Abstract
Atypical enteropathogenic Escherichia coli (aEPEC) strains are diarrheal pathogens that lack bundle-forming pilus production but possess the virulence-associated locus of enterocyte effacement. aEPEC strain 1551-2 produces localized adherence (LA) on HeLa cells; however, its isogenic intimin (eae) mutant produces a diffuse-adherence (DA) pattern. In this study, we aimed to identify the DA-associated adhesin of the 1551-2 eae mutant. Electron microscopy of 1551-2 identified rigid rod-like pili composed of an 18-kDa protein, which was identified as the major pilin subunit of type 1 pilus (T1P) by mass spectrometry analysis. Deletion of fimA in 1551-2 affected biofilm formation but had no effect on adherence properties. Analysis of secreted proteins in supernatants of this strain identified a 150-kDa protein corresponding to SslE, a type 2 secreted protein that was recently reported to be involved in biofilm formation of rabbit and human EPEC strains. However, neither adherence nor biofilm formation was affected in a 1551-2 sslE mutant. We then investigated the role of the EspA filament associated with the type 3 secretion system (T3SS) in DA by generating a double eae espA mutant. This strain was no longer adherent, strongly suggesting that the T3SS translocon is the DA adhesin. In agreement with these results, specific anti-EspA antibodies blocked adherence of the 1551-2 eae mutant. Our data support a role for intimin in LA, for the T3SS translocon in DA, and for T1P in biofilm formation, all of which may act in concert to facilitate host intestinal colonization by aEPEC strains.
INTRODUCTION
Enteropathogenic Escherichia coli (EPEC) is a common cause of infant diarrhea in both developing and developed countries (1, 2). The hallmark of EPEC pathogenesis is the formation of attaching and effacing (AE) lesions characterized by intimate bacterial adherence, microvillus effacement, and accumulation of polymerized actin and many other cytoskeleton elements, resulting in pedestal-like structures underneath adherent bacteria. The virulence factors necessary for the establishment of the AE lesion are encoded in a chromosomal pathogenicity island called the locus of enterocyte effacement (LEE) (3).
The EPEC pathotype is divided into typical (tEPEC) and atypical EPEC (aEPEC). This classification is based on the occurrence of the virulence-associated EAF (EPEC adherence factor) plasmid (pEAF) in tEPEC and its absence in aEPEC (4). Although tEPEC strains were major causative agents of acute diarrhea in very young children in Brazil until the 1990s (5), there is currently a clear decrease in their frequency (6, 7). Retrospective studies indicate that aEPEC strains have existed as important agents of diarrhea in developed countries since the 1960s and are currently emerging pathogens that produce acute and persistent diarrhea affecting children and adults worldwide (2, 8, 9).
The first step in the establishment of diarrheal disease caused by the various pathotypes of diarrheagenic E. coli (DEC) is the colonization of the gastrointestinal tract, which is mediated by specific fimbrial and nonfimbrial adherence factors (10). A feature of tEPEC is the production of a characteristic pattern of adherence, termed localized adherence (LA), in which bacteria bind to localized areas of the cultured epithelial cell surface, forming compact microcolonies that are visualized after 3 h of infection (11). Generally, most aEPEC strains are weakly adherent and display a LA-like (LAL) phenotype manifested as loose cell-associated bacterial microcolonies; less common are aEPEC strains that display the typical LA pattern. These patterns can be observed only in adherence assays performed for extended periods of infection (e.g., 6 h). Some strains however, do exhibit diffuse (DA) or aggregative (AA) adherence (12–15).
Upon infection of host cells EPEC translocates a range of effector proteins to the eukaryotic cell by means of the type 3 secretion system (T3SS) (16, 17). The EspA protein forms a long filamentous extension of the T3SS needle complex, and EspB and EspD are coupled to the end of the EspA filament to generate a pore in the host cell membrane through which effectors are injected into the host cell cytosol (18, 19).
The LEE-encoded intimin adhesin is required for intimate adhesion to epithelial cells and cytoskeletal reorganization (20). The translocated intimin receptor (Tir) (21) is injected via T3SS and inserted in the plasma membrane of eukaryotic cells and is crucial for development of AE lesions. So far, intimin has been recognized as the main adhesin in aEPEC strains (22, 23).
Our previous studies on the adherence mechanisms of aEPEC strains isolated from clinical cases in Brazil identified a strain (1551-2, ONT:H−) that forms LA on cultured epithelial cells. However, an isogenic intimin mutant strain adheres to HeLa cells exhibiting a diffuse-adherence (DA) pattern (24). Our main interest in the present study was to elucidate the nature of the aEPEC 1551-2 adhesin responsible for the DA phenotype.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1. aEPEC 1551-2 was isolated from a child with diarrhea in the absence of other recognized pathogens in an epidemiological study conducted in São Paulo, Brazil (13). Strains were grown aerobically in Luria-Bertani (LB) medium or Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) at 37°C. Recombinant DNA and molecular biology techniques were performed as previously described (25).
Table 1.
Escherichia coli strains and plasmids used in this study
| E. coli strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| 1551-2 | aEPEC (ONT:H−) isolated from a child with diarrhea in Brazil (Nalr) | 13 |
| 1551-2 eae::Km | 1551-2 harboring the suicide vector pJP5603 inserted into the eae gene (Nalr Kmr) | 24 |
| 1551-2 ΔfimA | fimA::zeo (Nalr Zeor) | This study |
| 1551-2 sslE::Km | 1551-2 harboring the suicide vector pJP5603 inserted into sslE gene (Nalr Kmr) | This study |
| 1551-2 escN::Km | 1551-2 harboring the suicide vector pJP5603 inserted into escN gene (Nalr Kmr) | This study |
| 1551-2 eae::Km Δtir | Double intimin/Tir mutant in aEPEC 1551-2 (Nalr Kmr Zeor) | This study |
| 1551-2 eae::Km ΔfimA | Double intimin/T1P mutant in aEPEC 1551-2 (Nalr Kmr Zeor) | This study |
| 1551-2 eae::Km ΔespA | Double intimin/EspA mutant in aEPEC 1551-2 (Nalr Kmr Zeor) | This study |
| Plasmids | ||
| pEscN | pACYC184 vector harboring the escN gene from EPEC prototype E2348/69 | 31 |
| pJP5603 | 3.1-kb R6K-based suicide vector, Kmr | 29 |
| pKOBEG-Apra | Red recombinase system plasmid, Aprar | 44 |
| 1711-4ΔfliC | Source of zeocin cassette | 44 |
| pGEM-T Easy | Cloning vector | Promega |
| pBAD/Myc-His A | Cloning vector | Invitrogen |
| pFimA | pBAD/Myc-His A vector harboring the fimA gene from aEPEC 1551-2 | This study |
| pInt | pBAD/Myc-His A vector harboring the eae gene from aEPEC 1551-2 | This study |
| pEspA | pBAD/Myc-His A vector harboring the espA gene from aEPEC 1551-2 | This study |
Nalr, nalidixic acid resistant; Kmr, kanamycin resistant; Zeor, zeocin resistant; Aprar, apramycin resistant.
Epithelial cell adherence.
The assay described by Cravioto et al. (26) was used, with some adaptations, to determine the adherence pattern of the strains. HeLa cells at 70% confluence were cultivated in 24-well tissue culture plates containing glass coverslips and DMEM supplemented with 10% bovine fetal serum (Gibco Invitrogen, USA) and 1% antibiotics (Gibco Invitrogen, USA). After one wash with phosphate-buffered saline (PBS), pH 7.4, 1.0 ml of fresh medium (DMEM supplemented with 2% fetal bovine serum) was added to the cell monolayers. E. coli strains were grown overnight in LB broth without shaking and then diluted 1:50 in the medium contained in the microplates. After an incubation period of 3 h at 37°C, the monolayers were washed once with PBS and fresh medium was added to the wells, followed by an additional incubation period of 3 h for a total of 6 h. After six washes with PBS, the preparations were fixed with methanol, stained with May Grünwald-Giemsa stain, and examined by light microscopy.
For quantitative experiments, after the adherence assay (performed as described above) the washed monolayers were lysed in 1% Triton X-100 for 30 min at room temperature. Following cell lysis, adherent bacteria were quantified by plating serial dilutions onto MacConkey agar plates to obtain the total number of cell-associated bacteria. Assays were carried out in triplicate, and the results represent the means ± standard errors from at least three independent experiments.
Adherence inhibition experiments with EspA antibody.
For adherence inhibition experiments, samples (approximately 107 CFU) of an overnight culture of the 1551-2 eae::Km mutant strain grown in LB broth were incubated with 1:100, 1:50, and 1:10 dilutions of anti-EspA antibody in DMEM at room temperature for 30 min and then added to the HeLa cell monolayers. After incubation for 6 h, the cells were lysed with PBS–0.1% Triton X-100, diluted, and plated onto MacConkey agar to estimate the number of bacteria adhering to the cell monolayers (27). All inhibition experiments were performed at least three times in triplicate on different days to ensure reproducibility.
Gene amplification.
For PCR, the GoTaq Green master mix (Promega, Madison, WI) was used with 1 μM each of the primers (Table 2).
Table 2.
Primers used for PCR amplification
| Purpose and designation | Primer sequence (5′ → 3′) |
|---|---|
| Allelic exchange | |
| tir-zeo(F) | ATGCCTATTGGTAATCTTGGTCATAATCCCAATGTGAGTGGTCATCGCTTGCATTAGAAAGG |
| tir-zeo(R) | TTAAACGAAACGATTGGATCCCGGCACTGGTGGGTTATTCGAATGATGCAGAGATGTAAG |
| fimA-zeo(F) | ATGAAAATTAAAACTCTGGCAATTGTTGTTCTGTCGGCTCGTCATCGCTTGCATTAGAAAGG |
| fimA-zeo(R) | TTATTGATACTGAACCTTGAAGGTTGCATCCGCGTTAGCTGAATGATGCAGAGATGTAAG |
| espA-zeo(F) | ATGGATACACCAAATGCAACATCCGTTGCTAGTGCGAGCGGTCATCGCTTGCATTAGAAAGG |
| espA-zeo(R) | TTATTTACCAAGGGATATTGCCGAAATAGTTCTGTATTGCGAATGATGCAGAGATGTAAG |
| Gene amplification by PCR | |
| espA-F | GGGGTACCTCGGTGTTTTTCAGGCTGC |
| espA-R | CGAGCTCATTTCAAGCTGGCTATTATT |
| fimA-F | GTCGGTTTTAACATTCAACTGAATG |
| fimA-R | AATAACGCGCCTGGAACGGAAT |
| fimA-NcoI-F | AGCAGCCCATGGAAATTAAAACTCTGG |
| fimA-HindIII-R | GGTAGGAAGCTTTTATTGATACTGAACCTT |
| eae-NcoI-F | ATACCATGGTTACTCATGGTTGTTATACCCGG |
| eae-HindIII-R | ATAAAGCTTTCTCACACAGACTCGATAGGCATAATTACC |
| espA-NcoI-F | ATACCATGGATACACCAAATGCAACATCCGTTGC |
| espA-HindIII-R | ATAAAGCTTTTTACCAAGGGATATTGCCGAAATAG |
| tir-F | CACCTATAAGGCAGTCTGTTGCTG |
| tir-R | GGTTTCAGTTGCACTTGCAGCAG |
| escN-F | CGACGACTATTGCAGAGT |
| escN-R | GCCTTATCTGCTTCAGGA |
| EscN-B-19-F | CGCGGATCCTCTAAGGGAATAATATCGAACTTAAAG |
| EscN-S-20-R | ACGCGTCGACTCAGGCAACCACTTTGAATAGGC |
| sslE-F | GATTCTGATGGCGCGTAATGATAA |
| sslE-R | CTGATACAGACGCTGAATGTTGG |
| sslE-F2 | AGGGCGTTATCAACAAGCTGTG |
| sslE-R2 | CAGCGGTTTTTCCATATAGTTGAG |
| PJP-F | CCCAGTCACGACGTTGTAAAACG |
| PJP-R | AGCGGATAACAATTTCACACAGG |
Mutant construction and complementation.
Derivative mutants were constructed by the one-step allelic exchange recombination method, using the thermosensitive plasmid pKOBEG-Apra, which carries lambda red recombination genes (28). Primers containing a 40-bp region homologous to the 5′ and 3′ extremities of the target genes (fimA, tir and espA) and specific sequence for the zeocin (Zeo) resistance-encoding gene were used to amplify the Zeo cassette. Amplicons were purified from the agarose gel and quantified (BioPhotometer; Eppendorf) before they were electroporated into competent aEPEC 1551-2 or 1551-2 eae::Km cells containing the pKOBEG-Apra plasmid. Recombinant bacteria were selected on Zeo-containing LB agar plates (60 μg/ml). Primers employed for mutagenesis are described in Table 2. The loss of the target gene in the isogenic mutants was confirmed by PCR, and all mutants were checked for susceptibility to apramycin, indicative of the loss of pKOBEG-Apra.
For directed insertional mutagenesis, internal segments of the escN or sslE gene were prepared by PCR and cloned into the pGEM-T Easy vector (Promega). This recombinant plasmid was digested with EcoRI, and the segment of interest was cloned into the corresponding sites in suicide vector pJP5603 (29). After transformation of recombinant plasmids into DH5α(λpir), transformants were selected on LB agar plates containing kanamycin (50 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 80 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 0.5 mM).
Plasmid DNA of transformants harboring the correct insert was transformed into strain S17-1(λpir) for mobilization into a nalidixic acid-resistant derivative of strain 1551-2 via filter mating on cellulose nitrate membranes. Transconjugants were selected on L agar plates containing kanamycin and nalidixic acid as previously described (30). The integration of suicide vector pJP5603 in the escN gene was confirmed by PCR analysis using primers EscN-B-19-F and PJP-R to verify the right junction and primers EscN-S-20-R and PJP-F to verify the left junction (Table 2). The escN mutant was complemented with the pEscN plasmid carrying the escN gene from the prototype tEPEC E2648/69 in the pACYC184 vector (31).
The integration of suicide vector pJP5603 in the sslE gene was also confirmed by PCR using primers sslE-F2 and PJP-R to verify the right junction and primers sslE-R2 and PJP-F to verify the left junction. For complementation, the fimA gene from aEPEC 1551-2 was amplified by PCR using primers fimA-NcoI-F and fimA-HindIII-R (Table 2). The PCR product was then digested with NcoI and HindIII and cloned in the respective site of the pBAD/Myc-His A vector, originating recombinant plasmid pFimA, and transformed into 1551-2 ΔfimA. T1P production in the complemented strain was induced with 0.01% l-arabinose. The eae and the espA genes from the 1551-2 strain were cloned in the pBAD/Myc-His A vector, in order to complement 1551-2 eae::Km and 1551-2 eae::Km ΔespA. Experiments with the complemented strains carrying the pInt (eae gene in pBAD/Myc-His A) or the pEspA (espA in pBAD/Myc-His A) plasmids were carried out in the presence of 0.01% l-arabinose.
All mutant derivatives obtained in this study were confirmed to grow at the same rate as the wild-type strain under the conditions tested in this work (data not shown).
Biofilm formation assay on abiotic surface.
Adhesion to abiotic surface (polystyrene) was analyzed using 24-well plates as described before (32). Overnight cultures of bacteria grown in LB broth were adjusted to an optical density at 600 nm (OD600) of 1.0 (corresponding to ca. 108 CFU/ml), and 40-μl aliquots were added to the wells containing 1 ml of LB and incubated at 37°C for 48 h. Unbound bacteria were removed after washing the cultures three times with PBS, and biofilms were fixed for 20 min with 2% formalin, washed, and then stained with 1% crystal violet (CV) for 20 min. Wells were thoroughly rinsed three times with PBS, and the dye was solubilized in 1 ml of methanol. Finally, the amount of extracted crystal violet was determined by measuring the OD600 by using an enzyme-linked immunosorbent assay (ELISA) Multiskan plate reader.
Pellicle formation.
Strains were grown for 48 h in LB at 37°C with shaking (225 rpm) in glass tubes. Following incubation, the pellicles formed at the air-liquid interface were washed three times with PBS and then stained with CV (1%) for 20 min. After staining, pellicles were washed thoroughly with distilled water and photographed (33).
Agglutination of yeast cells.
Surface-expressed T1P on aEPEC strains was assayed by the ability to agglutinate mannan-rich yeast (Saccharomyces cerevisiae) cells on glass slides as previously described (34). Equal volumes of bacterial and yeast suspensions were mixed on a glass slide and rocked for 1 to 2 min until the aggregates were visualized by naked eye.
Pilus purification and characterization.
To purify the pilus structures, aEPEC 1551-2 growing in 35 plates (150 by 15 mm) of MacConkey agar at 37°C was harvested, and the pili were purified by mechanical shearing and differential centrifugation in order to remove outer membranes and bacterial debris (35). Pilus purification was monitored by transmission electron microscopy (TEM) and SDS-PAGE, as previously described (36). The pilus-containing supernatant was denatured with HCl (37) and resolved by SDS-PAGE (38). An 18-kDa protein was excised from the gel and subjected to mass spectrometry analysis after trypsin digestion for protein identification.
Heat extraction of surface-associated proteins.
Toward identifying a putative adhesin, outer membrane proteins from aEPEC 1551-2 were heat extracted as follows. Briefly, 10 ml of an overnight bacterial culture in LB or DMEM was harvested, resuspended in 100 μl of PBS (pH 7.4), and heated at 65°C for 1 h (39). This treatment was followed by centrifugation at 10,000 × g at 4°C, and the supernatant was analyzed by SDS-PAGE in 12% polyacrylamide gels (38).
Analysis of aEPEC secreted proteins.
Determination of aEPEC secreted proteins was done as previously described, with minor modifications (40). Briefly, strains were grown overnight in LB at 37°C in a shaker at 225 rpm and then subcultured 1:50 into 5 ml of DMEM. After 6 h of growth, 1.5 ml of each culture was harvested by centrifugation at 16,000 × g for 5 min, and the supernatant was filtered through a 0.45-μm filter (Millipore). Trichloroacetic acid was added to a final concentration of 10% (vol/vol) to precipitate the secreted proteins. After overnight incubation at 4°C, the Eppendorf tubes were centrifuged for 20 min at 4°C, the supernatant was discarded, and the pellet was dissolved in 20 μl of SDS-PAGE sample buffer. Saturated Tris was added to neutralize the residual trichloroacetic acid. The secreted proteins were used for SDS-PAGE (12%) analysis.
Ultrastructural studies.
The presence of pili on 1551-2 was visualized by negative staining with 1% phosphotungstic acid and electron microscopy (EM) in a Philips CM12 electron microscope at 80 kV as previously described (27, 36). Glass coverslips containing fixed HeLa cells with adhering bacteria were prepared for scanning electron microcopy (SEM) as described previously (27) and visualized by using a Hitachi S-4500 scanning electron microscope (Hitachi, Tokyo, Japan).
Statistical analysis.
All analyses were performed by using Student's t test. A P value of ≤0.05 by two-tailed analysis was taken to indicate statistical significance.
RESULTS
aEPEC 1551-2 continues to adhere to HeLa cells in the absence of intimin.
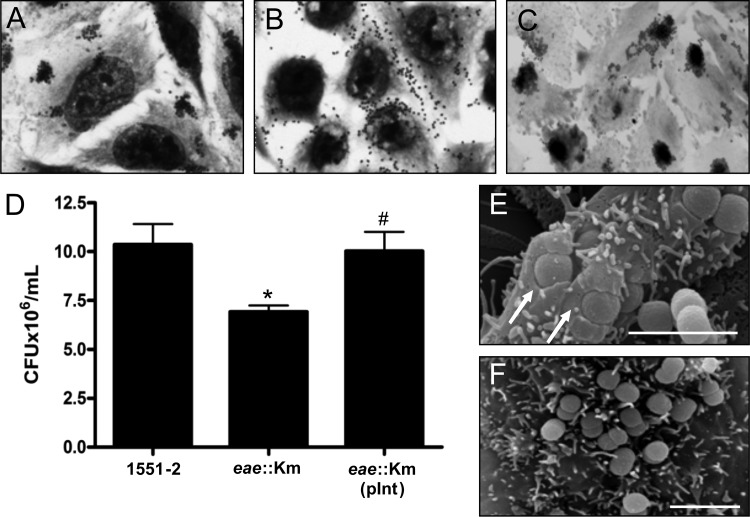
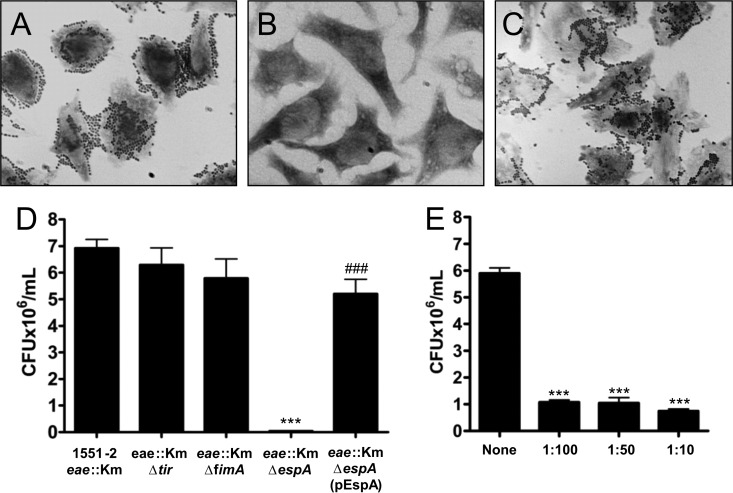
We have previously demonstrated that deletion of the eae gene completely abolished the ability of 1551-2 to display the LA pattern in HeLa cells (24). Surprisingly, 1551-2 eae::Km remained adherent to HeLa cells and no longer displayed LA but DA (Fig. 1A and B). Quantitatively, the eae mutant was approximately 33.7% less adherent than 1551-2 (P < 0.05) (Fig. 1D). Complementation of 1551-2 eae::Km in trans with eae in pInt restored the ability of this strain to produce the original LA pattern of 1551-2 (Fig. 1C) and at comparable adherence levels (P > 0.05) (Fig. 1D). The complemented strain was significantly more adherent than the eae mutant (P < 0.05). As expected, 1551-2 eae::Km lost the ability to produce the AE lesions observed in the wild-type strain (Fig. 1E and F).
Fig 1.
Atypical EPEC 1551-2 interactions with HeLa cells. (A, B, and C) Light microscopy images demonstrating the differences in the adherence patterns between 1551-2 (LA), the derivative eae mutant (DA), and the complemented strain (LA), respectively (original magnification, ×1,000). (D) Quantitative adherence assay demonstrating a reduction of approximately 33.7% in adherence of 1551-2 eae::Km compared with the wild-type strain. *, P < 0.05; #, significantly more adherent than 1551-2 eae::Km (P < 0.05). (E and F) SEM images showing the presence or absence of AE lesions (arrows) after 6 h of contact by 1551-2 and 1551-2 eae::Km, respectively. Bar = 2 μm.
Identification and role of T1P in cell adherence of aEPEC 1551-2.
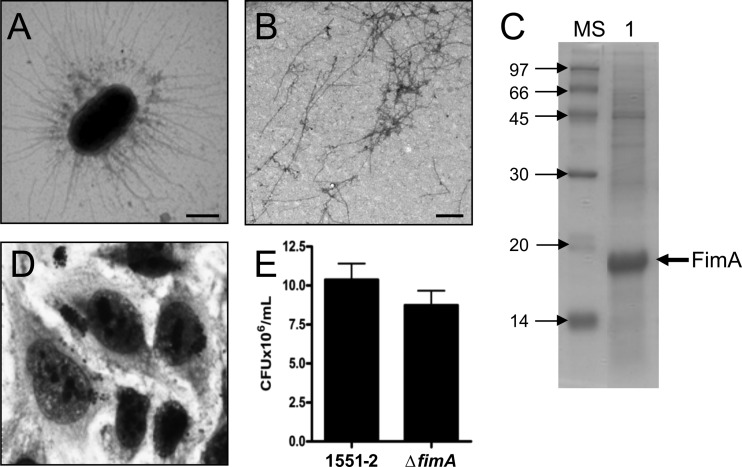
We set out to investigate the production of fimbrial adhesins in 1551-2 propagated at 37°C using different culture media (e.g., MacConkey, Luria-Bertani, Minca, 1% tryptone, colonization factor agar [CFA]) by electron microscopy analyses. Bacteria recovered from all the different media tested showed the presence of rigid rod-like pili (Fig. 2A), and MacConkey agar was selected for pilus purification (Fig. 2B). The purified pili dissociated into monomers of 18 kDa in 16% polyacrylamide gels under denaturing conditions with HCl treatment (Fig. 1C). Mass spectrometry analyses of the 18-kDa protein showed the presence of peptides whose amino acid sequence matched the sequence of the T1P major pilin subunit FimA (data not shown). In order to investigate the presence of functional T1P in 1551-2, the standard yeast agglutination assay was employed. 1551-2 recovered from the supernatant of infected HeLa cells after 6 h of incubation agglutinated yeast cells through recognition of mannosidic residues. This phenotype could be abolished in the presence of 3% d-mannose (data not shown).
Fig 2.
Identification and purification of T1P from 1551-2 and analysis of its role in HeLa cell adherence. (A) Micrograph of piliated 1551-2 obtained from the supernatant of infected cells after 6 h of incubation and visualized by TEM. (B) TEM micrograph showing pili purified from 1551-2. (C) Depolymerization of purified pili in a 16% SDS-PAGE gel showing an 18-kDa protein (lane 1) whose mass spectrometry analysis showed identity to the FimA pilin subunit protein. MS, mass standards. (D) Light microscopy micrograph showing the LA pattern exhibited by 1551-2 ΔfimA after 6 h of incubation with HeLa cells (original magnification, ×1,000). (E) Quantification of adhering bacteria showing only minor differences in the level of adherence between the wild type and the T1P mutant (P > 0.05). Bar = 500 nm.
The contribution of T1P in the adherence process of 1551-2 was investigated by mutating the fimA gene. The resulting fimA mutant lost its capacity to agglutinate yeasts, confirming the absence of T1P. However, the fimA mutant retained the ability to adhere to HeLa cells in an LA pattern (Fig. 2D). Although a reduction of approximately 15% could be observed between the fimA mutant and the wild-type strain (Fig. 2E), this difference could not be considered statistically significant (P > 0.05).
Identification and characterization of SslE, a type 2 secretion substrate, in aEPEC 1551-2.
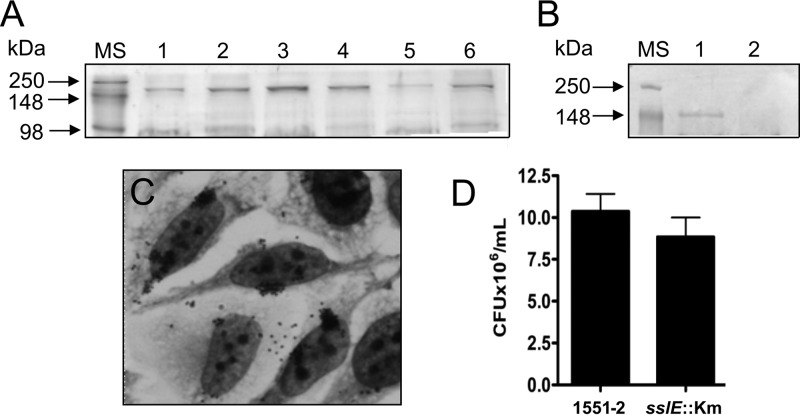
A protein of approximately 150 kDa was identified in the heat extracts and secreted protein fractions analyzed by SDS-PAGE (Fig. 3). Mass spectrometry analyses of this protein showed the presence of peptides whose amino acid sequence matched the sequence of a recently reported type 2 secreted protein termed SslE (secreted and surface-associated lipoprotein from E. coli) (41). We observed that this protein is produced in both LB medium and DMEM, at 37 and 28°C, and in an atmosphere supplemented with 5% CO2 (Fig. 3A). Given that SslE was suggested to be involved in biofilm formation in the tEPEC E2348/69 and rabbit EPEC strains, we investigated the contribution of this protein in the adherence of 1551-2 by deleting the sslE gene. Absence of the SslE corresponding band in the mutant strain can be observed in Fig. 3B. Qualitative and quantitative adherence assays did not indicate any contribution of this protein to the ability of 1551-2 to adhere to HeLa cells or to form LA (Fig. 3C and D).
Fig 3.
Identification and characterization of the T2SS substrate SslE in supernatants of 1551-2. (A) Heat extract of the 1551-2 strain grown in DMEM (lanes 1, 3, and 5) and LB (lanes 2, 4, and 6) at 37°C with shaking (lanes 1 and 2), at 28°C with shaking (lanes 3 and 4), and at 37°C under a 5% CO2 atmosphere without shaking (lanes 5 and 6). (B) Secreted protein extract showing the presence of the T2SS SslE in the wild type (lane 1) and absence in the sslE mutant (1551-2 sslE::Km) (lane 2). (C) Light microscopy image showing LA by the sslE mutant after 6 h of incubation with HeLa cells (original magnification, ×1,000). (D) Quantitative comparison of adherence between the wild type and sslE mutant showing no statistical difference (P > 0.05).
Contribution of the T3SS proteins to aEPEC 1551-2 adherence.
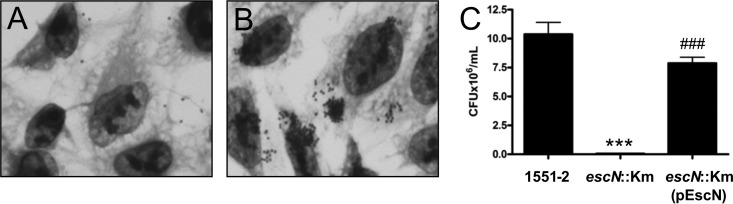
In order to evaluate if the DA phenotype observed in the 1551-2 eae::Km strain is dependent on proteins secreted by the T3SS, we deleted the escN gene in the aEPEC 1551-2 strain. This gene encodes a cytoplasmic ATPase protein responsible for energizing the secretion machinery (31). The adherence of the 1551-2 escN::Km strain was drastically reduced (approximately 99.3%) compared with wild-type 1551-2, with only a few isolated bacteria adhering to HeLa cells (P < 0.005), strongly supporting the involvement of the T3SS translocon in the adherence of 1551-2 (Fig. 4A). Trans-complementation of the 1551-2 escN::Km strain with pEscN restored the LA pattern after 6 h of incubation (Fig. 4B and C).
Fig 4.
Contribution of T3SS to the adherence of aEPEC 1551-2 to HeLa cells. (A and B) Adherence of the T3SS-ATPase-deficient mutant (1551-2 escN::Km) and the complemented strain after 6-h assay (original magnification, ×1,000). (C) Quantitative comparison of adherence between the indicated strains demonstrating that adherence of 1551-2 to HeLa is dependent on a functional T3SS. ***, significantly less adherent than 1551-2 (P < 0.005); ###, significantly more adherent than the 1551-2 escN::Km mutant (P < 0.001).
The DA phenotype is dependent on the T3SS needle complex.
Aiming to confirm the involvement of the T3SS needle complex in promoting DA, we constructed a double intimin/EspA mutant. In contrast to 1551-2 eae::Km, which displays the DA pattern in contact with HeLa cells, the intimin/EspA double mutant strain became nonadherent (Fig. 5A and B). Complementation of this mutant with the pEspA plasmid restored the ability of the mutant to adhere in a DA manner (Fig. 5C). Comparative quantitative assays demonstrated that adherence of the 1551-2 eae::Km ΔespA double mutant was reduced approximately by 99.4% with respect to the intimin mutant (P < 0.0001) (Fig. 5D). Similar results were observed in the adhesion inhibition assay with EspA antibodies (Fig. 5E), confirming the involvement of the T3SS translocon in the DA phenotype observed in the eae mutant strain. Complementation with pEspA plasmid quantitatively restored the adherence of 1551-2 eae::Km ΔespA compared with 1551-2 eae::Km (P < 0.005) (Fig. 5D). However, no statistical difference was observed between the 1551-2 eae::Km strain and the double mutant complemented strain (P > 0.05).
Fig 5.
The DA phenotype is dependent on the T3SS needle complex. (A, B, and C) Light microscopy images showing the adherence patterns observed in 1551-2 eae::Km, the double eae espA mutant, and the complemented strain, respectively (original magnification, ×1,000). (D) Effect of mutations in T3SS-related and fimA genes in the DA phenotype of 1551-2 eae::Km to HeLa cells. While mutations in tir and fimA genes did not affect the adherence ability of the eae mutant, deletion of espA significantly impaired its adherence. ***, significantly less adherent than 1551-2 eae::Km (P < 0.0001); ###, significantly more adherent than 1551-2eae::Km ΔespA (P < 0.005). (E) Quantification of adherence of the 1551-2eae::Km strain in the presence of anti-EspA antibodies. Bacteria were added to HeLa cells in the absence (none) or the presence of three dilutions of the anti-EspA (1:100, 1:50, and 1:10). ***, statistically significant with respect to the values obtained in the absence (none) of the anti-EspA antibody (P < 0.0001).
Additionally, two other double mutants were generated, intimin/Tir (1551-2 eae::Km Δtir) and intimin/T1P (1551-2 eae::Km ΔfimA) mutants, that were approximately 16.3 and 9.1% less adherent, respectively, than the intimin mutant (Fig. 5D). These mutants were still able to display DA on HeLa cells, discarding the involvement of T1P and the intimin-Tir interaction in the DA phenotype (data not shown).
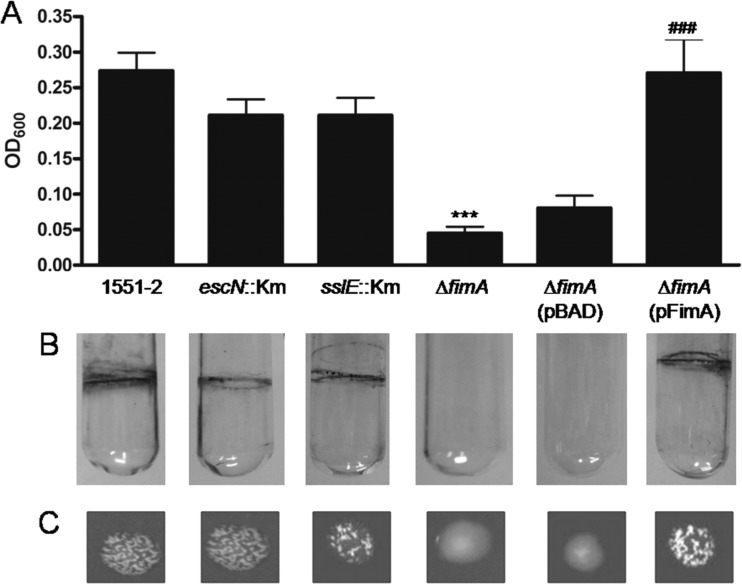
T1P mediated biofilm formation in aEPEC 1551-2.
The ability of 1551-2 and its derivate mutants to develop biofilm on an abiotic surface (polystyrene) was investigated. In comparison with the wild-type strain, the mutation in the fimA gene drastically affected the ability of 1551-2 to form biofilm on plastic (Fig. 6). The quantitative biofilm assay confirmed the difference in adherence efficiency between the wild-type and the fimA mutant strains (P < 0.0001). Our results indicate that T3SS proteins as well as the T2SS substrate SslE did not interfere with the biofilm formation phenotype in strain 1551-2 (Fig. 6A).
Fig 6.
Biofilm formation by aEPEC 1551-2 and derived mutants. (A) Quantitative determination of crystal violet adsorption upon biofilm formation by the indicated strains. Biofilm formation by 1551-2 is dependent on T1P production but not on T3SS or the T2SS substrate. (B) Pellicle formation at the air-liquid interface upon bacterial growth in glass tubes. This phenotype was dependent on T1P production. (C) Images comparing yeast agglutination by the indicated strains. ***, significantly less adherent than 1551-2 (P < 0.0001); ###, significantly more adherent than 1551-2 ΔfimA (P < 0.005).
aEPEC 1551-2 formed a pellicle at the air-liquid interface when grown with shaking in LB for 48 h at 37°C (Fig. 6B). Interestingly, the fimA mutant lost the ability to adhere to the glass tube wall in a ring-like format, while the escN and the sslE mutants preserved their biofilm-producing traits. The association of pellicle formation at the air-liquid interface with T1P production could be confirmed by demonstrating the close association between the presence of T1P, investigated by the ability to agglutinate yeast cells, and the ring-like formation in glass tube (Fig. 6C). Deletion of the fimA gene, confirmed by loss of the ability to agglutinate yeast, completely abolished the ability to form a pellicle at the air-liquid interface and to develop biofilm on a polystyrene surface.
The introduction of pFimA into the fimA mutant resulted in regaining of the capacity to adhere to abiotic surfaces (polystyrene and glass) and express T1P, as confirmed by the yeast agglutination assay (Fig. 6C). The pBADMyc-His A plasmid vector was employed as a negative control, confirming that the plasmid caused no alteration in the phenotype of the fimA mutated strain.
DISCUSSION
The importance of intimin as an adhesin has been extensively documented in tEPEC and aEPEC strains (20, 22, 23, 42). Although it is clear that intimin is an important adhesin of aEPEC, it is also evident that intimin is not the only adhesion factor present in this group of strains (24, 43, 44). We previously reported that aEPEC strain 1551-2 forms LA 6 h postinfection and that an isogenic eae mutant shows DA (24). These data indicated that the switch from LA to DA pattern was intimin dependent but that the DA phenotype was intimin-Tir independent. This prompted us to search in 1551-2, using a PCR-based approach, for adhesin genes previously shown to be involved in DA of other DEC strain adhesin genes, namely, the genes for F1845, AIDA-I, and LDA (locus for diffuse adherence). We also surveyed in 1551-2 many other fimbrial genes present in other pathogenic E. coli strains, for example, the genes for AAF/I-II, LPF, HCP, curli, T1P, ECP, and P and S fimbriae (8, 13, 45, 46). Except for fimE, none of the genes encoding these adhesins were found in 1551-2 (46). Thus, in this study, in order to identify the elusive DA adhesin, we undertook a different approach: using TEM, we sought novel pili upon growth of the bacteria in different growth conditions, we analyzed supernatants for new secreted proteins, and we studied the role of the T3SS translocon.
Much to our surprise, the pili produced by and purified from the 1551-2 strain turned out to be T1P. Of note, bacteria harvested from the 6-h adherence assay strongly agglutinated yeast cells, demonstrating that T1P is produced in the presence of HeLa cells. T1P are ubiquitous among the Enterobacteriaceae and have been vastly studied and implicated in various virulence-associated events of pathogenic E. coli strains, e.g., invasion and persistence in host cells, biofilm formation, induction of proinflammatory molecules, and cell signaling (reviewed in reference 47). T1P enable uropathogenic E. coli (UPEC) strains to attach to mannose residues on the surface of urinary epithelium cells and play a significant role in the colonization and invasion of bladder epithelial cells as the first critical step in the establishment of cystitis and urinary tract infections (48, 49). T1P of prototypic tEPEC strain E2348/69 were not associated with cell adherence (30) but were considered important for development of the AA pattern of enteroaggregative E. coli (EAEC) strain 042 (50). In the present study, 1551-2 ΔfimA showed the same level of adherence to HeLa cells as the wild-type strain, indicating that these pili were not the DA adhesins. Discrepancies in the role of T1P in cell adherence by different E. coli strains could be attributed to nucleotide variations in the tip adhesin fimH gene resulting in differences in their receptor-binding capabilities (51, 52). Studies showing variations in fimH and possible implications of these variations in aEPEC adherence are missing. Likewise, the fact that in our initial PCR 1551-2 was fimA negative could be explained by differences in the flanking regions of the gene that were not totally compatible with the sequence of the primers employed. Thus, it is possible that variants of fimA also exist among aEPEC.
The search for new potentially relevant surface-associated (heat extracts) or secreted proteins in supernatants of 1551-2 eae::Km cultures obtained in LB broth or DMEM led us to identify a large protein of approximately 150 kDa. Mass spectrometry analyses of this protein revealed that it was a product of the yghJ chromosomal gene, which encodes a predicted lipoprotein. YghJ production was demonstrated in bacteria grown at 28° and 37°C in an atmosphere supplemented or not with 5% CO2. The fact that YghJ is produced under conditions similar to those that induce production of the BFP and T3S effectors in tEPEC (e.g., 37°C in DMEM) might indicate an important biological significance.
While this work was in progress, a study by Baldi et al. (41) became available demonstrating that tEPEC E2348/69 secretes an outer membrane lipoprotein via T2SS, which they called SslE (for secreted and surface-associated lipoprotein from E. coli) and found to be important for biofilm maturation but not for cell adherence (41). SslE is an ortholog protein of the accessory colonization factor D (AcfD) of Vibrio cholerae, which is required for efficient colonization of infant mouse intestine (53). It was apparent that the YghJ protein of aEPEC 1551-2 and the SslE of E2348/69 were the same protein.
In order to establish the function of this outer membrane lipoprotein in the adherence processes of strain 1551-2, we constructed an sslE mutant. The mutation in the sslE gene led to the absence of SslE in supernatants but did not affect the secretion of EspA, EspB, or EspD, which are dependent on the T3SS (data not shown). The 1551-2 sslE::Km mutant did not lose the ability to produce LA on HeLa cells, did not show a significant reduction in the number of bacteria associated with the epithelial cells (P > 0.05), and was not associated with biofilm formation. Thus, the role of SslE in the pathogenicity of 1551-2 or other aEPEC strains in general remains unclear.
After ruling out the involvement of T1P and SslE as DA adhesin, we investigated if the DA phenotype would be dependent on a functional T3SS. We created an insertional mutation in escN; this gene encodes the ATPase required for secretion of the T3SS assembly components (31). 1551-2 escN::Km was no longer adherent in HeLa cells, providing compelling evidence of the role of the T3SS in DA of 1551-2 eae::Km. We also tested the escN mutant in differentiated Caco-2 cells. As observed in HeLa cells, the ability of 1551-2 escN::Km to adhere to this enterocyte cell line model was significantly reduced (approximately 91.4%) in comparison with that of the wild-type 1551-2 strain (P < 0.0001) (data not shown). The role of the T3SS in the virulence properties of EPEC and EHEC has been well established (54). In particular, the EspA filament, or translocon, was shown to link the bacteria to the host cell membrane and to be responsible for injection of proteins into the cytosol. Previous studies carried out with tEPEC E2348/69 and an EHEC strain 413/89-1 (serotype O26:H−) pointed to a possible involvement of the T3SS filaments in adherence to epithelial cells (42, 55, 56). The T3SS apparatus was also demonstrated to be responsible for attachment to leaves in EHEC O157 and O26 strains (57, 58). To specifically define the role of the EspA translocon in DA, we then constructed a double intimin/EspA mutant. As predicted, this mutant totally lost its adherence capabilities. Thus, this event is intimin-Tir independent, as the single eae and the double eae tir mutants displayed DA. Further, the DA pattern was inhibited with anti-EspA antibodies. All together, these data strongly indicated that the T3SS translocon is responsible for the DA displayed by 1551-2 eae::Km. Nonetheless, future studies are needed to elucidate whether the EspA filament per se or the tip-associated pore-forming proteins, EspB and EspD, are responsible for mediating host cell adherence.
Biofilm formation is a property not fully explored among aEPEC isolates. The ability of an aEPEC strain of serotype O55:H7 to produce biofilm at low temperatures was associated with curli production (33). While curli biogenesis genes are commonly found among aEPEC isolates, production of curli is a rare event (46). The EspA filament of tEPEC E2348/69 was suggested to be responsible for biofilm development on plastic under static conditions (59). In this study, we showed that the T3SS translocon is not associated with biofilm in 1551-2, since the escN mutant strain is still able to produce the same level of biofilm as the wild-type 1551-2 strain. In conclusion, our data support a role for intimin in LA, for the T3SS translocon in DA, and for T1P in biofilm formation, suggesting that all of these adhesins act in concert to facilitate host intestinal colonization by aEPEC strains.
ACKNOWLEDGMENTS
This work was supported by grants from FAPESP (2011/12664-5) and CNPq (304453/2011-0) to T.A.T.G. This work was also supported in part by NIAID, NIH award AI66012 to J.A.G.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
We are grateful to José L. Puente for providing the pEscN plasmid and to Inga Benz and M. Alexander Schmidt for providing AIDA-I antibodies. We also thank Jean-Marc Ghigo (Unité de Génétique des Biofilms, Departement de Microbiologie, Institut Pasteur) for providing the pKOBEG-Apra plasmid.
Footnotes
Published ahead of print 29 July 2013
REFERENCES
- 1.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trabulsi LR, Keller R, Gomes TAT. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaper JB. 1996. Defining EPEC. Rev. Microbiol. 27:130–133 [Google Scholar]
- 5.Gomes TAT, Rassi V, MacDonald KL, Ramos SR, Trabulsi LR, Vieira MAM, Guth BE, Candeias JA, Ivey C, Toledo MR, Blake PA. 1991. Enteropathogens associated with acute diarrheal disease in urban infants in São Paulo, Brazil. J. Infect. Dis. 164:331–337 [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues J, Thomazini CM, Morelli A, de Batista GC. 2004. Reduced etiological role for enteropathogenic Escherichia coli in cases of diarrhea in Brazilian infants. J. Clin. Microbiol. 42:398–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liebchen A, Benz I, Mellmann A, Karch H, Gomes TAT, Yamamoto D, Hernandes RT, Sampaio J, Sampaio SC, Fruth A, Schmidt MA. 2011. Characterization of Escherichia coli strains isolated from patients with diarrhea in São Paulo, Brazil: identification of intermediate virulence factor profiles by multiplex PCR. J. Clin. Microbiol. 49:2274–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes TAT, Irino K, Girão DM, Girão VB, Guth BE, Vaz TM, Moreira FC, Chinarelli SH, Vieira MAM. 2004. Emerging enteropathogenic Escherichia coli strains? Emerg. Infect. Dis. 10:1851–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandes RT, Elias WP, Vieira MA, Gomes TA. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297:137–149 [DOI] [PubMed] [Google Scholar]
- 10.Torres AG, Zhou X, Kaper JB. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scaletsky IC, Silva ML, Trabulsi LR. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues J, Scaletsky IC, Campos LC, Gomes TA, Whittam TS, Trabulsi LR. 1996. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 64:2680–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieira MAM, Andrade JR, Trabulsi LR, Rosa AC, Dias AM, Ramos SR, Frankel G, Gomes TAT. 2001. Phenotypic and genotypic characteristics of Escherichia coli strains of nonenteropathogenic E. coli (EPEC) serogroups that carry eae and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J. Infect. Dis. 183:762–772 [DOI] [PubMed] [Google Scholar]
- 14.Abe CM, Trabulsi LR, Blanco J, Blanco M, Dahbi G, Blanco JE, Mora A, Franzolin MR, Taddei CR, Martinez MB, Piazza RM, Elias WP. 2009. Virulence features of atypical enteropathogenic Escherichia coli identified by the eae+ EAF-negative stx− genetic profile. Diagn. Microbiol. Infect. Dis. 64:357–365 [DOI] [PubMed] [Google Scholar]
- 15.Mora A, Blanco M, Yamamoto D, Dahbi G, Blanco JE, López C, Alonso MP, Vieira MAM, Hernandes RT, Abe CM, Piazza RM, Lacher DW, Elias WP, Gomes TAT, Blanco J. 2009. HeLa-cell adherence patterns and actin aggregation of enteropathogenic Escherichia coli (EPEC) and Shiga-toxin-producing E. coli (STEC) strains carrying different eae and tir alleles. Int. Microbiol. 12:243–251 [PubMed] [Google Scholar]
- 16.Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911–921 [DOI] [PubMed] [Google Scholar]
- 17.Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438 [DOI] [PubMed] [Google Scholar]
- 18.Daniell SJ, Delahay RM, Shaw RK, Hartland EL, Pallen MJ, Booy F, Ebel F, Knutton S, Frankel G. 2001. Coiled-coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect. Immun. 69:4055–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiya K, Ohishi M, Ogino T, Tamano K, Sasakawa C, Abe A. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U. S. A. 98:11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 22.Pelayo JS, Scaletsky IC, Pedroso MZ, Sperandio V, Girón JA, Frankel G, Trabulsi LR. 1999. Virulence properties of atypical EPEC strains. J. Med. Microbiol. 48:41–49 [DOI] [PubMed] [Google Scholar]
- 23.Carvalho HM, Teel LD, Kokai-Kun JF, O'Brien AD. 2005. Antibody against the carboxyl terminus of intimin alpha reduces enteropathogenic Escherichia coli adherence to tissue culture cells and subsequent induction of actin polymerization. Infect. Immun. 73:2541–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandes RT, Silva RM, Carneiro SM, Salvador FA, Fernandes MC, Padovan AC, Yamamoto D, Mortara RA, Elias WP, Briones MRS, Gomes TAT. 2008. The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion. Cell. Microbiol. 10:415–425 [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Cravioto A, Gross RJ, Scotland S, Rowe B. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional enteropathogenic serotypes. Curr. Microbiol. 3:95–99 [Google Scholar]
- 27.Girón JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361–379 [DOI] [PubMed] [Google Scholar]
- 28.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. 10.1093/nar/28.22.e97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penfold RJ, Pemberton JM. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146 [DOI] [PubMed] [Google Scholar]
- 30.Elliott SJ, Kaper JB. 1997. Role of type 1 fimbriae in EPEC infections. Microb. Pathog. 23:113–118 [DOI] [PubMed] [Google Scholar]
- 31.Gauthier A, Puente JL, Finlay BB. 2003. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect. Immun. 71:3310–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Oliveira-Garcia D, Dall'Agnol M, Rosales M, Azzuz AC, Alcántara N, Martinez MB, Girón JA. 2003. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell. Microbiol. 5:625–636 [DOI] [PubMed] [Google Scholar]
- 33.Weiss-Muszkat M, Shakh D, Zhou Y, Pinto R, Belausov E, Chapman MR, Sela S. 2010. Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl. Environ. Microbiol. 76:1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock V, Witsø IL, Klemm P. 2011. Biofilm formation as a function of adhesin, growth medium, substratum and strain type. Int. J. Med. Microbiol. 301:570–576 [DOI] [PubMed] [Google Scholar]
- 35.de Oliveira-Garcia D, Dall'Agnol M, Rosales M, Azzuz AC, Martinez MB, Girón JA. 2002. Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8:918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girón JA, Ho AS, Schoolnik GK. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710–713 [DOI] [PubMed] [Google Scholar]
- 37.McMichael JC, Ou JT. 1979. Structure of common pili from Escherichia coli. J. Bacteriol. 138:969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 39.Girón JA, Lange M, Baseman JB. 1996. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 64:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC. 2007. Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14:131–137 [DOI] [PubMed] [Google Scholar]
- 41.Baldi DL, Higginson EE, Hocking DM, Praszkier J, Cavaliere R, James CE, Bennett-Wood V, Azzopardi KI, Turnbull L, Lithgow T, Robins-Browne RM, Whitchurch CB, Tauschek M. 2012. The type II secretion system and its ubiquitous lipoprotein substrate, SslE, are required for biofilm formation and virulence of enteropathogenic Escherichia coli. Infect. Immun. 80:2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleary J, Lai LC, Shaw RK, Straatman-Iwanowska A, Donnenberg MS, Frankel G, Knutton S. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150:527–538 [DOI] [PubMed] [Google Scholar]
- 43.Scaletsky IC, Michalski J, Torres AG, Dulguer MV, Kaper JB. 2005. Identification and characterization of the locus for diffuse adherence, which encodes a novel afimbrial adhesin found in atypical enteropathogenic Escherichia coli. Infect. Immun. 73:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampaio SC, Gomes TAT, Pichon C, du Merle L, Guadagnini S, Abe CM, Sampaio JL, Le Bouguénec C. 2009. The flagella of an atypical enteropathogenic Escherichia coli strain are required for efficient interaction with and stimulation of interleukin-8 production by enterocytes in vitro. Infect. Immun. 77:4406–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandes RT, Vieira MAM, Carneiro SM, Salvador FA, Gomes TAT. 2006. Characterization of atypical enteropathogenic Escherichia coli strains that express typical localized adherence in HeLa cells in the absence of the bundle-forming pilus. J. Clin. Microbiol. 44:4214–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandes RT, Velsko I, Sampaio SC, Elias WP, Robins-Browne RM, Gomes TAT, Girón JA. 2011. Fimbrial adhesins produced by atypical enteropathogenic Escherichia coli strains. Appl. Environ. Microbiol. 77:8391–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 48.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497 [DOI] [PubMed] [Google Scholar]
- 49.Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP. 2001. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J. Cell Sci. 114:4095–4103 [DOI] [PubMed] [Google Scholar]
- 50.Moreira CG, Carneiro SM, Nataro JP, Trabulsi LR, Elias WP. 2003. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 226:79–85 [DOI] [PubMed] [Google Scholar]
- 51.Sokurenko EV, Courtney HS, Maslow J, Siitonen A, Hasty DL. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 177:3680–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boddicker JD, Ledeboer NA, Jagnow J, Jones BD, Clegg S. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45:1255–1265 [DOI] [PubMed] [Google Scholar]
- 53.Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 55.Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ebel F, Podzadel T, Rohde M, Kresse AU, Krämer S, Deibel C, Guzmán CA, Chakraborty T. 1998. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147–161 [DOI] [PubMed] [Google Scholar]
- 57.Shaw RK, Berger CN, Feys B, Knutton S, Pallen MJ, Frankel G. 2008. Enterohemorrhagic Escherichia coli exploits EspA filaments for attachment to salad leaves. Appl. Environ. Microbiol. 74:2908–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saldaña Z, Sánchez E, Xicohtencatl-Cortes J, Puente JL, Girón JA. 2011. Surface structures involved in plant stomata and leaf colonization by shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:119. 10.3389/fmicb.2011.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreira CG, Palmer K, Whiteley M, Sircili MP, Trabulsi LR, Castro AF, Sperandio V. 2006. Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J. Bacteriol. 188:3952–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]