Abstract
Use of Foxp3-positive (Foxp3+) T-regulatory (Treg) cells as potential cellular therapy in patients with autoimmunity, or post-stem cell or -organ transplantation, requires a sound understanding of the transcriptional regulation of Foxp3. Conserved CpG dinucleotides in the Treg-specific demethylation region (TSDR) upstream of Foxp3 are demethylated only in stable, thymus-derived Foxp3+ Treg cells. Since methyl-binding domain (Mbd) proteins recruit histone-modifying and chromatin-remodeling complexes to methylated sites, we tested whether targeting of Mbd2 might promote demethylation of Foxp3 and thereby promote Treg numbers or function. Surprisingly, while chromatin immunoprecipitation (ChIP) analysis showed Mbd2 binding to the Foxp3-associated TSDR site in Treg cells, Mbd2 targeting by homologous recombination, or small interfering RNA (siRNA), decreased Treg numbers and impaired Treg-suppressive function in vitro and in vivo. Moreover, we found complete TSDR demethylation in wild-type (WT) Treg cells but >75% methylation in Mbd2−/− Treg cells, whereas reintroduction of Mbd2 into Mbd2-null Treg cells restored TSDR demethylation, Foxp3 gene expression, and Treg-suppressive function. Lastly, thymic Treg cells from Mbd2−/− mice had normal TSDR demethylation, but compared to WT Treg cells, peripheral Mbd2−/− Treg cells had a marked impairment of binding of Tet2, the DNA demethylase enzyme, at the TSDR site. These data show that Mbd2 has a key role in promoting TSDR demethylation, Foxp3 expression, and Treg-suppressive function.
INTRODUCTION
Methylation of regulatory DNA sequences controls gene transcription. The DNA methyltransferase enzymes, namely, Dnmt1, Dnmt3a, and Dnmt3b, methylate the 5 position of the cytosine (C) residue in cytosine-guanine dinucleotides (CpG) to form 5-methylcytosine (m5C). The presence of m5CpG dinucleotides in the first exon or promoter region can affect gene transcription by direct or indirect mechanisms. The direct mechanism involves interference of m5CpG dinucleotides with transcription factor binding to a promoter. The indirect mechanism of gene regulation typically involves the actions of methylated binding domain (Mbd) proteins that block the interaction of transcription factors with key DNA sequences (1). Mbd proteins form complexes with multiple proteins involved in the stabilization of heterochromatin structure, including histone/protein deacetylase (HDAC) enzymes and corepressor (Sin3a) and ATP-dependent chromatin-remodeling proteins (2, 3). Surprisingly, it has been proposed that one well-characterized 44-kDa Mbd protein, Mbd2, may act as a DNA demethylase by removing repressive methyl residues and thereby activating gene transcription (4). Overexpression of Mbd2 can activate gene expression as a result of demethylation of CpG islands within promoter regions (5), and correlations between high levels of Mbd2 and DNA demethylation were reported in several autoimmune diseases, including systemic lupus erythematosus (SLE), dermatomyositis and systemic sclerosis (6–8), rheumatoid arthritis (9), and psoriasis (10), as well as in breast cancer (11). However, other groups have failed to detect any demethylation activity of Mbd2 (12–14), and so it remains controversial as to whether Mbd2 itself has direct demethylase activity. Recently, members of the ten-eleven translocation (Tet) family of proteins were shown to be capable of mediating DNA demethylation (15).
T-regulatory (Treg) cells are important for the maintenance of immune homeostasis. Foxp3, a forkhead-containing transcription factor, is the master regulator of Treg function (16). Mutations of Foxp3 in scurfy mouse or in patients with immune dysregulation, polyendocrinopathy, enteropathy, or X-linked (IPEX) syndrome result in the development of severe autoimmunity (17, 18). Foxp3 expression, and thereby Treg function, is subject to epigenetic regulation through methylation of CpG islands within the promoter and enhancer regions of Foxp3. Naturally occurring, thymus-derived Treg cells show demethylation at the Treg-specific demethylation region (TSDR) (19), which is also known as CNS2 (20). Demethylation of this region in thymic Treg cells can result from the actions of one or more Tet enzymes (21), whereas the TSDR in conventional T cells remains heavily methylated.
We have shown that pan-histone deacetylase (HDAC) inhibitor therapy increases Treg numbers and suppressive function (22) and that targeting of selected HDAC isoforms, including HDAC6 (23), HDAC9 (24), and Sirt1 (25), can augment Treg function. The conventional view is that Mbd proteins bind to methylated DNA sequences at promoter or enhancer regions and recruit HDAC enzymes that deacetylate lysine residues within histone tails, thereby promoting a condensed, inactive chromatin structure (26). Given the important role of Mbd proteins in recruiting HDAC enzymes and promoting the functions of corepressor complexes, we sought to evaluate the role of Mbd proteins in Treg cells. In particular, based upon our preliminary findings of the expression of Mbd2 by Treg cells, we wished to determine whether deletion of Mbd2 would impair corepressor complex formation or, based upon the demethylase reports, leave the promoter or enhancer region methylated within Treg cells. Here, we report that Mbd2 deletion in Treg cells increases Foxp3 methylation at the TSDR and decreases Treg numbers and function in vitro and in vivo.
MATERIALS AND METHODS
Animals.
Six- to-8-week-old BALB/c (H-2d), BALB/c/Rag1−/− (H-2d), and C57BL/6 (H-2b) mice were purchased from The Jackson Laboratory, and Mbd2−/− mice (H-2d) were provided by Adrian Bird (27). Mice were housed under specific-pathogen-free conditions, with 12-h light/dark cycles and free access to food and tap water, and studied using a protocol approved by the Institutional Animal Care and Use Committee of the Children's Hospital of Philadelphia.
Flow cytometry.
Single-cell suspensions of cells from pooled lymph nodes (LN), spleen, or thymus were stained using monoclonal antibodies (MAbs) to Foxp3 (phycoerythrin [PE]-Cy5), CD4 (Pacific blue), CD8 (allophycocyanin [APC]), CD25 (fluorescein isothiocyanate [FITC]), CD44 (PE), and CD62L (APC-Cy7) and analyzed on a flow cytometer (Cyan; Dako). All MAbs were purchased from BD Pharmingen, except anti-Foxp3 MAb (clone FJK-16S; eBioscience).
In vitro assay of Treg function.
CD4+ CD25+ Treg cells, CD4+ CD25− T effector (Teff) cells, and antigen-presenting cells were isolated from pooled LN and spleen cells using magnetic beads (Miltenyi Bioscience) (22). Carboxyfluorescein succinimidyl ester (CFSE)-labeled Teff cells (0.05 × 106/well) and an equal number of γ-irradiated antigen-presenting cells were added to wells, along with serially diluted wild-type (WT) or Mbd2−/− Treg cells, starting at a 1:1 ratio. After 3 days, Teff cell proliferation was assessed according to CFSE dilution (23).
Gene expression.
RNA was isolated from fresh or activated Treg cells and Teff cells isolated using magnetic beads; activation was performed overnight using CD3 MAb (1 μg/ml) and γ-irradiated antigen-presenting cells. First-strand cDNA was created with TaqMan reverse transcription reagents (Applied Biosystems), and quantitative PCR (qPCR) was undertaken using master mix and indicated primers and probes (Applied Biosystems) (23).
Homeostatic proliferation.
Congenic Thy1.1+ Teff cells (1 × 106) purified using magnetic beads were adoptively transferred to Rag1−/− mice either alone or along with 0.5 × 106 Thy1.2+ WT or Mbd2−/− CD4+ CD25+ Treg cells (22). Spleen and lymph nodes were isolated after 7 days, and total numbers of Thy1.1+ CD4+ Teff cells were determined by flow cytometry.
In vitro and in vivo induction of Treg from Teff.
Teff cells isolated with magnetic beads were cultured for 5 days with a 1:1 ratio of CD3/CD28 MAb-coated beads, 10 U/ml interleukin-2 (IL-2), and differing levels of transforming growth factor β1 (TGF-β1). Foxp3+ cells were identified by flow cytometric staining with Foxp3 and CD4 MAbs. For in vivo conversion, 0.5 million Teff cells were injected intravenously (i.v.) into immunodeficient Rag1−/− mice. After 3 weeks, single-cell suspensions of cells from lymph nodes and spleens were prepared and CD4+ Foxp3+ cells were quantified by flow cytometry.
Treg function in cardiac allograft recipients.
In the first transplant model, hearts from C57BL/6 mice were engrafted heterotopically into the abdomens of Mbd2−/− or WT BALB/c recipients and 5 million donor splenocytes plus CD40L MAb (MR-1, 200 μg) were given i.v. immediately postsurgery (28). Allograft survival was monitored by palpation and verified by histologic examination. In a second model, Treg and Teff cells were isolated from BALB/c mice using magnetic beads, and 1 million Teff and 1.5 million WT or Mbd2−/− Treg cells were injected i.v. into Rag1−/− mice (BALB/c) that had received C57BL/6 cardiac allografts. Graft survival was monitored as a function of the ability of injected Treg cells to suppress Teff cell-dependent alloreactivity and allograft rejection (22).
ChIP analysis.
A chromatin immunoprecipitation (ChIP) assay for Mbd2 and Tet1, -2, and -3 binding to the Foxp3 TSDR site was undertaken using BALB/c Treg and Teff cells and a ChIP assay kit (Upstate). Quantitative PCR (qPCR) was performed using the same primers as those employed in methyl collector assays (29).
Mbd2 viral transduction.
Plasmid MinR-1 vector containing an Mbd2 construct (MinR1-Mbd2) was generated from pCMV-Sport6 expression vector containing murine Mbd2 (pCMV-Sport6-Mbd2) (MMM 1013-9200215; Openbiosystems). Mbd2 cDNA was cut from pCMV-Sport6-Mbd2 using NotI (R0189S; New England BioLabs) and SalI (R0138S; New England BioLabs), and blunt ends were added by the use of DNA polymerase I Klenow fragments (M0210S; New England BioLabs) in the presence of deoxynucleoside triphosphates (dNTPs). Mbd2 cDNA with blunt ends was ligated into MinR-1 vector using T4 ligase (203003; Stratagene), and the plasmid sequence was verified. Retroviruses were generated by cotransfection of MinR1-Mbd2 or parental MinR1 vector with pCLeco (Invitrogen) helper plasmid into the 293T Phoenix ecotropic packaging cell line, using Lipofectamine 2000 (11668-019; Invitrogen). Supernatants were collected and used to infect purified Teff cells or Treg cells isolated using magnetic beads. Teff cells were stimulated for 16 h using CD3 and CD28 MAbs (2 μg/ml) plus mouse IL-2 (10 U/ml), and isolated Treg cells were stimulated similarly except for the addition of 100 U/ml of mouse IL-2. Activated T cells were infected with 48-h viral supernatants harvested from the transfected Phoenix cells and cultured at 37°C with 5% CO2 for 24 h followed by use in suppression assays or by RNA extraction (22).
siRNA knockdown.
For small interfering RNA (siRNA) studies, CD4+ CD25+ Treg cells were transfected using Lipofectamine 2000 and siRNA (Santa Cruz Biotechnology) specific for Mbd2 or a negative-control siRNA without specificity for rat, human, or mouse RNA sequences. Transfected cells were evaluated using qPCR and Treg suppression assays.
Methylation studies.
Bisulfite-converted genomic DNA from purified Treg cells was subjected to nested primer PCR for the Foxp3 TSDR (30), and the PCR product was cloned and sequenced. We assessed general methylation of CpG islands in the Foxp3 first intron site, using purified DNA (DNA kit; Qiagen) from isolated Treg and Teff cells that was sonicated (15 s on, 5 min off), and fragment sizes were verified by agarose gels. Fragmented DNA was pulled down with Mbd2 protein, and quantitative Sybr green PCR was performed using forward 5′-GATAGACTAGCCACTTCTCG and reverse 5′-CCTGTTGTCACAACCTGAAC primers.
Western blotting.
Proteins isolated from Treg cells, separated by SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membranes were detected with antibodies to MBD2 (Millipore) and individual Tet1, -2, and -3 enzymes (Abiocode).
Microarrays.
Microarray experiments were performed using whole-mouse-genome oligoarrays (Mouse430a; Affymetrix), and array data were analyzed using MAYDAY 2.12 software (31). Array data were subjected to robust multiarray average normalization. For determination of differential expression, we calculated fold changes and tested for significance by significance analysis of microarrays (SAM) using 100 permutations with a false-discovery-rate (FDR; Storey)-adjusted P of <0.05. Data with >1.5× differential expression (log2 0.5849) were included in the analysis. Data underwent z-score transformation for display in heatmaps. We used DAVID v6.7 for functional annotation clustering (32).
Statistical analysis.
Data were analyzed using GraphPad Prism 5.0d software. All normally distributed data were displayed as means ± standard deviations (SD). Measurements between two groups were done with a Student t test or Mann-Whitney U test. Groups of three or more were analyzed by one-way analysis of variance (ANOVA) or the Kruskal-Wallis test.
Nucleotide sequence accession number.
We deposited our data in the NCBI Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession number GSE48653.
RESULTS
Mbd2 deletion alters Foxp3+ Treg numbers and gene expression.
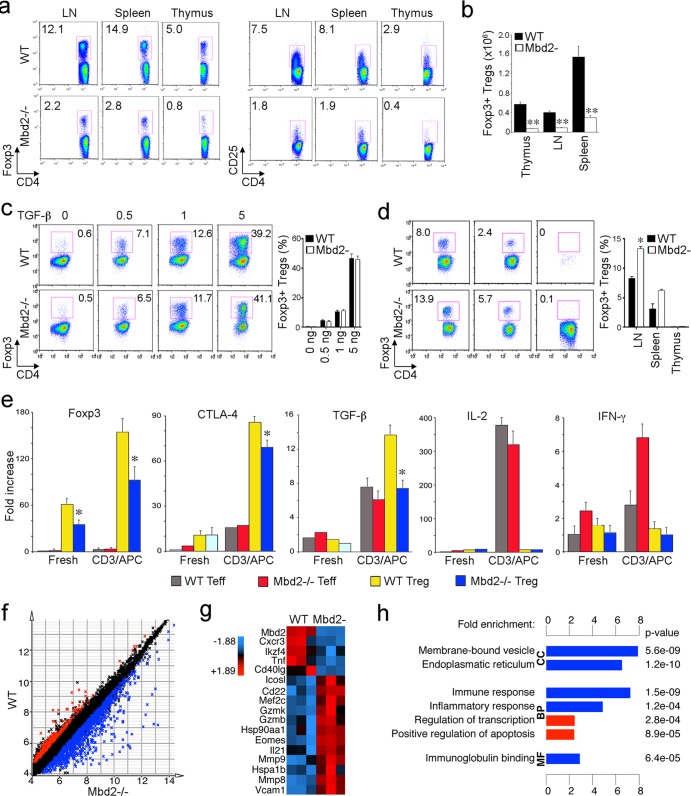
Mbd2-deficient mice were previously shown to be viable and to develop normally, despite a defect in maternal nurturing behavior (27). Likewise, the development of lymphoid organs and major lymphocyte subsets in Mbd2−/− mice was found to be normal, though under polarizing conditions, Th1 cells from Mbd2−/− mice were unable to silence expression of IL-4, indicating the normal repression of Gata-3 by Mbd2 (33). To date, no data have been reported on the effects of Mbd2 deletion in Treg cells. In preliminary studies, we confirmed the presence of normal proportions of CD4 and CD8 T cells in the thymus and peripheral lymphoid organs of Mbd2−/− mice (data not shown). However, we also found that compared to the WT controls, Mbd2−/− mice had significant decreases in the proportions of Treg cells in the thymus, LN, and spleen (Fig. 1a). For example, whereas WT mice showed 11% to 15% Foxp3+ CD4+ T cells, Mbd2−/− mice had <3% Foxp3+ CD4+ T cells in the periphery, and there was also a marked decrease in CD4+ CD25+ T cell numbers in lymphoid tissues of Mbd2−/− versus WT mice (Fig. 1a). These changes were reflected by correspondingly decreased absolute numbers of Foxp3+ Treg cells (Fig. 1b).
Fig 1.
Effects of Mbd2 deletion on Foxp3+ Treg cell numbers and gene expression. (a) Flow cytometric analysis of Foxp3+ CD4+ and CD4+ CD25+ cells within thymus, spleen, and pooled lymph nodes (LN) of WT and Mbd2−/− mice. The proportion of labeled cells is shown in each panel, and data are representative of 4 mice/group. (b) Absolute numbers of Foxp3+ CD4+ Treg cells (Tregs) in thymus, LN, and spleen as determined by flow cytometry (means ± SD, 4 mice/group); **, P < 0.01 for Mbd2−/− versus WT mice. (c) In vitro induction of Treg from Teff under conditions of increasing concentrations of TGF-β1; there was no significant difference in iTreg generation results between WT and MBD2−/− mice. (d) In vivo induction of Treg cells from Teff cells injected 3 weeks earlier into immunodeficient mice. Mbd2−/− Teff cells showed enhanced iTreg generation in the case of LN samples (*, P < 0.05), whereas no significant differences in in vivo Treg generation were detected in splenic or thymic samples between the 2 groups. (e) Expression of selected Treg-associated genes, plus the cytokines IL-2 and IFN-γ, by freshly isolated or TCR-activated Treg cells and Teff cells of WT and Mbd2−/− mice (qPCR, means ± SD, 4 mice/group); *, P < 0.05 for Mbd2−/− versus WT Treg cells. (f) Expression matrix scatter plot of microarray data showing 1,034 probes with increased (blue) and 266 probes with decreased (red) gene expression in MBD2−/− compared to WT Treg cells (1.5-fold expression; significance determined by SAM using 100 permutations and FDR-adjusted P < 0.05). (g) Genes of interest with respect to Treg function; data are shown after z-score transformation. (h) DAVID functional annotation clustering of probes with increased (blue) or decreased (red) expression in MBD2−/− versus WT Treg cells. Data are grouped by gene ontology terms CC (cellular component), BP (biological process), and MF (molecular function). Abbreviations: Cxcr, chemokine (c-x-c motif) receptor; Ikzf4, IKAROS family zinc finger 4 (Eos); Tnf, tumor necrosis factor; Icosl, Icos ligand; Mef2c, myocyte enhancer factor 2c; Gzm, granzyme; Hsp, heat shock protein; Eomes, eomesodermin; Mmp, matrix metallopeptidase; Vcam, vascular adhesion molecule.
In addition to their normal development within the thymus, Foxp3+ Treg cells can arise in the periphery by conversion of conventional CD4+ CD25− Teff cells into so-called inducible Treg cells (iTreg cells). This led us to check whether iTreg development in vitro or in vivo was normal when using Mbd2−/− Teff cells. We found that conversion of Mbd2−/− Teff cells into Foxp3+ Treg cells was equal to conversion of WT T cells when they were cultured under appropriate conditions in vitro (Fig. 1c). Likewise, analysis at 3 weeks after adoptive transfer of Mbd2−/− or WT Teff cells into Rag1−/− mice showed comparable or, in the case of lymph nodes, somewhat greater levels of iTreg development (Fig. 1d). Hence, the decrease in peripheral Treg cell numbers observed in Mbd2−/− mice was not likely to be due to impaired iTreg formation, at least under the in vitro or in vivo lymphopenic conditions analyzed to date.
Analysis by qPCR of gene expression in resting and activated Treg and Teff cells showed that Mbd2 deletion led to decreased Treg expression of Foxp3, cytotoxic-T lymphocyte-associated antigen 4 (CTLA-4), and TGF-β but did not affect the usual negligible expression of IL-2 or gamma interferon (IFN-γ) in Treg cells (Fig. 1e). We undertook Affymetrix microarray analysis to assess global gene expression by Mbd2−/− versus WT Treg cells. Differential expression of 1,300 (5.7%) genes was detected, with 1,034 genes upregulated and 266 genes downregulated in Mbd2−/− Treg cells (Fig. 1f). Mbd2 deletion led to broad changes in expression of multiple chemokine receptors and transcription factors (Fig. 1g). Functional annotation showed that Mbd2 deletion in Treg cells upregulated (blue) or downregulated (red) multiple pathways was associated with immune and inflammatory responses (Fig. 1h). Collectively, our data indicate that while conventional T cell development seems to be preserved in Mbd2−/− mice, these mice have a significant decrease in the overall number of peripheral Foxp3+ Treg cells. Mbd2−/− mice also have a markedly abnormal pattern of gene expression within residual Treg cells, including altered levels of Foxp3 and other prototypic Treg genes.
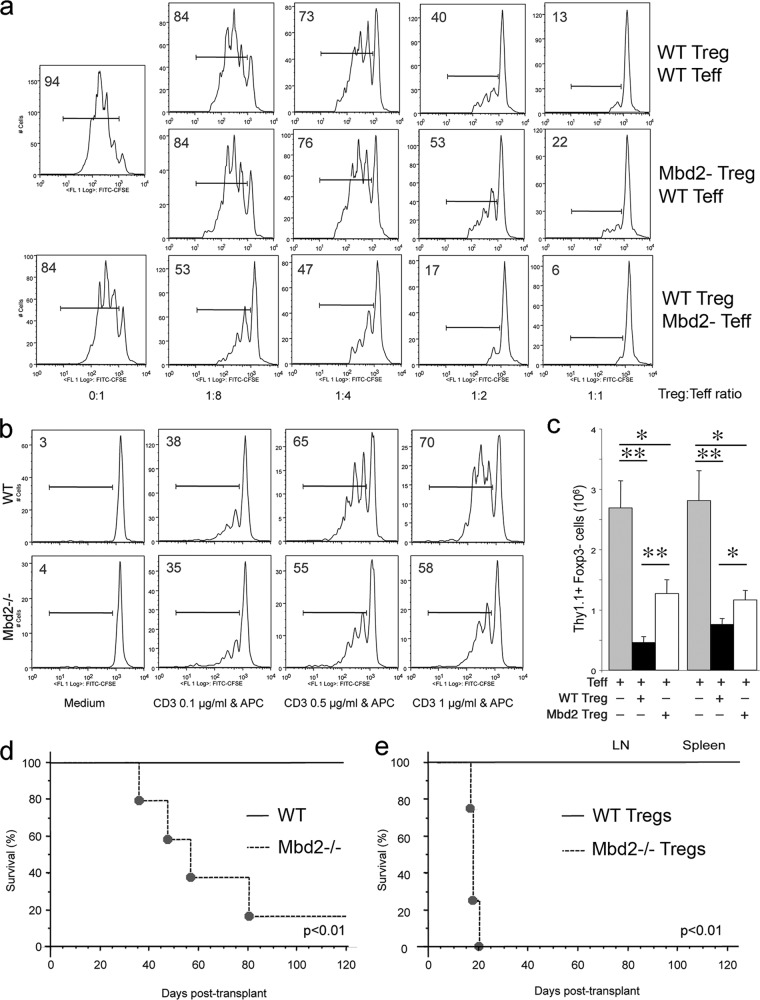
Mbd2 deletion significantly diminishes Foxp3+ Treg-suppressive functions.
In vitro studies using standard Treg suppression assays showed that Mbd2−/− Treg cells had decreased suppressive function compared to WT Treg cells (Fig. 2a, rows 1 and 2). To understand why Mbd2−/− mice, with decreased numbers of Foxp3+ Treg numbers (Fig. 1a) and decreased Treg function (Fig. 2a), do not develop autoimmunity, we assessed the responsiveness of Mbd2−/− Teff cells to Treg suppression. We found that Mbd2−/− Teff cells were more susceptible to Treg suppression than WT Teff cells (Fig. 2a, rows 1 and 3). Moreover, Mbd2−/− T cells underwent less T cell receptor (TCR)-induced proliferation than WT Teff cells receiving the same level of TCR stimulation (Fig. 2b). Hence, the normal phenotype of Mbd2−/− mice, despite reduced Treg numbers and function, likely reflects additional intrinsic Teff-associated factors that diminish T cell responses.
Fig 2.
Mbd2 deletion impairs Treg function in vitro and in vivo. (a) In vitro Treg suppression assays based on CFSE dilution by Teff cells proliferating in the presence of various ratios of WT or Mbd2−/− Treg cells; the proportion of proliferating cells is shown in each panel, and data are representative of 3 separate assays. (b) Mbd2−/− Teff cells display modestly impaired proliferative ability compared to WT Teff cells; the proportion of labeled cells is shown in each panel, and data are representative of 3 separate assays. (c) Mbd2−/− Treg cells displayed less ability than WT Treg cells to suppress the homeostatic expansion by 7 days of Thy1.1+ Foxp3− Teff cells that were adoptively transferred to Rag1−/− immunodeficient mice. Single-cell suspensions from LN or spleen samples were analyzed by flow cytometry for fluorescence-activated cell sorter (FACS) analyses (means ± SD, 4 mice/group). *, P < 0.05; **, P < 0.01 (for comparisons between groups, as shown). (d) Inability of a tolerance-inducing CD154 MAb/donor splenocyte transfusion protocol to prevent rejection in Mbd2−/− versus WT recipients of fully MHC-mismatched vascularized cardiac allografts (n = 5/group). (e) Differential survival of C57BL/6 cardiac allografts in BALB/c Rag1−/− immunodeficient hosts that were adoptively transferred with recipient Teff cells plus either WT or Mbd2−/− Treg cells (2:1 ratio of Teff to Treg cells, n = 4 mice/group).
We undertook 3 types of studies to assess if Mbd2 deletion affected Treg function in vivo. First, we used a model of homeostatic proliferation in which the adoptive transfer of 1 × 106 Thy1.1+ Teff cells into immunodeficient C57BL/6 Rag1−/− mice leads to rapid proliferation over 7 days, whereas cotransfer of 0.5 × 106 Treg cells normally markedly decreases Teff cell proliferation. Comparison of Teff proliferation in the presence of WT versus gene-targeted Treg cells in this model provides a quantitative test for evaluation of Treg suppression in vivo (22). We found that loss of Mbd2 significantly decreased the function of Treg cells, allowing increased Teff cell proliferation in the spleen and lymph nodes compared to the effects of transfer of WT Treg cells (Fig. 2c).
Second, we performed cardiac allografting across a full major histocompatibility complex (MHC) disparity, using C57BL/6 donors and Mbd2−/− or WT recipients. Each allograft recipient received 5 × 106 donor splenocytes plus 250 μg of CD40L MAb, intravenously, immediately after engraftment, to test the ability of Mbd2 deletion to curtail development of costimulation blockade-induced allograft tolerance (28). Whereas this well-established protocol led to long-term (>100-day) allograft survival in WT recipients, allografts were progressively rejected in Mbd2−/− recipients (P < 0.01) (Fig. 2d). As both induction and maintenance of long-term survival in this model are Treg dependent (34), the development of allograft rejection in Mbd2−/− mice pointed to a greater impact of Mbd deletion on Treg cells than on effector T cells.
As a third test of the effects of Mbd2 deletion on Treg function, we undertook cardiac allografting across the same fully MHC disparate strain combination but used immunodeficient Rag1−/− mice as recipients. Immediately postengraftment, recipients were injected intravenously with 1 × 106 WT Teff cells and 0.5 × 106 Treg cells from either WT or Mbd2−/− mice. Consistent with previous data (22), the adoptive transfer of WT Teff and WT Treg cells at a 2:1 ratio led to Treg-mediated suppression of effector T cell alloactivation and proliferation and resulted in long-term allograft survival (>100 days). In contrast, use of Mbd2−/− Treg cells in this model led to acute allograft rejection (P < 0.01) (Fig. 2e). Taken together, our in vitro and in vivo data demonstrate significant impairment of Foxp3+ Treg-suppressive functions as a result of Mbd2 deletion.
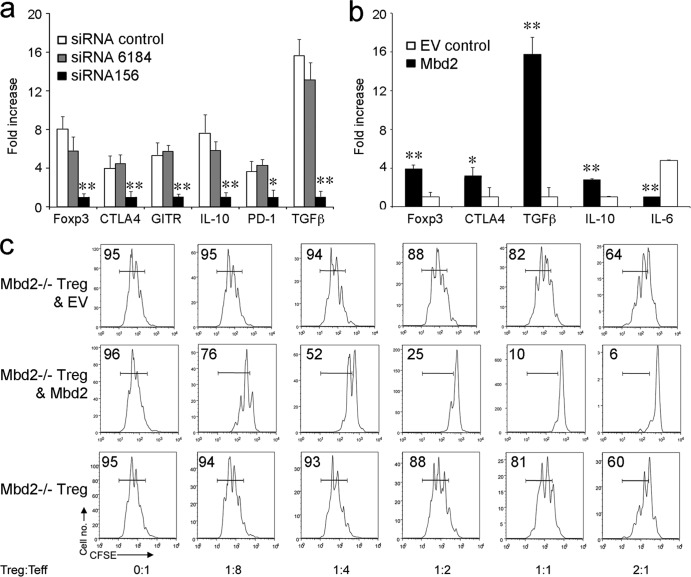
Knockdown or retroviral transduction of Mbd2 modulates Treg expression of Foxp3.
Our studies thus far showed an association between the deletion of Mbd2 by homologous recombination and decreased Treg numbers and suppressive function. We therefore tested the effects of siRNA-mediated knockdown of Mbd2 on gene expression in normal Treg cells and conversely whether viral transduction of Mbd2−/− Treg cells with Mbd2 would affect Treg numbers or function. We found that siRNA knockdown of Mbd2 in WT Treg cells significantly decreased Treg expression of Foxp3, as well as expression of multiple associated genes required for optimal Treg function, including CTLA-4, GITR, PD-1, IL-10, and TGF-β (Fig. 3a). In reciprocal studies, retroviral transduction of Mbd2−/− Treg cells with Mbd2 enhanced expression of Foxp3, CTLA-4, IL-10, and TGF-β and suppressed expression of IL-6 (Fig. 3b). Retroviral transduction also greatly increased Treg-suppressive function (Fig. 3c). Hence, alterations in the levels of Mbd2 can directly affect Foxp3 expression and Treg-suppressive function.
Fig 3.
Mbd2 knockdown or transduction modulates Foxp3 expression and Treg function. (a) Knockdown of Mbd2 decreased Treg expression of Foxp3 and Foxp3-dependent genes compared to the effects of scrambled siRNA or control lipofectamine. Data are shown as expression levels relative to that seen using specific Mbd2-targeting siRNA (means ± SD, *, P < 0.05; **, P < 0.01). (b) Viral transduction of Mbd2−/− Treg cells with Mbd2 increased expression of Foxp3, CTLA4, and TGF-β and IL-10 genes and decreased IL-6 gene expression. Data are shown as expression levels relative to empty vector (EV control) (means ± SD, *, P < 0.05; **, P < 0.01). (c) Effects of Mbd2 viral transduction on Treg-suppressive function in vitro; the proportion of proliferating cells is shown in each panel, and data are representative of 3 separate assays.
Mbd2 and Foxp3 methylation.
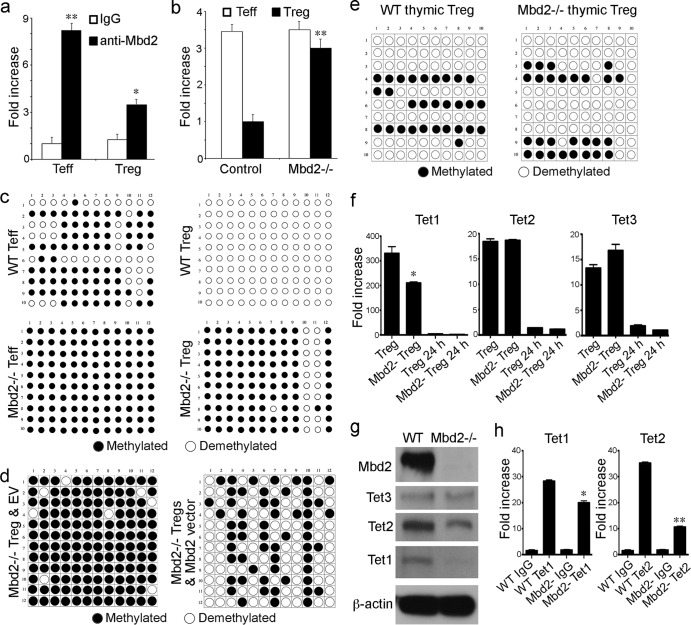
Since Foxp3 is a master regulator of Treg development and function and Mbd2 is known to bind methylated CpG islands, we assessed the effects of Mbd2 deletion on methylation of the best-characterized Foxp3 enhancer region, located at the TSDR site. We first tested whether Mbd2 was able to bind to this Foxp3 enhancer site by performing ChIP using an anti-Mbd2 antibody. We found that Mbd2 bound to the Foxp3 enhancer region in Teff cells and, to a lesser extent, Treg cells (Fig. 4a). Use of methyl collector (Mbd protein) to pull down methylated DNA showed more binding of Mbd2 to Foxp3 in the case of Mbd2−/− Treg cells; levels of binding now rose almost to the levels seen in Teff cells (Fig. 4b). To verify the methylation status of the Foxp3 TSDR site, we undertook bisulfite conversion, cloning, and sequencing. We found that the TSDR was fully demethylated in WT Treg cells but was largely methylated in Mbd2−/− Treg cells as well as in WT or Mbd2−/− Teff cells (Fig. 4c). However, retroviral transduction of Mbd2−/− Treg cells with Mbd2 but not empty vector (EV) control resulted in a marked increase in Foxp3 TSDR demethylation (Fig. 4d). These molecular data show that the presence or absence of Mbd2 has a critical effect on the extent of Foxp3 demethylation in Treg cells but not in Teff cells.
Fig 4.
Mbd2 associates with Foxp3 and affects Foxp3 methylation. (a) ChIP assay using anti-Mbd2 Ab to pull down Mbd2-associated chromatin, followed by Sybr real-time qPCR to detect the TSDR of Foxp3; data are shown as fold increase using purified Teff and Treg WT cells (*, P < 0.05, **, P < 0.01 [for anti-Mbd2 Ab versus control IgG]). (b) Methylation at the Foxp3 TSDR site was assessed using a methyl collector (Mbd2 protein) to pull down methylated DNA, followed by Sybr real-time qPCR; data are shown as fold increase using purified Teff and Treg cells (**, P < 0.01 [for Mbd2−/− versus WT Treg cells]). (c) Analysis of individual CpG sites within the Foxp3 TSDR by bisulfite conversion, cloning, and sequencing, using WT versus Mbd2−/− Teff and Treg cells. (d) Analysis of individual CpG sites within the Foxp3 TSDR by bisulfite conversion, cloning, and sequencing, using Mbd2−/− Treg cells retrovirally transduced with either empty vector (EV) control or vector containing Mbd2. (e) Bisulfite conversion, cloning, and sequencing showed that Mbd2−/− and WT thymic Treg cells had comparable levels of TSDR CpG site demethylation. (f) Expression of Tet genes in peripheral Treg cells (qPCR, 4 mice/group), showing a decrease in Tet1 but not Tet2 or Tet3 expression in freshly isolated Mbd2−/− versus WT Treg cells. (g) Western blots of Mbd2, Tet1, Tet2, and Tet3 in WT or Mbd2−/− Treg cells, confirming deletion of Mbd2 in Mbd2−/− Treg cells and showing that Tet1 protein levels in MBD2 Treg cells were markedly decreased whereas Tet2 and Tet3 levels were affected only modestly compared to WT Treg cell results. (h) ChiP assay of Tet1 and Tet2 binding to the TSDR of WT and Mbd2−/− peripheral Treg cells; data are shown as fold increase by qPCR (*, P < 0.05; **, P < 0.01 [for Tet Ab binding in Mbd2−/− versus WT Treg cells]).
To further explore the level at which Mbd2 expression affects Treg development, we tested whether methylation of the thymic Treg TSDR was affected by MBD2 deletion. Assessment of CD4+ CD25+ CD8− Treg cells, isolated by cell sorting from thymi of Mbd−/− and WT mice, showed comparable levels of and extensive TSDR demethylation in the two sets of mice (Fig. 4e). Hence, Mbd2−/− Treg cells have a normal pattern of almost complete TSDR demethylation during thymic development, but this is not maintained in the periphery. Since demethylation of DNA requires Tet1, -2, and -3, and especially the last two in the periphery (15), we checked Tet gene and protein expression in peripheral Treg cells. Compared to freshly isolated WT Treg cells, gene expression of Tet1 but not Tet2 or Tet3 was decreased in the corresponding peripheral Treg of Mbd2−/− mice (Fig. 4f). Levels of Tet1 protein were also markedly reduced in Mbd2−/− Treg cells, whereas levels of Tet2 and Tet3 proteins were decreased only slightly compared to levels in WT Treg cells (Fig. 4g). We also undertook ChIP assays to assess Tet protein binding to the Foxp3 TSDR. While no suitable anti-Tet3 Ab was available for ChIP studies, we did detect a slight decrease in Tet1 binding, and a marked decrease in Tet2 binding, to the TSDR of Mbd2−/− versus WT Treg cells (Fig. 4h). Collectively, these data suggest that Mbd2 may promote Foxp3 demethylation in the periphery at least in part by facilitating the recruitment of Tet enzymes to CpG island sites that affect Foxp3 gene expression.
DISCUSSION
In the fields of autoimmunity and transplantation, there is currently much interest in Treg cells either as potential cellular therapies or as targets for pharmacologic modulation. In the latter case, the requirement for acetylation of Foxp3 protein at certain lysines for optimal DNA binding and Treg function (35, 36) has spurred research into the use of HDAC inhibitors beyond the field of oncology (37). A broad understanding of the transcriptional control of Foxp3 expression is essential for efforts to rationally employ Foxp3+ Treg cells for therapeutic application in autoimmunity and posttransplantation. Much attention has been focused on the TSDR site that contains numerous CpG dinucleotides that are demethylated only in stable Foxp3+ Treg cells. The use of Dnmt enzyme inhibitors to reduce methylation in Treg cells is one possibility but is potentially too risky for application beyond the cancer field, given the potential mutagenic effects of these agents. Alternatively, since stable methylation typically requires the binding of Mbd proteins that recruit histone-modifying and chromatin-remodeling complexes to methylated sites, we reasoned that targeting of Mbd proteins might be a new approach to regulation of methylation in Treg cells. Thus, the repressive activity of Mbd2 is thought to be mediated by MeCP1, an ATP-dependent chromatin-remodeling complex consisting of about 10 proteins, including Mbd2, HDAC1, HDAC2, Mi-2/NuRD, and several Rb family and other proteins (38). However, the current data indicate that targeting Mbd2, at least, is also inadvisable in this context, given the resultant decreased Treg numbers and suppression function of Treg cells in Mbd2−/− mice. Indeed, in contrast to our initial expectations, it is possible that Mbd2 targeting is relevant to cancer therapy, at least in those cases in which cancers are associated with impaired host antitumor responses as a result of active suppression by Foxp3+ Treg cells (39, 40). Anti-sense targeting of Mbd2 in vitro and in vivo with immunodeficient mice bearing human cancer xenografts has proven effective (41, 42), and it is possible that this strategy will be even more useful in immunocompetent hosts. This mechanism may also contribute to the known resistance to tumorigenesis of Mbd2−/− mice when crossed to the ApcMin background (43).
The array of DNA demethylases now forms a complex, increasingly crowded field, with considerable attention recently being given to cytidine deaminases and members of the Tet family (44). The current studies point to a major role for Mbd2 in regulation of Foxp3+ Treg cells but do not address the controversy as to whether Mbd2 itself is a promoter-specific DNA demethylase. Rather, our data show that its loss leads to impaired Foxp3 expression and decreased Treg function and that its restoration promotes Foxp3 demethylation and restores Treg function to normal. A recent study showed that thymic Foxp3 TSDR demethylation is an active process involving the Tet family of proteins (21). Maintenance of TSDR demethylation in the periphery may also require the actions of one or more Tet enzymes, as suggested by our current studies. These data indicate that Mbd2 affects the expression of Tet proteins in Treg cells, though the details remain to be explored. Thus, the lack of TSDR demethylation in Mbd2−/− Treg cells may be due to decreased expression of Tet protein, resulting in decreased Tet enzyme binding to the Treg TSDR. However, Mbd2 might also recruit Tet proteins to methylated DNA. Without Mbd2, the binding of Tet proteins, especially Tet2, to the Treg TSDR was markedly impaired.
Additional studies are required to further tease out the importance and precise interactions of Mbd2 with Foxp3 and Tet enzymes. Why Mbd2 binding in Teff cells is not associated with TSDR demethylation whereas it appears key in Foxp3+ Treg cells is currently unknown. While many pathways function differently in Treg cells versus conventional T cells as a result of Foxp3-dependent events and additional epigenetic processes (16, 20), more distal actions of Mbd2 may also play a role. For example, loss of Mbd2 can affect rRNA stability and associated posttranscriptional modifications (45, 46), as well as protein-protein interactions. Thus, while Mbd2 binds to methylated DNA with high affinity via its Mbd region, it also has an N-terminal extension of 152 amino acids that can significantly affect its interactions with DNA as well as with other proteins (47), such as the zinc finger protein MIZF (48). Clearly, there are multiple levels at which Mbd2 may contribute to gene regulation, including in Foxp3+ Treg cells. Ultimately, unraveling this complexity will require identification of both the Foxp3 and Treg methylomes, and knowledge of their interactions may provide a fertile means for manipulation of the functions of these key cells in disease states ranging from autoimmunity to cancer.
ACKNOWLEDGMENTS
We declare that we have no competing financial interests.
This work was supported by National Institutes of Health awards K08AI095353 (to U.H.B.) and R01AI073938 and P01AI073489 (to W.W.H.).
Footnotes
Published ahead of print 26 August 2013
REFERENCES
- 1.Kass SU, Pruss D, Wolffe AP. 1997. How does DNA methylation repress transcription? Trends Genet. 13:444–449 [DOI] [PubMed] [Google Scholar]
- 2.Defossez PA, Stancheva I. 2011. Biological functions of methyl-CpG-binding proteins. Prog. Mol. Biol. Transl. Sci. 101:377–398 [DOI] [PubMed] [Google Scholar]
- 3.Dhasarathy A, Wade PA. 2008. The MBD protein family—reading an epigenetic mark? Mutat. Res. 647:39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. 1999. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397:579–583 [DOI] [PubMed] [Google Scholar]
- 5.Detich N, Theberge J, Szyf M. 2002. Promoter-specific activation and demethylation by MBD2/demethylase. J. Biol. Chem. 277:35791–35794 [DOI] [PubMed] [Google Scholar]
- 6.Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Vilardell-Tarres M. 2007. Transcript overexpression of the MBD2 and MBD4 genes in CD4+ T cells from systemic lupus erythematosus patients. J. Leukoc. Biol. 81:1609–1616 [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Ou TT, Wu CC, Li RN, Lin YC, Lin CH, Tsai WC, Liu HW, Yen JH. 2011. Global DNA methylation, DNMT1, and MBD2 in patients with systemic lupus erythematosus. Lupus 20:131–136 [DOI] [PubMed] [Google Scholar]
- 8.Lei W, Luo Y, Yan K, Zhao S, Li Y, Qiu X, Zhou Y, Long H, Zhao M, Liang Y, Su Y, Lu Q. 2009. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand. J. Rheumatol. 38:369–374 [DOI] [PubMed] [Google Scholar]
- 9.Liu CC, Fang TJ, Ou TT, Wu CC, Li RN, Lin YC, Lin CH, Tsai WC, Liu HW, Yen JH. 2011. Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunol. Lett. 135:96–99 [DOI] [PubMed] [Google Scholar]
- 10.Zhang P, Su Y, Chen H, Zhao M, Lu Q. 2010. Abnormal DNA methylation in skin lesions and PBMCs of patients with psoriasis vulgaris. J. Dermatol. Sci. 60:40–42 [DOI] [PubMed] [Google Scholar]
- 11.Vilain A, Vogt N, Dutrillaux B, Malfoy B. 1999. DNA methylation and chromosome instability in breast cancer cell lines. FEBS Lett. 460:231–234 [DOI] [PubMed] [Google Scholar]
- 12.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58–61 [DOI] [PubMed] [Google Scholar]
- 13.Boeke J, Ammerpohl O, Kegel S, Moehren U, Renkawitz R. 2000. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem. 275:34963–34967 [DOI] [PubMed] [Google Scholar]
- 14.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62–66 [DOI] [PubMed] [Google Scholar]
- 15.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Rudensky AY. 2007. Foxp3 in control of the regulatory T cell lineage. Nat. Immunol. 8:457–462 [DOI] [PubMed] [Google Scholar]
- 17.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73 [DOI] [PubMed] [Google Scholar]
- 18.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21 [DOI] [PubMed] [Google Scholar]
- 19.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38. 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463:808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toker A, Engelbert D, Garg G, Polansky JK, Floess S, Miyao T, Baron U, Duber S, Geffers R, Giehr P, Schallenberg S, Kretschmer K, Olek S, Walter J, Weiss S, Hori S, Hamann A, Huehn J. 2013. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J. Immunol. 190:3180–3188 [DOI] [PubMed] [Google Scholar]
- 22.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. 2007. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13:1299–1307 [DOI] [PubMed] [Google Scholar]
- 23.de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, Bradner JE, Mazitschek R, Kozikowski AP, Matthias P, Hancock WW. 2011. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3+ T-regulatory cells. Mol. Cell. Biol. 31:2066–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. 2010. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138:583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beier UH, Wang L, Bhatti TR, Liu Y, Han R, Ge G, Hancock WW. 2011. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell. Biol. 31:1022–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasai N, Defossez PA. 2009. Many paths to one goal? The proteins that recognize methylated DNA in eukaryotes. Int. J. Dev. Biol. 53:323–334 [DOI] [PubMed] [Google Scholar]
- 27.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. 2001. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock WW, Sayegh MH, Zheng XG, Peach R, Linsley PS, Turka LA. 1996. Costimulatory function and expression of CD40 ligand, CD80, and CD86 in vascularized murine cardiac allograft rejection. Proc. Natl. Acad. Sci. U. S. A. 93:13967–13972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Liu Y, Beier UH, Han R, Bhatti TR, Akimova T, Hancock WW. 2013. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood 121:3631–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas RM, Gao L, Wells AD. 2005. Signals from CD28 induce stable epigenetic modification of the IL-2 promoter. J. Immunol. 174:4639–4646 [DOI] [PubMed] [Google Scholar]
- 31.Battke F, Symons S, Nieselt K. 2010. Mayday—integrative analytics for expression data. BMC Bioinformatics 11:121. 10.1186/1471-2105-11-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 33.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. 2002. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell 10:81–91 [DOI] [PubMed] [Google Scholar]
- 34.Lee I, Wang L, Wells AD, Dorf ME, Ozkaynak E, Hancock WW. 2005. Recruitment of Foxp3+ T regulatory cells mediating allograft tolerance depends on the CCR4 chemokine receptor. J. Exp. Med. 201:1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. 2006. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J. Biol. Chem. 281:36828–36834 [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Wang L, Han R, Beier UH, Hancock WW. 2012. Two lysines in the forkhead domain of foxp3 are key to T regulatory cell function. PLoS One 7:e29035. 10.1371/journal.pone.0029035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, de Zoeten EF, Greene MI, Hancock WW. 2009. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat. Rev. Drug Discov. 8:969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Q, Zhang Y. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa H, Sakaguchi S. 2010. Regulatory T cells in tumor immunity. Int. J. Cancer 127:759–767 [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang L, Predina J, Han R, Beier UH, Wang LCS, Kapoor V, Bhatti TR, Akimova T, Singhal S, Brindle PK, Cole PA, Albelda SM, Hancock WW. Inhibition of p300 impairs Foxp3+ T-regulatory cell function and promotes anti-tumor immunity. Nat. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov MA, Lamrihi B, Szyf M, Scherman D, Bigey P. 2003. Enhanced antitumor activity of a combination of MBD2-antisense electrotransfer gene therapy and bleomycin electrochemotherapy. J. Gene Med. 5:893–899 [DOI] [PubMed] [Google Scholar]
- 42.Campbell PM, Bovenzi V, Szyf M. 2004. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis 25:499–507 [DOI] [PubMed] [Google Scholar]
- 43.Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. 2003. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat. Genet. 34:145–147 [DOI] [PubMed] [Google Scholar]
- 44.Carey N, Marques CJ, Reik W. 2011. DNA demethylases: a new epigenetic frontier in drug discovery. Drug Discov. Today 16:683–690 [DOI] [PubMed] [Google Scholar]
- 45.Swaminathan V, Reddy BA, Ruthrotha Selvi B, Sukanya MS, Kundu TK. 2007. Small molecule modulators in epigenetics: implications in gene expression and therapeutics. Subcell. Biochem. 41:397–428 [PubMed] [Google Scholar]
- 46.Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. 2004. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J. Biol. Chem. 279:6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. 2003. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31:1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekimata M, Takahashi A, Murakami-Sekimata A, Homma Y. 2001. Involvement of a novel zinc finger protein, MIZF, in transcriptional repression by interacting with a methyl-CpG-binding protein, MBD2. J. Biol. Chem. 276:42632–42638 [DOI] [PubMed] [Google Scholar]