Abstract
Succinoyl trehalose lipids (STLs) are promising glycolipid biosurfactants produced from n-alkanes that are secreted by Rhodococcus species bacteria. These compounds not only exhibit unique interfacial properties but also demonstrate versatile biochemical actions. In this study, three novel types of genes involved in the biosynthesis of STLs, including a putative acyl coenzyme A (acyl-CoA) transferase (tlsA), fructose-bisphosphate aldolase (fda), and alkane monooxygenase (alkB), were identified. The predicted functions of these genes indicate that alkane metabolism, sugar synthesis, and the addition of acyl groups are important for the biosynthesis of STLs. Based on these results, we propose a biosynthesis pathway for STLs from alkanes in Rhodococcus sp. strain SD-74. By overexpressing tlsA, we achieved a 2-fold increase in the production of STLs. This study advances our understanding of bacterial glycolipid production in Rhodococcus species.
INTRODUCTION
Biosurfactants (BSs) are surface-active compounds that are abundantly produced by a number of microorganisms. BSs are broadly classified into four types according to their structures: glycolipid, lipopeptide, high-molecular-weight polymer, and organic acid. Specific examples include rhamnolipids, which are glycolipid-type BSs produced by Pseudomonas aeruginosa, and surfactins, which are lipopeptide-type BSs produced by Bacillus subtilis (1). BSs have received increased attention due to their unique properties, which include higher biodegradability, lower toxicity, and more-versatile biological functions than are typical of synthetic surfactants (1–3). Glycolipid-type BSs have been especially well studied, as microorganisms that produce glycolipid-type BSs are known to have higher productivity than other types of BS-producing microorganisms.
Along with functional analyses of glycolipid-type BSs, their biosynthetic pathways have also been studied. Through the isolation of mutants and through genetic approaches, a clear understanding has emerged in studies of Pseudomonas bacteria (4). In P. aeruginosa, it is known that rhamnolipid is synthesized from dTDP-l-rhamnose and β-hydroxyalkanoyl-β-hydroxyalkanoyl (HAA)-acyl carrier protein (ACP) or -coenzyme A (CoA) (5, 6). The key genes of rhamnolipid synthesis are rhlA and rhlB, which catalyze the synthesis of HAA-ACP or -CoA and the formation of monorhamnolipid, respectively (5). Several types of glycolipids are produced by yeasts. Mannosylerythritol lipids (MELs), which are produced by Pseudozyma and Ustilago species, are representative BSs produced by yeast (7, 8). A number of essential genes for MEL synthesis have been discovered. The genes encoding erythritol/mannosyl transferase, which catalyzes the synthesis of the hydrophilic sugar moiety, and acyl-transferase, which transfers a hydrophilic acyl group to mannosylerythritol, are key genes for MEL biosynthesis (9, 10). As indicated above, it is known that the synthesis of the sugar moiety and the synthesis and transfer of the acyl chain are important reactions for glycolipid-type BS biosynthesis.
Trehalose lipids are glycolipid-type BSs that are primarily produced by bacterial Rhodococcus species. Recent studies revealed that trehalose lipids have many bioactivities, such as antitumor, antiviral and antimicrobial activities (11). Therefore, commercial production of trehalose lipids is anticipated. However, the biosynthesis pathway of trehalose lipids in Rhodococcus species is poorly understood. Succinoyl trehalose lipids (STLs), which are efficiently produced from n-alkanes, represent one promising type of trehalose lipids secreted by Rhodococcus sp. strain SD-74. These STLs are also recovered via precipitation (12). This bacterial strain primarily produces two types of STL homologues, STL-1 and STL-2, in addition to other minor components (13). Interestingly, the hydrophobic acyl groups of all STLs have the same carbon chain length as the n-alkane used as the substrate. In other words, SD-74 can produce STLs with carbon chains of a specific length with both defined unsaturated bonds and a defined substituent group. This property is highly useful for the commercial production of BSs, as it is difficult to produce specifically defined derivatives in most BS producers. STLs have one or two succinic acids and two fatty acids attached to a trehalose moiety, thereby allowing STLs to behave as an anionic surfactant depending on the pH of the aqueous solution. Ishigami et al. previously reported that a mixture of STLs exhibited potential emulsifying and dispersing activities that reflected their unique hydrophobic structures (14). Furthermore, we previously demonstrated that purified STL-1 has the highest surface activity among any reported sugar-based surfactants (15). These STLs were also shown to inhibit growth of the influenza virus and to induce cell differentiation (16, 17). In addition, among all tested trehalose lipid producers, Rhodococcus sp. SD-74 gave the best trehalose lipid yield (40 g/liter) from n-hexadecane under highly osmotic conditions (18). Despite these attractive properties, very little is known regarding whether the biosynthetic pathway of STLs is similar to that of other trehalose lipids produced by Rhodococcus species. Here, we identify the genes required for the production of STLs using a novel approach, i.e., the transposome mutagenesis system. The genes discovered using this approach had predicted functions in fatty acid metabolism, gluconeogenesis, and acyl group transfer, which are important processes in glycolipid production. Predicting the functions of these genes will allow us to better understand the biosynthesis pathway of STLs and the distinctive properties of SD-74. Based on the results of our study, we overexpressed STL-producing genes and successfully increased production 2-fold. This increased production rate corresponds to a 40-g/liter increase in theoretical yield. Therefore, our study will provide greater insight for studies of STLs.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Our strain of Rhodococcus sp. SD-74 was used as an STL producer in this study. Rhodococcus sp. SD-74 was grown on PMY medium (1.0% glycerol, 0.5% polypeptone, 0.3% yeast extract, 0.3% malt extract, 0.1% KH2PO4, 0.1% K2HPO4 [wt/vol, pH 7.0]) (20). FPY medium (2.0% fructose, 0.5% polypeptone, 0.5% yeast extract, 0.1% NaNO3, 0.1% KH2PO4, 0.1% MgSO4·7H2O [wt/vol, pH 7.0_) was used as the preculture medium for resting-cell conditions. The Escherichia coli strains were cultivated in LB medium. E. coli strains DH5α and JM109 were used for DNA manipulation, and BL21(DE3) was utilized for recombinant protein expression.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Rhodococcus sp. strains | ||
| SD-74 | Wild type | This study |
| 330 | STL productivity-deficient Tn mutant | This study |
| 1510 | STL productivity-deficient Tn mutant | This study |
| 1608 | STL productivity-deficient Tn mutant | This study |
| E. coli strains | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK+ mK+) supE44 λ− thi-1 gyrA relA1 | TaKaRa |
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15] | TaKaRa |
| BL21(DE3) | E. coli B, F− dcm ompT hsdSB(rB+ mB+) gal (DE3) | TaKaRa |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector, Ampr | Stratagene |
| pSMT3 | Rhodococcus-E. coli shuttle vector, Hgr | 19 |
| pSMT3-tlsA | pSMST3 derivative carrying traT | This study |
| pSMT3-fda | pSMST3 derivative carrying fda | This study |
| pSMT3-alkB | pSMST3 derivative carrying alkB | This study |
| pSMT3-traT-D191A | pSMT3 derivative carrying mutant tlsA(D191A) | This study |
| pSMT3-traT-H192A | pSMT3 derivative carrying mutant tlsA(H192A) | This study |
| pSMT3-traT-D196A | pSMT3 derivative carrying mutant tlsA(D196A) | This study |
| pSTM3-PalkB | pSMT3 derivative carrying alkB promoter | This study |
| pSMT3-PalkB::tlsA | pSMT3 derivative for traT overexpression | This study |
| pSMT3-PalkB::fda | pSMT3 derivative for fda overexpression | This study |
| pET21b | Apr, His tag, T7 promoter | Novagen |
| pET21b-fda | pET21b derivative, Fda expression vector | This study |
| pET21b-fda-His | pET21b derivative, Fda-His expression vector | This study |
Transformation of Rhodococcus sp. SD-74.
A 500-ml flask containing 100 ml PMY medium was inoculated with 1 ml of a stationary-phase culture of Rhodococcus sp. SD-74. The flask was incubated at 30°C with shaking at 200 rpm until the culture reached an optical density at 660 nm (OD660) of 0.5. The cells were pelleted via centrifugation at 3,000 rpm for 6 min at 4°C. Washes were performed three times with sterile water at 4°C. The cell pellet was diluted in 0.5 ml of sterile water, and 0.1-ml aliquots were used for electroporation.
Transposon mutagenesis of SD-74.
Electroporation was performed as previously reported (7, 21). The electrocompetent SD-74 cells were mixed with 20 ng of transposome (EZ::TN <KAN-2> Tnp Transposome kit; Epicentre, Madison, WI) and placed in a 0.1-cm electrode gap cuvette. Electroporation was performed using the following parameters: 2.0 kV, 25 μF, and 200 Ω. The cells were immediately resuspended in 1 ml of PMY medium and then incubated for 2 h at 26°C with shaking at 200 rpm. Next, the cells were overlaid onto PMY agar plates containing 20 μg/ml of kanamycin and were incubated at 30°C. Transposants appeared within 3 days.
Screening for non-STL-producing mutants.
The first screening was performed to identify mutants that did not produce STLs. The transposants were transferred to a 24-well microtiter plate (Marubishi Engineering) containing 0.9 ml of PMY medium and 0.1 ml of n-hexadecane, which was shaken at 500 rpm at 30°C for 6 days. The culture broths were collected and acidified to a pH of 3.0 with 6N HCl. The precipitated STLs were extracted with ethyl acetate and were analyzed via thin-layer chromatography (TLC) on silica plates (silica gel 60 aluminum sheets; Merck) with chloroform-methanol-water (65:25:4, vol/vol/vol). The compounds were located on the plates via charring at 110°C for 10 min after spraying with 0.1% anthrone–sulfuric acid (75% [vol/vol] sulfuric acid). The anthrone-sulfuric acid method was also used for STL quantification. During quantification of the STLs, trehalose was used for the standard compounds (22). We selected non-STL-producing mutants, which were then used for a second screening to eliminate mutants that were unable to produce STLs due to low growth. The mutants were cultivated in a 500-ml Erlenmeyer flask with FPY medium (50 ml) at 30°C with shaking at 200 rpm for 4 days. Whole cells were then washed with 0.05 M Tris buffer (pH 8.0) at 4°C and then transferred into a 500-ml Erlenmeyer flask containing 0.05 M Tris buffer (27 ml) and n-hexadecane (3 ml). For resting-cell conditions, the flasks were shaken at 200 rpm for 4 days at 30°C. After the aqueous layer was carefully collected, the production of STLs was confirmed using the methods described above.
DNA sequencing.
After cloning, the chromosomal DNA flanking the transposon insertions was sequenced. The chromosomal DNA from the mutants was extracted by using Isoplant II (Nippon Gene) following the manufacturer's instructions. The chromosomal DNA was then digested with ApaI, KpnI, or SacI (Toyobo) and was ligated into the respective pBluescript II SK(+) plasmid. The ligation mixtures were electroporated into DH5α, and the transformants were selected with kanamycin. After vector purification with the GenElute plasmid miniprep kit (Sigma), following the manufacturer's instructions, sequencing was performed using a genetic analyzer 310 (Applied Biosystems). The primers used for sequencing are shown in Table S1 in the supplemental material. The entire sequence of the open reading frame (ORF) was obtained via the primer walking method using the primers shown in Table S1.
Data analysis.
The DNA and protein homology searches were performed using NCBI BLAST (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/).
Complementation of STL-producing genes.
The chromosomal DNA of wild-type SD-74 was obtained using the method described above. For complementation of STL-producing genes, the ORFs were amplified and cloned into the Rhodococcus-E. coli shuttle vector pSMT3. The primers used for complementation are described in Table S1 in the supplemental material. The complementary and empty vectors were transformed in each STL-producing gene mutant via electroporation, and the productivity of these transformants was confirmed as described above (“Transformation of Rhodococcus sp. SD-74” and “Screening for non-STL-producing mutants”).
Construction of Fda expression plasmids.
The chromosomal DNA of wild-type SD-74 was obtained and amplified using the fda primers described in Table S1 in the supplemental material. The primer (exp-fda-Rv-His) fused a His tag-coding sequence followed by a HindIII restriction site to the fda gene. The PCR products were then cloned into a pET21b. To confirm whether the His tag affected Fda activity, a native Fda expression vector was also constructed.
Production and purification of recombinant protein from E. coli.
The production of Fda and Fda-His was performed using E. coli BL21(DE3). An overnight culture was used to inoculate 20 ml of LB medium that was supplemented with 50 μg/ml of carbenicillin. The cells were grown at 37°C, with shaking at 200 rpm, until an optical density of 0.8 at 600 nm was reached. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 0.1 mM. The cells were further grown at 20°C, with shaking at 200 rpm, for 3 h and harvested via centrifugation. The cells were resuspended in buffer I (50 mM Tris-HCl, pH 7.4, 1 mM 2-mercaptoethanol, 1 mM MgCl2) and then sonicated. The insoluble material was removed via centrifugation, and the supernatant was used for protein purification. The aldolase activity was assayed using a crude enzyme solution. Purification of the His-Fda fusion protein was performed using a MagExtractor His tag kit (Toyobo), following the manufacturer's instructions.
SDS-PAGE analysis.
The samples were treated with sodium dodecyl sulfate (SDS) loading buffer containing 5% 2-mercaptoethanol at 98°C for 3 min and then separated using polyacrylamide gel electrophoresis (PAGE) (23). After electrophoresis, the gels were stained using Bio-Safe Coomassie stain (Bio-Rad).
Aldolase activity assay.
The aldolase activity of Fda was measured in a coupled assay with fructose-1,6-bisphosphate as a substrate (24). The fructose-1,6-bisphosphates were cleaved to glyceraldehyde-3-phosphates and dihydroxyacetone phosphates by Fda or Fda-His. Next, the glyceraldehyde-3-phosphates were converted to dihydroxyacetone-phosphates by triosephosphate isomerase. As a result of this reaction, two molecules of dihydroxyacetone phosphates were produced from one molecule of fructose-1,6-bisphosphate. Thus, two molecules of dihydroxyacetone phosphates were converted to l-glycerol-3-phosphates by reacting with α-glycerolphosphate dehydrogenase. Because this reaction required NADH, the aldolase activity was measured by quantifying NADH, or the reduced absorbance at 320 nm. A unit of aldolase activity is defined as the amount of enzyme that catalyzes the aldol cleavage of 1 μmol of fructose-1,6-bisphosphate per minute.
Site-directed mutagenesis of HX3DX14Y motif of TlsA.
Site-directed mutagenesis of the HX3DX14Y motif of TlsA was performed via an overlap extension PCR method (23), using the primers described in Table S1 in the supplemental material to obtain the mutations H192A (where alanine was substituted for the histidine at position 192) and D196A within the motif and D191A just before the motif. The mutated sequences were confirmed using a genetic analyzer 310 (Applied Biosystems) and inserted into pSMT3. The constructed plasmids were transformed into strain 330, and the transformants' STL productivity was confirmed as previously described.
Total RNA extractions.
Wild-type SD-74 was grown in PMY medium or PMY medium containing 10% n-hexadecane (vol/vol) at 30°C with shaking at 200 rpm for 1 day. After centrifugation, RNA was extracted from each strain using Isogen (Nippon Gene) and then purified using RNeasy (Qiagen) following the manufacturer's instruction. The extracted RNA was treated twice with DNase according to the manufacturer's instructions (Qiagen). The RNA purity and concentration were determined using UV spectrophotometry and then visualized using agarose gel electrophoresis to assess the quality.
cDNA preparation.
To synthesize cDNA from the extracted RNA, which was treated with DNase, a reverse transcriptase (RT) reaction was performed with 1 μl of SuperScript III using random hexamers. Fifty nanograms of RNA was then added, and thus, the final volume of the reaction mixture was 20 μl.
Real-time RT-PCR analysis.
Real-time RT-PCR was performed using a LightCycler (Roche). The reaction mixture underwent 45 cycles of 95°C for 10 s, 60°C for 1 s, and 72°C for 12 s with a temperature transition rate of 20°C/s. A melting curve analysis was performed at 50°C to 95°C (temperature transition rate of 0.2°C/s), with stepwise fluorescence detection following amplification. The cycle threshold values were obtained, and the number of RNA copies per microgram of total RNA was calculated using a standard curve of known DNA amounts with target r2 coefficients larger than 0.997 in the range of 5 × 103 to 5 × 108 molecules per reaction. The 16S rRNA was used as a housekeeping gene to compare the amounts of RNA in each reaction (25). Three independent experiments were performed in duplicate for each RNA sample.
Nucleotide sequence accession numbers.
Newly determined gene sequences were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers AB847084, AB847085, and AB847086.
RESULTS
Three types of genes were essential for STL biosynthesis.
We performed a random insertion mutagenesis using a transposome system to identify the genes essential for STL production. We obtained 2,600 kanamycin-resistant mutants. In the first screening, we isolated 21 mutants that were unable to produce STLs in PMY medium that contained n-hexadecane. To exclude the mutants that were not able to produce STLs due to inability to grow in the PMY medium with n-hexadecane, we performed a second screening using a resting-cell reaction. As a result, 6 mutants were yielded that lacked STL production ability. By performing random insertion mutagenesis, in the first and second screenings, we obtained 6 mutants (strains 330, 1510, 1598, 1608, 1646, and 2120) that were not able to produce STLs in PMY medium containing n-hexadecane and under resting-cell conditions. We next identified the genes containing the Tn5 insertion through sequencing. From this sequence analysis, we identified three types of genes that were disrupted by this insertion. The mutant strains 1510 and 1598 had a Tn5 insertion in the same gene. Similarly, the mutant strains 1608, 1646, and 2120 also had the insertion in the same gene. Thus, we used mutant strains 330, 1510, and 1608 as representatives for the three types of non-STL-producing mutants (Fig. 1) to further study the genes required for production. We next addressed whether the transposon mutants' defects in STL productivity were caused by just one gene mutation. Complementation studies were performed on the predicted genes, which were rendered defective by a transposon; the STL productivity of the transformants was then verified (Fig. 1). The production of STLs was restored in all complementary strains, thereby indicating that the three types of genes were essential for this process.
Fig 1.
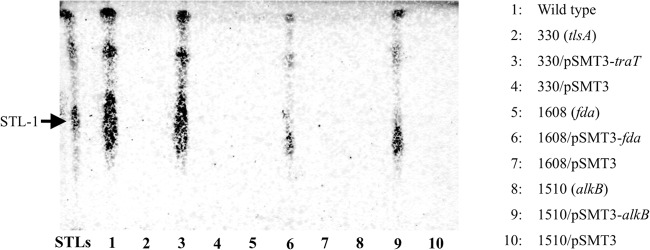
Analysis of STL production by the wild type, the non-STL-producing mutants, and the complemented strains. Ethyl acetate extracts of the resting-cell reaction solution were analyzed using thin-layer chromatography (chloroform-methanol-water, 65:25:4). The arrow indicates STL-1. STL detection was performed using the anthrone-sulfuric acid method. The wild-type strains produced STLs (lane 1). However, three types of mutant (strains 330, 1510, and 1608) lost STL production (lanes 2, 5, and 8, respectively). Complementation of the disrupted genes restored the STL productivity of the three transposon mutants (lanes 3, 6, and 9), though complementation with the empty vector did not restore STL productivity (lanes 4, 7, and 10).
Sugar synthesis involved in STL production.
Using sequencing, we next discovered an open reading frame flanking the insertion in strain 1608. We predicted through homology analysis that the 2,033-bp open reading frame was a fructose-bisphosphate aldolase gene (fda) (accession no. AB847085). The start codon of this ORF is GTG, and the predicted Shine-Dalgarno (SD) sequence (GGAGG) exists 9 bp upstream from the start codon. The amino acid sequence exhibited 76%, 71%, and 61% homology with the Fda sequences of M. tuberculosis H37Rv, Corynebacterium glutamicum, and Streptomyces glutamicum, respectively. Fructose-bisphosphate aldolase is responsible for the transformation of triose and hexose. It is assumed that strain 1608 was unable to produce STLs due to its inability to synthesize trehalose in the sugar biosynthesis pathway. Therefore, we performed complementation of the fda function via the addition of fructose. As a result, we demonstrated that STL productivity was recovered through the addition of fructose to the resting-cell medium (final concentration of 1%, wt/vol) (Fig. 2A).
Fig 2.
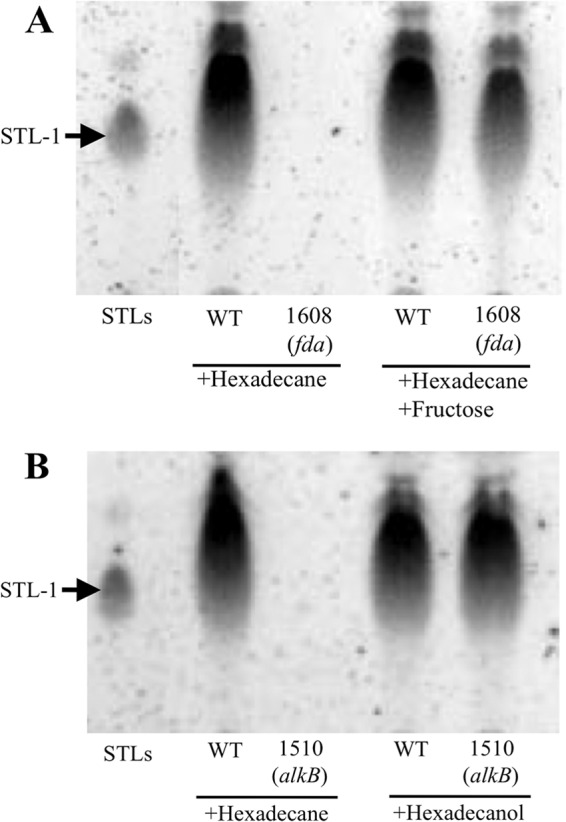
Complementation of STL productivity via the addition of the putative intermediates for STL production. STL production was analyzed via TLC using the anthrone-sulfuric acid method. The arrows indicate STL-1. (A) Additional fructose restored STL production in strain 1608 (fda mutant). (B) The STL production of strain 1510 (alkB mutant) was restored via the addition of hexadecanol.
Next, to characterize the fda encoding fructose-1,6-bisphosphate aldolase, we performed the expression and purification of Fda from Rhodococcus sp. SD-74. The expression of Fda from the SD-74 strain was performed in E. coli BL21(DE3). To confirm whether the expression of Fda and Fda-His was successful, a SDS-PAGE analysis was performed (Fig. 3). The molecular mass, which was estimated using the amino acid sequence of Fda from SD-74, was 36.7 kDa. The bands were located at approximately 36 and 38 kDa. These were the predicted sizes of Fda and Fda-His, respectively. The purification of the crude His-tagged Fda in solution was performed using the MagExtractor kit. The purified protein was confirmed via SDS-PAGE (Fig. 3), which was then used for aldolase activity analysis. The molecular mass of Fda-His was slightly higher than that of native Fda due to the fused His tag. The purified Fda-His showed aldolase activity (1,330 mU/mg protein), while the noninduced cell lysate of E. coli did not show the aldolase activity. Moreover, the crude extract of Fda-His has a lower aldolase activity (213 mU/mg protein) than purified Fda-His. The crude extract of native Fda has aldolase activity sufficiently similar (259 mU/mg protein) to that of crude Fda-His. This finding indicated that the Fda of SD-74 had aldolase activity and that the fused His tag did not affect the aldolase activity of Fda. Because Fda showed the aldolase activity and the lack of fda was restored by the addition of fructose, it is expected that Fda acts as a supplier of the trehalose moiety through gluconeogenesis from n-alkane metabolism.
Fig 3.
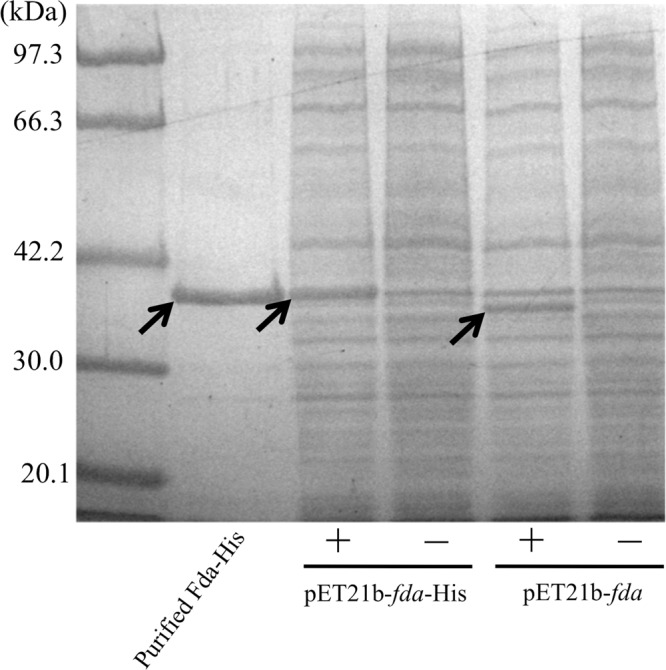
SDS-PAGE analysis of purified and crude His-tagged Fda and native Fda. The arrows indicate Fda or Fda-His. SDS-PAGE analysis was performed to confirm Fda and Fda-His expression and purification via the addition of 0.1 mM IPTG (+). −, no IPTG.
Alkane monooxygenase was required for alkane metabolism under STL-producing conditions.
We identified the open reading frame flanking the insertion of strain 1510 through sequencing and predicted that the 1,188-bp open reading frame was an alkane monooxygenase gene (alkB) (accession no. AB847086). This ORF used GTG as a start codon, and the SD sequence was predicted at 7 bp upstream from the start codon (GGAGG). We demonstrated that alkB contained a Hist-1 box [HE(L/M)XHK], a Hist-2 box [EHXXGHH], a Hist-3 box [LQRH(S/A)DHHA], and an HYG motif [NYXEHYG(L/M)]. These are strongly conserved in alkane monooxygenases and are also required for the enzyme's activity (Table 2). The amino acid sequence exhibited an 82% homology for both the alkane-1-monooxygenase (alkB1) of Rhodococcus sp. strain Q15 and the alkB1 of Rhodococcus erythropolis NRRLB-16531. We also identified, by amino acid sequence homology, three types of open reading frames that were predicted to encode rubredoxins (rubA1 and rubA2) and rubredoxin reductase (rubB). The alkB gene was located at the head of that gene cluster. The rubA1, rubA2, and rubB genes lined up downstream from the alkB in order. Moreover, the several ORFs of these genes included overlapping start and stop codons. The head-to-tail composition of the alkB gene cluster suggests that these genes may be transcribed as an operon. Generally, the n-alkane oxidation reaction requires reduced rubredoxin to oxidize n-alkane as a substrate. Therefore, we next determined that strain 1510 did not produce STLs because of its inability to convert alkane to alcohol. We cultured strain 1510 in PMY medium containing 10% hexadecanol instead of n-hexadecane under resting-cell conditions. This change resulted in the strain regaining the ability to produce STLs (Fig. 2B).
Table 2.
Amino acid alignments of selected alkane monooxygenases
| Proteina | 1st residue | Sequence (1st, 2nd, or 3rd segment)b | Last residue |
|---|---|---|---|
| AlkB1-Q15 | 131 | GVTAGIGINTAHELGHKKESVERWLSKIVLAQSAYGHFYLEHNRGHHVRVSTPEDPATSRFGETLYGFWP | 198 |
| AlkB2-Q15 | 147 | GCVAGIGINTAHELGHKKDDLERWLSKITLAQSFYGHFYIEHNRGHHVRVATPEDPASSRFGESFWTFLP | 216 |
| AlkB1-16531 | 131 | GVTAGIGINTAHELGHKKESVERWLSKIVLAQSAYGHFYLEHNRGHHVRVSTPEDPATSRFGETLYGFWP | 198 |
| AlkB2-16531 | 147 | GCVAGIGINTAHELGHKKDDLERWLSKITLAQSFYGHFYIEHNRGHHVRVATPEDPASSRFGESFWTFLP | 216 |
| AlkB-H37Rv | 150 | GVLGGVGINTAHEMGHKKDSLERWLSKITLAQTCYGHFYIEHNRGHHVRVSTPEDPASARFGETLWEFLP | 219 |
| AlkM-ADP1 | 143 | GAINGIAVNTAHELSHKADRLDHILSHLALVPTGYNHFRIEHPYGHHKRAATPEDPASSQMGETFYEFWP | 208 |
| AlkB-GPo1 | 136 | GIVNGLALNTGHELGHKKETFDRWMAKIVLAVVGYGHFFIEHNKGHHRDVATPMDPATSRMGESIYKFSI | 196 |
| AlkB-SD74 | 131 | GVTAGIGINTAHELGHKKENLERWLSKIVLAQSAYGHFYIEHNRGHHVRVSTPEDPASSRFGESFYRFWP | 198 |
| AlkB1-Q15 | 199 | RSVGGGLKSAWHLEKTRFERLGTTHWSIKN----DVLNAWLMSVVLFGVALAVFGIGIAPYLVIQAFIGF | 264 |
| AlkB2-Q15 | 217 | RSVWGSLRSSWSLEKARLDRLGKKPWT---I-RNGVLHSWLMSVVLFGVLVAVFGLSVLPFLVLQAVFGF | 282 |
| AlkB1-16531 | 199 | RSVGGGLKSAWHLEKTRFERLGTTHWSIKN----DVLNAWLMSVVLFGVALAVFGIGIAPYLVIQAFIGF | 264 |
| AlkB2-16531 | 217 | RSVWGSLRSSWSLEKARLDRLGKKPWT---I-RNDVLHSWLMSVVLFGVLVAVFGLSVLPFLVLQAVFGF | 282 |
| AlkB-H37Rv | 220 | RSVIGGLRSAVHLEAQRLRRLGVSPWNPMTYLRNDVLNAWLMSVVLWGGLIAVFGPALIPFVIIQAVFGF | 289 |
| AlkM-ADP1 | 209 | RTVFGSLKSAIEIETHRLKRKGKKFWSKDNEL----LQGWGMSAAFHSSIIAIFGKGTIPYLVTQAFYGI | 274 |
| AlkB-GPo1 | 197 | REIPGAFIRAWGLEEQRLSRRGQSVWSFDNEILQPMI----ITVILYAVLLALFGPKMLVFLPIQMAFGW | 262 |
| AlkB-SD74 | 199 | RSVAGGLKSAWHLEKARFERLGTTHWSIKN----DVLNAWLMTLVLFGAVVAAFGIGILPYVLIQAVIGF | 264 |
| AlkB1-Q15 | 265 | SLLEAINYLEHYGLLRQKTASGRYERCTPAHSWNSDRICTNVFLYHLQRHSDHHANPTRRYQALRSMEEA | 334 |
| AlkB2-Q15 | 283 | CLLETVNYLEHYGLKRRRLDSGRYERAAPEHSWNSDHICTNIFLYHLQRHSDHHANPTRRYQTLRSMDGA | 352 |
| AlkB1-16531 | 265 | SLLEAINYLEHYGLLRQKTASGRYERCTPAHSWNSDRICTNVFLYHLQRHSDHHANPTRRYQALRSMEEA | 334 |
| AlkB2-16531 | 283 | CLLETVNYLEHYGLKRRRLDSGRYERAAPEHSWNSDHICTNIFLYHLQRHSDHHANPTRRYQTLRSMDGA | 352 |
| AlkB-H37Rv | 290 | SLLEAVNYLEHYGLLRQKSANGRYERCAPVHSWNSDHIVTNLFLYHLQRHSDHHANPTRRYQTLRSMAGA | 359 |
| AlkM-ADP1 | 275 | SLFEIINYIEHYGLKRQKRADGNYERTMPEHSWNNNNIVTNLFLYQLQRHSDHHAYPTRPFQALRHFDEA | 344 |
| AlkB-GPo1 | 263 | WQLTSANYIEHYGLLRQKMEDGRYEHQKPHHSWNSNHIVSNLVLFHLQRHSDHHAHPTRSYQSLRDFPGL | 332 |
| AlkB-SD74 | 265 | SLLEAINYLEHYGLRRQKTASGRYERCTPAHSWNSDRLCTNVFLYHLQRHSDHHANPTRRYQTLRSMDQA | 334 |
AlkB-SD74, putative alkane monooxygenase of SD-74; AlkB1-16531 and AlkB2-16531, alkane monooxygenases of Rhodococcus sp. strain Q15 (31); AlkB1-Q15 and AlkB2-Q15, alkane monooxygenases of R. erythropolis NRRLB-16531 (30); AlkB-H37, putative AlkB of M. tuberculosis H37Rv (accession no. NP_217769); Rv AlkM-ADPI, alkane monooxygenase of Acinetobacter sp. ADP1 (accession no. AJ002316); AlkB-GPo1, alkane monooxygenase of Pseudomonas putida GPo1 (accession no. AJ245436).
Underlines indicate conserved Hist-1, Hist-2, and Hist-3 boxes and HYG motifs.
The Rhodococcus strains R. erythropolis NRRL B-16531 and Rhodococcus sp. Q15 have multiple genes that encode alkane oxygenases (26, 27). We therefore hypothesized that SD-74 also had multiple alkane oxygenases and that strain 1510 would grow under conditions where n-hexadecane was the only carbon source. We then cultured strain 1510 in minimal medium (0.1% NH4NO3, 0.01% MgSO4, 0.4% K2HPO4, 0.04% KH2PO4 [wt/vol]) with 2% n-hexadecane. These results revealed that strain 1510 could grow with n-hexadecane. However, the growth rate of the alkB mutant (3.33 × 105 CFU/ml/day) was lower than that of the wild-type (5.67 × 106 CFU/ml/day). Moreover, STL production of the alkB mutant was not restored under those conditions (data not shown). This result suggested the possibilities that SD-74 had at least one other alkane oxygenase gene, that alkB was essential for STL production, and that the lack of alkB did not completely affect the growth activity; these possibilities suggest that AlkB has other functions that are essential for STL production.
A putative acyl-CoA transferase was essential for STL production.
By sequencing the DNA flanking the insertion of the transposon in strain 330, we identified a 1,491-bp open reading frame (accession no. AB847084). The start codon of orf1 is ATG, and a predicted SD sequence is present 9 bp upstream from the start codon. The predicted protein sequence of ORF1 had no amino acid sequence homology with the reported proteins. However, ORF1 included an amino acid motif common in acyl-CoA transferases, the HX3DX14Y motif (28, 29). Therefore, the gene orf1 was named trehalose lipid synthase A (tlsA). This motif is conserved in papA1, which is found in Mycobacterium tuberculosis, the product of which was characterized as an acyl-CoA transferase, an enzyme that transfers palmitoyl-CoA to trehalose-2-sulfate (30). This type of acyl-CoA transferase, containing an HX3DX14Y motif, is widely conserved among Gram-positive bacteria (Table 3). Considering this fact, the TlsA of strain SD-74 may function similarly to papA1 by transferring acyl-CoA to trehalose or its derivatives. This is the final step of STL production. Due to the presence of the HX3DX14Y motif, TlsA was predicted to be an acyltransferase. To confirm that the HX3DX14Y motif was essential for TlsA activity, we constructed TlsA mutants in which histidine and aspartic acid in the HX3DX14Y motif were replaced with alanines (H192A and D196A), as well as the aspartic acid just before the motif (D191A). We analyzed the STL production in the transformants of strain 330 with the mutated tlsA gene. All of the mutants with one of the ORF1 mutations (D191A, H192A, or D196A) exhibited defective STL production (Fig. 4), indicating that the HX3DX14Y motif is essential for the activity of TlsA. This result partly suggests that TlsA has acyltransferase activity and acts as an STL synthase using trehalose and acyl-CoA as substrates.
Table 3.
Amino acid alignments of acyltransferases
| Protein | Species | H positionb | HX3DX14Y motif regiona | No. of residuesc |
|---|---|---|---|---|
| PapA1 | Mycobacterium bovis | 171 | SIDHLHADGQFVGVGLMEFQSMYTAL | 511 |
| PapA1 | Mycobacterium tuberculosis | 171 | SIDHLHADGQFVGVGLMEFQSMYTAL | 511 |
| PapA2 | Mycobacterium avium | 177 | SADHLVIDGMSVGVIFLEIHLTYAAL | 503 |
| PapA2 | Mycobacterium smegmatis | 167 | SVDHVHVDATFLGLMLIEIHLMYAAL | 480 |
| PapA2 | Streptomyces coelicolor | 172 | AFDHSNVDAYSIYRIPAEVGELYAAG | 473 |
| TlsA | Rhodococcus sp. SD-74 | 192 | AIDHAHTDMQSMILMFAEIRISYQAE | 499 |
Fig 4.
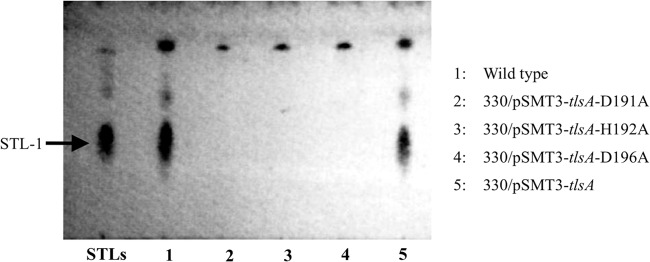
STL production by the TlsA mutants in which histidine and aspartate in the HX3DX14Y motif were replaced with alanines. STLs were produced by the wild type, as shown by the results in lane 1. While the TlsA-complemented strain recovered its STL production (lane 5), transformants that contained one of three mutations in TlsA, D191A, H192A, or D196A, lost STL production (lanes 2, 3, and 4, respectively). The analysis of STL production was performed using TLC, and detection of STLs used the anthrone-sulfuric acid method.
Construction of an alkane-inducible expression vector and overexpression of STL-producing genes.
Using the results of the genetic analysis, we increased the production of STLs using a genetic engineering procedure. First, we analyzed the transcript levels of three genes to find a promoter that is induced by alkanes (the first substrate of STLs) and that has strong promoter activity under STL-producing conditions. We cultured a wild-type strain of SD-74 in PMY medium or PMY medium with 10% n-hexadecane (vol/vol) to create non-STL-producing and STL-producing conditions, respectively. The total RNA was extracted to compare the fda, alkB, and tlsA transcript levels under each condition. Quantification of the transcripts was performed via RT-PCR using 16S rRNA as a housekeeping gene. The fda transcripts remained at a nearly constant level under each condition (Fig. 5A). The levels of alkB transcripts increased under the STL-producing conditions (Fig. 5B). Under the STL-producing conditions, the expression levels of tlsA were approximately 10-fold greater than the levels under non-STL-producing conditions (Fig. 5C).
Fig 5.
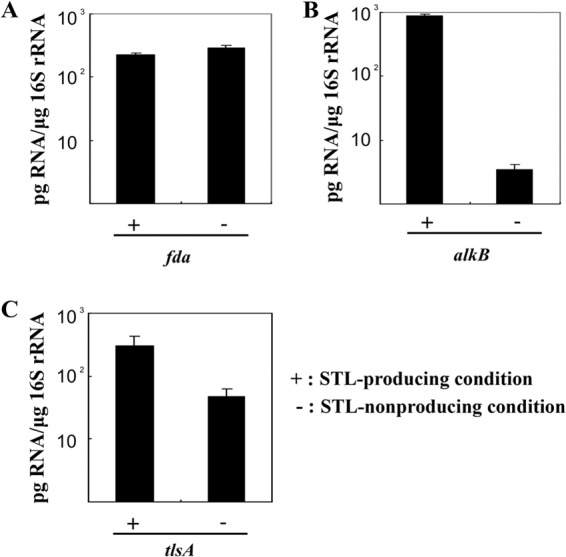
Real-time RT-PCR analysis of STL production-related gene expression. Rhodococcus sp. SD-74 was grown under non-STL-producing (−) and STL-producing conditions (+). The mRNA level of the gene in each sample was normalized to that of 16S rRNA. (A) Expression of fda. The expression level of fda did not change under either condition. (B) Expression of alkB. The expression level of alkB increased significantly under STL-producing conditions. (C) Expression of tlsA. The expression level of tlsA was slightly increased under STL-producing conditions. The results are averages based on three or more independent determinations, and the standard errors are indicated.
The RT-PCR results showed that alkB transcription was notably increased under the STL-producing conditions and in the presence of n-hexadecane (Fig. 5B). Therefore, we constructed an overexpression plasmid using the promoter region of alkB to increase STL production. We cloned the putative promoter region of alkB and inserted the promoter region into pSMT3. The overexpression plasmid pSMT3-PalkB was used for overexpression of the STL-producing genes tlsA and fda. We transformed strain SD-74 with the overexpression plasmids pSMT3-PalkB::tlsA and pSMT3-PalkB::fda and then analyzed STL production in tlsA- and fda-overexpressing mutants. After production, the STLs were extracted with ethyl acetate and quantified using the anthrone-sulfuric acid method. The STL production volume of the tlsA-overexpressing mutant was 2-fold greater than that of the nonoverexpressing strain. This increasing production volume represents 40 g/liter when converted into grams at theoretical yields. The fda-overexpressing mutant produced slightly fewer STLs than the nonoverexpressing strain (Fig. 6). These results suggest that TlsA is the rate-determining enzyme and that the putative last step of STL synthesis is the rate-limiting step of STL production.
Fig 6.
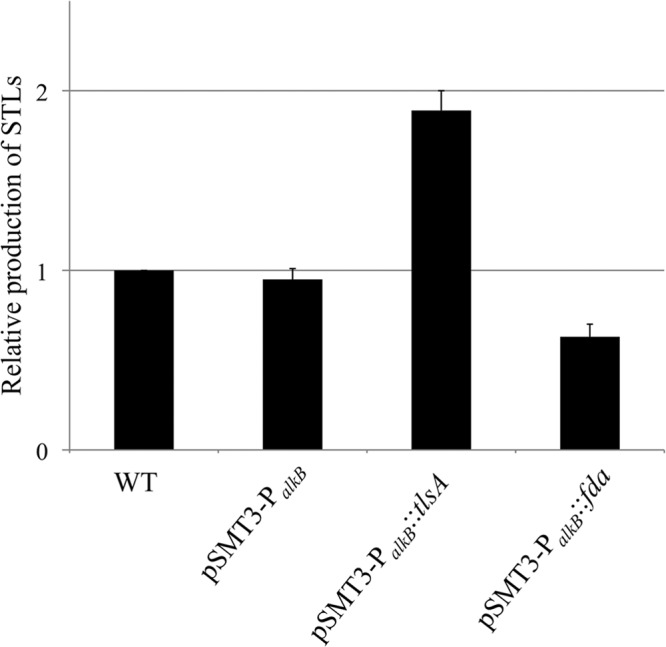
Overproduction of STLs by overexpression of the STL-producing genes tlsA and fda. STLs were produced under the resting-cell conditions and then extracted by ethyl acetate. The quantification of STLs was performed using the anthrone-sulfuric acid method, and trehalose was used for the standard curve. STL production nearly doubled with overexpression of tlsA compared to the level of production by the nonoverexpressing strain. The results are averages based on three or more independent determinations, and the standard errors are indicated.
DISCUSSION
In this study, we identified the three types of genes required for the biosynthesis of STLs and determined their functions. In addition, we succeeded in increasing the production of STLs by 40 g/liter using genetic engineering methods. STLs are of great importance because of their unique properties, which include high surface activity, growth inhibition of the influenza virus, and induction of cell differentiation in human leukemia cells. However, the STL biosynthesis pathway has not yet been elucidated. Using a transposome mutagenesis system, we described the STL biosynthesis pathway for the first time. By screening for strains that could not produce STLs and analyzing the sequences of DNA flanking the insertion, we identified three types of genes required for STL production.
First, we determined that the gene fda was essential for STL production and that it was predicted to encode fructose-bisphosphate aldolase. This enzyme is involved in the conversion of sugars from triose to hexose or vice versa. To support this conclusion, we purified Fda, which was expressed in E. coli. It exhibited high aldolase activity with fructose-1,6-bisphosphate as a substrate. Therefore, the gene fda encodes fructose-1,6-bisphosphate aldolase and Fda has aldolase activity. This means that fda is important in the biosynthesis of trehalose from n-hexadecane. Trehalose is a fundamental disaccharide that is abundant in mycobacteria in a free form or in glycoconjugates in the cytosol and the cell wall (31). In mycobacteria, which are related to Rhodococcus spp., the trehalose accumulates in the cell during growth under nutrient-limited conditions (32). Thus, it may be predicted that fda transcripts involved in sugar biosynthesis are maintained at similar levels under STL-producing conditions. Indeed, fda transcripts remained at a nearly constant level in both STL-producing and -nonproducing conditions. Our results indicate that fda is essential for de novo trehalose synthesis from n-alkane and, therefore, that trehalose synthesis through alkane metabolism and gluconeogenesis is an important pathway for glycolipid synthesis in SD-74.
Second, we identified alkB, which is predicted to be involved in the first step of alkane metabolism. Common motifs encoded by alkB are strongly conserved in alkane monooxygenases and are required for the enzyme's activity (Table 2). Therefore, alkB encodes alkane monooxygenase and is required for the conversion of alkanes to alcohols. To support this hypothesis, the STL-producing activity of an alkB mutant was recovered via the addition of hexadecanol, which is a product of the n-hexadecane oxidizing reaction. According to these results, the oxidation of alkanes is an essential part of the initial steps in the STL biosynthesis pathway from alkanes. Moreover, the results indicated that the transcriptional level of alkB was significantly upregulated by the addition of n-hexadecane. Interestingly, despite the defect in alkane monooxygenase activity of AlkB, an alkB mutant was still able to grow with n-hexadecane as the only carbon source. However, this mutant was unable to produce STLs (data not shown). These findings suggest that AlkB is essential for STL production and that Rhodococcus spp. have at least two genes that encode alkane monooxygenases that are not involved in STL production. In the alkB disruption mutant, a product accumulated that was presumably trehalose or succinoyltrehalose (data not shown). Therefore, it is possible that AlkB is involved not only in alkane oxidation but also in the regulation of STL production.
Third, we showed that the gene tlsA was essential for STL production. Using sequence analysis, TlsA was predicted to be an acyl-CoA transferase because of the HX3DX14Y motif, which is commonly found in acyl-CoA transferases. The HX3DX14Y motif contributes greatly to the acyl transferase activity of Pap in M. tuberculosis. Pap functions by transferring acyl groups, which are activated by CoA, to the hydroxyl groups of alcohols (29). Site-directed mutagenesis of the HX3DX14Y motif demonstrated that this motif was also important for TlsA and suggested that the function of TlsA was similar to that of Pap in M. tuberculosis. Thus, TlsA likely functions by transferring fatty acids to trehalose or its derivatives. In support of this hypothesis, putative STL precursors accumulated in the tlsA mutant strain (data not shown). These findings suggest that TlsA works during the final step of the STL biosynthesis pathway and that it plays an important role in trehalose lipid or succinoyl trehalose lipid synthesis. The expression level of tlsA increased approximately 10-fold under STL-producing conditions. This result suggested that the transcription of tlsA is promoted by the presence of n-hexadecane, similar to alkB transcription. Our findings suggested that tlsA is essential for direct synthesis of the STLs. Therefore, it is possible that STL production was increased by the upregulation of tlsA transcription.
Next, we attempted to increase STL production by overexpressing STL-producing genes. The putative alkB promoter was used for overexpression because alkB was highly induced under STL-producing conditions (Fig. 5C). Although the STL production of the fda-overexpressing mutant was similar to that in the control, STL production in the tlsA-overexpressing mutant was 2-fold that of the other strains (Fig. 6). Therefore, we succeeded in increasing STL production. These findings further suggest the possibility that the enzymatic activity of TlsA is the rate-limiting step of STL biosynthesis.
We propose a hypothetical biosynthesis pathway for STLs from n-alkanes based on the identification of three gene types involved in STL production (Fig. 7). In this proposed biosynthesis pathway, alkane metabolism, sugar synthesis, and the generation of STLs involved alkB, fda, and tlsA, respectively. Initially, alkanes are oxidized by AlkB and converted to acyl-CoA or acetyl-CoA by alkane metabolic enzymes. Subsequently, acetyl-CoA is converted to succinate through the tricarboxylic acid cycle or converted into trehalose by gluconeogenic enzymes, which include fda and trehalose-synthesizing enzymes. Finally, acyl-CoA, succinate, and trehalose are converted to STLs by STL-producing enzymes, such as those encoded by tlsA. It is possible that this final step is the rate-limiting step of STL biosynthesis. In this hypothetical pathway, STL production was involved in β-oxidation and gluconeogenesis, which are important for growth. In fact, fda and alkB mutants were confirmed to show lower growth rates than the wild type with n-hexadecane as the only carbon source (data not shown). Interestingly, the tlsA mutant also showed lower growth than the wild type in minimal medium (data not shown). Therefore, it is suggested that STLs are required for effective utilization of n-alkanes in nature.
Fig 7.
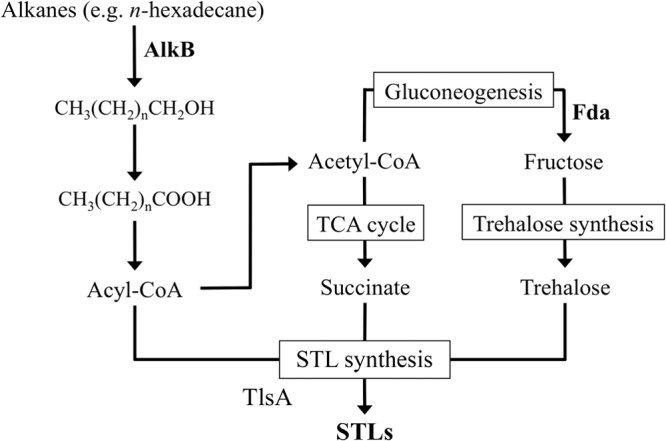
The proposed biosynthesis pathway of succinoyl trehalose lipids from alkanes. There are three general phases in STL production from alkanes. The alkane metabolism phase requires AlkB as the initial enzyme, and Fda works as a gluconeogenesis enzyme for trehalose backbone synthesis. In the final step of STL biosynthesis, TlsA transfers acyl groups to trehalose or its derivatives. TCA, tricarboxylic acid.
In this study, a detailed analysis of STL-producing genes revealed a novel pathway for trehalose lipid biosynthesis that was unknown in other Rhodococcus spp. The elucidation of the STL biosynthesis pathway was expected to allow increased production, and we achieved a 2-fold increase in production by upregulating tlsA transcription using the alkB promoter (Fig. 6). This is the first report of successfully increased production of trehalose lipids via genetic manipulation. It is possible that further analysis of the STL biosynthesis pathway will allow further increases in STL production. In addition, different types of trehalose lipids are produced by various Rhodococcus spp. (11), and it is supposed that trehalose lipid biosynthesis pathways do not vary greatly between species. Therefore, knowledge of STL biosynthesis contributes to a better understanding of trehalose lipid biosynthesis in Rhodococcus spp. This knowledge is very important not only for commercial purposes but also for elucidating the ecological purpose of trehalose lipid production in Rhodococcus spp.
Supplementary Material
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01664-13.
REFERENCES
- 1.Banat IM, Makkar RS, Cameotra SS. 2000. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 53:495–508 [DOI] [PubMed] [Google Scholar]
- 2.Imura T, Ohta N, Inoue K, Yagi N, Negishi H, Yanagishita H, Kitamoto D. 2006. Naturally engineered glycolipid biosurfactants leading to distinctive self-assembled structures. Chem. Eur. J. 12:2434–2440 [DOI] [PubMed] [Google Scholar]
- 3.Kitamoto D, Isoda H, Nakahara T. 2002. Functions and potential applications of glycolipid biosurfactants—from energy-saving materials to gene delivery carriers. J. Biosci. Bioeng. 94:187–201 [DOI] [PubMed] [Google Scholar]
- 4.Desai JD, Banat IM. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller MM, Hausmann R. 2011. Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production. Appl. Microbiol. Biotechnol. 91:251–264 [DOI] [PubMed] [Google Scholar]
- 6.Reis RS, Pereira AG, Neves BC, Freire DMG. 2011. Gene regulation of rhamnolipid production in Pseudomonas aeruginosa—a review. Bioresource Technol. 102:6377–6384 [DOI] [PubMed] [Google Scholar]
- 7.Kitamoto D, Morita T, Fukuoka T, Konishi M, Imura T. 2009. Self-assembling properties of glycolipid biosurfactants and their potential applications. Curr. Opin. Colloid Interface Sci. 14:315–328 [Google Scholar]
- 8.Lang S. 2002. Biological amphiphiles (microbial biosurfactants). Curr. Opin. Colloid Interface Sci. 7:12–20 [Google Scholar]
- 9.Hewald S, Linne U, Scherer M, Marahiel MA, Kamper J, Bolker M. 2006. Identification of a gene cluster for biosynthesis of mannosylerythritol lipids in the basidiomycetous fungus Ustilago maydis. Appl. Environ. Microbiol. 72:5469–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita T, Ito E, Kitamoto HK, Takegawa K, Fukuoka T, Imura T, Kitamoto D. 2010. Identification of the gene PaEMT1 for biosynthesis of mannosylerythritol lipids in the basidiomycetous yeast Pseudozyma antarctica. Yeast 27:905–917 [DOI] [PubMed] [Google Scholar]
- 11.Franzetti A, Gandolfi I, Bestetti G, Smyth TJP, Banat IM. 2010. Production and applications of trehalose lipid biosurfactants. Eur. J. Lipid Sci. Technol. 112:617–627 [Google Scholar]
- 12.Lang S, Philp JC. 1998. Surface-active lipids in rhodococci. Antonie van Leeuwenhoek 74:59–70 [DOI] [PubMed] [Google Scholar]
- 13.Uchida Y, Tsuchiya R, Chino M, Hirano J, Tabuchi T. 1989. Extracellular accumulation of mono-succinoyl and di-succinoyl trehalose lipids by a strain of Rhodococcus erythropolis grown on n-alkanes. Agric. Biol. Chem. 53:757–763 [Google Scholar]
- 14.Ishigami Y, Suzuki S, Funada T, Chino M, Uchida Y, Tabuchi T. 1987. Surface-active properties of succinoyl trehalose lipids as microbial biosurfactants. J. Jpn. Oil Chem. Soc. 36:847 [Google Scholar]
- 15.Tokumoto Y, Nomura N, Uchiyama H, Imura T, Morita T, Fukuoka T, Kitamoto D. 2009. Structural characterization and surface-active properties of a succinoyl trehalose lipid produced by Rhodococcus sp. SD-74. J. Oleo Sci. 58:97–102 [DOI] [PubMed] [Google Scholar]
- 16.Isoda H, Kitamoto D, Shinmoto H, Matsumura M, Nakahara T. 1997. Microbial extracellular glycolipid induction of differentiation and inhibition of the protein kinase C activity of human promyelocytic leukemia cell line HL60. Biosci. Biotechnol. Biochem. 61:609–614 [DOI] [PubMed] [Google Scholar]
- 17.Sudo T, Zhao XX, Wakamatsu Y, Shibahara M, Nomura N, Nakahara T, Suzuki A, Kobayashi Y, Jin CY, Murata T, Yokoyama KK. 2000. Induction of the differentiation of human HL-60 promyelocytic leukemia cell line by succinoyl trehalose lipids. Cytotechnology 33:259–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida Y, Misawa S, Nakahara T, Tabuchi T. 1989. Factors affecting the production of succinoyl trehalose lipids by Rhodococcus erythropolis SD-74 grown on n-alkanes. Agric. Biol. Chem. 53:765–769 [Google Scholar]
- 19.Garbe TR, Barathi J, Barnini S, Zhang Y, Abouzeid C, Tang D, Mukherjee R, Young DB. 1994. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140:133–138 [DOI] [PubMed] [Google Scholar]
- 20.Komeda H, Hori Y, Kobayashi M, Shimizu S. 1996. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc. Natl. Acad. Sci. U. S. A. 93:10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes PJ, Powell JAC, Archer JAC. 2001. Construction of Rhodococcus random mutagenesis libraries using Tn5 transposition complexes. Microbiology 147:2529–2536 [DOI] [PubMed] [Google Scholar]
- 22.Morris DL. 1948. Quantitative determination of carbohydrates with Dreywood's anthrone reagent. Science 107:254–255 [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Rutter WJ, Hunsley JR, Groves WE, Calder J, Rajkumar TV, Woodfin BM. 1966. Fructose diphosphate aldolase. Methods Enzymol. 9:479–498 [Google Scholar]
- 25.Miranda-Casoluengo R, Duffy PS, O'Connell EP, Graham BJ, Mangan MW, Prescott JF, Meijer WG. 2005. The iron-regulated iupABC operon is required for saprophytic growth of the intracellular pathogen Rhodococcus equi at low iron concentrations. J. Bacteriol. 187:3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Beilen JB, Smits THM, Whyte LG, Schorcht S, Rothlisberger M, Plaggemeier T, Engesser KH, Witholt B. 2002. Alkane hydroxylase homologues in gram-positive strains. Environ. Microbiol. 4:676–682 [DOI] [PubMed] [Google Scholar]
- 27.Whyte LG, Smits THM, Labbe D, Witholt B, Greer CW, van Beilen JB. 2002. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 68:5933–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buglino J, Onwueme KC, Ferreras JA, Quadri LEN, Lima CD. 2004. Crystal structure of PapA5, a phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis. J. Biol. Chem. 279:30634–30642 [DOI] [PubMed] [Google Scholar]
- 29.Onwueme KC, Ferreras JA, Buglino J, Lima CD, Quadri LEN. 2004. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc. Natl. Acad. Sci. U. S. A. 101:4608–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P, Schelle MW, Jain M, Lin FL, Petzold CJ, Leavell MD, Leary JA, Cox JS, Bertozzi CR. 2007. PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis virulence factor Sulfolipid-1. Proc. Natl. Acad. Sci. U. S. A. 104:11221–11226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Smet KAL, Weston A, Brown IN, Young DB, Robertson BD. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199–208 [DOI] [PubMed] [Google Scholar]
- 32.Elbein AD, Mitchell M. 1973. Levels of glycogen and trehalose in Mycobacterium smegmatis and the purification and properties of the glycogen synthetase. J. Bacteriol. 113:863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


