Abstract
Resistance of Acinetobacter baumannii clinical isolates to carbapenems is on the rise worldwide mainly in association with the production of OXA-23. Until recently, however, OXA-23 was absent in Spain. In this work, we report the molecular characterization of a hospital outbreak of OXA-23-producing A. baumannii in Barcelona caused by a multidrug-resistant (MDR) clone belonging to international clone IC-II/sequence type ST85 between October 2010 and May 2011. blaOXA-23 was carried in a plasmid of 90 kb and located within the composite transposon Tn2006.
TEXT
Acinetobacter baumannii is considered an important opportunistic pathogen causing pneumonia, bacteremia, and other respiratory and urinary tract infections (1, 2), and it is also responsible for nosocomial outbreaks worldwide, especially in intensive care units (ICUs) (3). Eradication of nosocomial infections caused by A. baumannii is particularly troublesome due to the ability of this bacterium to rapidly acquire antimicrobial resistance (4) and persist in the environment for long periods of time (5). Increased resistance to carbapenems in A. baumannii has raised special concerns during the last decade, especially since it is associated mostly with the production of acquired carbapenemases belonging to either class B metallo-β-lactamases or carbapenem-hydrolyzing OXA-type class D β-lactamases (2).
Outbreak multidrug-resistant (MDR) isolates causing hospital infections across Europe usually belong to one of the three main international clones or lineages (IC-I, IC-II, and IC-III) (6), with recent studies reporting the spread of genetically related epidemic clones of A. baumannii producing OXA-23 and belonging to IC-II within the Mediterranean region (7–11). Until very recently, however, OXA-23-producing A. baumannii was absent in Spain (12). In this work, we report the molecular characterization of an OXA-23-producing A. baumannii strain causing a hospital outbreak in Barcelona between October 2010 and May 2011.
(Some of this work was presented at the 23rd European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], 27 to 30 April 2013, Berlin, Germany.)
In October 2010, the first carbapenem-resistant A. baumannii isolate associated with this outbreak was identified by Vitek (bioMérieux, Marcy l'Etoile, France) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics GmbH, Leipzig, Germany) (13) from a bronchial aspirate from a 36-year-old male patient admitted to the ICU of the Hospital del Mar, Barcelona, Spain, because of cardiac arrest due to cocaine overdose. The patient had a record of multiple psychiatric stays during the previous 2 years and was already colonized upon ICU admission. Until May 2011, subsequent screenings recovered carbapenem-resistant A. baumannii isolates from 17 different patients and 3 environmental dry surfaces within the ICU. In 10 of the 17 patients (59%), A. baumannii was recovered from rectal samples and its presence was considered colonization, whereas in the remaining patients (41%) it was considered infection and was recovered from different sites/sources (bronchial aspirates, blood culture, urine, or samples from skin and soft tissue infections). The implementation of infection control measures, such as reinforcement of hand hygiene, environmental and equipment cleaning, and cohort isolation of patients, led to outbreak containment, and no additional isolates were recovered after May 2011.
Antimicrobial susceptibility testing performed by Vitek (bioMérieux, Marcy l'Etoile, France) and Etest (AB bioMérieux, Solna, Sweden) interpreted according to CLSI guidelines (14) showed all isolates to be resistant to ceftazidime, cefotaxime, (MIC > 256 μg/ml), cefepime, imipenem, meropenem, ciprofloxacin (MIC > 32 μg/ml), and amikacin (MIC > 64 μg/ml), intermediate to tetracycline (MIC, 8 μg/ml), and susceptible to tobramycin (MIC, 1 μg/ml), gentamicin (MIC, 0.38 μg/ml), and colistin (MIC, 0.125 μg/ml). MIC values of kanamycin, aztreonam and chloramphenicol were >256 μg/ml, and those of tigecycline were 1 to 2 μg/ml.
The analysis of the isolates by pulsed-field gel electrophoresis (PFGE) profiles of ApaI-digested genomic DNA (15) revealed that all isolates were clonally related (Fig. 1). Multilocus sequence typing (MLST) following the Pasteur scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html) assigned all isolates to sequence type ST85, and multiplex PCR to identify clonal lineages was positive for group 1 (IC-II) (16). Additional typing following the 3ST scheme (http://www.hpa-bioinformatics.org.uk/AB/ab_type1.php) provided allele sequences matching those of ST85 isolates from France (allele sequence numbers for ompA, csuE, and blaOXA-51 being 8, 11, and 10, respectively) (17).
Fig 1.
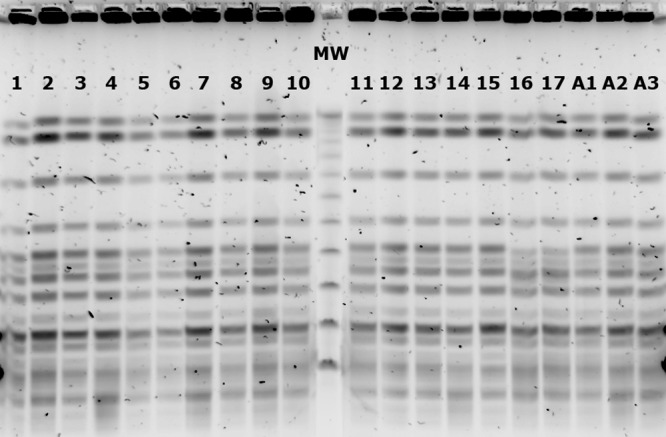
ApaI PFGE patterns of OXA-23-producing A. baumannii isolates. Lanes 1 to 17, isolates recovered from patients; lanes A1 to A3, environmental isolates; MW, lambda ladder PFGE molecular weight marker.
PCR detection of metallo- and carbapenem-hydrolyzing OXA-type β-lactamase genes (18–20) was positive only for blaOXA-51 and blaOXA-23. The insertion sequence ISAba1 was found upstream of both blaOXA-23 and the chromosomal blaADC gene but not upstream of blaOXA-51. DNA sequencing confirmed that the blaOXA-23 sequences displayed 100% identity to that of the original blaOXA-23 (GenBank accession number AJ132105.1). S1 nuclease digestion followed by pulsed-field gel electrophoresis and Southern hybridization with a digoxigenin (DIG)-labeled probe against the blaOXA-23 gene demonstrated the genetic location of blaOXA-23 within a plasmid of circa 90 kb. Plasmid replicon typing performed according to the scheme devised by Bertini et al. (21) was positive for the repAci6 gene, in agreement with previous studies showing a positive correlation between the presence of this replicon group (GR6) and blaOXA-23 carriage (22). Further characterization of the genetic environment of blaOXA-23 by inverse PCR (12) revealed its inclusion within a canonical Tn2006 composite transposon (23).
PCR amplification and sequencing of the quinolone resistance-determining region (QRDR) also showed missense mutations in both the gyrA (Ser83→Leu) and parC (Ser80→Leu) genes, concomitant with high-level resistance to ciprofloxacin, as previously described (24). PCR detection of aminoglycoside-modifying genes and 16S rRNA methylases was positive only for aphA6, which is commonly associated with high-level resistance to amikacin (25).
Plasmid transfer to a rifampin-resistant A. baumannii ATCC 17978 strain was successfully achieved by both transformation (26) and conjugation (27) upon selection on LB agar plates containing 8 μg/ml imipenem and 75 μg/ml rifampin. Selected colonies were positive for both blaOXA-23 and aphA6, showing coexistence of the two genes in the same plasmid, as well as for the repAci6 gene. Acquisition of the plasmid containing the blaOXA-23 and aphA6 genes turned the ATCC 17978 strain resistant to carbapenems (imipenem and meropenem MICs increased from 0.38 and 0.5 μg/ml, respectively, to >32 μg/ml), kanamycin (MIC increased from 6 μg/ml to >256 μg/ml), and amikacin (MIC increased from 3 μg/ml to 48 μg/ml).
Although outbreak MDR A. baumannii isolates are usually included within one of the three major international clones (IC-I, IC-II, and IC-III), published data seem to indicate that isolates belonging to IC-II possess a considerable ability for epidemic clonal spread (28). During the last decade, genetically related OXA-23-producing A. baumannii isolates belonging to IC-II have also been shown to disseminate within the Mediterranean region and other European countries, with a high percentage of isolates also being included within the widespread clonal complex (CC) CC92/CC2 (7, 9–11, 29–32). It has been suggested that such isolates are progressively replacing previously predominant clones carrying alternative OXA-type genes (7, 9, 10, 29). Until recently, however, carbapenem-resistant A. baumannii in Spain was mostly attributed to OXA-40 and, to a lesser extent, OXA-58 (33), whereas the presence of OXA-23-producing A. baumannii was limited to a single IC-II/ST2 isolate described in Mallorca and most likely imported from Portugal (12).
In the present study, we have described an outbreak caused by a single A. baumannii MDR clone producing OXA-23 and belonging to IC-II that rapidly disseminated among inpatients and environmental samples within one hospital in Barcelona. MLST typing assigned this clone to ST85, which is unrelated to CC92/CC2 but included in a different CC together with ST6 (17) and, therefore, cannot be associated with the OXA-23-producing ST2 epidemic clones emerging in Portugal, Italy, or Greece. ST85 has previously been linked to NDM-1 (34) and OXA-23 in France (35), the latter enzyme being responsible for a small outbreak in Marseille, but it has also been reported in carbapenem-susceptible isolates in Greece (17).
Of note, the composite transposon Tn2006, characteristically harboring the blaOXA-23 gene in A. baumannii isolates belonging to CC92/CC2, has consistently been found in the chromosome, where it might be contained within AbaR4-like resistance islands (8, 10, 12, 23, 31), although IC-II isolates bearing both a chromosomal copy and a plasmid-encoded copy and IC-I isolates containing only a plasmid-encoded OXA-23 have also been described (10, 23). The presence of a plasmid-carried Tn2006 in the present study reflects the potential dissemination of blaOXA-23 into epidemic clones other than those within CC92/CC2.
These findings reveal that the emergence of blaOXA-23-positive isolates in the Mediterranean region could be attributed to the horizontal mobilization of Tn2006 among several genetically related outbreak MDR clones belonging to IC-II with an increased capability to disseminate and persist within the hospital setting. The identification of such outbreak lineages in Spain might represent the final expansion of OXA-23 throughout the Mediterranean region and, perhaps, the end of OXA-40 hegemony within the Iberian Peninsula as well.
ACKNOWLEDGMENTS
This study was supported by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, cofinanced by European Regional Development Fund (ERDF) “A Way to Achieve Europe,” the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015), and the Spanish Ministry of Health (grant number FIS 11/02024). This study was also supported by grant 2009 SGR 1256 from the Departament d'Universitats, Recerca i Societat de la Informació, of the Generalitat de Catalunya and by funding from the European Community (Saturn, contract HEALTH-F3-2009241796, and AntiPathoGN, contract HEALTH-F3-2008-223101).
Footnotes
Published ahead of print 22 July 2013
REFERENCES
- 1.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/journal.pone.0022674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Espinal P, Vila-Farrés X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durante-Mangoni E, Zarrilli R. 2011. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6:407–422 [DOI] [PubMed] [Google Scholar]
- 4.Zhao WH, Hu ZQ. 2012. Acinetobacter: a potential reservoir and dispenser for β-lactamases. Crit. Rev. Microbiol. 38:30–51 [DOI] [PubMed] [Google Scholar]
- 5.Espinal P, Martí S, Vila J. 2012. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J. Hosp. Infect. 80:56–60 [DOI] [PubMed] [Google Scholar]
- 6.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Arezzo S, Principe L, Capone A, Petrosillo N, Petrucca A, Visca P. 2011. Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J. Antimicrob. Chemother. 66:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grosso F, Quinteira S, Peixe L. 2011. Understanding the dynamics of imipenem-resistant Acinetobacter baumannii lineages within Portugal. Clin. Microbiol. Infect. 17:1275–1279 [DOI] [PubMed] [Google Scholar]
- 9.Liakopoulos A, Miriagou V, Katsifas EA, Karagouni AD, Daikos GL, Tzouvelekis LS, Petinaki E. 2012. Identification of OXA-23-producing Acinetobacter baumannii in Greece, 2010 to 2011. Euro Surveill. 17:pii:20117 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20117 [PubMed] [Google Scholar]
- 10.Minandri F, D'Arezzo S, Antunes LC, Pourcel C, Principe L, Petrosillo N, Visca P. 2012. Evidence of diversity among epidemiologically related carbapenemase-producing Acinetobacter baumannii strains belonging to international clonal lineage II. J. Clin. Microbiol. 50:590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Touati M, Diene SM, Racherache A, Dekhil M, Djahoudi A, Rolain JM. 2012. Emergence of blaOXA-23 and blaOXA-58 carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii isolates from University Hospital of Annaba, Algeria. Int. J. Antimicrob. Agents 40:89–91 [DOI] [PubMed] [Google Scholar]
- 12.Espinal P, Macia MD, Roca I, Gato E, Ruiz E, Fernandez-Cuenca F, Oliver A, Rodriguez-Bano J, Bou G, Tomas M, Vila J. 2013. First report of an OXA-23 carbapenemase-producing Acinetobacter baumannii clinical isolate related to Tn2006 in Spain. Antimicrob. Agents Chemother. 57:589–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin. Microbiol. Infect. 18:1097–1103 [DOI] [PubMed] [Google Scholar]
- 14.CLSI 2013. Performance standards for antimicrobial susceptibility testing, 23rd informational supplement, M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815 [DOI] [PubMed] [Google Scholar]
- 17.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, Zarrilli R, Tsakris A. 2011. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000–09). J. Antimicrob. Chemother. 66:2767–2772 [DOI] [PubMed] [Google Scholar]
- 18.Higgins PG, Lehmann M, Seifert H. 2010. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 35:305. [DOI] [PubMed] [Google Scholar]
- 19.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 20.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]
- 21.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:4168–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Towner KJ, Evans B, Villa L, Levi K, Hamouda A, Amyes SG, Carattoli A. 2011. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D β-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:2154–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vila J, Ruíz J, Goñi P, Jiménez de Anta T. 1997. Quinolone-resistance mutations in the topoisomerase IV parC gene of Acinetobacter baumannii. J. Antimicrob. Chemother. 39:757–762 [DOI] [PubMed] [Google Scholar]
- 25.Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233–1240 [DOI] [PubMed] [Google Scholar]
- 26.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 27.Poirel L, Guibert M, Bellais S, Naas T, Nordmann P. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karah N, Sundsfjord A, Towner K, Samuelsen O. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247 [DOI] [PubMed] [Google Scholar]
- 29.Manageiro V, Jones-Dias D, Ferreira E, Louro D, Canica M. 2012. Genetic diversity and clonal evolution of carbapenem-resistant Acinetobacter baumannii isolates from Portugal and the dissemination of ST118. Int. J. Antimicrob. Agents 40:398–403 [DOI] [PubMed] [Google Scholar]
- 30.Mezzatesta ML, D'Andrea MM, Migliavacca R, Giani T, Gona F, Nucleo E, Fugazza G, Pagani L, Rossolini GM, Stefani S. 2012. Epidemiological characterization and distribution of carbapenem-resistant Acinetobacter baumannii clinical isolates in Italy. Clin. Microbiol. Infect. 18:160–166 [DOI] [PubMed] [Google Scholar]
- 31.Saule M, Samuelsen O, Dumpis U, Sundsfjord A, Karlsone A, Balode A, Miklasevics E, Karah N. 2013. Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob. Agents Chemother. 57:1069–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schleicher X, Higgins PG, Wisplinghoff H, Korber-Irrgang B, Kresken M, Seifert H. 2013. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005–2009). Clin. Microbiol. Infect. 19:737–742 [DOI] [PubMed] [Google Scholar]
- 33.Villalón P, Valdezate S, Medina-Pascual MJ, Carrasco G, Vindel A, Saez-Nieto JA. 2013. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenem-hydrolysing oxacillinase and metallo-β-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J. Antimicrob. Chemother. 68:550–553 [DOI] [PubMed] [Google Scholar]
- 34.Bonnin RA, Poirel L, Naas T, Pirs M, Seme K, Schrenzel J, Nordmann P. 2012. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 18:E362–E365 [DOI] [PubMed] [Google Scholar]
- 35.Kempf M, Rolain JM, Azza S, Diene S, Joly-Guillou ML, Dubourg G, Colson P, Papazian L, Richet H, Fournier PE, Ribeiro A, Raoult D. 2013. Investigation of Acinetobacter baumannii resistance to carbapenems in Marseille hospitals, south of France: a transition from an epidemic to an endemic situation. APMIS 121:64–71 [DOI] [PubMed] [Google Scholar]


