Abstract
The ability of chemoautotrophic ammonia-oxidizing archaea to compete for ammonia among marine microorganisms at low ambient concentrations has been in part attributed to their extremely high affinity for ammonia, but as yet there is no mechanistic understanding of supporting metabolism. We examined transcription of selected genes for anabolic functions (CO2 fixation, ammonia transport, and cell wall synthesis) and a central catabolic function (ammonia oxidation) in the thaumarchaeon Nitrosopumilus maritimus SCM1 growing at two ammonia concentrations, as measured by combined ammonia and ammonium, one well above the Km for ammonia oxidation (∼500 μM) and the other well below the Km (<10 nM). Transcript levels were generally immediately and differentially repressed when cells transitioned from ammonia-replete to ammonia-limiting conditions. Transcript levels for ammonia oxidation, CO2 fixation, and one of the ammonia transport genes were approximately the same at high and low ammonia availability. Transcripts for all analyzed genes decreased with time in the complete absence of ammonia, but with various rates of decay. The new steady-state mRNA levels established are presumably more reflective of the natural physiological state of ammonia-oxidizing archaea and offer a reference for interpreting message abundance patterns in the natural environment.
INTRODUCTION
The generally extremely low availability of ammonia in the open ocean, from below detection to low micromolar concentrations, suggests that free ammonia is a limiting resource for microbial growth (1–6). Thus, competition for ammonia among marine microorganisms is intense. However, the generally high numbers of ammonia-oxidizing archaea (AOA) in these environments indicate that they are not limited by ammonia (here defined as combined ammonia and ammonium) concentrations in the low nanomolar range. This inference is supported by the exceptionally low Km for ammonia oxidation of Nitrosopumilus maritimus (Km ≈ 133 nM for combined NH4+ + NH3) (7), by the direct determination of the Km for ammonia oxidation in a marine system dominated by AOA (Km ≈ 100 nM) (8), and by environmental metatranscriptomic analyses showing significant abundance of transcripts for genes coding for the thaumarchaeal ammonia monooxygenase (amoA), ammonia transport (amt), and cell wall S-layer proteins (slp) (9–14). Thus, the AOA may exert primary control over ammonia availability throughout the marine water column, including the control of nitrification in marine oxygen minimum zones (1, 15–18).
However, there is as yet no reference for relating environmental measures of transcript abundance to the physiological state of the contributing populations. The constantly low concentrations of ammonia typical of the marine environment cannot be maintained using standard batch culture conditions. We therefore developed a simple system, based on growing N. maritimus in dialysis bags, as a mechanism to establish quasi-steady-state growth at ammonia concentrations in the nanomolar range.
Once growth conditions were established, we examined transcript abundance patterns for genes encoding key functions in ammonia acquisition and cell synthesis. Selected genes encompassed those coding for one subunit of the ammonia monooxygenase (amoA), two ammonia transporters (amt1 and amt2), two S-layer proteins (slp1 and slp2), and the 4-hydroxybutyryl-coenzyme A (CoA) dehydratase (hcd) in the pathway for CO2 fixation (19). The transcripts for these genes in N. maritimus were examined under conditions of ammonia excess (batch culture), ammonia limitation (dialysis bag growth), extended ammonia starvation, and short-term fluctuations in ammonia availability.
MATERIALS AND METHODS
Growth conditions.
All studies were performed with N. maritimus SCM1 (20). The strain was maintained in 500 ml of HEPES-buffered synthetic Crenarchaeota medium (7) in 2-liter glass bottles at 30°C in the dark without shaking and transferred (1% inoculum) to fresh medium at late exponential phase. The concentration of nitrite was determined colorimetrically with the Griess-Ilosvay reagent (quantitative limit, 1.0 μM) (21). The concentration of NH4+ was determined using a fluorescence microplate reader and the o-phthaldialdehyde (OPA) reagent (quantitative limit, 10 nM) (22).
Comparative analyses of alternative growth states: batch, dialysis bag, and starvation.
Batch culture experiments (three biological replicates) were carried out in 2-liter glass bottles without shaking in the dark (see Fig. S1 in the supplemental material). One milliliter of medium was immediately quenched on ice for determinations of transcript abundance (n = 2), cell numbers (n = 1), and NH4+ and NO2− concentrations (as described below).
The dialysis bag experiments were conducted using a 10-liter glass bottle containing approximately 9 liters of medium, without added NH4Cl, preincubated overnight at 30°C (see Fig. S1 in the supplemental material). This medium contains approximately ∼1.6 μM ammonia determined by the OPA reagent as described above, originating from the trace levels of ammonia present in the reagent grade chemicals used to prepare the synthetic medium. Dialysis tubing (12,000- to 14,000-Da molecular-mass cutoff Spectra/Pro4, 6.4-mm diameter; Spectrum Labs, Rancho Dominguez, CA) cut to approximately 15.5-cm lengths was boiled in 800 ml of sodium bicarbonate buffer (790 ml of ultrapure water, 16 g NaHCO3, 1.6 ml of 500 mM EDTA, pH 8.0) for 10 min. After the dialysis tubes were rinsed in 800 ml of ultrapure water twice, they were boiled in 800 ml of sterile ultrapure water for 10 min and then washed in 50 ml of sterile preincubated SCM1 medium without NH4Cl.
The rinsed dialysis tubing was filled with 3.5 ml of batch culture SCM1 at late exponential growth (day 6) and clamped at both ends. Dialysis bag cultures were transferred into a preincubated 10-liter glass bottle containing approximately 9 liters of medium, incubated at 30°C, and stirred with a stir bar in the dark. As shown in Fig. S1 in the supplemental material, replicate dialysis bag cultures were removed at 1, 2, 3, and 6 days of incubation, for a total of three biological replicates. Culture material was recovered from the dialysis bag with a 3-ml syringe, immediately quenched on ice, and then aliquoted for determinations of transcript abundance (1 ml), amoA gene abundance (1 ml), and cell counts (1 ml). Cells were harvested by centrifugation (20,000 × g) for 10 min at 4°C in 1.5-ml PCR microcentrifuge tubes (Eppendorf, Hamburg, Germany). The supernatant was removed, immediately filtered with Millex-GV (pore size, 0.22 μm; Millipore, Billerica, MA, USA), and stored at −20°C for later determination of NH4+ and NO2− concentrations. One milliliter of RNALater (Qiagen, Valencia, CA) was added to each cell pellet, mixed well, and then centrifuged at 4°C, 20,000 × g, for 10 min. After discarding 0.95 ml of supernatant, the fixed cells were stored at −80°C until used for quantitative reverse transcription-PCR (qRT-PCR) analysis (as described below).
Dialysis bag cultures (n = 6, for biological triplicates) for starvation experiments were established from 3.5 ml of mid-exponential-growth SCM1 and placed in 10-liter glass bottles containing approximately 9 liters medium without added NH4Cl (see Fig. S1 in the supplemental material). After 1 day of incubation, 7 ml of medium recovered from two of the dialysis bag cultures was transferred to a sterile 15-ml conical tube (BD, Franklin Lakes, NJ) to initiate conditions of NH4+ starvation. The starvation experiments were carried out in the 15-ml tubes incubated at 30°C without shaking in the dark (see Fig. S1 in the supplemental material), removing 1-ml samples at different times (n = 2) to monitor transcript abundance and cell numbers (n = 1).
RNA extraction.
Total RNA was extracted using an RNeasy Minikit (Qiagen) in combination with an RNase-Free DNase set (Qiagen) according to the manufacturer's instructions. The extracted RNA solutions were treated with DNase (Turbo DNA-free kit; Life Technologies, Carlsbad, CA), again according to the manufacturer's instructions. The absence of DNA contamination was confirmed by PCR.
qRT-PCR.
The mRNAs of amoA (Nmar_1500), amt1 (Nmar_0588), amt2 (Nmar_1698), hcd (Nmar_0207), slp1 (Nmar_1201), and slp2 (Nmar_1547) genes were quantified by real-time PCR in a LightCycler system (Roche Diagnostics, Mannheim, Germany) with LightCycler RNA Master SYBR green I kit (Roche Diagnostics). Each reaction of the one-step reverse transcription-PCR was conducted in a 10-μl volume containing 0.5 μl of template RNA solution, 0.5 μM each primer, 3.75 μl of LightCycler RNA Master SYBR green I, and 3.25 mM Mn(OAc)2. The PCR primer sets in Table S1 in the supplemental material and PCR cycling in Table S2 in the supplemental material were used for quantitative RT-PCR. The standard curves for each mRNA were generated using an RNA standard in a 10-fold dilution series of 102 to 108 copies per reaction. The amplification efficiencies of quantitative RT-PCRs averaged 70.8% (r2 = 0.999), 80.3% (r2 = 1.000), 72.2% (r2 = 0.999), 76.8% (r2 = 0.999), 74.1% (r2 = 0.999), and 81.0% (r2 = 0.999) for amoA, amt1, amt2, hcd, slp1, and slp2 transcripts, respectively.
Cell counts.
Cells were fixed in 2% paraformaldehyde for at least 1 h. The fixed cells were diluted in phosphate-buffered saline to adjust the density from 10 to 100 cells per field for the cell counting, filtered on 0.2-μm-pore-size polycarbonate GTBP membrane filters (Millipore, Billerica, MA, USA), and then stained with 2 μl of Abnova 4′,6′-diamidino-2-phenylindole (DAPI) (1.5 mg ml−1; Abnova, Taipei, Taiwan). The cell numbers were determined for each sample by epifluorescence microscopy, counting cells in each of 20 random fields.
Response to short-term ammonia starvation and ammonia readdition.
Rinsed dialysis tubing was filled with 3.5 ml batch culture SCM1 at late exponential growth (day 6), clamped at both ends, and used for studies of growth under conditions of low ammonia availability and in response to short-term changes in ammonia availability. Conditions of ammonia limitation were established by transferring the dialysis bag cultures to 10-liter glass bottles containing 9 liters synthetic medium at 1.6 μM NH4+. This concentration of ammonium originates from the trace levels present in the reagent grade chemicals used to prepare the synthetic medium. After incubation for 8 h in low ammonia medium, replicate dialysis bag cultures were transferred to a 10-liter bottle containing approximately 9 liters of medium without added NH4Cl (see Fig. S2 in the supplemental material). Ammonia starvation was initiated at specified times by transferring individual replicates to a sterile petri dish without medium in a plastic bag containing a wetted filter strip and incubating at 30°C. Dialysis bags were transferred between bottles containing medium without ammonia and sterile petri dishes without medium for analyses of transcriptional response to short-term fluctuations in ammonia availability (see Fig. S2 in the supplemental material). Culture material was removed using a 3-ml syringe and immediately quenched on ice prior to analysis of transcript abundance and NH4+ and NO2− concentrations.
RESULTS AND DISCUSSION
N. maritimus retained within dialysis bags suspended in a large volume of medium poised at ∼2 μM total ammonia grew at near-maximum growth rates while maintaining the concentration of ammonia at or below 10 nM, the analytical limit of detection (Fig. 1). Since cells showed no tendency to attach to the dialysis bag walls under these growth conditions, the cells on average were experiencing ammonia concentrations in the very low nanomolar range. At these low ammonium concentrations, oxygen was not a limiting resource. Ammonia in batch cultures was depleted below the detection limit of 10 nM after N. maritimus entered stationary phase as described previously (7). The concentration of ammonia in the dialysis bags decreased following transfer to the ammonia-limited medium (Fig. 1A and B) and was maintained below 10 nM by active consumption by N. maritimus throughout the experiments. In contrast, the NO2− concentration within the dialysis bags was slighter higher than in the bulk medium, reflecting a production rate exceeding the rate of equilibration between compartments. N. maritimus cell numbers within the dialysis bags increased throughout the experimental period, confirming their ability for continuous growth at low nanomolar concentrations of ammonia (Fig. 1C and Table 1). The abundances of amoA genes determined by quantitative PCR were smaller than those obtained by direct cell count (Table 1). The inefficiencies of cell harvesting and extraction likely caused this discrepancy. However, since this loss is relatively low and is expected to be comparable for DNA and RNA, this does not alter our conclusions.
Fig 1.
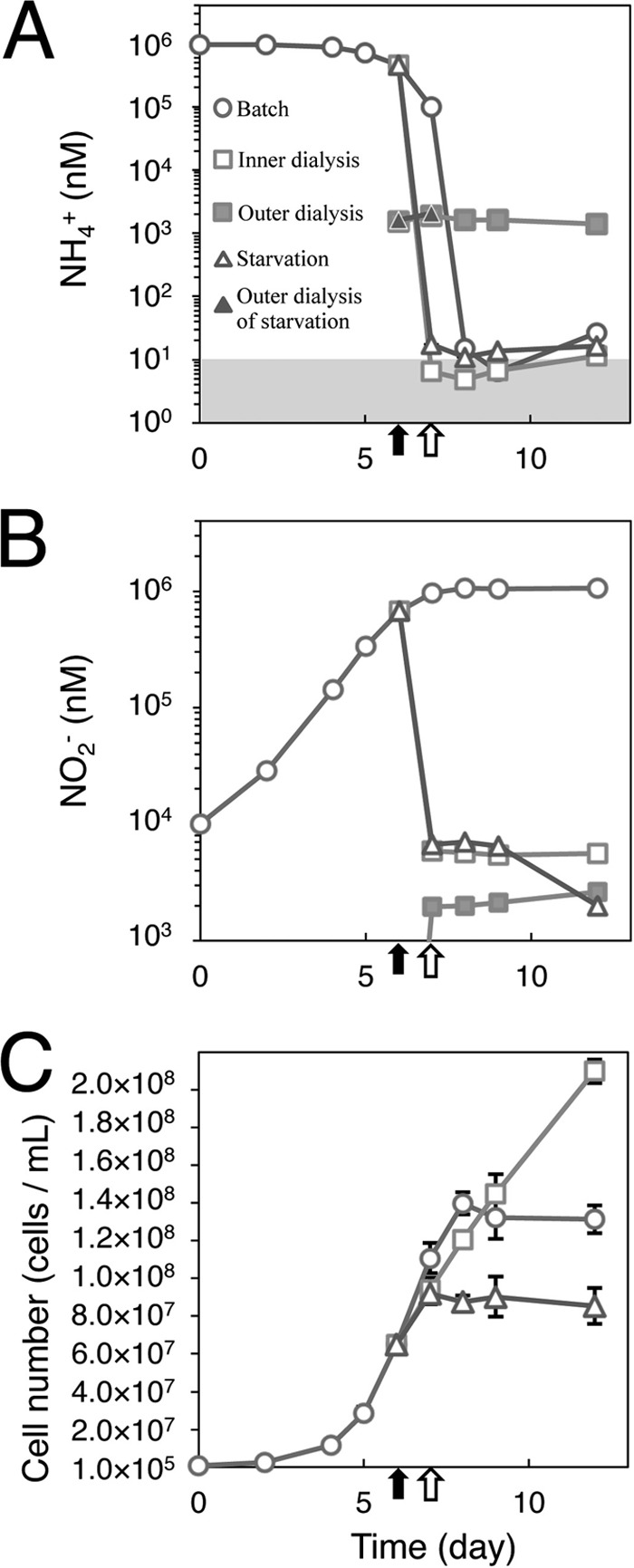
Changes in concentration of NH4+ (A) and NO2− (B) and in cell numbers of N. maritimus SCM1 (C) during batch culture (circles) and dialysis bag growth (open squares, inner dialysis bag; filled squares, outer dialysis bag) and in response to ammonia starvation (open triangles, inner dialysis bag and batch culture under ammonia starvation; filled triangles, outer dialysis bag for ammonia starvation). The gray zone shows the ammonia detection limit (∼10 nM). Black arrows indicate times when cells were transferred to dialysis bags. Open black arrows show times when cells were removed from dialysis bags for starvation experiments. Bars are standard errors (n = 3).
Table 1.
Changes in cell numbers, growth rates, and amoA gene abundances of N. maritimus SCM1 during batch culture and dialysis bag growth and in response to ammonia starvation
| Characteristic measured and process | Value at day: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 5 | 6 | 7 | 8 | 9 | 12 | |
| No. of cells/ml | |||||||||
| Batch culture | 8.2 × 105 | 2.6 × 106 | 1.2 × 107 | 2.9 × 107 | 6.5 × 107 | 1.1 × 108 | 1.4 × 108 | 1.3 × 108 | 1.3 × 108 |
| Dialysis bag growth | 6.5 × 107 | 9.4 × 107 | 1.2 × 108 | 1.5 × 108 | 2.1 × 108 | ||||
| Ammonia starvation | 6.5 × 107 | 9.2 × 107 | 8.8 × 107 | 9.0 × 107 | 8.6 × 107 | ||||
| Growth rate (no. of cells/ml/day) | |||||||||
| Batch culture | 8.9 × 105 | 4.5 × 106 | 1.7 × 107 | 3.6 × 107 | 4.6 × 107 | 2.9 × 107 | 0 | 0 | |
| Dialysis bag growth | 2.9 × 107 | 2.7 × 107 | 2.4 × 107 | 2.2 × 107 | |||||
| Ammonia starvation | 2.7 × 107 | 0 | 2.6 × 106 | 0 | |||||
| No. of amoA gene copies/ml | |||||||||
| Batch culture | 4.9 × 105 | 1.2 × 106 | 9.6 × 106 | 1.8 × 107 | 2.7 × 107 | 4.6 × 107 | 5.5 × 107 | 4.9 × 107 | 8.0 × 107 |
| Dialysis bag growth | 2.7 × 107 | 4.1 × 107 | 4.6 × 107 | 4.8 × 107 | 1.1 × 108 | ||||
| No. of amoA gene copies/cell | |||||||||
| Batch culture | 0.6 | 0.5 | 0.8 | 0.6 | 0.4 | 0.4 | 0.4 | 0.4 | 0.6 |
| Dialysis bag growth | 0.4 | 0.4 | 0.4 | 0.3 | 0.5 | ||||
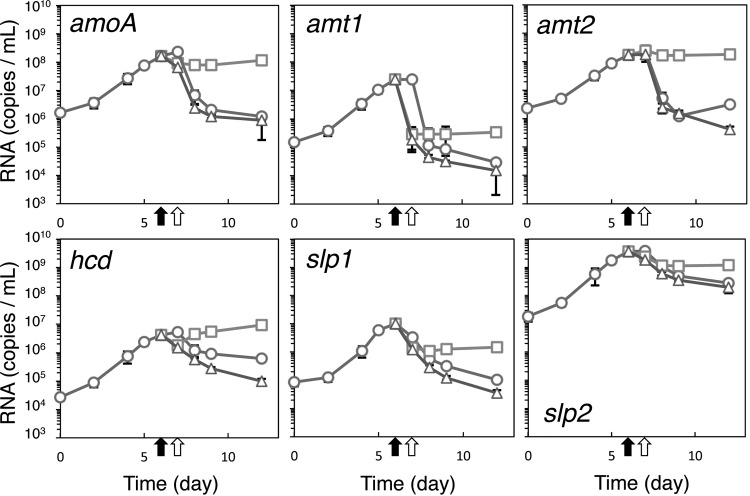
Prior to transfer to the dialysis bags, the highest per-cell transcript values for those genes evaluated were for those coding for ammonia oxidation (amoA), ammonia uptake (amt2), and one of two closely related S-layer proteins (slp2) (Table 2). This is consistent with published metatranscriptomic analyses of natural marine systems, which found higher-level transcripts for amoA, amt2, and slp2. Thus, transcripts of these genes, coding for central energy-generating and anabolic functions, are expected to provide useful signatures for active cell growth in environmental samples (9–14). The per-cell values for these transcripts varied between a high value for slp2 (∼60 transcripts/cell) and a low value for hcd (∼0.08 transcripts/cell). Since the amoA gene counts were comparable to cell counts (Table 1), the low abundances of the per-cell transcripts except for slp2 likely reflect a physiological state in which some cells in a population are not actively transcribing those genes. The per-cell abundance of all transcripts decreased during dialysis bag incubation, approaching new steady-state values by day 12. Relatively small changes in transcript abundance for amoA, amt2, hcd, and slp2 genes were observed following transition to growth at the low ammonia concentration maintained in the dialysis bags (Fig. 2). Reduced per-cell abundances were about 54% (hcd), 28% (amt2), 21% (amoA), and 10% (slp2) of levels determined prior to transfer to the dialysis bags. These data indicate that the cellular investment in central systems for energy generation and anabolism does not vary greatly over a wide range of ammonia concentrations, in this study varying from below 10 nM to 500 μM.
Table 2.
Changes in the per-cell transcript and gene abundance of N. maritimus SCM1 during batch culture and dialysis bag growth and in response to ammonia starvation
| Gene and process | Transcript copies per cell on day: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 5 | 6 | 7 | 8 | 9 | 12 | |
| amoA | |||||||||
| Batch culture | 1.9918 | 1.4173 | 2.3362 | 2.6137 | 2.4978 | 2.0328 | 0.0473 | 0.0155 | 0.0090 |
| Dialysis bag growth | 2.4978 | 0.9762 | 0.6374 | 0.5351 | 0.5429 | ||||
| Ammonia starvation | 2.4978 | 0.7141 | 0.0271 | 0.0135 | 0.0102 | ||||
| amt1 | |||||||||
| Batch culture | 0.1832 | 0.1444 | 0.2926 | 0.3619 | 0.3749 | 0.2232 | 0.0008 | 0.0006 | 0.0002 |
| Dialysis bag growth | 0.3749 | 0.0031 | 0.0024 | 0.0020 | 0.0016 | ||||
| Ammonia starvation | 0.3749 | 0.0020 | 0.0005 | 0.0003 | 0.0002 | ||||
| amt2 | |||||||||
| Batch culture | 2.8194 | 1.9220 | 2.7713 | 3.0709 | 2.7285 | 2.1475 | 0.0373 | 0.0093 | 0.0238 |
| Dialysis bag growth | 2.7285 | 2.6570 | 1.3680 | 1.1728 | 0.8749 | ||||
| Ammonia starvation | 2.7285 | 1.9577 | 0.0277 | 0.0165 | 0.0049 | ||||
| hcd | |||||||||
| Batch culture | 0.0326 | 0.0343 | 0.0660 | 0.0837 | 0.0635 | 0.0472 | 0.0087 | 0.0068 | 0.0047 |
| Dialysis bag growth | 0.0635 | 0.0196 | 0.0378 | 0.0376 | 0.0448 | ||||
| Ammonia starvation | 0.0635 | 0.0165 | 0.0065 | 0.0030 | 0.0011 | ||||
| slp1 | |||||||||
| Batch culture | 0.1048 | 0.0487 | 0.0963 | 0.2033 | 0.1539 | 0.0301 | 0.0040 | 0.0024 | 0.0008 |
| Dialysis bag growth | 0.1539 | 0.0152 | 0.0089 | 0.0086 | 0.0070 | ||||
| Ammonia starvation | 0.1539 | 0.0131 | 0.0032 | 0.0014 | 0.0004 | ||||
| slp2 | |||||||||
| Batch culture | 22.1288 | 21.4997 | 49.6467 | 60.4198 | 55.9631 | 33.6562 | 7.4532 | 3.6854 | 2.0885 |
| Dialysis bag growth | 55.9631 | 24.9313 | 9.3454 | 7.7554 | 5.6251 | ||||
| Ammonia starvation | 55.9631 | 19.5799 | 6.8197 | 3.8748 | 2.2453 | ||||
Fig 2.
Changes in transcript abundance of N. maritimus SCM1 genes amoA, amt1, amt2, hcd, slp1, and slp2 associated with changing ammonia availability during batch culture (circles) and dialysis bag growth (squares) and in response to ammonia starvation (triangles). Black arrows show times of transfer from batch culture to dialysis bags. Open black arrows show times of transfer of cells from dialysis bags for starvation analyses. Bars are standard errors (n = 3).
In contrast to those genes for which transcript numbers remained relatively high in a low-ammonia environment, transition to nanomolar-ammonia growth conditions resulted in a much greater reduction in transcripts for an alternative S-layer gene (slp1) and ammonia transporter gene (amt1), reduced to 3% and 0.4% of initial abundances, respectively. Such a rapid decrease is suggestive of active message decay. Also, since these genes code for variants of the ammonia transporter and S-layer proteins, they may function primarily during growth at higher ammonia concentrations, Amt2 possibly functioning as a lower-affinity transporter and Slp2 supporting cell wall synthesis associated with higher growth rates. It is also possible that the varying stoichiometry of S-layer proteins with varying ammonia availability may be of functional significance. For example, recent structural analyses of the S-layer protein of the methanogenic archaeon Methanosarcina acetivorans revealed a highly negatively charged pore surface, suggesting a possible role of this S-layer in selective ion uptake (23). Therefore, it is essential to investigate the structure of S-layer of N. maritimus in order to evaluate if it serves a similar function in the collection of ammonia, or ammonium, at extremely low concentrations.
The dual role of ammonia—assimilated for cell synthesis or oxidized as a source of energy and reductant—also presents a possible conflict for this organism. Ammonia/ammonium collected at the cell surface must be partitioned between anabolic and catabolic pathways, necessitating a balance in the affinities of enzyme systems controlling the flux of ammonia through these alternative pathways. Studies of ammonia assimilation by Escherichia coli have shown that active ammonia transport by its AmtB ammonia permease is essential for growth below ∼20 μM NH4+ (24). At higher ammonia concentrations, diffusion of ammonia through the membrane is a major source of ammonia for biosynthesis. In N. maritimus, transcripts for two ammonia permease genes are abundant at the higher ammonia concentration, with transcripts for one (amt2) present in about 10-fold-higher abundance than the second (amt1). The rapid depletion of amt1 transcripts with transition from high to low ammonia concentrations (Fig. 2, top middle panel) suggests that this permease has a higher Km (lower affinity) than that of the second (amt2). Thus, the Amt2 permease presumably supports ammonia assimilation at concentrations in the very low nanomolar range.
All transcripts decreased following short-term elimination of ammonia, albeit having significantly different rates of decay, with transcripts for amoA and amt1 being rapidly depleted in the absence of ammonia (Fig. 3). However, cells were also immediately responsive to the resupply of nanomolar amounts of ammonia and within 1 to 2 h returned to near prestarvation transcript levels. Thus, these results point to an extremely rapid adaptive response to changing ammonia availability and further enforce the importance of rapid preservation of cells harvested for metatranscriptomic analyses (10).
Fig 3.
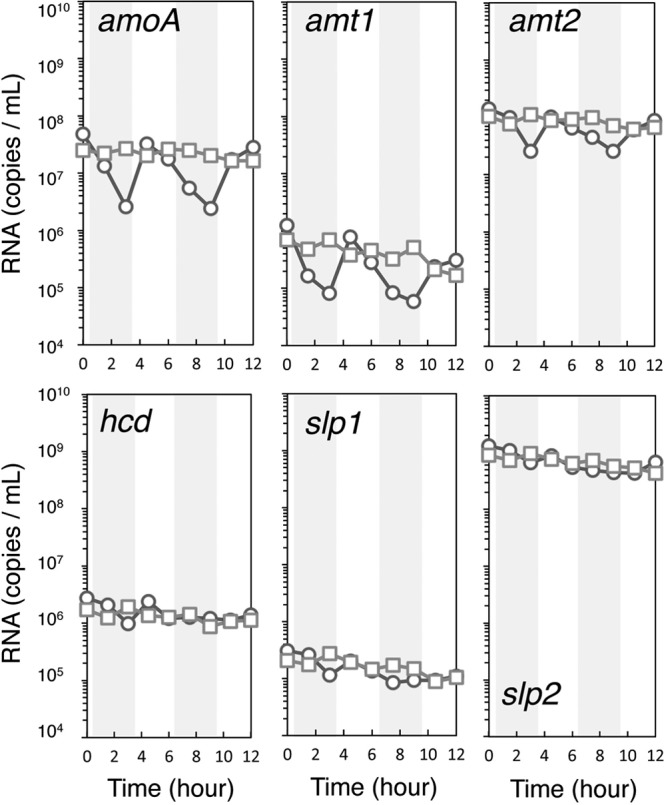
Changes in transcript abundance of N. maritimus genes amoA, amt1, amt2, hcd, slp1, and slp2 associated with short-term ammonia starvation under short-term ammonia starvation followed by readdition (circles) or continuous dialysis bag growth (squares). Gray bars correspond to periods of ammonia starvation.
A similar, rapid decay of amoA transcripts in Nitrosomonas europaea was observed under short-term ammonium starvation (25). In contrast, amoC transcript numbers remained elevated under these conditions (26). The response of ammonia-starved N. europaea and Nitrosospira briensis to ammonia addition was also similar to that of N. maritimus, as evidenced by a rapid increase in amoA transcripts following readdition of 15 or 50 mM ammonia to N. europaea and 5 mM ammonia to N. briensis (27, 28). Thus, the AOA and ammonia-oxidizing bacteria (AOB) may share similar regulatory strategies to cope with changing ammonia concentrations, although differing vastly in the range of ammonia concentrations to which they are responsive. The Km of N. maritimus for ammonia oxidation (133 nM) is much lower than that of AOB N. europaea (Km = 553 μM) (7), Nitrosococcus oceani (Km = 101 μM) (7), and N. briensis (Km = 3 μM) (28).
Although additional studies are essential to establish differences in the ammonia affinity of the two permeases and to evaluate the suggested variation in S-layer protein stoichiometry associated with changing ammonia concentrations, our results provide an important context for interpreting transcript abundance patterns observed in analyses of natural marine populations. More generally, the strategy used in this study for growing marine oligotrophs under nutrient limitation may assist in the study of other microorganisms living at the extremes of energy limitation common to much of the biosphere (29, 30).
Supplementary Material
ACKNOWLEDGMENTS
We thank Willm Martens-Habbena, Tony D. Bertagnolli, Wei Qin, and Liu He for discussions. We thank two anonymous reviewers for helpful comments.
This work was supported by NSF grants MCB-0920741 and NSF Dimensions program grant OCE-1046017 to D.A.S and JSPS to T.N. (21770028).
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02028-13.
REFERENCES
- 1.Beman JM, Popp BN, Francis CA. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441 [DOI] [PubMed] [Google Scholar]
- 2.Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. 2011. Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 77:2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herfort L, Schouten S, Abbas B, Veldhuis MJW, Coolen MJL, Wuchter C, Boon JP, Herndl GJ, Sinninghe Damsté JS. 2007. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol. Ecol. 62:242–257 [DOI] [PubMed] [Google Scholar]
- 4.Rees AP, Woodward EMS, Joint I. 2006. Concentrations and uptake of nitrate and ammonium in the Atlantic ocean between 60 degrees N and 50 degrees S. Deep Sea Res. Part II Top. Stud. Oceanogr. 53:1649–1665 [Google Scholar]
- 5.Santoro AE, Casciotti KL. 2011. Enrichment and characterization of ammonia oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. 5:1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro AE, Casciotti KL, Francis CA. 2010. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12:1989–2006 [DOI] [PubMed] [Google Scholar]
- 7.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979 [DOI] [PubMed] [Google Scholar]
- 8.Horak REA, Qin W, Schauer A, Armbrust EV, Ingalls AE, Moffett JW, Stahl DA, Devol AH. 9 May 2013. Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. [Epub ahead of print.] 10.1038/ismej.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker BJ, Lesniewski RA, Dick GJ. 2012. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 6:2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feike J, Jürgens Hollibaugh KJT, Krüger S, Jost G, Labrenz M. 2012. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 6:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. 2011. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 5:866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radax R, Rattei T, Lanzen A, Bayer C, Tore Rapp H, Urich T, Schleper C. 2012. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. 14:1308–1324 [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Tyson GW, Eppley JM, DeLong EF. 2011. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. Environ. Microbiol. 5:999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart FJ, Ulloa O, DeLong EF. 2012. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ. Microbiol. 14:23–40 [DOI] [PubMed] [Google Scholar]
- 15.Beman JM, Popp BN, Alford SE. 2012. Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol. Oceanogr. 57:711–726 [Google Scholar]
- 16.Church MJ, Wai B, Karl DM, DeLong EF. 2010. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ. Microbiol. 12:679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labrenz M, Sintes E, Toetzke F, Zumsteg A, Herndl GJ, Seidler M, Jürgens K. 2010. Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J. 4:1496–1508 [DOI] [PubMed] [Google Scholar]
- 18.Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart GJ, Sinninghe Damsté JS. 2011. Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J. 5:1896–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PSG, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 21.Hewitt EJ, Nicholas DJD. 1964. Enzymes of inorganic nitrogen metabolism, p 167–172 In Linskens HF, Sanwal BD, Tracey MV. (ed), Modern methods of plant analysis, vol 7 Springer, Heidelberg, Germany [Google Scholar]
- 22.Holmes RM, Aminot A, Kerouel R, Hooker BA, Peterson BJ. 1999. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can. J. Fish. Aquat. Sci. 56:1801–1808 [Google Scholar]
- 23.Arbing MA, Chan S, Shin A, Phan T, Ahn CJ, Rohlin L, Gunsalus RP. 2012. Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans. Proc. Natl. Acad. Sci. U. S. A. 109:11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T. 2012. Need-based activation of ammonium uptake in Escherichia coli. Mol. Syst. Biol. 8:616. 10.1038/msb.2012.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei X, Sayavedra-Soto LA, Arp DJ. 2004. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150:1869–1879 [DOI] [PubMed] [Google Scholar]
- 26.Berube PM, Samudrala R, Stahl DA. 2007. Transcription of all amoC copies is associated with recovery of Nitrosomonas europaea from ammonia starvation. J. Bacteriol. 189:3935–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein LY, Arp DJ. 1998. Ammonium limitation results in the loss of ammonia-oxidizing activity in Nitrosomonas europaea. Appl. Environ. Microbiol. 64:1514–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bollmann A, Schmidt I, Saunders AM, Nicolaisen MH. 2005. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–220 [DOI] [PubMed] [Google Scholar]
- 30.Hoehler TM, Jørgensen BB. 2013. Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11:83–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.