Abstract
The eukaryotic flagellum (or cilium) is a broadly conserved organelle that provides motility for many pathogenic protozoa and is critical for normal development and physiology in humans. Therefore, defining core components of motile axonemes enhances understanding of eukaryotic biology and provides insight into mechanisms of inherited and infectious diseases in humans. In this study, we show that component of motile flagella 22 (CMF22) is tightly associated with the flagellar axoneme and is likely to have been present in the last eukaryotic common ancestor. The CMF22 amino acid sequence contains predicted IQ and ATPase associated with a variety of cellular activities (AAA) motifs that are conserved among CMF22 orthologues in diverse organisms, hinting at the importance of these domains in CMF22 function. Knockdown by RNA interference (RNAi) and rescue with an RNAi-immune mRNA demonstrated that CMF22 is required for propulsive cell motility in Trypanosoma brucei. Loss of propulsive motility in CMF22-knockdown cells was due to altered flagellar beating patterns, rather than flagellar paralysis, indicating that CMF22 is essential for motility regulation and likely functions as a fundamental regulatory component of motile axonemes. CMF22 association with the axoneme is weakened in mutants that disrupt the nexin-dynein regulatory complex, suggesting potential interaction with this complex. Our results provide insight into the core machinery required for motility of eukaryotic flagella.
INTRODUCTION
The flagellum (also called cilium) is a prominent eukaryotic organelle that functions in motility, environmental sensing, adhesion, and mating (1, 2). In humans, flagellar motility is essential for normal development and physiology (3). Defective motility, due to flagellar paralysis or dysregulated beating, results in a variety of diseases, including primary ciliary dyskinesia, situs inversus, hydrocephalus, respiratory malfunction, and male factor infertility (4). In addition, motile flagella provide the primary mode of locomotion for many pathogenic protozoa, including Trypanosoma spp., Leishmania spp., Giardia lamblia, and Trichomonas vaginalis, which cause immense morbidity throughout the developing world (5–7). For example, Trypanosoma brucei strains depend on a motile flagellum to complete their life cycle, enabling infections of humans and livestock that result in a significant loss of life and economic hardship in sub-Saharan Africa (8, 9). Therefore, the motile flagellum is a critical component in a range of heritable and infectious diseases.
Flagella are present in all major eukaryotic groups and are built upon a microtubule axoneme that may be motile or nonmotile (10–12). Motile flagella exhibit structural elaborations that are not seen in immotile flagella and in most organisms share a canonical 9 + 2 axoneme structure, which consists of nine outer doublet microtubules arranged symmetrically around a pair of singlet microtubules. Outer doublets provide a scaffold for assembly of dynein motors and regulatory complexes, with adjacent doublets connected via nexin links. Projections extending outward from each of the central pair microtubules form a sheath-like structure that envelopes the central pair (13). Radial spokes extend inward from each outer doublet microtubule and terminate adjacent to the sheath of the central pair microtubules. Recent cryoelectron tomography studies have defined additional axonemal substructures, such as microtubule inner proteins (14) and novel interdoublet linkages (15). Structural variations are present in some organisms, but the general architecture of the 9 + 2 axoneme described above is conserved across broad evolutionary distances, consistent with the idea that the last eukaryotic common ancestor had a motile axoneme (16).
Because of the importance of flagellum motility to human health and disease, there is great interest in identifying essential functional and structural components of these organelles, as these represent candidate disease genes and/or therapeutic targets. Moreover, such efforts offer insight into flagellum biology and motility mechanisms by defining fundamental building blocks needed for assembly and operation of motile axonemes. The deep evolutionary origins of the eukaryotic axoneme, together with the availability of complete genome sequences from many divergent eukaryotes, have made it possible to identify candidate motility genes based on phylogenetic distribution (17–20). These approaches take advantage of the fact that proteins crucial for axoneme motility are conserved in most organisms with motile flagella but are absent in organisms that lack motile flagella. Proteins identified may be conserved across diverse taxa or may represent lineage-specific elaborations, depending upon how the analysis is developed (21). As with any genomic approach, direct analyses of resultant candidates must then be conducted to interrogate the predicted role in axoneme motility.
Previously, we performed an in silico screen to identify genes that are broadly conserved in organisms with motile flagella and absent in organisms that lack a motile flagellum (19). We identified a cohort of 50 genes postulated to serve as core components of motile flagella (CMF), i.e., having axonemal motility functions conserved across diverse taxa. Several CMF genes encode proteins determined to have known or implicated functions in axoneme motility through independent studies. These include subunits of the nexin-dynein regulatory complex (NDRC; trypanin, CMF44, CMF46, CMF70) (22–26), protofilament ribbon proteins (CMF2, CMF3, CMF4, CMF19) (27–29), dynein subunits (CMF39, CMF73) (30, 31), and the trypanosome orthologue of the move backwards only 2 (MBO2) gene product (CMF8) (32, 33). However, a large number of CMF proteins have not yet been subjected to direct, in-depth analysis. Among the cohort of CMF proteins, CMF22 piqued our interest because its amino acid sequence contains IQ and ATPase associated with a variety of cellular activities (AAA) sequence motifs, which have potential to confer regulatory roles via Ca2+ and nucleotide signaling, respectively (34, 35). Each of these is of interest in the context of flagellum function, because calcium ions and nucleotides both function in regulation of flagellum beating (36, 37). Here we report an in-depth analysis of CMF22 in the African trypanosome T. brucei, which has emerged as a powerful experimental system for functional analysis of flagellar proteins (38). We employ expanded phylogenetic analysis, together with biochemical and gene-knockdown approaches, to demonstrate that CMF22 is an axonemal protein required for regulation of flagellar beating.
MATERIALS AND METHODS
Trypanosome transfection and cell maintenance.
Procyclic 29-13 cells (39) were used for all experiments. Cultures were maintained in Cunningham's semidefined medium (SM) supplemented with 10% heat-inactivated fetal calf serum, 15 mg/ml G418, and 50 mg/ml hygromycin (23). Transfection and selection were done as previously described (23). Clonal lines were established through limiting dilution. RNA interference (RNAi) was induced by adding 1 μg/ml tetracycline (Tet).
Database searches.
T. brucei CMF22 (GenBank accession number XP_828418.1) orthologues were identified in genome databases as reciprocal best BLAST hits using the NCBI BLAST portal as well as species-specific portals when necessary and the T. brucei sequence as the original query. Hits were confirmed by individual alignments where there was ambiguity. An organism was listed as having an orthologue if it had a hit and returned T. brucei CMF22 as the top hit in a BLAST search against T. brucei. Domain identification was done using the SMART database (40) and individual alignments where there was ambiguity. A summary of the organisms used and the corresponding accession numbers of the CMF22 orthologues is provided in Table S1 in the supplemental material.
In situ tagging and RNAi knockdown.
In situ tagging of CMF22 was performed using the pMOTag2H vector (41) for direct integration of a tagging cassette at the C terminus of endogenous CMF22. The final 515 bp of the CMF22 open reading frame (ORF) was amplified from 29-13 procyclic genomic DNA with forward (5′-ATGGTACCTGAAGAAAGAGGTACTTAAGGGC-3′) and reverse (5′-ATCTCGAGTTCCTTCTTCTTGGGTCTCTTCTTAG-3′) primers containing a KpnI site (underlined) and an XhoI site (underlined), respectively. This fragment was ligated into the corresponding sites of pMOTag2H directly upstream of the 3× hemagglutinin (HA) cassette. Subsequently, 655 bp of the 3′ untranslated region (UTR) immediately downstream of the CMF22 stop codon was amplified with forward (5′-ATGGATCCAGCCCGGAATGTTCCTCAC-3′) and reverse (5′-ATACTAGTGACGCCGGTCGACCTCGAAG-3′) primers containing a BamHI site (underlined) and a SpeI site (underlined), respectively. This fragment was then cloned into the corresponding sites directly downstream of the puromycin resistance (PuroR) cassette. The entire tagging cassette (CMF22ORF-3×HA-igr-PuroR-CMF22 3′ UTR) was excised from the resulting plasmid and stably transfected into 29-13 cells.
RNAi plasmids were constructed in the p2T7TiB plasmid (42). A 405-bp fragment of the CMF22 3′ UTR was selected using parameters described previously (43, 44) and designed using the RNAit algorithm (45) as described previously (44, 46). This region was amplified with forward (5′-ATGGATCCAGCCCGGAATGTTCCTCAC-3′) and reverse (5′-ATAAGCTTGCTTCGCACCTGTACAACG-3′) primers containing a BamHI site (underlined) and a HindIII site (underlined), respectively. The resulting amplicon was cloned into the corresponding sites of p2T7TiB between opposing, tetracycline-inducible T7 promoters. Following linearization with NotI, this plasmid was stably transfected into 29-13 cells to generate a CMF22 3′ UTR-knockdown (CMF22-UKD) line. All DNA constructs were verified by direct sequencing. The rescue UTR-knockdown RNAi-immune (UKD-Ri) cell line was generated by stably transfecting the CMF22 C-terminal in situ tagging cassette (see above) into the CMF22-UKD line. The same CMF22 C-terminal in situ tagging cassette (see above) was stably transfected into the trypanin-UKD (46) and CKF70-knockdown (CMF70-KD) (25) cell lines to generate trypanin and CMF70 knockdowns that express tagged CMF22. These cell lines were grown in medium without puromycin for a week prior to tetracycline induction.
Cytoskeletal fractionation, Western blotting, and immunofluorescence and electron microscopy.
Procyclic 29-13 cells were subjected to detergent fractionation using a two-step fractionation method as previously described (25, 38). Nonionic 1% Nonidet P-40 (NP-40) was used to solubilize the cell, and the pellet fraction was subsequently extracted in 0.5 M and 1 M NaCl PMN (10 mM NaPO4, pH 7.4, 1 mM MgCl2, 150 mM NaCl) buffer. Cell lysates and detergent fractionations were analyzed by Western blotting as previously described (23). Primary antibody dilutions were as follows: monoclonal anti-HA antibody (HA11.1; Covance), 1:1,000; and monoclonal antitrypanin antibody (monoclonal antibody 37.2), 1:5,000 (23); monoclonal antitubulin antibody E7 supernatant, 1:2,500. Monoclonal antibody E7, directed against beta-tubulin, was developed previously (47) and was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa Department of Biological Sciences.
For immunofluorescence, cells were extracted in PEME buffer [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.9, 2 mM EGTA, 0.1 mM EDTA, 1 mM MgSO4, 25 μg/ml aprotinin, 25 μg/ml leupeptin] containing 1% NP-40 (48). Cell cytoskeletons were fixed in 2% paraformaldehyde, quenched in 0.1 M glycine, and subjected to indirect immunofluorescence microscopy as previously described (38). Procyclic 2913 cytoskeletons were used as a negative control and showed no staining (data not shown). The following primary antibody dilutions were used: monoclonal anti-HA antibody (HA11.1; Covance), 1:200; monoclonal anti-PFR antibody (rat anti-PFR2; a gift from Thomas Seebeck), 1:2,500; mouse monoclonal antitrypanin antibody (9C7), 1:50; rat anti-HA antibody clone 3F10 (Roche Applied Science), 1:100; and monoclonal antitubulin clone YL 1/2 (Millipore), 1:2,500. Cytoskeletons were mounted after staining with Vectashield containing DAPI (4′,6-diamidino-2-phenylindole).
For transmission electron microscopy (TEM), log-phase CMF22-UKD cells were uninduced or induced with tetracycline for 72 h, washed twice with phosphate-buffered saline (PBS), resuspended in 1 ml fixative (3% paraformaldehyde and 3% glutaraldehyde in 0.1 M Na cacodylate containing 1% filtered tannic acid), and incubated for 1 h. For detergent-extracted cytoskeletons, cells were washed twice in PBS and resuspended in PEME buffer plus 1% NP-40 for 10 min. Cells were then pelleted at 4°C and resuspended in 1 ml fixative for 1 h (as described above). Fixed cells were pelleted and resuspended in fixative without tannic acid, treated with 1% osmium tetroxide, dehydrated in ethanol, washed three times in Eponate resin, and embedded in Eponate 182 resin. After polymerization, cells were sectioned into 80-nm-thick sections and analyzed using a JEOL 1230 transmission electron microscope at the University of Iowa Central Microscopy Research Facility (49).
Northern blotting.
Total RNA was extracted from cells using a Qiagen RNeasy miniprep kit. Northern blotting was performed on RNA samples (5 μg/lane) from each cell line as previously described (50), with the exception that digoxigenin (DIG)-labeled probes (DIG nucleic acid detection kit [Roche]) were used in place of 32P-labeled probes to detect RNA levels. A unique 523-bp probe corresponding to nucleotides 1398 to 1920 of the CMF22 ORF was used in accordance with the manufacturer's instructions. rRNA was cross-linked and visualized under UV light.
Growth curves and motility assays/videos.
Cell growth was monitored using a Z1 Coulter particle counter (Beckman Coulter), with curves plotted as cumulative growth, and data points are reported as the averages obtained in three independent experiments with duplicate samples taken at each time point. Error bars indicate the standard deviation of the three independent experiments. For motility assays, cells at a density of 3 × 106 cells/ml were pipetted into polyglutamate-coated motility chambers (51) at room temperature and imaged on a Zeiss Axioskop II microscope within 10 min after removal from the incubator. For motility traces, videos were collected using dark-field optics with a ×20 objective as described previously (51). For tetracycline-inducible conditions, 1 μg/ml tetracycline was added to cultures for 72 h before video capture. Motility traces were performed as described previously (51), and average curvilinear velocity was determined using Metamorph software (Molecular Dynamics). A Student's unpaired t test was also used to determine significance. To quantify parasite propulsive motility, traces were used to calculate the mean squared displacement (MSD) of individual cells in the x and y dimensions according to the formula 〈ri(t)2〉 = 〈[pi(t) − pi(0)]2〉, where ri(t) is the distance traveled by the parasite over time interval t, pi(t) is the position of the parasite at any given time t, and pi(0) is the position of the parasite at the start of time interval t. The time scale of t ranged from 1 to 30 s in increments of 1 s. MSD is calculated for each instance i of a given time interval. Several cell MSDs were then averaged to obtain an ensemble average. Error bars indicate the standard deviation. For CMF22-UKD, 20 uninduced parasites were compared to 20 parasites that were induced with tetracycline for 72 h. For CMF22-UKD-Ri, 20 uninduced parasites were compared to 20 parasites that were tetracycline induced for 72 h. For the tetracycline removal experiment, CMF22-UKD cells were induced with tetracycline for 72 h and tetracycline was removed by diluting cells to 1 × 106 cells/ml in fresh medium without tetracycline, with continuous dilution to maintain log-phase growth for 5 days. After 5 days of tetracycline removal, motility traces were performed and MSD was determined as described above. For single-cell motility videos, cells were imaged using differential interference contrast optics with a ×100 oil objective. Videos were either captured at 30 frames per second with a COHU charge-coupled-device camera and imported using Adobe Premiere Elements software as described previously (51) or captured at 1,000 frames/second using an X-PRI mono 2s high-speed camera (AOS Technologies AG) and the AOS Imaging Studio Light (v2) software from the manufacturer.
RESULTS
The original identification of CMF genes surveyed 10 organisms, and these represented a limited number of eukaryotic clades (19). We therefore expanded our analysis with CMF22 to include 115 organisms and incorporate representatives from each of the five major clades that encompass most eukaryotic diversity (52, 53). Within this set, we included members of the chromalveolate and rhizaria clades that were not represented previously. Using reciprocal best BLAST, we identified CMF22 orthologues in 85 out of 86 organisms with motile flagella, 0 out of 4 with only immotile flagella, and 3 out of 25 with no flagella (see Table S1 in the supplemental material). Notably, a CMF22 orthologue was identified in representatives from all five clades for which there is a complete genome sequence of a ciliated organism available (Fig. 1A). Together, these results indicate that CMF22 was likely present in the last eukaryotic common ancestor and are consistent with a central role for CMF22 in axonemal motility.
Fig 1.
CMF22 is broadly conserved among organisms with motile flagella. (A) Representative organisms from each of the indicated eukaryotic clades are shown along with a depiction indicating whether these organisms have a motile flagellum, an immotile flagellum, or no flagellum. The presence or absence of CMF22 orthologues is noted in the right-most column. Only organisms for which the complete genome sequence is available were considered. Groups are based on the work of Keeling et al. (53). (B) The CMF22 domain architecture is conserved in diverse organisms. The schematic shows the predicted domain architecture for CMF22 orthologues from the indicated organisms. The IQ and AAA motifs were identified using SMART (black and gray boxes) or individual alignments (hash-marked box). The organisms (with their GenBank accession numbers in parentheses) used were as follows: Trypanosoma brucei (XP_828418.1), Homo sapiens (NP_001257513.1), Chlamydomonas reinhardtii (XP_001690665.1), Paramecium tetraurelia (XP_001429179.1), and Bigelowiella natans (jgi|Bigna1|142761). aa, amino acids.
Analysis of the CMF22 primary amino acid sequence revealed the presence of sequence motifs with the potential for regulatory input/output, namely, an IQ motif near the N terminus and an AAA motif near the C terminus (Fig. 1B). The presence and position of the IQ and AAA domains are conserved in CMF22 orthologues from diverse organisms (Fig. 1B), supporting a requirement for these sequence motifs for CMF22 function. Although the SMART predicting algorithm detected a weak hit for the IQ motif in the Bigelowiella natans orthologue, individual alignment showed that all the conserved residues (IQXXXRGXXXR) are present (data not shown). The combined phylogenetic distribution and domain structure of CMF22 support a role in axonemal motility, with potential for regulatory function. We therefore investigated this idea directly through biochemical and functional analysis of the CMF22 protein in T. brucei.
CMF22 is an axonemal protein.
To determine the location of CMF22, we utilized in situ tagging (41) to place a 3× HA epitope at the protein's C terminus. Anti-HA antibodies specifically recognized a single protein of approximately 110 kDa in total cell lysates from the CMF22-HA-tagged cell line (Fig. 2A and B). This size is consistent with the size predicted for the CMF22 protein (102 kDa). To determine if CMF22-HA is associated with the flagellum, cells were extracted with detergent to separate detergent-soluble proteins (S1) from insoluble cytoskeletons (P1). Cytoskeletons were further extracted with 0.5 M NaCl to solubilize the subpellicular cytoskeleton (S2), leaving an insoluble flagellum skeleton (P2) that includes the axoneme, paraflagellar rod (PFR), basal body, and flagellum attachment zone (FAZ) (48). The entire cellular pool of CMF22 fractionated with the axonemal marker trypanin in the flagellum skeleton fraction, even when using high-salt (1 M NaCl) extraction (Fig. 2B). Notably, these high-salt-extraction conditions solubilize most outer arm dyneins as well as much of the central pair apparatus and portions of the PFR (38).
Fig 2.
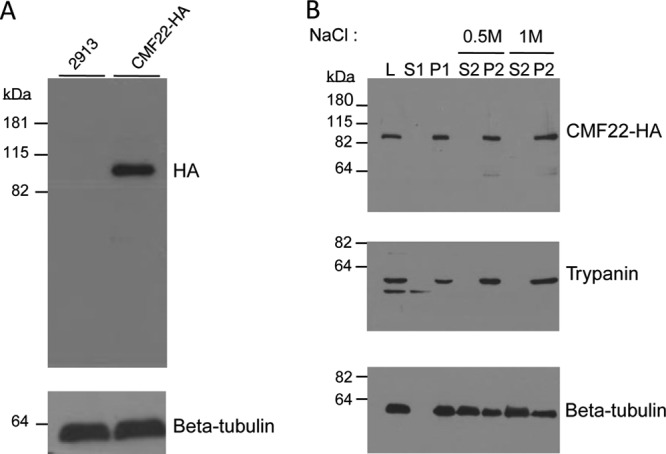
CMF22 is associated with the flagellum skeleton. (A) Western blot of total cell lysates from control (2913) or CMF22-HA-tagged (CMF22-HA) cells. Blots were probed with antibodies against the HA epitope or loading control (tubulin). (B) Western blot of whole-cell lysates (lane L) and the indicated subcellular fractions from CMF22-HA cells. Blots were probed with antibodies against the indicated proteins. Fractions correspond to nonionic detergent-soluble (lane S1) and insoluble (lane P1) fractions, as well as soluble (lane S2) and insoluble (lane P2) fractions obtained by extraction of P1 with NaCl at the concentrations indicated at the top. The lower band in the trypanin blot is likely a degradation product.
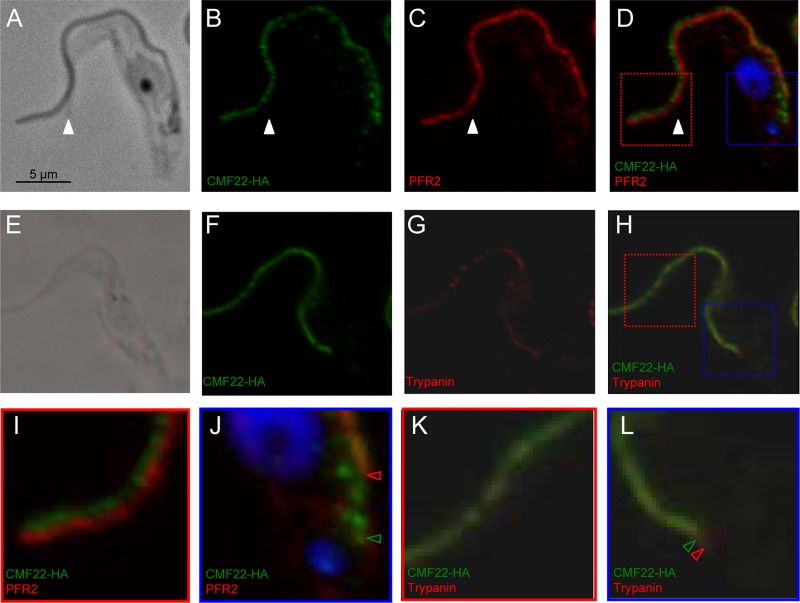
The biochemical fractionation pattern observed for CMF22 is consistent with an axonemal protein but would also be observed for protein components of the basal body, PFR, or FAZ. To distinguish between these possibilities, we used immunofluorescence to examine the distribution of CMF22 in the cell. CMF22-HA was found to be distributed along the entire length of the flagellum (Fig. 3) and clearly distinct from the basal body (see Fig. S1 in the supplemental material). CMF22-HA staining was adjacent to, but not coincident with, PFR staining along most of the flagellum (Fig. 3A to D and I). Moreover, at the proximal end of the flagellum, where the flagellum emerges from the cell body, CMF22-HA staining extended beyond the end of the PFR and nearly all the way to the kinetoplast (Fig. 3J, open arrowheads). Thus, CMF22 is not part of the PFR. At the distal end of the flagellum, CMF22-HA extended all the way to the flagellum tip, beyond the cell body (Fig. 3A to D, white arrowheads, and I), demonstrating that CMF22 is not part of the FAZ, which stops at the end of the cell body (54). Finally, CMF22-HA overlapped and was interspersed with the axonemal marker trypanin along the entire length of the flagellum, including the proximal end of the axoneme (Fig. 3H, K, and L and 4). This staining pattern differs from the side-by-side staining pattern observed for CMF22 and PFR. A similar pattern was also observed in dividing cells, where the newly forming flagellum is seen posterior to the old flagellum (Fig. 4). Therefore, the combined biochemical, immunofluorescence, and phylogenetic data demonstrate that CMF22 is an axonemal protein.
Fig 3.
CMF22 is localized to the axoneme. Indirect immunofluorescence was performed on detergent-extracted cytoskeletons from CMF22-HA cells. Samples were stained with anti-HA antibodies to detect CMF22-HA and costained with antibodies against markers for the PFR (PFR2) (A to D, I, and J) or axoneme (trypanin) (E to H, K, and L), as indicated. DNA was visualized with DAPI (blue). Phase-contrast, individual fluorescence, and merged fluorescence images are shown. Regions boxed in red and blue in panel D are enlarged in panels I and J, respectively. Regions boxed in red and blue in panel H are enlarged in panels K and L, respectively. The white arrowheads in panels A to D mark the end of the cell body. Red and green arrowheads in panel J mark the proximal end of PFR and CMF22 staining, respectively. Red and green arrowheads in panel L mark the proximal end of trypanin and CMF22 staining, respectively.
Fig 4.
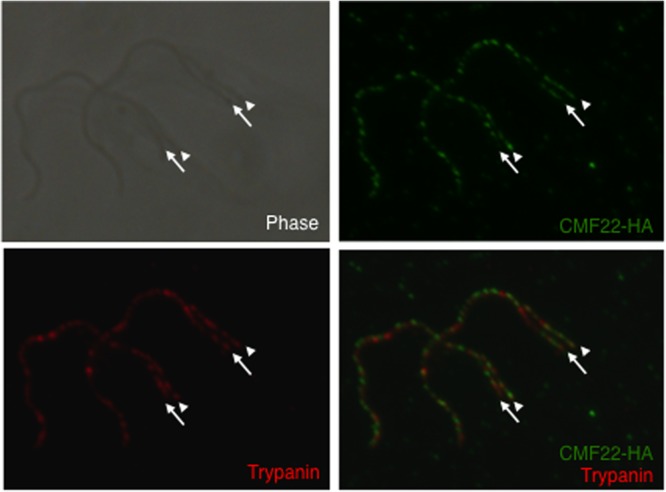
CMF22 localizes to the axoneme in nascent flagella. Indirect immunofluorescence was performed on detergent-extracted cytoskeletons from dividing CMF22-HA cells. Samples were stained with anti-HA (green) and antitrypanin (red) antibodies. The newly emergent daughter flagellum (arrows) is posterior to the old flagellum (arrowheads). Phase-contrast, individual fluorescence, and merged fluorescence images are shown.
CMF22 is required for propulsive motility.
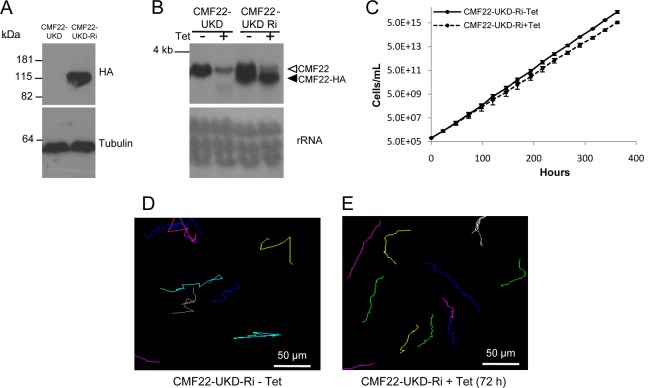
To investigate the CMF22 function, we used Tet-inducible RNAi to target the CMF22 3′ UTR, so as to allow subsequent rescue experiments (see below) (44, 46). Northern blotting using RNA from CMF22-UTR-knockdown (CMF22-UKD) parasites showed a single CMF22 mRNA of approximately 3.7 kb that was dramatically reduced within 72 h of Tet induction (Fig. 5A). Knockdown of CMF22 resulted in a modest growth defect (Fig. 5B), with a doubling time of 14.6 h for the knockdown in comparison to a doubling time of 10.1 h for control cells. This is consistent with prior preliminary analysis of a CMF22 open reading frame knockdown (19). We did not observe any obvious morphological or flagellum defects in the knockdown, based on light microscopy, nor did we observe any obvious ultrastructural defect in the flagellum, based on transmission electron microscopy (see Fig. S2 in the supplemental material). To test the requirement of CMF22 for cell motility, CMF22-UKD parasites were analyzed using high-resolution, single-cell video microscopy, as well as automated particle tracking and motility trace analysis (46). A motility defect was immediately evident in CMF22-UKD cells when analyzed using automated particle tracking (Fig. 5C and D) and high-resolution video microscopy (see Movies S1 and S2 in the supplemental material). Knockdown cells retained a vigorously beating flagellum, but beating was unproductive, as the cells were incapable of translocation, while uninduced controls were motile and readily moved in and out of the field of view (see Movies S1 and S2 in the supplemental material).
Fig 5.
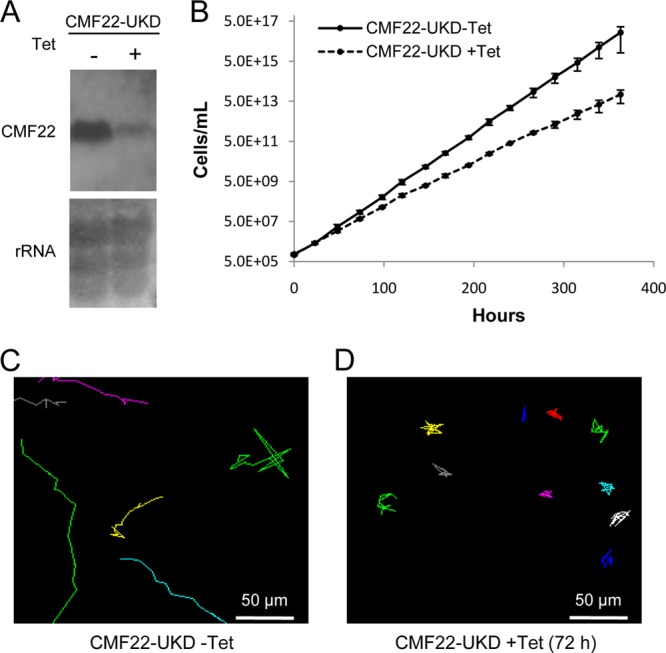
CMF22 knockdown disrupts motility. (A) Northern blot of RNA prepared from CMF22-UKD cells grown in the absence (−) or presence (+) of Tet, as indicated. (Top) Probe with a DNA fragment unique to the CMF22 ORF; (bottom) rRNA as a loading control. (B) Growth curve of CMF22-UKD grown in the absence or presence of Tet for 2 weeks, as indicated. Data are averages of three independent experiments. (C and D) Motility traces of CMF22-UKD grown in the absence or presence of Tet for 72 h, as indicated. Each line traces the movement of an individual trypanosome over a 30-s time interval.
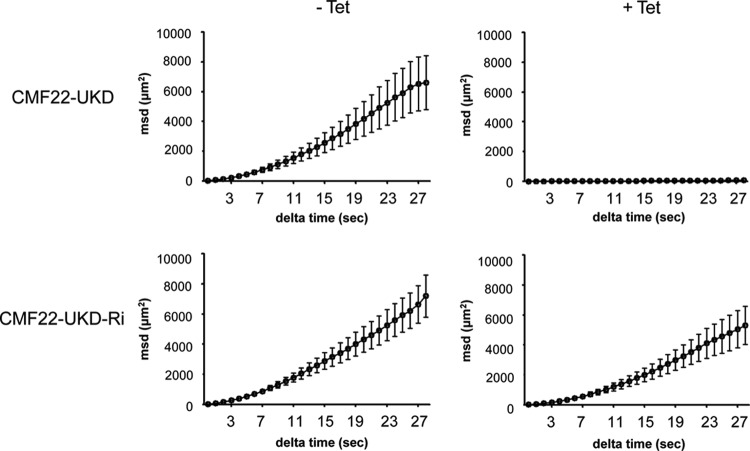
To determine the penetrance of the motility phenotype and quantify the impact on cell movement, we employed motility trace analysis and automated particle tracking (46, 51). CMF22-UKD parasites were significantly slower than the uninduced controls (2.15 μm/s for the knockdown versus 4.48 μm/s for the controls; P < 0.0001). Cell speed provides a reasonable assessment of cell motility but has limitations because it does not filter out Brownian motion, which contributes to the speed calculation. We therefore measured mean squared displacement as a function of the time interval to assess the impact of CMF22 knockdown on propulsive cell movement. Using this approach, we found propulsive cell movement to be essentially absent in the CMF22 knockdown (Fig. 8, top). Thus, CMF22 knockdown resulted in a defective flagellar beat that was incapable of driving propulsive motility. The defect was reversed following removal of tetracycline (see Fig. S3 in the supplemental material).
Fig 8.
CMF22 is required for propulsive motility. MSD is plotted as a function of the time interval (delta time) for CMF22-UKD cells and for CMF22-UKD-Ri cells grown in the absence or presence of Tet, as indicated.
Despite the block in propulsive motility, the flagellum of CMF22-knockdown cells continued to beat. Close examination of flagellum beating using high-speed video microscopy revealed defective coordination of axonemal beating along the length of the flagellum. Beating was erratic and variable in any given cell and from cell to cell, making it difficult to assign a uniform description, but prominent features included erratic movement of the flagellum tip, coupled with sharp bending in the cell's anterior end (see Movies S2 and S4 in the supplemental material). Base-to-tip beats were common (Fig. 6, +Tet; see Movie S4 in the supplemental material), which is in contrast to the findings for control cells, where the dominant beat was tip to base (55, 56) (Fig. 6, −Tet; see Movies S1 and S3 in the supplemental material). Tip-to-base beats could be observed in knockdown cells (see Fig. S4 in the supplemental material), although not as clearly or as consistently as in control cells, and generally did not appear to propagate along the whole cell (see Movies S5 and S8 in the supplemental material). Thus, it appears that erratic and reverse flagellar beating combine to block propulsive cell movement.
Fig 6.

Time-lapse series showing the reverse beat in CMF22-UKD following tetracycline induction. (Top) Control cells (CMF22-UKD cells in the absence of Tet) retain a wild-type tip-to-base beat; time-lapse image series taken from frames at 100 to 300 ms of Movie S3 in the supplemental material are shown. (Bottom) After Tet induction for 72 h, CMF22-UKD cells display a flagellar beat that originates at the base of the flagellum and propagates toward the flagellum tip (reverse beat); frames at 150 to 350 ms of Movie S4 in the supplemental material are shown. White arrows, first waveform; black arrows, second waveform; filled arrows, position of the waveform at each time point; unfilled arrows; approximate position where the waveform originates; P, posterior end of the cell; A anterior end of the cell.
Maintaining CMF22 expression in the knockdown restores motility.
We next asked whether the motility defect in the knockdown was specific to loss of CMF22 expression. For this, we took advantage of the fact that RNAi targets the 3′ UTR of CMF22, allowing us to express CMF22 under inducing conditions by changing the 3′ UTR (44, 46). We used in situ tagging (41) to replace the CMF22 3′ UTR with the alpha-tubulin 3′ UTR, while simultaneously incorporating an HA tag at one CMF22 allele in the knockdown. With this method, an HA-tagged copy of the gene that is immune to RNAi and that is expressed from the endogenous CMF22 locus is generated (46). We refer to this cell line as UTR-knockdown RNAi immune (UKD-Ri). Immunoblotting with anti-HA antibodies showed a single band in lysates from UKD-Ri cells (Fig. 7A). The alpha-tubulin 3′ UTR is smaller than the CMF22 3′ UTR; therefore, CMF22-HA mRNA is smaller than endogenous CMF22 mRNA, and this size difference enabled us to distinguish between the endogenous and RNAi-immune CMF22 mRNA in Northern blots. A probe specific for the CMF22 ORF hybridized to a single mRNA in the UKD parental line and two mRNAs in the UKD-Ri cell line, corresponding to endogenous and HA-tagged mRNAs (Fig. 7B). The abundance of the endogenous mRNA (Fig. 7B, open arrowhead) was dramatically reduced upon RNAi induction, while the HA-tagged mRNA (Fig. 7B, filled arrowhead) was unaffected. Thus, CMF22-UKD-Ri cells retain CMF22 expression even under RNAi induction, and this cell line was used to test for rescue of the motility phenotype.
Fig 7.
HA-tagged CMF22 rescues the motility defect of CMF22-knockdown cells. (A) Western blot of total protein prepared from CMF22-UKD cells or CMF22-UKD-Ri cells probed with anti-HA or antitubulin antibody, as indicated. UKD-Ri cells contain an RNAi-immune HA-tagged CMF22 gene; see the text for details. (B) Northern blot of total RNA prepared from CMF22-UKD or CMF22-UKD-Ri cells grown in the absence or presence of Tet, as indicated. Open arrowhead, endogenous CMF22 mRNA; filled arrowhead, HA-tagged mRNA. Membranes were probed with a DNA fragment specific to the CMF22 ORF (top), and rRNA is shown as a loading control (bottom). (C) Growth curve of CMF22-UKD-Ri cells grown in the absence or presence of Tet, as indicated. (D and E) Motility traces of CMF22-UKD-Ri cells grown in the absence or presence of Tet, as indicated.
RNAi knockdown of endogenous CMF22 had a minimal effect on the growth of CMF22-UKD-Ri cells (Fig. 7C), with a doubling time of 11.5 h compared to a doubling time of 10.5 h for the controls without tetracycline (−Tet). Importantly, UKD-Ri cells retained propulsive motility even upon knockdown of endogenous CMF22, as demonstrated by motility trace (Fig. 7D and E), movies of individual cells (see Movies S6 and S7 in the supplemental material), and mean squared displacement analyses (Fig. 8, bottom). The slope of the MSD curve was slightly less under induced conditions, but propulsive motility was clearly restored. Therefore, the motility defect in CMF22-KD cells is attributed specifically to the loss of CMF22. Moreover, the ability of HA-tagged CMF22 to support propulsive motility demonstrates that the localization of the HA-tagged protein reflects the location of the endogenous protein.
The stability of the CMF22-HA interaction with the axoneme is reduced in NDRC knockdowns.
The phylogenetic distribution, biochemical fractionation, and localization of CMF22, together with the phenotype of CMF22 knockdowns, demonstrate that CMF22 plays an important role in axonemal motility. Consistent with these findings, a recent study found that the CMF22 orthologue in Chlamydomonas reinhardtii showed reduced abundance in axonemes from NDRC mutants, suggesting that it may be associated with the NDRC (57). The fractionation and localization of CMF22, together with the phenotype of CMF22 knockdowns, are consistent with this idea. To test for a potential interaction of CMF22 with the NDRC, we asked whether cofractionation of CMF22 with axonemes was altered in trypanosomes deficient in the NDRC subunit trypanin or CMF70. In control cells, CMF22 fractionated exclusively with 1% NP-40-extracted cytoskeleton pellets and 1 M NaCl-extracted axoneme pellets (Fig. 2). As shown in Fig. 9, Tet-inducible knockdown of trypanin or CMF70 resulted in nearly half of the cellular pool of CMF22 being solubilized with 1% NP-40 (Fig. 9). Therefore, an intact NDRC is required for the stable association of CMF22 with the axoneme. In the reciprocal experiment, the stable association of trypanin with 1 M NaCl-extracted axonemes was not altered by CMF22 knockdown (Fig. 9).
Fig 9.
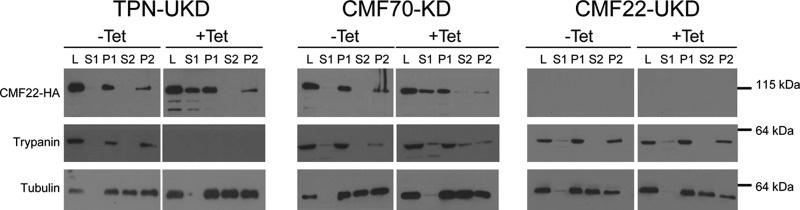
CMF22-HA is less stably associated with the axoneme in NDRC mutants. (A) Western blot of the indicated subcellular fractions from trypanin-UTR-knockdown, CMF70-knockdown, or CMF22-ORF-knockdown cells probed with anti-HA, antitrypanin, or antitubulin antibody, as indicated. Fractions correspond to whole-cell lysate (lanes L), nonionic detergent-soluble (lanes S1) and insoluble (lanes P1) fractions, as well as soluble (lanes S2) and insoluble (lanes P2) fractions obtained by extraction of P1 with 1 M NaCl. The lower-molecular-mass bands in the anti-HA and antitrypanin blots are likely degradation products.
DISCUSSION
Axonemal motility is essential for human development and physiology, as well as for the motility of pathogens that cause tremendous human suffering worldwide (4, 5). Therefore, defining core components of motile axonemes enhances understanding of eukaryotic biology and provides insight into mechanisms of inherited and infectious diseases in humans. In this study, we used phylogenetic, biochemical, and functional analysis to demonstrate that CMF22 is a broadly conserved component of the motile axoneme and is required for wild-type flagellum beating in T. brucei. Ablation of CMF22 expression by RNAi resulted in altered flagellar beating, including erratic and reverse beating, and completely blocked directional cell motility.
CMF22 is represented in each of the five major eukaryotic clades for which there is a representative having a motile flagellum and a completely sequenced genome (Fig. 1). This distribution indicates that CMF22 arose early in eukaryotic evolution as a core component of the machinery that drives flagellar motility. CMF22 function has not been directly studied in other organisms. However, CMF22 mRNA is among the most abundant transcripts in human sperm (96th percentile) and exhibits 5-fold reduced expression in patients with male factor infertility, which is characterized by abnormal sperm morphology and motility (58). These findings support the phylogenetic analyses by indicating a role for CMF22 in flagellar motility in organisms as diverse as trypanosomes and humans. We did not identify a CMF22 orthologue in Plasmodium falciparum, which elaborates a motile flagellum during gametocytogenesis (59). P. falciparum axonemes are not well studied but exhibit unusual features relative to the canonical structure (60). Our analysis suggests either that P. falciparum has dispensed with the need for CMF22 or that the gene sequence has diverged sufficiently to preclude detection on the basis of sequence similarity.
Identification of CMF22 orthologues in three organisms reported to lack a flagellum, Chlorella variabilis, Ostreococcus tauri, and Aureococcus anophagefferens, at first seems contrary to its overall restriction to organisms with motile flagella, but further analysis indicates otherwise. Ostreococcus and Chlorella are both green algae with no flagellated stage yet described. However, as noted previously (20, 61), these organisms retain genes for several flagellum proteins, including NDRC subunits trypanin and CMF70, the dynein subunit LC1, the dynein-NDRC assembly factor CCDC39, and MBO2. Aureococcus is in a different lineage (Chromalveolates), and no flagellated stage is yet described. Again, however, orthologues of flagellar genes are present, such as centriolar Pix proteins (62), and in our own analysis, we found LC1, IFT88, BBS5, and trypanin (data not shown). The presence of flagellar genes in these organisms has been suggested to indicate that they might possess a cryptic flagellated stage, for example, a cryptic gamete stage (63), that flagellar genes have been retained for other functions, or even that they have recently lost flagella and there has not been enough time for genomic loss (20). In any case, given the presence of several genes for flagellum proteins, it is not entirely surprising to find that CMF22 is retained in these organisms.
CMF22 knockdown completely blocks propulsive cell motility but does not cause flagellar paralysis. Rather, flagella with the CMF22 knockdown exhibit an abnormal beating pattern characterized by erratic beating and frequent reversals of waveform propagation. The organism with the knockdown does not show any indication of defective flagellum assembly or major defects in growth that accompany gross disruptions of axoneme structure in T. brucei (23, 64–67). Indeed, TEM analysis did not reveal any obvious defect in axoneme ultrastructure. These data argue for a role of CMF22 in flagellar beat regulation, rather than a structural role or a direct role in generating forces that power microtubule sliding. Two proteins previously shown to function in beat regulation in T. brucei are trypanin and CMF70, which function as part of the NDRC (26). The organism with the CMF22 knockdown exhibits erratic flagellar beating, as observed in the CMF70-knockdown cells, but does not exhibit the continuous cell tumbling that characterizes organisms with the trypanin knockdown (23, 25). Also, despite frequent reversals of flagellar wave propagation, the CMF22-knockdown cells do not exhibit significant backward cell motility, such as that described in outer arm dynein mutants with reverse wave propagation (51, 66). The absence of backward cell locomotion in the CMF22-knockdown cells may be due to reverse (base-to-tip) beats occurring simultaneously with forward (tip-to-base) beats, which counter each other and block propulsive motility. Thus, CMF22 appears to function in beat regulation, but its precise role is distinct from that of previously described flagellar mutations that lead to altered flagellum beating in T. brucei.
Given the flagellum beating defect of the CMF22-knockdown cells, together with biochemical and immunofluorescence data, we expect CMF22 to reside within an axonemal subcomplex that is crucial to motility regulation. Canonical subcomplexes of motile axonemes are inner and outer arm dynein motors, radial spokes, the NDRC, and the central pair apparatus (68). The entire cellular pool of CMF22 remains with insoluble axonemes after extraction with 1 M NaCl, which solubilizes outer arm dyneins and much of the central pair apparatus (38). Thus, it is unlikely that CMF22 is a subunit of outer arm dynein complexes or the central pair apparatus. In support of this conclusion, CMF22 is retained in organisms that lack outer dyneins, Physcomitrella, and in organisms that lack the central pair apparatus, Thalasiossira (20, 69, 70). Notably, NDRC subunits trypanin and CMF70 are likewise conserved in Physcomitrella and Thalasiossira, the latter of which also lacks radial spokes and inner arm dyneins (20). Altogether, these data suggest that CMF22 can function independently of the central pair apparatus, radial spokes, or any single subset of axonemal dyneins and point to the NDRC or one of the novel axonemal subcomplexes recently identified by cryoelectron microscopy (14) to be the site of CMF22 action. Current efforts are aimed at distinguishing between these possibilities. During preparation of the manuscript, Bower et al. (57) reported that the CMF22 orthologue in Chlamydomonas reinhardtii is a candidate NDRC subunit, because it shows reduced abundance in axonemes from drc mutants. Our data support and extend the important Chlamydomonas studies through fractionation, localization, and functional analysis of the T. brucei CMF22 protein and are consistent with an NDRC function for CMF22. As reported for C. reinhardtii, we observed that CMF22 is less stably associated with the axoneme in cells deficient in other NDRC components, which is a hallmark of NDRC subunits (71, 72). The availability of an organism with the CMF22 knockdown allowed us to also ask the reciprocal question, namely, whether or not NDRC subunits are impacted by the loss of CMF22. We found that association of trypanin with salt-extracted axonemes is not affected by CMF22 knockdown. Bower and colleagues (57) suggested that FAP82, the C. reinhardtii CMF22 orthologue, may correspond to one of the NDRC distal densities that contact the B tubule of the adjacent outer doublet (73). The finding that CMF22 knockdown does not alter trypanin fractionation supports this model, and structural studies are under way to directly test this idea.
A notable feature of CMF22 is the protein's domain architecture, which includes an N-terminal IQ motif and a C-terminal AAA domain. IQ motifs function as binding sites for calmodulin and other EF-hand proteins (74). AAA domains are nucleotide binding domains that function in regulation and ATP hydrolysis in a variety of cellular contexts (75). The presence and position of each of these domains are conserved in CMF22 orthologues from diverse organisms (Fig. 1B), indicating that there is selective pressure to retain them and supporting the notion that they are important for protein function. The CMF22 AAA domain lacks the glutamate residue in the Walker B motif that is critical for catalysis (76), so it probably does not hydrolyze ATP. However, it might still function in regulation of motility through nucleotide binding.
The CMF22 IQ motif is of particular interest because it links CMF22 to potential functions in Ca2+ regulation of flagellar motility, which is conserved across diverse phyla and likely has roots in regulatory mechanisms that appeared early in eukaryotic evolution. Chemotaxis of mammalian sperm as well as flagellated protists is dependent upon modulation of the motility apparatus in response to extracellular cues, using Ca2+ as the second messenger (12, 77–81). Targets of Ca2+ are not well understood, but pioneering studies in Chlamydomonas recently identified three axonemal protein complexes that bind calmodulin and are predicted to function in Ca2+ regulation (82–84). The Chlamydomonas CMF22 orthologue, FAP82, was identified in the flagellar proteome (85) but was not identified as part of calmodulin-binding complexes in Chlamydomonas (86). Nonetheless, Ca2+-regulated motility is conserved in another trypanosome species, Crithidia oncopelti, where the direction of flagellar wave propagation is regulated by the concentration of calcium. Forward (tip-to-base) beating predominates at low Ca2+ concentrations (<0.1 mM), and reverse (base-to-tip) beating dominates at higher Ca2+ concentrations (87, 88). The Ca2+ response is observed in demembranated flagella of C. oncopelti (36), indicating that at least some of the targets are axonemal proteins. The axonemal location of CMF22 and frequent reversals of flagellar beating in the organism with the CMF22 knockdown are consistent with a role for CMF22 in flagellar beat regulation.
T. brucei and related trypanosomatids are deadly pathogens that cause tremendous human suffering worldwide, and the flagellum is central to trypanosome biology, transmission, and pathogenesis (89). Distinctive characteristics of the T. brucei flagellum structure and motility suggest specialized mechanisms for controlling flagellar beating and parasite motility (55, 90–93). In addition to providing insight into the fundamental biology of flagella, understanding beat regulation in these organisms thus offers opportunities for discovering novel therapeutic or transmission-blocking strategies. Of the 50 original CMF genes identified in T. brucei (19), 5 have now been demonstrated or implicated to function as part of the NDRC: trypanin, CMF70, CMF46, and CMF44 (22, 23, 25, 26, 64) and, as discussed above, CMF22. While CMF proteins themselves are broadly conserved, they interact with conserved as well as organism-specific proteins (94, 95). Continued analysis of CMF22 and other CMF proteins therefore offers opportunities to enhance understanding of flagellum motility with relevance to the pathogenesis of trypanosomes and related human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
Funding for the work was provided by grants from the NIH (R01AI052348) and the Burroughs Wellcome Fund to K.L.H. H.T.N. is the recipient of an NIH-NRSA fellowship (GM007185). J.S. is the recipient of a Beckman Scholars fellowship. G.L. is the recipient of the Warsaw Fellowship.
We thank Jose Rodriguez and Neville Kisalu for assistance with MSD analysis and Alvaro Sagasti for use of the X-PRI mono 2s high-speed camera. We also thank Edwin Saada and Pius Kabututu for technical assistance. We thank Chantal Allamargot (University of Iowa) for electron microscopy. We are grateful to Stephen King for helpful comments and the members of the Kent Hill laboratory for critical reading of the manuscript and comments on the work.
Footnotes
Published ahead of print 12 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00068-13.
REFERENCES
- 1.Takeda S, Narita K. 2012. Structure and function of vertebrate cilia, towards a new taxonomy. Differentiation 83:S4–S1122118931 [Google Scholar]
- 2.Wilson NF. 2008. Gametic cell adhesion and fusion in the unicellular alga Chlamydomonas. Methods Mol. Biol. 475:39–51 [DOI] [PubMed] [Google Scholar]
- 3.Snell WJ, Pan J, Wang Q. 2004. Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117:693–697 [DOI] [PubMed] [Google Scholar]
- 4.Fliegauf M, Benzing T, Omran H. 2007. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8:880–893 [DOI] [PubMed] [Google Scholar]
- 5.Ginger ML, Portman N, McKean PG. 2008. Swimming with protists: perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6:838–850 [DOI] [PubMed] [Google Scholar]
- 6.Bastin P, Pullen TJ, Moreira-Leite FF, Gull K. 2000. Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes Infect. 2:1865–1874 [DOI] [PubMed] [Google Scholar]
- 7.Vincensini L, Blisnick T, Bastin P. 2011. 1001 model organisms to study cilia and flagella. Biol. Cell 103:109–130 [DOI] [PubMed] [Google Scholar]
- 8.Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. 2008. Kinetoplastids: related protozoan pathogens, different diseases. J. Clin. Invest. 118:1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill KL. 2003. Biology and mechanism of trypanosome cell motility. Eukaryot. Cell 2:200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642 [DOI] [PubMed] [Google Scholar]
- 11.Satir P, Christensen ST. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400 [DOI] [PubMed] [Google Scholar]
- 12.Salathe M. 2007. Regulation of mammalian ciliary beating. Annu. Rev. Physiol. 69:401–422 [DOI] [PubMed] [Google Scholar]
- 13.Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 70:101–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–948 [DOI] [PubMed] [Google Scholar]
- 15.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2009. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell Biol. 186:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607:130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 [DOI] [PubMed] [Google Scholar]
- 18.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117:527–539 [DOI] [PubMed] [Google Scholar]
- 19.Baron DM, Ralston KS, Kabututu ZP, Hill KL. 2007. Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 120:478–491 [DOI] [PubMed] [Google Scholar]
- 20.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickstead B, Gull K. 2007. Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8:1708–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupp G, Porter ME. 2003. A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J. Cell Biol. 162:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ralston KS, Hill KL. 2006. Trypanin, a component of the flagellar dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colantonio JR, Vermot J, Wu D, Langenbacher AD, Fraser S, Chen JN, Hill KL. 2009. The dynein regulatory complex is required for ciliary motility and otolith biogenesis in the inner ear. Nature 457:205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabututu ZP, Thayer M, Melehani JH, Hill KL. 2010. CMF70 is a subunit of the dynein regulatory complex. J. Cell Sci. 123:3587–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. 2011. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286:29175–29191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. 2003. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J. Biol. Chem. 278:7725–7734 [DOI] [PubMed] [Google Scholar]
- 28.Norrander JM, deCathelineau AM, Brown JA, Porter ME, Linck RW. 2000. The Rib43a protein is associated with forming the specialized protofilament ribbons of flagellar microtubules in Chlamydomonas. Mol. Biol. Cell 11:201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linck RW, Norrander JM. 2003. Protofilament ribbon compartments of ciliary and flagellar microtubules. Protist 154:299–311 [DOI] [PubMed] [Google Scholar]
- 30.Bowman AB, Patel-King RS, Benashski SE, McCaffery JM, Goldstein LS, King SM. 1999. Drosophila roadblock and Chlamydomonas LC7: a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146:165–180 [PMC free article] [PubMed] [Google Scholar]
- 31.Yang P, Sale WS. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9:3335–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiBella LM, Sakato M, Patel-King RS, Pazour GJ, King SM. 2004. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol. Biol. Cell 15:4633–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam LW, Lefebvre PA. 2002. The Chlamydomonas MBO2 locus encodes a conserved coiled-coil protein important for flagellar waveform conversion. Cell Motil. Cytoskeleton 51:197–212 [DOI] [PubMed] [Google Scholar]
- 34.Erzberger JP, Berger JM. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93–114 [DOI] [PubMed] [Google Scholar]
- 35.Cheney RE, Mooseker MS. 1992. Unconventional myosins. Curr Opin. Cell Biol. 4:27–35 [DOI] [PubMed] [Google Scholar]
- 36.Sugrue P, Hirons MR, Adam JU, Holwill ME. 1988. Flagellar wave reversal in the kinetoplastid flagellate Crithidia oncopelti. Biol. Cell 63:127–131 [DOI] [PubMed] [Google Scholar]
- 37.Sakato M, King SM. 2004. Design and regulation of the AAA+ microtubule motor dynein. J. Struct. Biol. 146:58–71 [DOI] [PubMed] [Google Scholar]
- 38.Oberholzer M, Lopez MA, Ralston KS, Hill KL. 2009. Approaches for functional analysis of flagellar proteins in African trypanosomes. Methods Cell Biol. 93:21–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 40.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberholzer M, Morand S, Kunz S, Seebeck T. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145:117–120 [DOI] [PubMed] [Google Scholar]
- 42.LaCount DJ, Bruse S, Hill KL, Donelson JE. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol. 111:67–76 [DOI] [PubMed] [Google Scholar]
- 43.Benz C, Nilsson D, Andersson B, Clayton C, Guilbride DL. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125–134 [DOI] [PubMed] [Google Scholar]
- 44.Rusconi F, Durand-Dubief M, Bastin P. 2005. Functional complementation of RNA interference mutants in trypanosomes. BMC Biotechnol. 5:6. 10.1186/1472-6750-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redmond S, Vadivelu J, Field MC. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol. 128:115–118 [DOI] [PubMed] [Google Scholar]
- 46.Ralston KS, Kisalu NK, Hill KL. 2011. Structure-function analysis of dynein light chain 1 identifies viable motility mutants in bloodstream-form Trypanosoma brucei. Eukaryot. Cell 10:884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu DT, Klymkowsky MW. 1989. The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Dev. Biol. 136:104–117 [DOI] [PubMed] [Google Scholar]
- 48.Robinson D, Beattie P, Sherwin T, Gull K. 1991. Microtubules, tubulin, and microtubule-associated proteins of trypanosomes. Methods Enzymol. 196:285–299 [DOI] [PubMed] [Google Scholar]
- 49.Hutchings NR, Donelson JE, Hill KL. 2002. Trypanin is a cytoskeletal linker protein and is required for cell motility in African trypanosomes. J. Cell Biol. 156:867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merchant S, Bogorad L. 1986. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell. Biol. 6:462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baron DM, Kabututu ZP, Hill KL. 2007. Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J. Cell Sci. 120:1513–1520 [DOI] [PubMed] [Google Scholar]
- 52.Koonin EV. 2010. The incredible expanding ancestor of eukaryotes. Cell 40:606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. 2005. The tree of eukaryotes. Trends Ecol. Evol. 20:670–676 [DOI] [PubMed] [Google Scholar]
- 54.Kohl L, Sherwin T, Gull K. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105–109 [DOI] [PubMed] [Google Scholar]
- 55.Walker PJ, Walker JC. 1963. Movement of trypanosome flagella. J. Protozool. 10(Suppl):32 [Google Scholar]
- 56.Kohl L, Bastin P. 2005. The flagellum of trypanosomes. Int. Rev. Cytol. 244:227–285 [DOI] [PubMed] [Google Scholar]
- 57.Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. 2013. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol. Biol. Cell 24:1134–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF, Krawetz SA. 2007. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum. Mol. Genet. 16:763–773 [DOI] [PubMed] [Google Scholar]
- 59.Sinden RE. 1983. Sexual development of malarial parasites. Adv. Parasitol. 22:153–216 [DOI] [PubMed] [Google Scholar]
- 60.Sinden RE, Talman A, Marques SR, Wass MN, Sternberg MJ. 2010. The flagellum in malarial parasites. Curr. Opin. Microbiol. 13:491–500 [DOI] [PubMed] [Google Scholar]
- 61.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL. 2010. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woodland HR, Fry AM. 2008. Pix proteins and the evolution of centrioles. PLoS One 3:e3778. 10.1371/journal.pone.0003778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimsley N, Pequin B, Bachy C, Moreau H, Piganeau G. 2010. Cryptic sex in the smallest eukaryotic marine green alga. Mol. Biol. Evol. 27:47–54 [DOI] [PubMed] [Google Scholar]
- 64.Ralston KS, Lerner AG, Diener DR, Hill KL. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell 5:696–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440:224–227 [DOI] [PubMed] [Google Scholar]
- 66.Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P. 2006. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 119:3443–3455 [DOI] [PubMed] [Google Scholar]
- 67.Ralston KS, Hill KL. 2008. The flagellum of Trypanosoma brucei: new tricks from an old dog. Int. J. Parasitol. 38:869–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porter ME, Sale WS. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37–F42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manton I, Kowallik K, von Stosch HA. 1970. Observations on the fine structure and development of the spindle at mitosis and meiosis in a marine centric diatom (Lithodesmium undulatum). IV. The second meiotic division and conclusion. J. Cell Sci. 7:407–443 [DOI] [PubMed] [Google Scholar]
- 70.Bernhard D, Leipe DD, Sogin ML, Schlegel KM. 1995. Phylogenetic relationships of the Nassulida within the phylum Ciliophora inferred from the complete small subunit rRNA gene sequences of Furgasonia blochmanni, Obertrumia georgiana, and Pseudomicrothorax dubius. J. Eukaryot. Microbiol. 42:126–131 [DOI] [PubMed] [Google Scholar]
- 71.Huang B, Ramanis Z, Luck DJ. 1982. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28:115–124 [DOI] [PubMed] [Google Scholar]
- 72.Piperno G, Mead K, Shestak W. 1992. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118:1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187:921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamniuk AP, Vogel HJ. 2004. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27:33–57 [DOI] [PubMed] [Google Scholar]
- 75.White SR, Lauring B. 2007. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8:1657–1667 [DOI] [PubMed] [Google Scholar]
- 76.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizuno N, Taschner M, Engel BD, Lorentzen E. 2012. Structural studies of ciliary components. J. Mol. Biol. 422:163–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brokaw CJ. 1987. Regulation of sperm flagellar motility by calcium and cAMP-dependent phosphorylation. J. Cell. Biochem. 35:175–184 [DOI] [PubMed] [Google Scholar]
- 79.Gibbons IR, Cosson MP, Evans JA, Gibbons BH, Houck B, Martinson KH, Sale WS, Tang WJ. 1978. Potent inhibition of dynein adenosinetriphosphatase and of the motility of cilia and sperm flagella by vanadate. Proc. Natl. Acad. Sci. U. S. A. 75:2220–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.King SM. 2012. Integrated control of axonemal dynein AAA(+) motors. J. Struct. Biol. 179:222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Satir P. 2011. Coiled-coils and motile cilia. Nat. Genet. 43:10–11 [DOI] [PubMed] [Google Scholar]
- 82.Wargo MJ, Dymek EE, Smith EF. 2005. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 118:4655–4665 [DOI] [PubMed] [Google Scholar]
- 83.Dymek EE, Goduti D, Kramer T, Smith EF. 2006. A kinesin-like calmodulin-binding protein in Chlamydomonas: evidence for a role in cell division and flagellar functions. J. Cell Sci. 119:3107–3116 [DOI] [PubMed] [Google Scholar]
- 84.DiPetrillo CG, Smith EF. 2010. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J. Cell Biol. 189:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pazour GJ, Agrin N, Leszyk J, Witman GB. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heuser T, Dymek EE, Lin J, Smith EF, Nicastro D. 2012. The CSC connects three major axonemal complexes involved in dynein regulation. Mol. Biol. Cell 23:3143–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holwill ME, McGregor JL. 1975. Control of flagellar wave movement in Crithidia oncopelti. Nature 255:157–158 [DOI] [PubMed] [Google Scholar]
- 88.Holwill ME, McGregor JL. 1976. Effects of calcium on flagellar movement in the trypanosome Crithidia oncopelti. J. Exp. Biol. 65:229–242 [DOI] [PubMed] [Google Scholar]
- 89.Hill KL. 2010. Parasites in motion: flagellum-driven cell motility in African trypanosomes. Curr. Opin. Microbiol. 13:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hughes LC, Ralston KS, Hill KL, Zhou ZH. 2012. Three-dimensional structure of the Trypanosome flagellum suggests that the paraflagellar rod functions as a biomechanical spring. PLoS One 7:e25700. 10.1371/journal.pone.0025700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodriguez JA, Lopez MA, Thayer MC, Zhao Y, Oberholzer M, Chang DD, Kisalu NK, Penichet ML, Helguera G, Bruinsma R, Hill KL, Miao J. 2009. Propulsion of African trypanosomes is driven by bihelical waves with alternating chirality separated by kinks. Proc. Natl. Acad. Sci. U. S. A. 106:19322–19327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koyfman AY, Schmid MF, Gheiratmand L, Fu CJ, Khant HA, Huang D, He CY, Chiu W. 2011. Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc. Natl. Acad. Sci. U. S. A. 108:11105–11108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heddergott N, Kruger T, Babu SB, Wei A, Stellamanns E, Uppaluri S, Pfohl T, Stark H, Engstler M. 2012. Trypanosome motion represents an adaptation to the crowded environment of the vertebrate bloodstream. PLoS Pathog. 8:e1003023. 10.1371/journal.ppat.1003023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ralston KS, Kabututu ZP, Melehani JH, Oberholzer M, Hill KL. 2009. The Trypanosoma brucei flagellum: moving parasites in new directions. Annu. Rev. Microbiol. 63:335–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin J, Heuser T, Carbajal-Gonzalez BI, Song K, Nicastro D. 2012. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 69:88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.