 and 28.8 ± 0.3 nmol min−1 mg−1 of protein, respectively. The kcat values were 47.6 ± 4.3, 0.0273 ± 0.0037, and 0.0411 ± 0.00123 s−1, respectively, measured using the method described by Cleland (50).
The catalytic efficiencies (kcat/Km) for PNPGlc, Re, and Rg1 decreased in this order: PNPGlc (1.23 ± 0.13 μM−1 S−1) > Rg1 (1.64 ± 0.22 mM−1 min−1) > Re (0.51 ± 0.07 mM−1 min−1). BglQM has higher catalytic efficiency for Rg1 than Re; the attached rhamnose moiety of Re may decrease the affinity. The catalytic efficiencies for ginsenoside Rg1 is higher than that of β-glycosidase from Sulfolobus acidocaldarius (29) for ginsenosides Rd (4.8 mM−1 min−1) and Rb2 (0.77 mM−1 min−1) but lower than that for Rb1 (4.8 mM−1 min−1) and Rc (4.5 mM−1 min−1). No catalytic efficiencies of other recombinant glycoside hydrolases for Re and Rg1 have been reported before BglQM.
Optimization of substrate concentration.
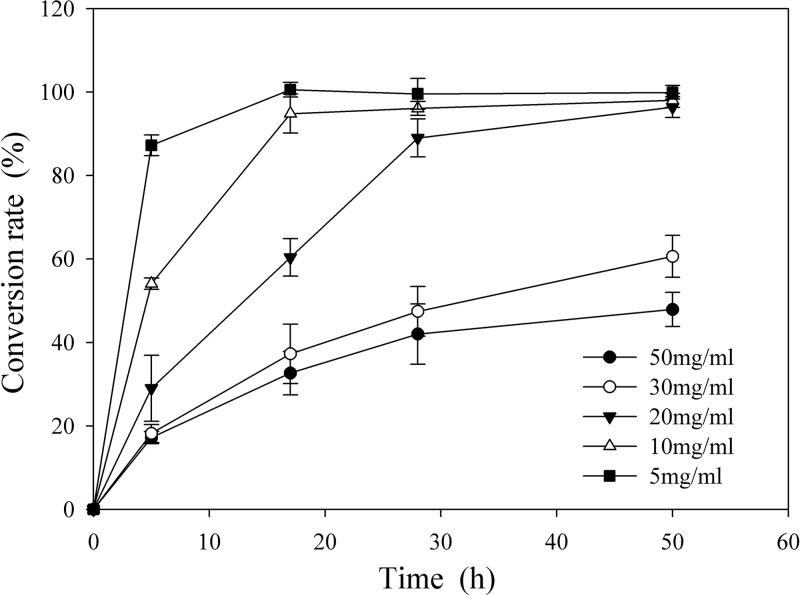
To determine an economical substrate and facilitate scaling up of the reaction, six substrate concentrations (final concentrations of PPTGM of 50, 30, 20, 10, 5, and 1 mg/ml) were reacted with crude recombinant BglQM having 0.50 U/ml PNPGlc activity, and the time courses of (S)-Rg2 and (S)-Rh1 production were determined by TLC and HPLC (Fig. 7). In experiments with moderate substrate concentrations (5 and 10 mg/ml), Re and Rg1 were completely converted to (S)-Rg2 and (S)-Rh1 within 20 h, which was around twice as fast as the bioconversion of 20 mg/ml PPTGM. The other two substrate concentrations failed to show complete conversion within 50 h and were excluded. However, the higher concentration of substrate (20 mg/ml) was adopted for the subsequent scaling up of biotransformation, allowing us to use a smaller, more economical reactor.
Fig 7.
Effects of PPTGM concentration on the production of (S)-Rg2 and (S)-Rh1 by BglQM. PPTGM was hydrolyzed by recombinant BglQM under optimum conditions for 50 h.
Scaling up the biotransformation of Rg1 and Re by crude recombinant BglQM.
Bacterial cells harboring pMAL-bglQM were induced, incubated for 24 h at 18°C, and harvested via centrifugation when the culture reached an OD600 of 28. Cell pellets (72 g of wet cells) were resuspended in 10 volumes (wt/vol) of 100 mM sodium phosphate buffer (pH 7.0) and disrupted by ultrasonication, and the supernatant was used as the crude enzyme for ginsenoside biotransformation. Almost half of the expressed pMAL-BglQM was in soluble form (Fig. 5). After sonication, the β-glucosidase activity of crude recombinant BglQM was measured using PNPGlc at 0.25 U/ml.
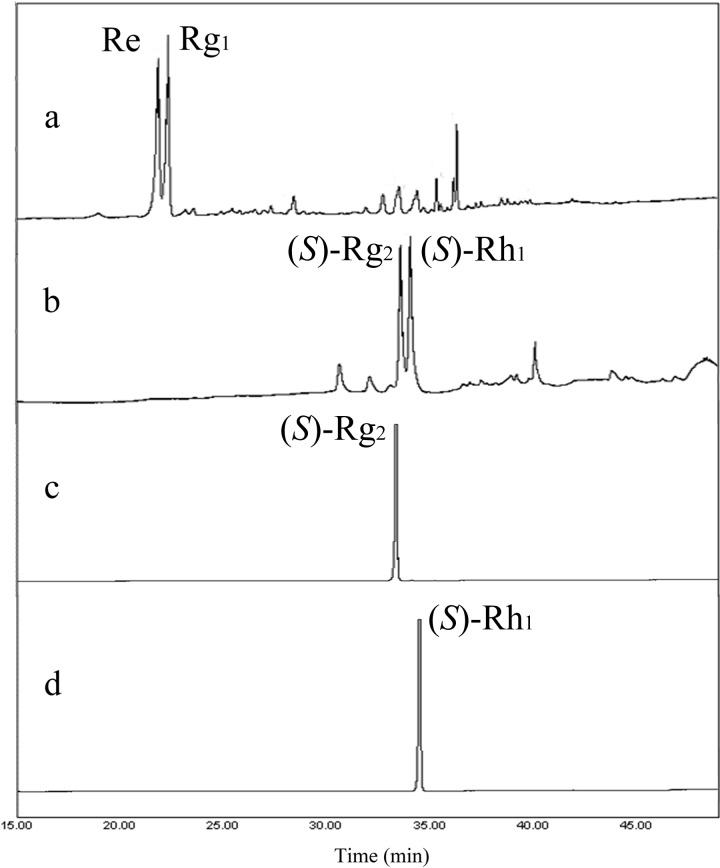
The reaction consisted of 1.5 liters crude recombinant BglQM and 60 g of PPTGM (substrate; 20 mg/ml) in 3 liters of 100 mM sodium phosphate buffer. As shown in Fig. 8, Re and Rg1 were completely converted to (S)-Rg2 and (S)-Rh1, respectively, within 50 h; at this point, Re and Rg1 were no longer detectable by HPLC. Chromatographic images of samples drawn from different time points and analyzed by HPLC are shown in Fig. 8.
Fig 8.
Transformation of PPTGM by recombinant BglQM, as assessed by HPLC. (a) The ginsenoside peaks of PPTGM; (b) the reaction mixture after treatment with BglQM for 50 h; (c) (S)-Rg2 after silica resin purification; and (d) (S)-Rh1 after silica resin purification. PPTGM, protopanaxatriol-type ginsenoside mixture.
Purification of biotransformed (S)-Rg2 and (S)-Rh1.
After n-butanol extraction and evaporation, we obtained 32.7 g of biotransformed ginsenosides mixture. Half of the crude mixture containing (S)-Rg2 and (S)-Rh1 was loaded into a Biotage SNAP flash chromatography cartridge, and gradient chloroform-methanol elution (9:1→8:2→7:3) was used to separate and purify (S)-Rg2 and (S)-Rh1. TLC analysis revealed that (S)-Rh1 was eluted at 14 to 16 BVs, while (S)-Rg2 was eluted at 20 to 23 BVs. This step was applied again for the residual crude mixture. Pure fractions were collected and evaporated to yield 5.4 and 4.6 g of (S)-Rg2 and (S)-Rh1, respectively, with 98% ± 0.5% and 97% ± 1.2% purity (as assessed by HPLC) (Fig. 8). The negative-ion ESI MS-MS spectra of the final products confirmed that we had obtained (S)-Rg2 and (S)-Rh1 (see Fig. S3 in the supplemental material).
DISCUSSION
Although (S)-Rg2 and (S)-Rh1 have antiallergic, anti-inflammatory, and antimetastatic activities, the lack of a selective mass production technology has hampered their commercial use. Red ginseng, which is a very popular health-promoting food in oriental herbal medicine, contains approximately 1% (S)-Rg2 and (S)-Rh1 based on dry weight. In total, red ginseng products account for 52.8% of the total health food market in South Korea, which was estimated to be worth $625 million in 2011 (58). The popularity of red ginseng appears to indirectly prove its effects. In the laboratory, a number of researchers have sought to biotransform major ginsenosides into larger quantities of (S)-Rg2 or (S)-Rh1 using microorganisms (36, 59) and enzymes (32, 37). Ko et al. (37) characterized the abilities of crude β-galactosidase from Aspergillus oryzae and crude lactase from Penicillium spp. to biotransform PPT-type ginsenosides to (S)-Rg2 and (S)-Rh1 but only on a 10-mg scale. Recently, a recombinant β-glucosidase belonging to glycoside hydrolase family 3 (subfamily 5) from Microbacterium esteraromaticum was shown to transform Re and Rg1 into (S)-Rg2 and (S)-Rh1, respectively, but without sufficient efficiency for gram-scale applications (32). Here, we report for the first time that BglQM can transform up to 20 mg/ml PPTGE (containing Re and Rg1) into multiple grams of crude (S)-Rg2 and (S)-Rh1 within 50 h. To the best of our knowledge, this is the first recombinant enzyme that has been reported to produce gram-scale (S)-Rg2 and (S)-Rh1 from PPTGM.
While surveying the ginsenoside-hydrolyzing enzymes, our research team has identified several ginsenoside-hydrolyzing bacteria (60, 61) and constructed several ginsenoside-hydrolyzing recombinant enzymes (23–26, 33, 52). Our novel identification of BglQM as an enzyme capable of converting Re and Rg1 into (S)-Rg2 and (S)-Rh1 should critically facilitate the mass production of (S)-Rg2 and (S)-Rh1 from protopanaxatriol-type ginsenoside mixtures derived from Panax quinquefolius (American ginseng) or Panax ginseng C. A. Meyer (Korean ginseng). Since ginsenosides were first isolated in the 1960s (62, 63), hundreds of publications have focused on ginseng and ginsenosides, but the lack of a technology for producing large quantities of pure ginsenoside compounds has limited the research into their physiological functions (64). Therefore, this paper may mark the beginning of new era in which (S)-Rg2 and (S)-Rh1 are used as core functional biomaterials in the cosmetic, health food, and medical industries.
In conclusion, we describe here the isolation of a ginsenoside-hydrolyzing β-glucosidase (BglQM) belonging to glycoside hydrolase family 3 from Mucilaginibacter sp. strain QM49 and the production of a recombinant enzyme for the biotransformation of the major ginsenosides Re and Rg1 into the pharmacologically active rare ginsenosides (S)-Rg2 and (S)-Rh1, respectively. This enzyme was expressed in E. coli BL21(DE3) in a soluble form. Characterization revealed that its optimum reaction conditions were 30°C and pH 8.0, and that this could be used for gram-scale production. In terms of yield, 5.4 and 4.6 g of (S)-Rg2 and (S)-Rh1, respectively, with 98% ± 0.5% and 97% ± 1.2% chromatographic purity were obtained via the biotransformation of 60 g of a PPT-type ginsenoside mixture followed by purification with silica resin. The conversion took place in a 10-liter jar fermenter in 100 mM sodium phosphate buffer (pH 8.0) at 30°C for 50 h, with an initial substrate concentration of 20 mg/ml. This is the first report of a cloned enzyme that is capable gram-scale production of (S)-Rg2 and (S)-Rh1 through biotransformation of the major ginsenosides Re and Rg1.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Intelligent Synthetic Biology Center of Global Frontier Project funded by the Ministry of Education, Science and Technology (2011-0031967), Republic of Korea.
Footnotes
Published ahead of print 28 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01150-13.
REFERENCES
- 1.Ang-Lee MK, Moss J, Yuan CS. 2001. Herbal medicines and perioperative care. JAMA 286:208–216 [DOI] [PubMed] [Google Scholar]
- 2.Hu C, Wei H, Kong H, Bouwman J, Gonzalez-Covarrubias V, van der Heijden R, Reijmers TH, Bao X, Verheij ER, Hankemeier T, Xu G, van der Greef J, Wang M. 2011. Linking biological activity with herbal constituents by systems biology-based approaches: effects of Panax ginseng in type 2 diabetic Goto-Kakizaki rats. Mol. Biosyst. 7:3094–3103 [DOI] [PubMed] [Google Scholar]
- 3.Attele AS, Wu JA, Yuan CS. 1999. Ginseng pharmacology: multiple constituents and multiple actions. Biochem. Pharmacol. 58:1685–1693 [DOI] [PubMed] [Google Scholar]
- 4.Christensen LP. 2009. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv. Food Nutr. Res. 55:1–99 [DOI] [PubMed] [Google Scholar]
- 5.Jia L, Zhao Y, Liang XJ. 2009. Current evaluation of the millennium phytomedicine- ginseng II: collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Curr. Med. Chem. 16:2924–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SK, Park JH. 2011. Trends in ginseng research in 2010. J. Ginseng. Res. 35:389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun LQ. 2004. Information on research and application of ginseng, the king of traditional and herbal medicines. Asian J. Drug Metab. Pharmacokinet. 4:261–284 [Google Scholar]
- 8.Lee JH, Hyun Kim YJDH. 2011. Cloning and characterization of alpha-L-arabinofuranosidase and bifunctional alpha-L-arabinopyranosidase/beta-D-galactopyranosidase from Bifidobacterium longum H-1. J. Appl. Microbiol. 111:1097–1107 [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Jin Y, Lim W, Ji S, Choi S, Jang S, Lee S. 2003. A ginsenoside-Rh1, a component of ginseng saponin, activates estrogen receptor in human breast carcinoma MCF-7 cells. J. Steroid. Biochem. Mol. Biol. 84:463–468 [DOI] [PubMed] [Google Scholar]
- 10.Park EK, Choo MK, Han MJ, Kim DH. 2004. Ginsenoside Rh1 possesses antiallergic and anti-inflammatory activities. Int. Arch. Allergy Immunol. 133:113–120 [DOI] [PubMed] [Google Scholar]
- 11.Wang YZ, Chen J, Chu SF, Wang YS, Wang XY, Chen NH, Zhang JT. 2009. Improvement of memory in mice and increase of hippocampal excitability in rats by ginsenoside Rg1's metabolites ginsenoside Rh1 and protopanaxatriol. J. Pharmacol. Sci. 109:504–510 [DOI] [PubMed] [Google Scholar]
- 12.Yoon JH, Choi YJ, Lee SG. 2012. Ginsenoside Rh1 suppresses matrix metalloproteinase-1 expression through inhibition of activator protein-1 and mitogen-activated protein kinase signaling pathway in human hepatocellular carcinoma cells. Eur. J. Pharmacol. 679:24–33 [DOI] [PubMed] [Google Scholar]
- 13.Li GX, Liu ZQ. 2008. The protective effects of ginsenosides on human erythrocytes against hemin-induced hemolysis. Food. Chem. Toxicol. 46:886–892 [DOI] [PubMed] [Google Scholar]
- 14.Liu ZQ, Luo XY, Sun YX, Chen YP, Wang ZC. 2002. Can ginsenosides protect human erythrocytes against free-radical-induced hemolysis? Biochim. Biophys. Acta 1572:58–66 [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Hu S, Song X. 2007. Adjuvant effects of protopanaxadiol and protopanaxatriol saponins from ginseng roots on the immune responses to ovalbumin in mice. Vaccine 25:1114–1120 [DOI] [PubMed] [Google Scholar]
- 16.Liu ZQ, Luo XY, Liu GZ, Chen YP, Wang ZC, Sun YX. 2003. In vitro study of the relationship between the structure of ginsenoside and its antioxidative or prooxidative activity in free radical induced hemolysis of human erythrocytes. J. Agric. Food. Chem. 51:2555–2558 [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Liu A, Zhou Y, San X, Jin T, Jin Y. 2008. Panax ginseng ginsenoside-Rg2 protects memory impairment via anti-apoptosis in a rat model with vascular dementia. J. Ethnopharmacol. 115:441–448 [DOI] [PubMed] [Google Scholar]
- 18.Choi KT. 2008. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C. A. Meyer. Acta Pharmacol. Sin. 29:1109–1118 [DOI] [PubMed] [Google Scholar]
- 19.Wang CZ, Aung HH, Ni M, Wu JA, Tong R, Wicks S, He TC, Yuan CS. 2007. Red American ginseng: ginsenoside constituents and antiproliferative activities of heat-processed Panax quinquefolius roots. Planta Med. 73:669–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim WY, Kim JM, Han SB, Lee SK, Kim ND, Park MK, Kim CK, Park JH. 2000. Steaming of ginseng at high temperature enhances biological activity. J. Nat. Prod. 63:1702–1704 [DOI] [PubMed] [Google Scholar]
- 21.Bae EA, Han MJ, Kim EJ, Kim DH. 2004. Transformation of ginseng saponins to ginsenoside Rh2 by acids and human intestinal bacteria and biological activities of their transformants. Arch. Pharm. Res. 27:61–67 [DOI] [PubMed] [Google Scholar]
- 22.Yun TK, Lee YS, Lee YH, Kim SI, Yun HY. 2001. Anticarcinogenic effect of Panax ginseng CA Meyer and identification of active compounds. J. Korean Med. Sci. 16(Suppl.:S6–S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.An DS, Cui CH, Lee HG, Wang L, Kim SC, Lee ST, Jin F, Yu H, Chin YW, Lee HK, Im WT, Kim SG. 2010. Identification and characterization of a novel Terrabacter ginsenosidimutans sp. nov. beta-glucosidase that transforms ginsenoside Rb1 into the rare gypenosides XVII and LXXV. Appl. Environ. Microbiol. 76:5827–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui CH, Kim SC, Im WT. 2013. Characterization of the ginsenoside-transforming recombinant beta-glucosidase from Actinosynnema mirum and bioconversion of major ginsenosides into minor ginsenosides. Appl. Microbiol. Biotechnol. 97:649–659 [DOI] [PubMed] [Google Scholar]
- 25.Hong H, Cui CH, Kim JK, Jin FX, Kim SC, Im WT. 2012. Enzymatic biotransformation of ginsenoside Rb1 and gypenoside XVII into ginsenosides Rd and F2 by recombinant β-glucosidase from Flavobacterium johnsoniae. J. Ginseng Res. 36:418–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JK, Cui CH, Yoon MH, Kim SC, Im WT. 2012. Bioconversion of major ginsenosides Rg1 to minor ginsenoside F1 using novel recombinant ginsenoside hydrolyzing glycosidase cloned from Sanguibacter keddieii and enzyme characterization. J. Biotechnol. 161:294–301 [DOI] [PubMed] [Google Scholar]
- 27.Kim SJ, Lee CM, Kim MY, Yeo YS, Yoon SH, Kang HC, Koo BS. 2007. Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J. Microbiol. Biotechnol. 17:905–912 [PubMed] [Google Scholar]
- 28.Ko SR, Suzuki Y, Choi KJ, Kim YH. 2000. Enzymatic preparation of genuine prosapogenin, 20(S)-ginsenoside Rh1, from ginsenosides Re and Rg1. Biosci. Biotechnol. Biochem. 64:2739–2743 [DOI] [PubMed] [Google Scholar]
- 29.Noh KH, Son JW, Kim HJ, Oh DK. 2009. Ginsenoside compound K production from ginseng root extract by a thermostable beta-glycosidase from Sulfolobus solfataricus. Biosci. Biotechnol. Biochem. 73:316–321 [DOI] [PubMed] [Google Scholar]
- 30.Quan LH, Jin Y, Wang C, Min JW, Kim YJ, Yang DC. 2012. Enzymatic transformation of the major ginsenoside Rb2 to minor compound Y and compound K by a ginsenoside-hydrolyzing beta-glycosidase from Microbacterium esteraromaticum. J. Ind. Microbiol. Biotechnol. 39:1557–1562 [DOI] [PubMed] [Google Scholar]
- 31.Quan LH, Min JW, Jin Y, Wang C, Kim YJ, Yang DC. 2012. Enzymatic biotransformation of ginsenoside Rb1 to compound K by recombinant beta-glucosidase from Microbacterium esteraromaticum. J. Agric. Food. Chem. 60:3776–3781 [DOI] [PubMed] [Google Scholar]
- 32.Quan LH, Min JW, Sathiyamoorthy S, Yang DU, Kim YJ, Yang DC. 2012. Biotransformation of ginsenosides Re and Rg1 into ginsenosides Rg2 and Rh1 by recombinant beta-glucosidase. Biotechnol. Lett. 34:913–917 [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Liu QM, Sung BH, An DS, Lee HG, Kim SG, Kim SC, Lee ST, Im WT. 2011. Bioconversion of ginsenosides Rb1, Rb2, Rc and Rd by novel beta-glucosidase hydrolyzing outer 3-O glycoside from Sphingomonas sp. 2F2: cloning, expression, and enzyme characterization. J. Biotechnol. 156:125–133 [DOI] [PubMed] [Google Scholar]
- 34.Wan JB, Zhang QW, Ye WC, Wang YT. 2008. Quantification and separation of protopanaxatriol and protopanaxadiol type saponins from Panax notoginseng with macroporous resins. Sep. Purif. Technol. 60:198–205 [Google Scholar]
- 35.Zhao Y, Chen B, Yao SZ. 2007. Separation of 20(S)-protopanaxdiol type ginsenosides and 20(S)-protopanaxtriol type ginsenosides with the help of macroporous resin adsorption and microwave assisted desorption. Sep. Purif. Technol. 52:533–538 [Google Scholar]
- 36.Bae EA, Shin JE, Kim DH. 2005. Metabolism of ginsenoside Re by human intestinal microflora and its estrogenic effect. Biol. Pharm. Bull. 28:1903–1908 [DOI] [PubMed] [Google Scholar]
- 37.Ko SR, Choi KJ, Uchida K, Suzuki Y. 2003. Enzymatic preparation of ginsenosides Rg2, Rh1, and F1 from protopanaxatriol-type ginseng saponin mixture. Planta Med. 69:285–286 [DOI] [PubMed] [Google Scholar]
- 38.Buck JD. 1982. Nonstaining KOH method for determination of gram reactions of marine bacteria. Appl. Environ. Microbiol. 44:992–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smibert RM, Krieg NR. 1994. Phenotypic characterization. Methods for general and molecular bacteriology, p 607–654 In Gerhardt P, Murray RGE, Wood WA, Krieg NR. (ed), Methods for general and molecular microbiology. ASM Press, Washington, DC [Google Scholar]
- 40.Collins MD. 1985. Analysis of isoprenoid quinones. Methods Microbiol. 18:329–366 [Google Scholar]
- 41.Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. Technical note 101. Microbial ID, Newark, DE [Google Scholar]
- 42.Im WT, Kim SY, Liu QM, Yang JE, Lee ST, Yi TH. 2010. Nocardioides ginsengisegetis sp. nov., isolated from soil of a ginseng field. J. Microbiol. 48:623–628 [DOI] [PubMed] [Google Scholar]
- 43.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. (Oxford) 41:95–98 [Google Scholar]
- 45.Kimura M. 1983. The neutral theory of molecular evolution. Cambridge University Press, New York, NY [Google Scholar]
- 46.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 48.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 49.Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167 [Google Scholar]
- 50.Cleland WW. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103–138 [DOI] [PubMed] [Google Scholar]
- 51.Riesenberg D, Menzel K, Schulz V, Schumann K, Veith G, Zuber G, Knorre WA. 1990. High cell density fermentation of recombinant Escherichia coli expressing human interferon alpha 1. Appl. Microbiol. Biotechnol. 34:77–82 [DOI] [PubMed] [Google Scholar]
- 52.An DS, Cui CH, Sung BH, Yang HC, Kim SC, Lee ST, Im WT, Kim SG. 2012. Characterization of a novel ginsenoside-hydrolyzing alpha-L-arabinofuranosidase, AbfA, from Rhodanobacter ginsenosidimutans Gsoil 3054T. Appl. Microbiol. Biotechnol. 94:673–682 [DOI] [PubMed] [Google Scholar]
- 53.Shipkowski S, Brenchley JE. 2005. Characterization of an unusual cold-active beta-glucosidase belonging to family 3 of the glycoside hydrolases from the psychrophilic isolate Paenibacillus sp. strain C7. Appl. Environ. Microbiol. 71:4225–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harvey AJ, Hrmova M, De Gori Varghese NJ, Fincher GB. 2000. Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 41:257–269 [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto W, Miki H, Nankai H, Sato N, Kawai S, Murata K. 1998. Molecular cloning of two genes for beta-D-glucosidase in Bacillus sp. GL1 and identification of one as a gellan-degrading enzyme. Arch. Biochem. Biophys. 360:1–9 [DOI] [PubMed] [Google Scholar]
- 56.Ruan CC, Zhang H, Zhang LX, Liu Z, Sun GZ, Lei J, Qin YX, Zheng YN, Li X, Pan HY. 2009. Biotransformation of ginsenoside Rf to Rh1 by recombinant beta-glucosidase. Molecules 14:2043–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quan LH, Min JW, Yang DU, Kim YJ, Yang DC. 2012. Enzymatic biotransformation of ginsenoside Rb1 to 20S-Rg3 by recombinant beta-glucosidase from Microbacterium esteraromaticum. Appl. Microbiol. Biotechnol. 94:377–384 [DOI] [PubMed] [Google Scholar]
- 58.Korea Food & Drug Administration (KFDA) 2011. Production record of health functional food and analysis result of that in 2011, Seoul, South Korea [Google Scholar]
- 59.Chi H, Ji GE. 2005. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol. Lett. 27:765–771 [DOI] [PubMed] [Google Scholar]
- 60.An DS, Wang L, Kim MS, Bae HM, Lee ST, Im WT. 2011. Solirubrobacter ginsenosidimutans sp. nov, isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 61:2606–2609 [DOI] [PubMed] [Google Scholar]
- 61.Wang L, An DS, Kim SG, Jin FX, Kim SC, Lee ST, Im WT. 2012. Ramlibacter ginsenosidimutans sp nov, with ginsenoside-converting activity. J. Microbiol. Biotechnol. 22:311–315 [DOI] [PubMed] [Google Scholar]
- 62.Shibata S, Fujita M, Itokawa H, Tanaka O, Ishii T. 1963. Studies on the constituents of Japanese and Chinese crude drugs XI. Panaxadiol, a sapogenin of ginseng roots. Chem. Pharm. Bull. Tokyo 11:759–761 [DOI] [PubMed] [Google Scholar]
- 63.Shibata S, Tanaka O, Soma K, Ando T, Iida Y, Nakamura H. 1965. Studies on saponins and sapogenins of ginseng. The structure of panaxatriol. Tetrahedron Lett. 42:207–213 [DOI] [PubMed] [Google Scholar]
- 64.Leung KW, Wong AS. 2010. Pharmacology of ginsenosides: a literature review. Chin. Med. 5:20–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.