Abstract
A 69-year-old patient presented with a tender, thickly crusted skin lesion of 1 week's duration. A bacterial culture swab taken from the underlying granular tissue yielded a pure isolate of a Gram-negative coccobacillus, presumptively identified as a novel Francisella species via 16S rRNA and multilocus gene sequence analysis.
CASE REPORT
On 5 January 2011, a 69-year-old white male presented to a dermatology clinic with a tender lesion on his left ankle. The patient had been well until the previous week when he cut down an Arizona ash tree near his home in southern Louisiana, close to the Mississippi River. Branches from the tree scratched his legs and abraded his left ankle. He applied mupirocin ointment to the ankle lesion, but it became increasingly tender and erythematous. His underlying medical conditions included non-insulin-dependent diabetes, hypertension, hyperlipidemia, and a history of a squamous cell carcinoma excised from his right arm in March 2010. The patient denied fever, chills, or other systemic symptoms.
On initial examination, the patient was afebrile with a 4- by 6-cm thickly crusted erythematous nodule on the medial aspect of the left ankle, with surrounding erythema and tenderness to palpation (Fig. 1A). Incision and drainage of the nodule was attempted; however, no pus or purulent drainage was noted. Tissue underlying the eschar was pink and granular. A biopsy specimen of the underlying granular tissue was submitted for histopathology, deep fungal culture, and atypical mycobacterial culture; a swab was also taken for routine bacterial culture.
Fig 1.
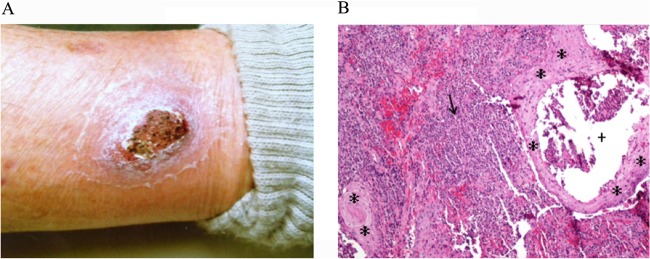
(A) The cutaneous lesion from which Francisella sp. LA11-2445 was isolated. (B) Histopathology (100× magnification) of a fragmented punch biopsy specimen. The arrow marks a dense, mixed dermal infiltrate of neutrophils, histiocytes, and lymphocytes. An intraepidermal neutrophilic microabscess is shown (+). The asterisks (*) indicate pseudoepitheliomatous epidermal hyperplasia.
Initial histopathology of the biopsied tissue was interpreted as squamous cell carcinoma. The patient was referred to a general surgeon for excision of the lesion but, because of the surrounding erythema, was also prescribed 500 mg ciprofloxacin twice daily (BID) for 7 days. The patient noted improvement in the lesion after 24 h. Deep fungal and atypical mycobacterial cultures were both negative. A Gram-negative coccobacillus grew from the bacterial swab taken from the underlying granular tissue and was presumptively identified as Francisella tularensis (described below). Based on the isolation of a Francisella species from the lesion, an additional 7 days of ciprofloxacin was prescribed, and the pathologist who performed histopathology was contacted to review the initial biopsy specimen. The pathologist's amended report noted probable pseudoepitheliomatous hyperplasia overlying a dense suppurative granulomatous dermatitis, rather than squamous cell carcinoma. However, significant atypia was noted, and the remaining lesion was excised in the operating room on 4 March 2011. The area healed well without further sequelae. Subsequent independent histopathology of the biopsy tissue similarly indicated no evidence of squamous cell carcinoma but rather marked inflammation and pseudoepitheliomatous epidermal hyperplasia with a dense, mixed dermal infiltrate including scattered small neutrophilic microabscesses, histiocytes, lymphocytes, and plasma cells (Fig. 1B). Intraepidermal neutrophilic microabscesses were also seen (Fig. 1B).
Testing of the bacterial culture swab was performed at a commercial laboratory, where a pure culture of a small Gram-negative coccobacillus was recovered on both blood and chocolate agars but not MacConkey agar. The organism was forwarded to a second commercial laboratory, where it was presumptively identified via growth characteristics, colony morphology, Gram stain, and biochemical testing (catalase and oxidase) as F. tularensis and subsequently submitted to the nearby Orange County Public Health Laboratory (OCPHL) in Santa Ana, CA, on 19 January 2011. Characterization performed at OCPHL indicated the organism grew well on chocolate, cysteine heart, and buffered charcoal yeast extract agars after 24 h incubation at 35°C in 5% CO2, whereas only minimal growth was observed on blood agar after 24 h. Biochemical testing revealed the bacterium was o-nitrophenyl-β-d-galactopyranoside (ONPG) positive and oxidase, catalase, indole, urease, gelatinase, and nitrate negative. It fermented maltose and glucose but did not utilize xylose, mannitol, lactose, sucrose, or fructose when grown on 1% cystine tryptic agar in the presence of the individual carbohydrates. Similarly, there was no fermentation of glycerol after 5 days in phenol red nitrate broth. The organism grew better in nutrient broth in the presence of 3% and 6% NaCl than in the presence of 0% NaCl. When the NaCl concentration was increased to 10%, there was no growth.
F. tularensis-specific testing indicated that the isolate was negative by the direct fluorescent-antibody assay (1) and by the Laboratory Response Network (LRN) real-time PCR for F. tularensis (3 of 3 PCR assays were negative). For 16S rRNA gene sequencing, the MicroSEQ microbial identification system (Life Technologies, Grand Island, NY) and associated protocols were used. A 475-bp region of the 16S rRNA gene sequence showed the highest similarity to Francisella spp., specifically 95% to Francisella philomiragia and 93% to F. tularensis when using the MicroSEQ ID 16S rDNA 500 Library and 96% to F. noatunensis subsp. noatunensis GM2212 (DQ309246) when using GenBank for comparison.
For identification of the Francisella species, the isolate was sent to the Centers for Disease Control and Prevention (CDC), Fort Collins, CO. Amplification and sequencing of a ∼1,300-bp segment of the 16S rRNA gene from the isolate (designated LA11-2445) was performed using primers and PCR conditions described previously (2). Segments of the pgm, rpoA, rpoB, tpiA, groEL, sdhA, and atpA genes were amplified and sequenced using PCR primers and conditions described in references 2, 3, and 4. Previous studies have shown that concatenation and analysis of the housekeeping genes chosen for this study yield phylogenies congruent with those based on whole-genome sequences of Francisella species (3, 5, 6). Available homologous sequences from Francisella species and other gammaproteobacteria were obtained from GenBank for nucleotide comparison. All sequences were trimmed in Lasergene v9.0 (DNASTAR, Madison, WI) and then exported into MEGA v5 (7) to generate alignments, distance comparisons, average nucleotide identity (ANI), and phylogenetic trees using maximum likelihood (ML). The generalized time-reversible nucleotide substitution model with gamma distribution (4 gamma categories) and invariant sites (GTR+I+G) was used for ML analyses. Statistical reliability of nodes was evaluated through bootstrap analysis with 1,000 replicates.
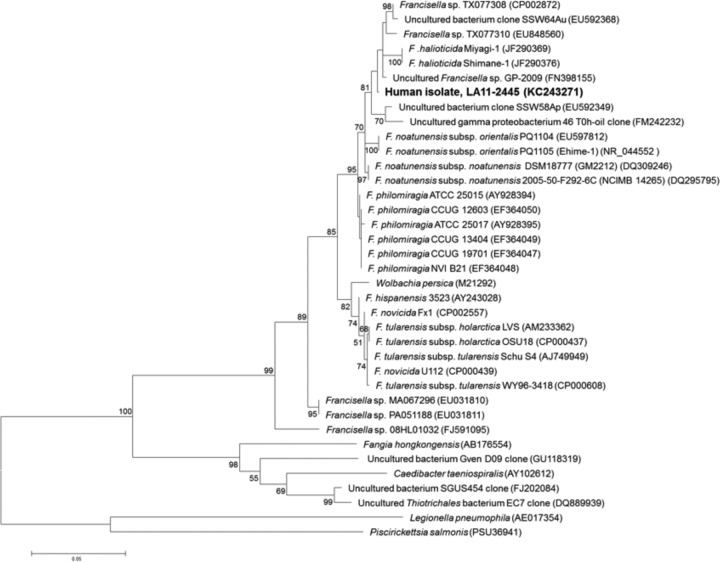
Distance comparison of a 1,205-bp fragment of the 16S rRNA gene sequence revealed that isolate LA11-2445 shared ≥95.5% sequence identity with all other Francisella spp. analyzed, compared to only 89.2% and 88.1% with Fangia hongkongensis and Caedibacter taeniospiralis, respectively. Maximum 16S rRNA sequence identity (99.56%) was observed between isolate LA11-2445 and Francisella sp. GP-2009. Phylogenetic analysis of 16S rRNA gene sequences revealed that LA11-2445 fell into a clade (statistical support of 81%) consisting of Francisella organisms associated with marine environments, including (i) GP-2009, an uncultured endosymbiont of a marine ciliate from the Adriatic Sea, (ii) TX077310 and TX077308, two isolates cultured from Gulf of Mexico seawater samples, (iii) Shimane-1 and Miyagi-1, two F. halioticida strains isolated from moribund giant abalone in Japan, and (iv) SSW64Au, SSW58Ap, and 46 T0h-oil, three uncultured bacteria from either the Salton Sea or the Berre lagoon, France (Fig. 2).
Fig 2.
Maximum-likelihood (ML) analysis showing the relationship of the clinical isolate, LA11-2445, to other Francisellaceae members based on a 1,205-bp region of the 16S rRNA gene. Bootstrap support values >50% are indicated. The scale bar corresponds to 0.05 substitutions per nucleotide position. GenBank accession numbers are indicated following strain designations.
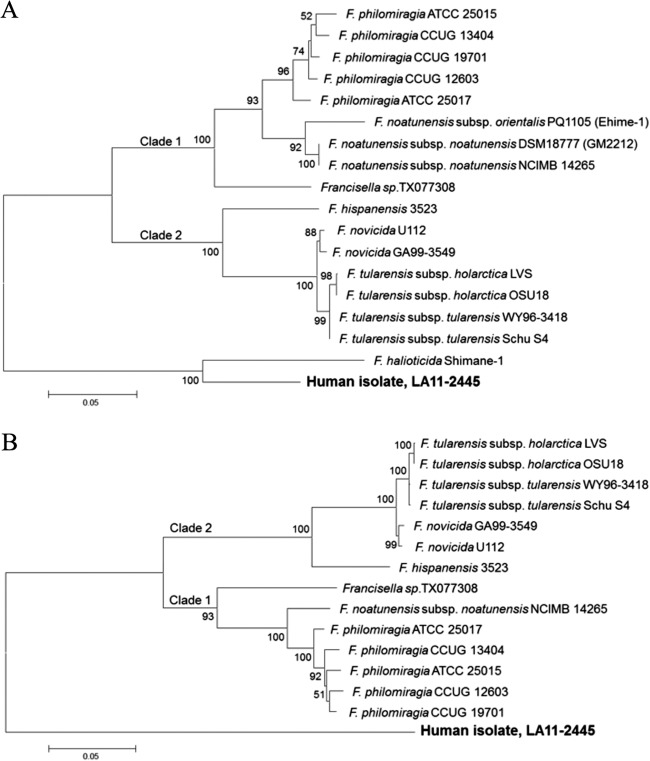
Incongruent phylogenetic topologies for Francisellaceae members have previously been shown for 16S rRNA compared to whole-genome sequence data (5). Therefore, in-frame gene sequences amplified from the housekeeping genes, pgm (567 bp), rpoA (264 bp), rpoB (255 bp), tpiA (321 bp), groEL (837 bp), sdhA (444 bp), and atpA (318 bp), were analyzed as concatenated sequences. Analysis of both 4- and 7-locus concatenated sequences was performed, as sequence data for all 7 genes of the most closely related Francisellaceae members, including F. halioticida, were not available in GenBank. Uncultured Francisella spp. also could not be included due to the absence of sequence information in GenBank for genes other than 16S rRNA genes. Concatenation of the 4 loci, in the order pgm, rpoA, rpoB, and tpiA, generated a 1,596-bp sequence, which was compared to homologous sequences from 17 other Francisella strains. Phylogenetic analysis indicated strong statistical support (100%) for the resulting topology, which consisted of two previously recognized clades, clade 1 (F. philomiragia, F. noatunensis, and Francisella sp. TX077308) and clade 2 (F. tularensis, F. novicida, and F. hispaniensis), and F. halioticida and LA11-2445 formed a separate clade (Fig. 3A). The ANI between LA11-2445 and F. halioticida was 89.3% across the four genes, compared to an ANI of 81 to 82% with all other Francisella spp. in clade 1 and clade 2.
Fig 3.
Maximum-likelihood (ML) analysis showing the relationship of the clinical isolate, LA11-2445, to other Francisellaceae members based on concatenated pgm, rpoA, rpoB, and tpiA gene sequences (A) and concatenated pgm, rpoA, rpoB, tpiA, groEL, sdhA, and atpA gene sequences (B). Bootstrap support values of >50% are shown. The scale bars correspond to 0.05 substitutions per nucleotide position. GenBank accession numbers for the pgm, rpoA, rpoB, tpiA, groEL, sdhA, and atpA gene sequences from LA11-2445 are KC243274, KC243275, KC243276, KC243278, KC243272, KC243273, and KC243277, respectively.
All 7 loci linked in the order pgm, rpoA, rpoB, tpiA, groEL, sdhA, and atpA yielded a 3,014-bp sequence. The same overall tree topology was observed for the 7-locus data set compared with the 4-locus data set, with 93 to 100% bootstrap support for clades 1 and 2 and with LA11-2445 separate from both clades 1 and 2 (Fig. 3B). The topologies observed for both the 4-locus and 7-locus phylogenetic trees (clade 1 and clade 2), with the exception of F. noatunensis subsp. orientalis, are also consistent with topologies generated using whole-genome sequence data (8), suggesting accurate placement of LA11-2445 into the Francisella genus via these analyses. These results indicate that Francisella sp. LA11-2445 falls into a clade with F. halioticida but represents a unique species, as the ANI observed between LA11-2445 and F. halioticida was 89.9% across 4 loci, below the 95% cutoff for bacterial species. Comparative analyses have shown that an ANI of 95% corresponds to a 70% reassociation value via DNA-DNA hybridization, the cutoff and method historically used to delineate bacterial species (9).
To look at the susceptibility of LA11-2445 to antibiotics used for treatment of Francisella infections, broth microdilution was performed with ciprofloxacin, chloramphenicol, doxycycline, gentamicin, levofloxacin, streptomycin, and tetracycline, utilizing the Clinical and Laboratory Standards Institute approved susceptibility method and breakpoints for F. tularensis (10, 11). MICs to the aforementioned antibiotics were as follows: 0.004 μg/ml for ciprofloxacin, 1.0 μg/ml for chloramphenicol, 0.25 μg/ml for doxycycline, 0.06 μg/ml for gentamicin, 0.008 μg/ml for levofloxacin, 0.5 μg/ml for streptomycin, and 1.0 μg/ml for tetracycline. All fell within the susceptible range, consistent with the patient's positive response to treatment with ciprofloxacin.
Members of the Francisella genus range from intracellular pathogens with different host preferences (F. tularensis, F. noatunensis, and F. halioticida) to specialized endosymbionts (Francisella-like endosymbionts) to generalists believed capable of a free-living existence (F. philomiragia and F. novicida) (8), with only a subset of Francisella spp. linked to human illness. The intracellular pathogen F. tularensis is the causative agent of tularemia, an acute and fatal illness in animals and humans, whereas F. noatunensis and F. halioticida infect and cause death in fish and abalone, respectively (3, 6, 12). F. novicida and F. philomiragia are infrequent causes of opportunistic infections in compromised individuals or saltwater near-drowning victims and are associated with saltwater or brackish water (13, 14). In recent years, several unclassified Francisella spp., also linked to marine and brackish environments, have been identified based on 16S rRNA gene analysis of uncultured samples or by isolation and characterization of the organism (15–18). To date, none of these Francisella species have been associated with human infections.
Here, we identify a Francisella species recovered from a human cutaneous lesion. Results of phylogenetic analyses, including an ANI of only 89.8% across 4 housekeeping genes to F. halioticida, the closest Francisellaceae member for which sequence is available, are consistent with this organism representing a novel Francisella species. Further description of this Francisella sp. is limited by the lack of similar strains and the availability of gene sequences, other than 16S rRNA gene sequences, for closely related uncultured Francisella species. F. halioticida and the uncultured Francisella spp. are all associated with marine environments. Consistent with this, Francisella sp. LA11-2445 was determined to be slightly halophilic, growing better in the presence of 6% NaCl than in the absence of NaCl.
Our findings are consistent with the bacterium being responsible for the patient's infection, as this Francisella sp. was the only organism isolated from the ankle lesion, the lesion showed marked inflammation, and the patient responded positively to treatment with ciprofloxacin. The mode of transmission is unclear. Given the relatedness of this organism with other Francisella spp. associated with marine environments and the patient's residing near brackish waterways, we hypothesize that the bacterium was introduced from the environment into an open wound caused by the patient's tree-cutting activities.
The phylogenetic landscape of Francisella continues to expand with the availability of genomic sequencing and publicly available sequence repositories. Although Francisella was once thought to be a fairly small genus comprised of only three species (F. tularensis, F. novicida, and F. philomiragia), it has become increasingly clear in the last decade that additional Francisella spp. are widespread in the environment, with some of these species having the potential to cause infections in humans.
Nucleotide sequence accession numbers.
The 16S rRNA, pgm, rpoA, rpoB, tpiA, groEL, sdhA, and atpA gene sequences from LA11-2445 were deposited in GenBank under accession numbers KC243271, KC243274, KC243275, KC243276, KC243278, KC243272, KC243273, and KC243277, respectively.
ACKNOWLEDGMENTS
We thank Sandy Urich and John Young for technical assistance.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
Footnotes
Published ahead of print 31 July 2013
REFERENCES
- 1.Johansson A, Petersen J, Sjöstedt A. 2007. Laboratory diagnostics and discrimination of subspecies and strains, p 27–34 In Tärnvik A. (ed), WHO guidelines on tularemia, WHO/CDS/EPR/2007.7 World Health Organization, Geneva, Switzerland [Google Scholar]
- 2.Kugeler KJ, Mead PS, McGowan KL, Burnham JM, Hogarty MD, Ruchelli E, Pollard K, Husband B, Conley C, Rivera T, Kelesidis T, Lee WM, Mabey W, Winchell JM, Stang HL, Staples JE, Chalcraft LJ, Petersen JM. 2008. Isolation and characterization of a novel Francisella sp. from human cerebrospinal fluid and blood. J. Clin. Microbiol. 46:2428–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikalsen J, Colquhoun DJ. Francisella asiatica sp. nov. isolated from farmed tilapia (Oreochromis sp.) and elevation of Francisella philomiragia subsp. noatunensis to species rank as Francisella noatunensis comb. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2009 doi: 10.1099/ijs.0.002139-0. [DOI] [PubMed] [Google Scholar]
- 4.Nübel U, Reissbrodt R, Weller A, Grunow R, Porsch-Ozcürümez M, Tomaso H, Hofer E, Splettstoesser W, Finke E-J, Tschäpe H, Witte W. 2006. Population structure of Francisella tularensis. J. Bacteriol. 188:5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjödin A, Svensson K, Öhrman C, Ahlinder J, Lindgren P, Duodu S, Johansson A, Colquhoun DJ, Larsson P, Forsman M. 2012. Genome characterisation of the genus Francisella reveals similar paths of host adaptation in pathogens of mammals and fish. BMC Genomics 13:268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brevik OJ, Ottem KF, Kamaishi T, Watanabe K, Nylund A. 2011. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J. Appl. Microbiol. 111:1044–1056 [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Methods Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahlinder J, Öhrman C, Svensson K, Lindgren P, Johansson A, Forsman M, Larsson P, Sjödin A. 2012. Increased knowledge of Francisella genus diversity highlights the benefits of optimized DNA-based assays. BMC Microbiol. 12:220–235. 10.1186/1471-2180-12-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute 2010. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria: approved guideline, 2nd ed, CLSI document M45-A2 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11.Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard, 8th ed, CLSI document M07-A8 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12.Ellis J, Oyston PC, Green M, Titball RW. 2002. Tularemia. Clin. Microbiol. Rev. 15:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CV, Brenner DJ. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger JD, Hollis DG, Weaver RE, Baker CN, Brown GR, Brenner DJ, Broome CV. 1989. Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognized human pathogen. Ann. Intern. Med. 110:888–892 [DOI] [PubMed] [Google Scholar]
- 15.Schrallhammer M, Schweikert M, Vallesi A, Verni F, Petroni G. 2011. Detection of a novel subspecies of Francisella noatunensis as endosymbiont of the ciliate Euplotes raikovi. Microb. Ecol. 61:455–464 [DOI] [PubMed] [Google Scholar]
- 16.Petersen JM, Carlson J, Yockey B, Pillai S, Kuske C, Garbalena G, Pottumarthy S, Chalcraft L. 2009. Direct isolation of Francisella spp. from environmental samples. Lett. Appl. Microbiol. 48:663–667 [DOI] [PubMed] [Google Scholar]
- 17.Dillon JG, McMath LM, Trout AL. 2009. Seasonal changes in bacterial diversity in the Salton Sea. Hydrobiologia 632:49–64 [Google Scholar]
- 18.Païssé S, Goñi-Urriza M, Coulon F, Duran R. 2010. How a bacterial community originating from a contaminated coastal sediment responds to an oil input. Microb. Ecol. 60:394–405 [DOI] [PubMed] [Google Scholar]