Abstract
Protein turnover is a key process for bacterial survival mediated by intracellular proteases. Proteolytic degradation reduces the levels of unfolded and misfolded peptides that accumulate in the cell during stress conditions. Three intracellular proteases, ClpP, HslV, and FtsH, have been identified in the Gram-positive bacterium Staphylococcus aureus, a pathogen responsible for significant morbidity and mortality worldwide. Consistent with their crucial role in protein turnover, ClpP, HslV, and FtsH affect a number of cellular processes, including metabolism, stress responses, and virulence. The ClpP protease is believed to be the principal degradation machinery in S. aureus. This study sought to identify the effect of the Clp protease on the iron-regulated surface determinant (Isd) system, which extracts heme-iron from host hemoglobin during infection and is critical to S. aureus pathogenesis. Inactivation of components of the Clp protease alters abundance of several Isd proteins, including the hemoglobin receptor IsdB. Furthermore, the observed changes in IsdB abundance are the result of transcriptional regulation, since transcription of isdB is decreased by clpP or clpX inactivation. In contrast, inactivation of clpC enhances isdB transcription and protein abundance. Loss of clpP or clpX impairs host hemoglobin binding and utilization and results in severe virulence defects in a systemic mouse model of infection. These findings suggest that the Clp proteolytic system is important for regulating nutrient iron acquisition in S. aureus. The Clp protease and Isd complex are widely conserved in bacteria; therefore, these data reveal a novel Clp-dependent regulation pathway that may be present in other bacterial pathogens.
INTRODUCTION
Staphylococcus aureus is a highly virulent pathogen responsible for considerable morbidity and mortality worldwide. Diseases caused by this organism encompass a wide range of organ involvement and severity. S. aureus can cause relatively mild skin and soft tissue infections as well as devastating invasive infections, including pneumonia, bacteremia, and osteomyelitis (1). S. aureus is the leading cause of superficial skin infections as well as infective endocarditis (2). Development of novel therapeutics to treat S. aureus infection is of paramount importance as the incidence of antibiotic resistance continues to surge and disease in individuals without known risk factors for staphylococcal infection increases (1, 3, 4).
Pathogens must acquire essential nutrients to cause disease. With few exceptions, all living organisms require iron for propagation and survival (5). In the vertebrate host, S. aureus can obtain iron through the acquisition of heme, a molecule consisting of an iron atom coordinated within a tetrapyrrole ring (6). A rich source of heme within the mammalian body is hemoglobin, the tetrameric oxygen-transporting protein. Approximately 80% of the body's iron is bound to hemoglobin and contained within circulating erythrocytes. Hemoglobin is composed of four globin peptides, each of which binds one molecule of heme (7). S. aureus exploits host hemoglobin as an iron source during infection through the activity of the iron-regulated surface determinant (Isd) system, which is composed of several proteins that capture hemoglobin, extract heme, and facilitate its transport across the cell wall and membrane into the cytoplasm (8). Host hemoglobin is primarily targeted through specific recognition by the staphylococcal hemoglobin receptor IsdB (9). IsdB has evolved to preferentially recognize human hemoglobin over hemoglobin from other animal species, indicating that interaction with this molecule may contribute to the species specificity of this pathogen (10). Upon hemoglobin binding, IsdB extracts heme and transfers it to IsdA or IsdC, which shuttle the molecule through the thick peptidoglycan layer to the membrane transport system, IsdDEF (11). Finally, IsdDEF pumps heme into the bacterial cytoplasm, where it is utilized as an intact molecule or is further degraded by the heme oxygenases IsdGI to release iron (12).
The Isd system is tightly regulated by the ferric uptake regulator, Fur. Under conditions of high intracellular iron, Fur dimerizes and interacts with DNA at sites containing its recognition motif, known as Fur boxes. Occupation of Fur boxes effectively inhibits transcription of downstream genes (13). Low iron conditions result in derepression of Fur-regulated genes, as the repressor dissociates from Fur boxes and allows transcription to proceed, promoting uptake of extracellular heme-iron in iron-limiting environments (8). However, an additional level of regulation exists to control levels of the heme oxygenase IsdG. While transcription of isdG is upregulated during iron deprivation, the IsdG protein is stable only in the presence of heme (14). In contrast, IsdI is produced upon iron starvation and is stable in the absence of intracellular heme. This posttranslational regulation of IsdG is the result of rapid degradation of apo-IsdG, a process that depends on a flexible loop within IsdG that is the most divergent region between IsdG and IsdI (15). Previous work has shown that degradation of IsdG requires ATP, suggesting the involvement of an ATP-dependent protease (15). S. aureus expresses three intracellular, ATP-dependent proteases, known as ClpP, FtsH, and HslV (ClpQ), that are important for managing protein levels and directing stress responses. The ClpP protease is believed to be the principal degradation machinery in S. aureus, while FtsH and HslV play less critical roles in the bacterial stress response (16–19). Experiments using strains inactivated for clpP, ftsH, or hslV demonstrated that none of these proteases are responsible for ensuring the heme-dependent stability of IsdG (15).
The Clp proteolytic system is critical for maintaining appropriate levels of unfolded, misfolded, and aggregated proteins within the cell in response to stress (20, 21). S. aureus encodes additional members of the Clp protease system, including the Hsp100/Clp ATPases ClpB, ClpC, ClpL, and ClpX. The Clp ATPases exhibit chaperone-like functions, recognizing and refolding misfolded or aggregated proteins (22). ClpC and ClpX can associate with the ClpP proteolytic subunit to compose the degradation machinery (23, 24). In this capacity, ClpC and ClpX provide the energy for degradation as well as specificity for targeted proteins. Proteins are targeted for degradation by the Clp ATPases through recognition of specific degrons, peptide sequences that tag or are exposed on proteins destined for destruction (25–28). For example, an SsrA tag is added to the C terminus of a protein upon stalling of the ribosome during translation (29, 30). In addition, proteins can be targeted by the N-end rule, which results in degradation of polypeptides upon exposure of a destabilizing residue (F, L, W, or Y) at the amino terminus of the protein (31). Finally, exposure of internal protein sequences following cleavage or activation can also promote degradation (32, 33). Upon substrate recognition, the Clp ATPases unfold and translocate proteins into the proteolytic chamber of ClpP (23, 24, 34, 35). This occurs through conserved motifs that provide ATP binding (Walker A motif) and ATP hydrolysis (Walker B motif) functions (36). As well as their role in proteolytic degradation, Clp ATPases can refold misfolded or aggregated proteins (22, 24, 37, 38). The Clp ATPases therefore provide both the energy required for degradation and the specificity for targeted proteins.
While a major function of proteolytic machinery is to maintain appropriate levels of misfolded or aggregated proteins that accumulate in response to various stressors, it also allows invading pathogens to quickly adapt to changing environments during infection (39). Alteration of entire genetic programs can be inefficient, whereas targeted protein degradation can rapidly alter the phenotypic profile of the organism in response to new environments or insults. Thus, direct targeting and degradation of specific proteins, including transcriptional regulators, allow the pathogen to quickly adjust its proteome and respond to environmental cues (19, 40, 41).
The data presented herein demonstrate that the Clp proteolytic system affects IsdB-dependent hemoglobin binding in S. aureus. Inactivation of clpP or its associated ATPase gene, clpX, dramatically influences abundance of the hemoglobin receptor IsdB, which subsequently affects hemoglobin capture and heme utilization by S. aureus. Components of the Clp system are required for efficient transcription of isdB, though this regulation does not involve Fur. Because of the critical role of host heme acquisition in staphylococcal pathogenesis (14, 42), we tested the contributions of individual Clp components to virulence in a systemic mouse model. Inactivation of Clp genes significantly reduced virulence in a systemic model of staphylococcal infection. Based on these data, we propose that the Clp proteolytic system regulates an additional, unknown factor that influences transcription of isdB. These findings could represent a novel Clp-dependent mechanism of regulating nutrient acquisition that may be relevant to the pathogenic mechanisms of other microbes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All experiments were performed with S. aureus Newman as the wild-type strain, and all mutants were generated in this background (43). A list of strains used in these studies is included in Table S1 in the supplemental material. The Newman clpP knockout with the clpP gene stably integrated into the chromosome was generated as described previously (44). Mutants for hslUV (clpYQ), ftsH, and fur have been reported previously (15, 45). Strains inactivated for clpB, clpC, clpL, and clpX were generated by allelic replacement as described previously (46). The ΔclpP Δfur double mutant was created by transducing Δfur::tetM into the ΔclpP knockout background as described previously (12). Triple mutants were generated by transducing ΔcodY::ermC, ΔagrA::ermC, ΔsaeR::ermC, or Δrot::tetM into the ΔclpP Δfur double mutant background. The ΔcodY::ermC and ΔagrA::ermC isolates were acquired from the Network of Antimicrobial Resistance in Staphylococcus aureus (NARSA) program. The ΔsaeR::ermC and Δrot::tetM mutants were generous gifts from C. Lee and V. Torres, respectively.
Iron-rich conditions were achieved by growing S. aureus strains in tryptic soy broth (TSB) (Difco), whereas iron restriction was generated by the addition of a 100 to 500 μM concentration of the iron chelator 2,2-dipyridyl (DIP) to TSB or RPMI 1640 medium (Invitrogen) supplemented with 1% Casamino Acids (RPMI + CAS).
Medium devoid of iron was generated by treating RPMI + CAS with 7% Chelex. The resulting medium, NRPMI, was then supplemented with 25 μM ZnCl2, 25 μM MnCl2, 100 μM CaCl2, and 1 mM MgCl2. To analyze growth in iron-deplete media supplemented with various iron sources, overnight cultures were grown in 0.5 mM EDDHA (iron chelator) and subcultured 1:50 into NRPMI with 1.5 mM EDDHA with either 300 μM FeSO4, 2 μM heme, or 2.5 μg/ml purified human hemoglobin. Bacterial growth was monitored over 24 h by measuring the optical density of the cultures at 600 nm (OD600).
Immunoblotting.
Bacteria were grown in TSB for 8 h and then subcultured 1:100 into medium containing TSB with or without DIP and incubated at 37°C with agitation for approximately 15 h. Cultures were harvested by centrifugation, and cell wall fractions were obtained following digestion with lysostaphin and subsequent centrifugation. Total protein content was assessed by bicinchoninic acid (BCA) analysis and normalized. Approximately 40 μg of total protein from each sample was separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with polyclonal rabbit serum against IsdB and with secondary goat anti-rabbit conjugated to Alexa Fluor 680 (Invitrogen). Imaging of blots and densitometric analyses were achieved using an Odyssey infrared imaging system (Li-Cor) at 700 nm. Student's t test was used to determine statistical significance between samples.
IsdB immunofluorescence.
Fluorescent labeling of surface-exposed IsdB was performed using a modification of a previous protocol (42). Briefly, overnight bacterial cultures grown in iron-rich or iron-poor medium were washed three times with cold phosphate-buffered saline (PBS; pH 7.4) and incubated with glass coverslips that had been precoated with poly-l-lysine. Bacteria were fixed to the slides by use of 2% formaldehyde. Coverslips were washed with PBS and blocked using a 1:50 goat anti-protein A polyclonal antibody in 3% bovine serum albumin (BSA) for 1 h to reduce protein A-dependent, nonspecific antibody binding. Coverslips were washed with PBS and incubated with polyclonal rabbit anti-IsdB for 2 h. Following incubation with primary antibody, the coverslips were washed three times with PBS and incubated with goat anti-rabbit Alexa Fluor 488 for 1 h. Coverslips were washed three final times and sealed face down on glass slides with nail polish. Imaging of coverslips was achieved using a fluorescence microscope. ImageJ software was used to quantify the fluorescence signal in each field (47).
Hemoglobin cosedimentation.
Hemoglobin binding by S. aureus strains was assessed as described previously (10). Briefly, hemoglobin was purified from human blood obtained from anonymous donors. S. aureus strains were grown overnight in RPMI + CAS and supplemented with 0.5 mM DIP to generate iron-restricted conditions. Bacterial cultures were normalized based on the optical density at 600 nm, resuspended in 1 ml PBS containing 10 μg/ml hemoglobin, and incubated at 37°C for 30 min with shaking at 180 rpm. Bacteria were then washed three times with ice-cold PBS, resuspended in 4% sodium dodecyl sulfate (SDS)–0.5 M Tris-HCl (pH 8.0), and boiled to release bound hemoglobin. Hemoglobin was separated from bacteria by centrifugation and subjected to 15% SDS-PAGE. Gels were silver stained (GE Healthcare), and the relative abundance of bound hemoglobin was determined by densitometric analysis using ImageJ software (47). Student's t test was used to determine statistical significance between samples.
RNA isolation.
Bacterial RNA was harvested using a modification of a previously described protocol (48). Overnight cultures of wild-type and mutant S. aureus were subcultured (1:100) in triplicate into TSB alone or supplemented with 1 mM DIP. Cultures were harvested at late exponential phase (OD600 = 1.0), mixed with an equal volume of ice-cold ethanol-acetone (1:1), and stored at −80°C. For RNA isolation, samples were thawed on ice and centrifuged at 5,000 × g for 10 min. Pellets were resuspended in LETS buffer (1 M LiCl, 0.5 M EDTA, 1 M Tris-HCl, pH 7.4, 10% SDS) and lysed in lysing matrix B tubes (MP Bio) by using a FastPrep Instrument (MP Bio) set at 6 m/s for 45 s. Samples were heated at 55°C for 5 min and centrifuged for 10 min at 16,000 × g. RNA was extracted by collecting the supernatant, mixing it with 1 ml TRIzol (TRI reagent; Sigma), and incubating the mixture for 5 min at room temperature. Chloroform (200 μl) was added to the samples, which were mixed vigorously for 15 s, incubated for 2 min at room temperature, and then centrifuged at 16,000 × g for 15 min. RNA was precipitated by collecting the top aqueous phase, mixing it with 1 ml isopropyl alcohol, and incubating the mixture for 10 min at room temperature. Samples were centrifuged, washed with 70% ethanol, and air dried for 1 min. The RNA pellet was then dissolved in 100 μl distilled water (dH2O). Contaminating DNA was removed by adding 10 units of DNase I (Amersham Biosciences) to RNA samples and incubating them in RQ1 buffer (Promega) at 37°C for 30 min. Following DNase treatment, the samples were cleaned using an RNeasy minikit according to the manufacturer's protocol (Qiagen). RNA concentration and purity were measured by determining the optical densities at 260 nm and 280 nm, respectively.
Real-time reverse transcription-PCR (RT-PCR).
For cDNA synthesis, 2 μg of total cellular RNA was reverse transcribed with Moloney murine leukemia virus (M-MLV) reverse transcriptase according to the manufacturer's recommendations (Promega). cDNA was amplified in triplicate by using iQ SYBR green Supermix (Bio-Rad) in an iCycler iQ instrument (Bio-Rad). Student's t test was used to determine statistical significance between samples.
Systemic mouse infection.
Animal protocols were approved by the Vanderbilt University IACUC. Seven-week-old female BALB/c mice (Jackson Laboratories) were infected with 107 CFU of S. aureus wild-type bacteria or strains inactivated for clpP, clpC, or clpX. Cultures were normalized, suspended in PBS, and inoculated into mice by retro-orbital injection. Mice were euthanized 96 h after infection, at which time kidneys, livers, and hearts were removed and homogenized in sterile PBS. The bacterial burden in each organ was assessed by enumerating CFU on tryptic soy agar. Student's t test was used to determine statistical significance between samples.
RESULTS
Inactivation of the S. aureus ClpP protease affects binding of host hemoglobin.
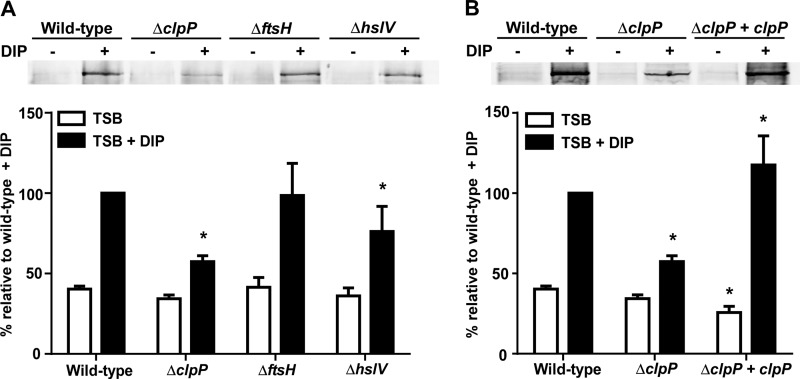
In an effort to identify the protease that degrades the apo-IsdG heme oxygenase, we performed immunoblot analyses to examine the effects of protease mutants on the abundance of Isd proteins under iron-rich and iron-poor conditions. While inactivation of clpP, ftsH, or hslV did not affect the abundance of IsdG (15), protein levels of the hemoglobin receptor IsdB were significantly decreased in strains inactivated for clpP or hslV (Fig. 1A). Loss of clpP also resulted in decreased abundance of several other Isd proteins, although the proteins were not affected to the same extent (see Fig. S1 in the supplemental material). Notably, restoration of wild-type IsdB protein levels was achieved by stably integrating clpP into the chromosome of a clpP-null strain, which demonstrated that the reduction in IsdB abundance was due to the loss of clpP (Fig. 1B). Loss of ftsH did not significantly alter IsdB abundance.
Fig 1.
Disruption of the S. aureus clpP proteolytic subunit reduces abundance of the hemoglobin receptor IsdB. (A) IsdB protein levels analyzed by immunoblotting. Wild-type S. aureus or strains inactivated for clpP, ftsH, or hslV were grown in TSB or TSB supplemented with the iron chelator DIP (1 mM). Total protein in cell wall fractions was normalized and separated by 15% SDS-PAGE. Proteins were transferred to nitrocellulose and probed with an anti-IsdB antibody. (Top) Blots are representative of at least 4 independent experiments. (Bottom) Graphical representation of IsdB protein abundance in TSB (white bars) or TSB plus DIP (black bars) as assessed by densitometry analysis of immunoblots. (B) Restoration of IsdB protein levels through complementation of clpP, assessed by immunoblotting (top) and densitometry (bottom). Data represent 4 independent experiments. Error bars represent standard errors of the means (SEM). *, P <0.025 relative to the wild type under the respective conditions, as calculated by Student's t test.
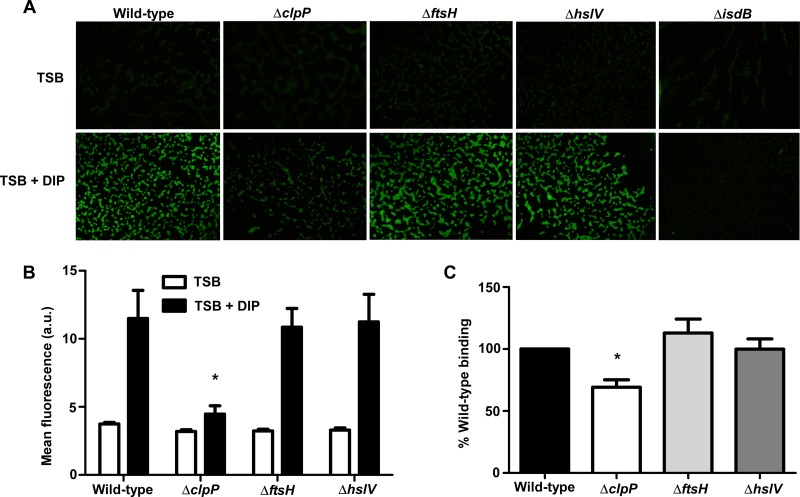
Recognition and binding of host hemoglobin by the IsdB receptor are the initiating steps of hemoglobin-dependent heme acquisition in S. aureus (9). As such, IsdB plays a critical role in obtaining exogenous heme-iron and promoting staphylococcal pathogenesis (42, 49). Since reduced IsdB protein levels could drastically affect hemoglobin recognition, the effects of protease knockouts on IsdB surface exposure and hemoglobin binding were analyzed. Localization of IsdB on the bacterial surface of protease mutants was monitored using immunofluorescence. These experiments revealed considerably less IsdB presented on the surface of the clpP mutant, as evidenced by a decrease in fluorescence labeling of ΔclpP cells incubated with anti-IsdB antibody. No difference was observed in IsdB surface localization in strains inactivated for ftsH or hslV compared to wild-type S. aureus (Fig. 2A and B). The reduction in IsdB levels in the ΔclpP strain corresponded with significantly decreased hemoglobin binding in a cosedimentation assay (Fig. 2C). In contrast, hemoglobin binding by strains inactivated for ftsH or hslV was similar to that of wild-type S. aureus. The immunofluorescence and hemoglobin binding data suggest that the reduction in IsdB abundance in the hslV mutant observed by immunoblotting did not significantly impair IsdB surface exposure or hemoglobin recognition. These results indicate that ClpP is required for effective hemoglobin capture by S. aureus.
Fig 2.
Disruption of the S. aureus clpP proteolytic subunit impairs binding of host hemoglobin. (A) Surface exposure of IsdB as analyzed using immunofluorescence microscopy. Wild-type S. aureus or protease knockouts were grown in iron-rich (TSB) or iron-poor (TSB + DIP) medium, fixed on coverslips, and probed with anti-IsdB and a secondary antibody conjugated to Alexa Fluor 488. Images are representative of at least 2 experiments. (B) Graphical representation of IsdB immunofluorescence. a.u., arbitrary units. *, P < 0.001. (C) Hemoglobin capture by wild-type or mutant S. aureus strains was determined using a whole-cell cosedimentation assay. Iron-starved cells were incubated with human hemoglobin, and captured hemoglobin was removed by pelleting cells, treating them with SDS, and boiling the samples. Bound hemoglobin was separated by SDS-PAGE, silver stained, and analyzed by densitometry. Data represent at least 5 experiments. Error bars represent SEM. *, P < 0.0001 relative to the wild type, as calculated by Student's t test.
ClpP is required for hemoglobin-dependent heme acquisition.
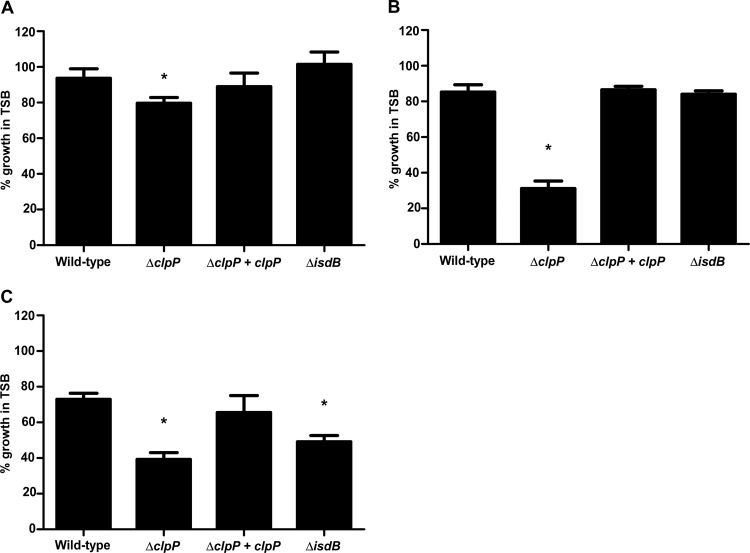
Since loss of clpP caused a significant reduction in IsdB protein levels and hemoglobin binding, we hypothesized that heme acquisition would also be substantially impeded in this mutant. To test this hypothesis, we measured growth of the ΔclpP strain under iron-depleted conditions supplemented with various iron sources and compared it to that of wild-type bacteria or a strain inactivated for isdB. Growth of the ΔclpP strain was significantly reduced compared to that of the wild type when iron sulfate, heme, or hemoglobin was added as the sole iron source; however, the defect was most notable with heme or hemoglobin (Fig. 3). Loss of clpP is known to reduce bacterial growth, which is consistent with the observed growth defect in the presence of iron sulfate (see Fig. S2 in the supplemental material) (40). The reduced growth in the presence of heme likely reflects the reduced abundance of heme transport proteins such as IsdA (see Fig. S1). Finally, the inability of the clpP knockout to grow using hemoglobin as an iron source is similar to that of an isdB mutant and suggests that clpP is required for hemoglobin acquisition due to its effects on IsdB. Importantly, all growth defects of the clpP mutant were rescued upon expression of a wild-type copy of clpP in trans (Fig. 3).
Fig 3.
ClpP is required for hemoglobin-dependent heme acquisition. Wild-type S. aureus, knockouts of clpP or isdB, and the complemented clpP mutant were grown in iron-depleted medium and supplemented with iron sulfate (A), heme (B), or hemoglobin (C). Growth was determined by measuring the optical density at 600 nm, and data are expressed as percent growth compared to that of each strain in iron-rich medium at 24 h. Data shown are means and SEM for three separate experiments. *, P < 0.0004 relative to the wild type, as calculated by Student's t test.
Inactivation of Clp ATPases affects binding of host hemoglobin.
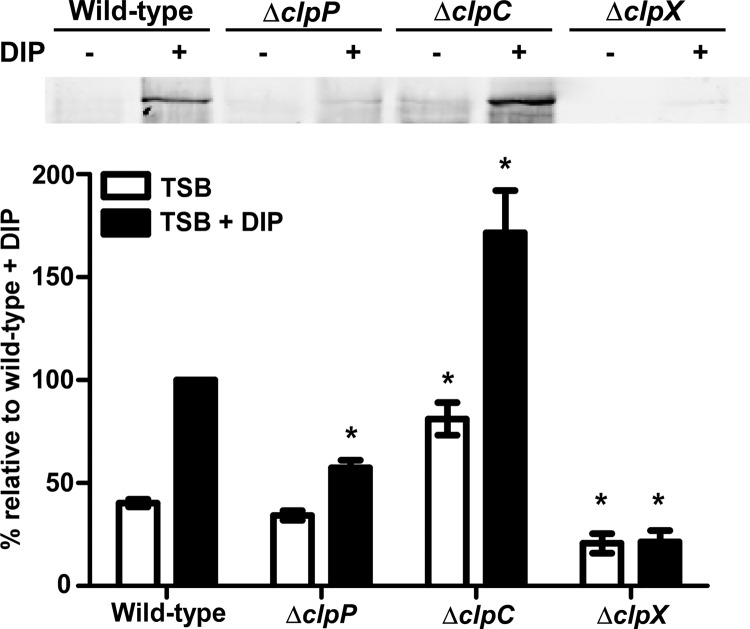
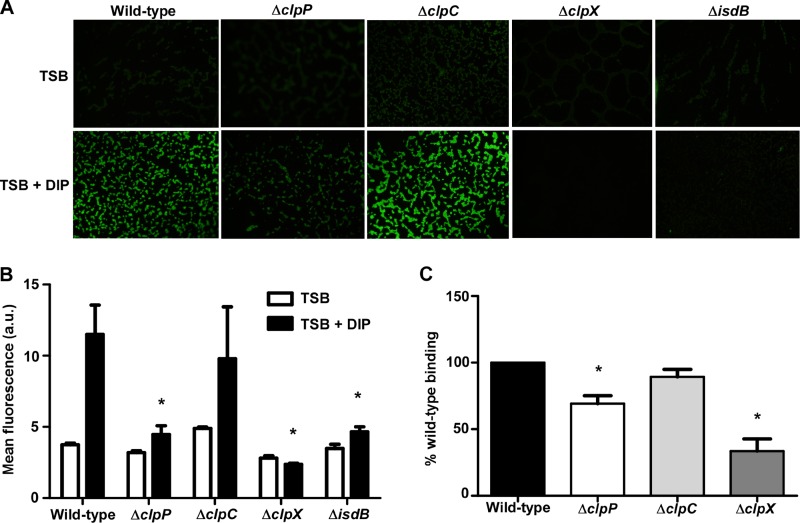
To determine if additional components of the Clp proteolytic system affect IsdB protein levels, immunoblots on cell wall fractions from strains inactivated for individual Clp ATPases were performed. IsdB levels were unchanged in the absence of clpB or clpL (see Fig. S3 in the supplemental material) but were significantly increased and decreased in the clpC and clpX mutants, respectively (Fig. 4). Strikingly, IsdB protein levels in the clpX mutant were almost undetectable by immunoblotting. IsdB surface exposure and hemoglobin binding were also significantly reduced in this strain (Fig. 5). These results demonstrate that multiple components of the ClpP proteolytic system alter the abundance of IsdB.
Fig 4.
Inactivation of the Clp ATPases drastically alters IsdB protein levels. (Top) IsdB protein levels were analyzed by Western blotting. Wild-type S. aureus or strains inactivated for clpP, clpC, or clpX were grown in iron-rich (TSB) or iron-poor (TSB + DIP) medium. Total protein in cell wall fractions was normalized and separated by 15% SDS-PAGE. Proteins were transferred to nitrocellulose and probed with antibody specific to IsdB. Blots are representative of at least 4 independent experiments. (Bottom) Graphical representation of IsdB protein abundance in TSB (white bars) or TSB plus DIP (black bars) as assessed by densitometry analysis of Western blots. Error bars represent SEM. *, P < 0.0002 relative to the wild type under the respective conditions, as calculated by Student's t test.
Fig 5.
Inactivation of clpX ATPase significantly impairs host hemoglobin binding. (A) Surface exposure of IsdB as determined by immunofluorescence microscopy. Wild-type S. aureus or strains inactivated for components of the Clp proteolytic system were grown in iron-rich or iron-poor medium, fixed on coverslips, and probed with anti-IsdB and a secondary antibody conjugated to Alexa Fluor 488. Images are representative of at least 2 experiments. (B) Graphical representation of IsdB immunofluorescence. *, P < 0.003. (C) Hemoglobin capture of wild-type or mutant S. aureus strains was determined using a whole-cell cosedimentation assay. Iron-starved cells were incubated with human hemoglobin, and captured hemoglobin was removed by pelleting cells, treating them with SDS, and boiling the samples. Bound hemoglobin was separated by SDS-PAGE, and gels were silver stained and analyzed by densitometry. Data represent at least 5 experiments. Error bars represent SEM. *, P < 0.0001 relative to the wild type, as calculated by Student's t test.
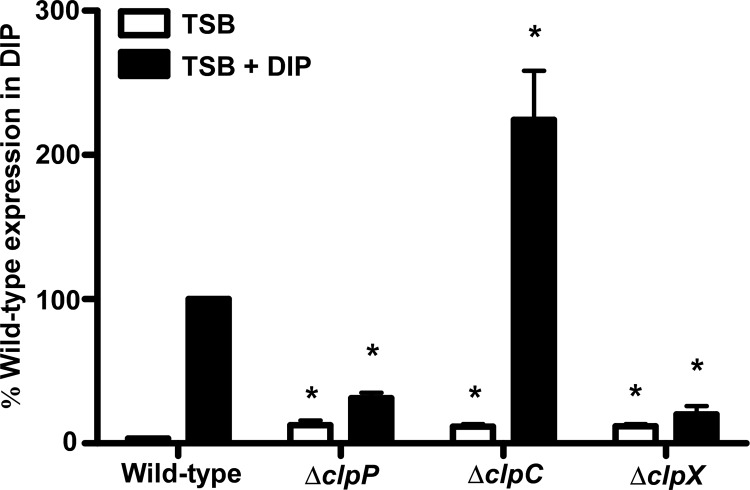
Disruption of Clp genes alters isdB abundance at the transcriptional level.
If the ClpXP proteolytic system is responsible for degrading IsdB, then an abundance of the IsdB receptor should increase following clpP or clpX inactivation. However, the reduction in IsdB levels observed in the absence of ClpP or ClpX indicates that this is not the case. While an increase in IsdB was observed upon inactivation of clpC, this does not imply that ClpC degrades IsdB, since this ATPase is not known to have protease activity alone but instead must complex with ClpP to promote proteolysis (50). Therefore, we hypothesized that the Clp system may indirectly affect IsdB via proteolysis of an isdB regulatory protein. To examine this possibility, expression of isdB was analyzed in the clp knockout backgrounds under iron-rich and iron-poor conditions. Quantitative real-time PCR analysis demonstrated that isdB expression was significantly decreased in both the clpP and clpX knockouts under iron-poor conditions compared to the expression in wild-type S. aureus (Fig. 6). In contrast, isdB expression was enhanced in the clpC mutant, which corresponded to increased IsdB protein levels observed by immunoblotting in this strain. The functional consequence of the ClpC-dependent increase in IsdB is not entirely clear, since hemoglobin binding did not appear to be affected in this mutant (Fig. 5C). These results support the notion that the Clp system alters IsdB abundance indirectly via an additional effector molecule that affects transcription of isdB.
Fig 6.
Inactivation of clp genes alters isdB expression. Expression of isdB was measured using qRT-PCR. Wild-type S. aureus or strains inactivated for clpP, clpC, or clpX were grown in TSB (white bars) or TSB with 1 mM DIP (black bars) to late exponential phase. RNAs were isolated from samples and converted to cDNAs. Expression of isdB was analyzed using primers specific to the isdB coding sequence. Data are expressed as fold transcript levels relative to wild-type isdB expression in TSB, which was set at 1. The graph represents data from 3 individual experiments. Error bars represent SEM. *, P < 0.007 relative to the wild type under the respective conditions, as calculated by Student's t test.
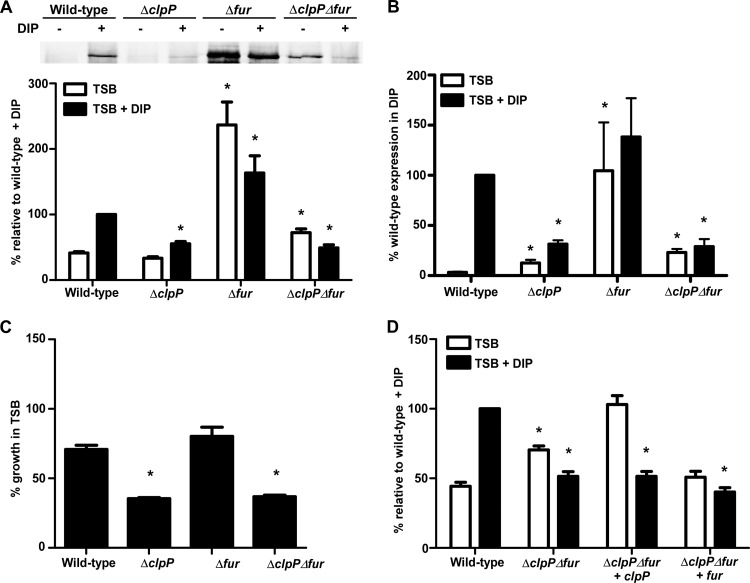
Clp-dependent regulation of isdB expression occurs independently of Fur.
Since Fur is the only known regulator of the Isd system, we hypothesized that the Clp proteins affected isdB transcription by altering Fur-mediated regulation. To address this hypothesis, IsdB protein levels were examined in a strain inactivated for both fur and clpP and compared to single Δfur and ΔclpP knockouts. As expected, high IsdB protein levels were observed in the Δfur knockout, irrespective of iron levels, demonstrating derepression of isdB in the absence of Fur (Fig. 7A and B). Strikingly, the double mutant lacking both fur and clpP produced significantly less IsdB than did the single Δfur mutant (Fig. 7A and B). Appreciable IsdB levels were observed in the ΔclpP Δfur double mutant under iron-rich conditions, although these levels were significantly reduced compared to those with inactivation of fur alone. IsdB levels in the ΔclpP Δfur double knockout under iron-poor conditions were significantly lower than those of the single Δfur mutant and similar to levels observed in the ΔclpP single mutant. Analysis of IsdB transcripts by quantitative RT-PCR (qRT-PCR) revealed that changes in IsdB protein levels in the ΔclpP Δfur double mutant occurred via transcriptional regulation (Fig. 7B). Furthermore, repression of isdB in the ΔclpP Δfur double mutant correlated with a significant growth defect when hemoglobin was the sole iron source (Fig. 7C). Complementation of the double mutant with clpP, which effectively generated a single fur mutant, resulted in alleviation of isdB repression under iron-rich conditions, whereas the IsdB protein was not produced under conditions of iron restriction (Fig. 7D). In contrast, complementation of the double mutant with fur resulted in reduced IsdB protein levels consistent with those of a single clpP mutant. These data suggest that either the Clp protease degrades an additional factor that inhibits expression of isdB in the absence of Fur or the ClpP protease activates or stabilizes an additional factor that is required to promote isdB expression.
Fig 7.
Clp proteins influence IsdB independently of Fur. (A) (Top) IsdB protein levels were analyzed by immunoblotting. Wild-type S. aureus or strains inactivated for fur alone or in combination with the knockout of clpP were grown in TSB alone or in the presence of DIP (1 mM), and cell wall fractions were blotted for IsdB. Blots are representative of 4 independent experiments. (Bottom) Graphical representation of IsdB protein abundance in TSB (white bars) or TSB plus DIP (black bars) as determined by densitometry analysis of Western blots. (B) Quantitative RT-PCR analysis of isdB expression in fur mutants. Error bars represent SEM. *, P < 0.015 relative to the wild type under the respective conditions, as calculated by Student's t test. (C) Wild-type or mutant strains were assessed for growth with hemoglobin as the sole iron source. *, P < 0.001 relative to the wild type at 24 h. (D) IsdB protein levels in ΔclpP Δfur mutants complemented with either clpP or fur on a plasmid were determined by Western blotting.
S. aureus Clp proteins are required for pathogenesis in a systemic staphylococcal infection.
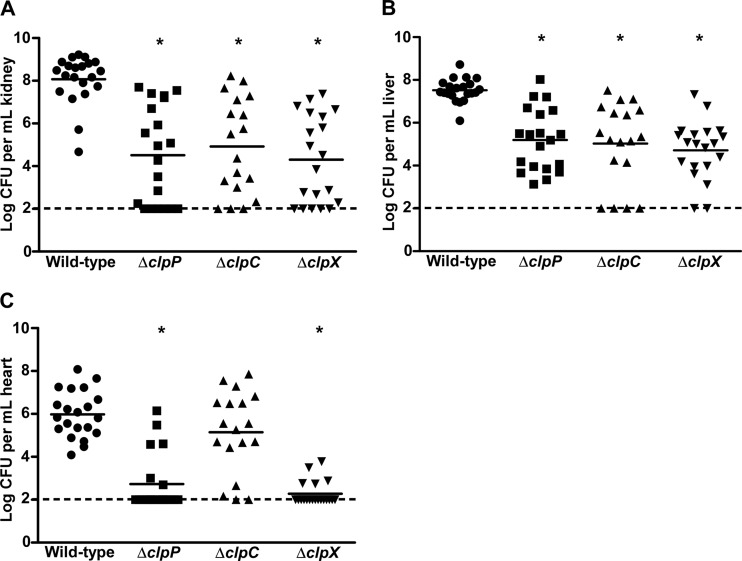
Isd-dependent heme acquisition is an important survival mechanism that S. aureus utilizes to combat iron restriction in the host. Previous work has shown that isd genes are expressed in vivo, and inactivation of individual components of the Isd system significantly reduces pathogenicity (14, 42). Loss of isdB alone leads to a 2-log decrease in colonization of murine hearts following systemic infection (42). In addition, IsdB is required for the formation of deep-seated abscesses within infected organs in a systemic infection model (49). Thus, based on data demonstrating a significant decrease in IsdB abundance and hemoglobin binding in the clpP and clpX knockouts, it was hypothesized that inactivation of individual Clp components would impede staphylococcal pathogenesis in an invasive infection model where hemoglobin-dependent heme acquisition is known to occur.
Systemic infection following retro-orbital inoculation of 7-week-old female BALB/c mice with clpP or clpX knockouts resulted in significantly reduced bacterial burdens in all organs tested (Fig. 8). The reduced growth rates of clpP and clpX mutants compared to wild-type S. aureus in liquid culture likely contributed to the observed virulence defects (see Fig. S2 in the supplemental material) (40). In addition, loss of clpC led to a reduction in bacterial burdens in the kidneys and livers, but the burdens in the hearts of infected animals were not different from those for the wild type (Fig. 8). Other groups have reported virulence defects in clp knockouts, using different S. aureus strains and alternate infection models (17, 51). Taken together, these data demonstrate that a loss of proteases is critical for the stress response and that protein degradation drastically affects infection. Thus, although the reduction in pathogenicity in our experiments cannot be attributed solely to the impact of the Clp system on heme acquisition, our data suggest that the reduced capacity to acquire host heme may contribute to the mutants' virulence defects during invasive infection.
Fig 8.
Clp proteolytic system is required for staphylococcal pathogenesis in a systemic infection model. Seven-week-old female BALB/c mice were retro-orbitally infected with 107 CFU wild-type S. aureus Newman or strains inactivated for clpP, clpC, or clpX. Infection progressed for 96 h, at which time mice were sacrificed, organs harvested, and bacterial burdens determined by serial dilution and plating. Numbers of recovered CFU from infected kidneys (A), livers (B), and hearts (C) are shown. Graphs include data from two individual experiments with a total of 20 mice. The dashed lines denote the limit of detection. Error bars represent SEM. *, P < 0.0001 relative to the wild type, as calculated by Student's t test.
DISCUSSION
Heme acquisition is a critical mechanism of S. aureus pathogenesis and is mediated by the Isd system, which strips heme from host hemoglobin and imports it into the bacterial cell (8, 52). Recognition of host hemoglobin during infection is the crucial initiating step of this process and occurs via the staphylococcal hemoglobin receptor IsdB (9). Moreover, the IsdB-hemoglobin interaction is critical to staphylococcal pathogenesis in a murine model of infection (10, 42, 49). Consistent with this, IsdB is highly immunogenic and has garnered attention as a potential staphylococcal vaccine target (53, 54). Data presented here suggest that hemoglobin-dependent heme acquisition through IsdB is driven not only by Fur-mediated repression but also by an additional level of proteolytic regulation.
Our results demonstrate that inactivation of components of the S. aureus ClpP protease drastically reduces abundance of IsdB. The fact that inactivation of the other intracellular proteases in S. aureus, including FtsH and HslV, did not affect hemoglobin binding through IsdB is not surprising, since the Clp protease has been demonstrated to be the main degradation system within this organism (16). Although HslV appears to be important for heat tolerance, inactivation of this protease did not affect susceptibility to other stressors or virulence in a murine skin abscess infection model (16). The membrane-bound FtsH protease is thought to act predominantly on misfolded, membrane-attached proteins (55). The significant reductions in IsdB protein and expression were observed in strains inactivated for clpP or clpX, suggesting that the ClpXP protease is the main proteolytic complex involved in regulating heme acquisition proteins.
The S. aureus Clp protease components initiate responses required to withstand numerous external pressures, including extreme temperature changes, alterations in osmolarity, and oxidative stress (17, 56, 57). Furthermore, the Clp system exerts profound effects on cellular physiology (58). The data reported here demonstrate that the Clp protease also regulates host hemoglobin binding and utilization through IsdB and suggest that this system may be important for nutrient acquisition in S. aureus.
Our results demonstrate that Clp proteins alter expression of isdB, which is also regulated in an iron-responsive manner by Fur. Previous studies indicated that ClpP may influence Fur-mediated repression, as six genes with putative Fur boxes were altered in expression in response to clpP inactivation, though the Isd system was not present within this list (19). The reported impact of ClpP on Fur-regulated genes is believed to contribute chiefly to defense against oxidative stress (19). Interestingly, expression of the Isd system has been reported to be upregulated in response to oxidative stress (59). Moreover, inactivation of clpC results in a 2.6-fold decrease in fur expression compared to that in wild-type cells (60). Despite the observed alterations in expression of fur or some Fur-regulated genes, there have been no reports of the Clp system acting directly on expression or degradation of Fur (60).
Repression of isdB expression in the absence of both fur and clpP suggests that an additional molecule is capable of regulating transcription at this locus. The Clp system has been implicated in regulating a number of virulence factors by altering the abundance of transcriptional regulators that control their expression, including Agr, Sae, Rot, and CodY (17, 19, 40, 41, 61, 62). While the impact of the Clp system on transcriptional regulation is evident, there have been no reports of these regulators affecting Isd-dependent heme acquisition. Western blot analysis of triple mutants inactivated for clpP, fur, and agrA, saeR, codY, or rot demonstrated that none of these regulators appear to repress IsdB protein levels in a clp-dependent manner (see Fig. S4 in the supplemental material). Thus, of the transcriptional regulators that are known to be affected by the ClpP system, there is no clear candidate for the observed transcriptional regulation of isdB.
Due to its influence on virulence factor production, the Clp proteolytic system is critical to the pathogenesis of a number of organisms, including Listeria monocytogenes, Streptococcus pneumoniae, Bacillus anthracis, Helicobacter pylori, Staphylococcus epidermidis, Salmonella enterica serovar Typhimurium, and Brucella suis (63–73). Likewise, loss of Clp proteins profoundly affects S. aureus pathogenesis. Lack of clpP or clpX results in decreased virulence in a skin abscess model of S. aureus infection (17). Inactivation of clpX in S. aureus results in attenuated virulence in a murine bacteremia model (51). Our results add to the growing importance of the Clp system by demonstrating that ClpP and ClpX are also required for pathogenesis in a systemic model of staphylococcal infection. In addition, our data show for the first time that ClpC is critical for survival and propagation in kidneys and livers of infected animals during S. aureus infection. The reason for the organ-specific virulence defects observed in the clpC mutant is not entirely clear. IsdB is expressed in the heart during infection and is critical for colonization of this organ, while it does not affect bacterial burdens in infected livers (42). It is intriguing to suggest that the enhanced IsdB expression and abundance observed in the clpC mutant in vitro may contribute to its ability to colonize the heart in vivo.
In summary, the data presented here implicate the Clp proteolytic complex in the control of an unknown transcriptional regulator that influences expression of the Isd system in S. aureus. Ongoing experiments in our lab aim to identify this factor and its mechanism of Isd regulation. Disruption of the Isd system by inactivation of the Clp proteolytic system has profound effects on hemoglobin-dependent heme acquisition, a critical mechanism of obtaining nutrient iron in S. aureus that is required for virulence in a systemic model of infection. Targeting the Clp protease with acyl-depsipeptides, which cause uncontrolled degradation by the Clp system, or with β-lactones, which inhibit ClpP, has proven to be highly effective in bacterial killing (74, 75). Our data lend support to the proposal that the Clp protease is an excellent target for antimicrobials. Since the Clp protease and Isd complexes are widely conserved in bacteria, these data reveal a novel Clp-dependent regulation pathway that may be relevant to pathogenic mechanisms of other bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Skaar laboratory for critically reading the manuscript.
This research was supported by NIH grants AI069233 and AI073843 to E.P.S. and F32 AI098380 to A.J.F., all funded by the NIAID, as well as by the Vanderbilt Mechanisms of Vascular Disease training grant T32 HL007751-16A2 (to A.J.F.). E.P.S. is a Burroughs Wellcome Fellow in the Pathogenesis of Infectious Diseases. The ΔcodY::ermC and ΔagrA::ermC isolates were acquired from the NARSA program, which is supported by NIAID/NIH contract HHSN272200700055C.
Footnotes
Published ahead of print 30 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00505-13.
REFERENCES
- 1.Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 2.Miro JM, Anguera I, Cabell CH, Chen AY, Stafford JA, Corey GR, Olaison L, Eykyn S, Hoen B, Abrutyn E, Raoult D, Bayer A, Fowler VG, International Collaboration on Endocarditis Merged Database Group 2005. Staphylococcus aureus native valve infective endocarditis: report of 566 episodes from the International Collaboration on Endocarditis Merged Database. Clin. Infect. Dis. 41:507–514 [DOI] [PubMed] [Google Scholar]
- 3.Purcell K, Fergie J. 2005. Epidemic of community-acquired methicillin-resistant Staphylococcus aureus infections: a 14-year study at Driscoll Children's Hospital. Arch. Pediatr. Adolesc. Med. 159:980–985 [DOI] [PubMed] [Google Scholar]
- 4.Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boyle-Vavra S, Leitch CD, Daum RS. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598 [DOI] [PubMed] [Google Scholar]
- 5.Crosa J. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61:319–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626–1628 [DOI] [PubMed] [Google Scholar]
- 7.Bullen JJ, Griffiths E. 1999. Iron and infection: molecular, physiological and clinical aspects. John Wiley and Sons, New York, NY [Google Scholar]
- 8.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906–909 [DOI] [PubMed] [Google Scholar]
- 9.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. 2006. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188:8421–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pishchany G, McCoy AL, Torres VJ, Krause JC, Crowe JE, Jr, Fabry ME, Skaar EP. 2010. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe 8:544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiedemann MT, Heinrichs DE, Stillman MJ. 2012. Multiprotein heme shuttle pathway in Staphylococcus aureus: iron-regulated surface determinant cog-wheel kinetics. J. Am. Chem. Soc. 134:16578–16585 [DOI] [PubMed] [Google Scholar]
- 12.Skaar EP, Gaspar AH, Schneewind O. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436–443 [DOI] [PubMed] [Google Scholar]
- 13.Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477 [DOI] [PubMed] [Google Scholar]
- 14.Reniere ML, Skaar EP. 2008. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 69:1304–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reniere ML, Haley KP, Skaar EP. 2011. The flexible loop of Staphylococcus aureus IsdG is required for its degradation in the absence of heme. Biochemistry 50:6730–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frees D, Thomsen L, Ingmer H. 2005. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch. Microbiol. 183:286–291 [DOI] [PubMed] [Google Scholar]
- 17.Frees D, Qazi SNA, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565–1578 [DOI] [PubMed] [Google Scholar]
- 18.Donegan NP, Thompson ET, Fu Z, Cheung AL. 2010. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J. Bacteriol. 192:1416–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, Hacker J, Ohlsen K. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188:5783–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katayama-Fujimura Y, Gottesman S, Maurizi MR. 1987. A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262:4477–4485 [PubMed] [Google Scholar]
- 21.Wickner S, Maurizi MR, Gottesman S. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888–1893 [DOI] [PubMed] [Google Scholar]
- 22.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. 1995. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 14:1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. U. S. A. 97:8898–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y-I, Burton RE, Burton BM, Sauer RT, Baker TA. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5:639–648 [DOI] [PubMed] [Google Scholar]
- 25.Flynn JM, Neher SB, Kim Y-I, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683 [DOI] [PubMed] [Google Scholar]
- 26.Baker TA, Sauer RT. 2006. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31:647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoskins JR, Pak M, Maurizi MR, Wickner S. 1998. The role of the ClpA chaperone in proteolysis by ClpAP. Proc. Natl. Acad. Sci. U. S. A. 95:12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoskins JR, Singh SK, Maurizi MR, Wickner S. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. U. S. A. 97:8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiler KC, Waller PRH, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993 [DOI] [PubMed] [Google Scholar]
- 31.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. 2006. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439:753–756 [DOI] [PubMed] [Google Scholar]
- 32.Neher SB, Flynn JM, Sauer RT, Baker TA. 2003. Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev. 17:1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoskins JR, Yanagihara K, Mizuuchi K, Wickner S. 2002. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. U. S. A. 99:11037–11042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63:1285–1295 [DOI] [PubMed] [Google Scholar]
- 35.Kirstein J, Moliere N, Dougan DA, Turgay K. 2009. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7:589–599 [DOI] [PubMed] [Google Scholar]
- 36.Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9:27–43 [PubMed] [Google Scholar]
- 37.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. 1999. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401:90–93 [DOI] [PubMed] [Google Scholar]
- 38.Levchenko I, Luo L, Baker TA. 1995. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 9:2399–2408 [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S, Maurizi MR. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frees D, Andersen JH, Hemmingsen L, Koskenniemi K, Bæk KT, Muhammed MK, Gudeta DD, Nyman TA, Sukura A, Varmanen P, Savijoki K. 2012. New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J. Proteome Res. 11:95–108 [DOI] [PubMed] [Google Scholar]
- 41.Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ESC, Siddiqui SM, Wah DA, Baker TA. 2004. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pishchany G, Dickey SE, Skaar EP. 2009. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 77:2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 44.Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70:186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. 2010. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78:1618–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 47.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 48.Collins JA, Irnov I, Baker S, Winkler WC. 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 21:3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23:3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wawrzynow A, Banecki B, Zylicz M. 1996. The Clp ATPases define a novel class of molecular chaperones. Mol. Microbiol. 21:895–899 [DOI] [PubMed] [Google Scholar]
- 51.Mei J-M, Nourbakhsh F, Ford CW, Holden DW. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399–407 [DOI] [PubMed] [Google Scholar]
- 52.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, Stillman MJ. 2008. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283:28125–28136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harro C, Betts R, Orenstein W, Kwak E-J, Greenberg HE, Onorato MT, Hartzel J, Lipka J, DiNubile MJ, Kartsonis N. 2010. Safety and immunogenicity of a novel Staphylococcus aureus vaccine: results from the first study of the vaccine dose range in humans. Clin. Vaccine Immunol. 17:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, Missiakas DM, Schneewind O. 2010. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28:6382–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akiyama Y, Kihara A, Tokuda H, Ito K. 1996. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J. Biol. Chem. 271:31196–31201 [DOI] [PubMed] [Google Scholar]
- 56.Frees D, Chastanet A, Qazi S, Sørensen K, Hill P, Msadek T, Ingmer H. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445–1462 [DOI] [PubMed] [Google Scholar]
- 57.Chatterjee I, Becker P, Grundmeier M, Bischoff M, Somerville GA, Peters G, Sinha B, Harraghy N, Proctor RA, Herrmann M. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187:4488–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hecker M, Schumann W, Völker U. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417–428 [DOI] [PubMed] [Google Scholar]
- 59.Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, Skaar EP, DeLeo FR. 2008. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180:500–509 [DOI] [PubMed] [Google Scholar]
- 60.Chatterjee I, Schmitt S, Batzilla CF, Engelmann S, Keller A, Ring MW, Kautenburger R, Ziebuhr W, Hecker M, Preissner KT, Bischoff M, Proctor RA, Beck HP, Lenhof H-P, Somerville GA, Herrmann M. 2009. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9:1152–1176 [DOI] [PubMed] [Google Scholar]
- 61.Frees D, Sørensen K, Ingmer H. 2005. Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect. Immun. 73:8100–8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, Ingmer H, Frees D. 2013. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 12:547–558 [DOI] [PubMed] [Google Scholar]
- 63.Butler SM, Festa RA, Pearce MJ, Darwin KH. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol. Microbiol. 60:553–562 [DOI] [PubMed] [Google Scholar]
- 64.Rouquette C, Ripio M-T, Pellegrini E, Bolla J-M, Tascon RI, Vázquez-Boland J-A, Berche P. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977–987 [DOI] [PubMed] [Google Scholar]
- 65.Nair S, Milohanic E, Berche P. 2000. ClpC ATPase is required for cell adhesion and invasion of Listeria monocytogenes. Infect. Immun. 68:7061–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286–1294 [DOI] [PubMed] [Google Scholar]
- 67.Rouquette C, De Chastellier C, Nair S, Berche P. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235–1245 [DOI] [PubMed] [Google Scholar]
- 68.Robertson GT, Ng W-L, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGillivray SM, Ebrahimi CM, Fisher N, Sabet M, Zhang DX, Chen Y, Haste NM, Aroian RV, Gallo RL, Guiney DG, Friedlander AM, Koehler TM, Nizet V. 2009. ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J. Innate Immun. 1:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loughlin MF, Arandhara V, Okolie C, Aldsworth TG, Jenks PJ. 2009. Helicobacter pylori mutants defective in the clpP ATP-dependent protease and the chaperone clpA display reduced macrophage and murine survival. Microb. Pathog. 46:53–57 [DOI] [PubMed] [Google Scholar]
- 71.Wang C, Li M, Dong D, Wang J, Ren J, Otto M, Gao Q. 2007. Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microbes Infect. 9:1376–1383 [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto T, Sashinami H, Takaya A, Tomoyasu T, Matsui H, Kikuchi Y, Hanawa T, Kamiya S, Nakane A. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ekaza E, Guilloteau L, Teyssier J, Liautard J-P, Köhler S. 2000. Functional analysis of the ClpATPase ClpA of Brucella suis, and persistence of a knockout mutant in BALB/c mice. Microbiology 146:1605–1616 [DOI] [PubMed] [Google Scholar]
- 74.Bottcher T, Sieber SA. 2008. β-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 130:14400–14401 [DOI] [PubMed] [Google Scholar]
- 75.Brotz-Oesterhelt H, Beyer D, Kroll H-P, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, Bandow JE, Sahl H-G, Labischinski H. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082–1087 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.