Abstract
We have investigated the response of primary human meningothelial cells to Neisseria meningitidis. Through a transcriptome analysis, we provide a comprehensive examination of the response of meningothelial cells to bacterial infection. A wide range of chemokines are elicited which act to attract and activate the main players of innate and adaptive immunity. We showed that meningothelial cells expressed a high level of Toll-like receptor 4 (TLR4), and, using a gene silencing strategy, we demonstrated the contribution of this pathogen recognition receptor in meningothelial cell activation. Secretion of interleukin-6 (IL-6), CXCL10, and CCL5 was almost exclusively TLR4 dependent and relied on MyD88 and TRIF adaptor cooperation. In contrast, IL-8 induction was independent of the presence of TLR4, MyD88, and TRIF. Transcription factors NF-κB p65, p38 mitogen-activated protein kinase (MAPK), Jun N-terminal protein kinase (JNK1), IRF3, and IRF7 were activated after contact with bacteria. Interestingly, the protein kinase IRAK4 was found to play a minor role in the meningothelial cell response to Neisseria infection. Our work highlights the role of meningothelial cells in the development of an immune response and inflammation in the central nervous system (CNS) in response to meningococcal infection. It also sheds light on the complexity of intracellular signaling after TLR triggering.
INTRODUCTION
Neisseria meningitidis (the meningococcus) is an obligate human commensal that normally inhabits the mucus membranes of the nasopharynx of around 10% of most human populations (1). Occasionally, however, it invades the bloodstream, where it can cause septicemia; it is also one of the few bacterial species that are capable of disrupting and penetrating the blood-brain barrier (BBB) and initiating meningitis. Interaction of the meningococcus with cells of the meninges results in an intense inflammatory response, which is the primary cause of pathology. The meninges are three protective membranes surrounding the brain and the spinal cord. They comprise the thick outer dura mater and the leptomeninges, composed of the arachnoid and the pia mater in close contact with the brain. The subarachnoid space is located between the arachnoid and the pia mater and is filled with a nutritive and protective liquid, the cerebrospinal fluid (CSF). The arachnoid and the pia mater are lined with specialized epithelial cells called meningothelial cells. They are able to establish cell-cell junctions building a firm layer restricting the passage of cells or substances between the CSF and brain tissues or CSF and blood (2–4). Thus, meningothelial cells, along with endothelial cells of the blood-brain barrier, participate in isolating the central nervous system (CNS) from the rest of the body.
The cellular and molecular processes leading to meningococcal meningitis are gradually being elucidated. In susceptible persons, bacteria cross the epithelial layer of the nasopharynx, invade the bloodstream, and reach the cerebral vascular endothelium. Meningococci bind to the laminin receptor expressed by the endothelium (5) and can cross the blood-brain barrier via a paracellular route, after disruption of cell junction components (6, 7).
Penetration of the CSF by meningococci results in the recruitment of immune cells and the development of a massive inflammatory response (8). Meningothelial cells are among the major cell types exposed to meningococci during meningitis, where they may constitute an important source of proinflammatory cytokines, chemokines, or antibacterial peptides (9) and may thus play an important role in the recruitment of leukocytes in the CSF (10–13). The modality of pathogen recognition by these cells is poorly understood, however. Although they express Toll-like receptor 2 (TLR2) and -4, the role of these pathogen recognition receptors in sensing of bacterial pathogens has been challenged recently by Humphries and colleagues, who described a TLR2- and 4-independent activation by Neisseria outer membrane vesicles (11). While the massive inflammation and leukocyte infiltration observed in the CSF after bacterial penetration may be protective, they can also result in damage to neuronal tissues and lead to neurological sequelae (14). Thus, it is important to characterize the meningothelial cell response to N. meningitidis to better understand their contribution in the immune response observed in the CSF.
Here we investigated the modulation of gene expression after exposure of primary meningothelial cells to serogroup B N. meningitidis isolate MC58. Our report provides a comprehensive description of the meningothelial cell response to meningococcal infection. Using gene silencing and specific inhibitors, we characterized the signaling processes leading to meningothelial cell activation. In particular, we demonstrate the major contribution of TLR4 in the meningothelial cell response to a meningococcal challenge. Our work supports the idea of meningothelial cells as major players in the immune response in the CNS. It also provides new insights with respect to proposed alternative therapeutic strategies to sustain bacterial elimination and to reduce or prevent the deleterious consequences of the immune response.
MATERIALS AND METHODS
Meningothelial cell isolation and culture.
Meningothelial cells were isolated from surgically removed tumors and cultured as described previously (13). Cells were propagated in 75-cm2 culture flasks in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS; Invitrogen), 1% l-glutamine, and 1% antibiotic-antimycotic solution (both from Sigma). Flasks were incubated at 37°C in an atmosphere of 5% CO2. Cells were split on average every 10 to 14 days using nonenzymatic cell dissociation solution (Sigma); the medium was changed every 5 to 7 days.
Production of human monocyte-derived macrophages (hMDM).
Heparinized blood from healthy donors was obtained after prior consent and ethical committee approval. Peripheral blood mononuclear cells were separated on a Histopaque density gradient (Sigma). After monocyte purification by plastic adherence, differentiation into macrophages was carried out for 6 days in RPMI 1640 medium (Sigma) supplemented with 1% l-glutamine and 1% antibiotic-antimycotic solution (both from Sigma) and 10% HAB serum (PAA) in the presence of 50 ng/ml of macrophage colony-stimulating factor (M-CSF) (R&D Systems).
Infection of meningothelial cells.
Meningothelial cells (passages 4 to 7) were cultured to confluence in 24-well plates. Before stimulation, cells were washed three times in antibiotic- and serum-free DMEM and resuspended in 300 μl of antibiotic-free DMEM supplemented with 2% heat-inactivated FCS. Cells were stimulated with approximately 1 × 105 CFU of meningococci (serogroup B strain MC58) per well (bacterial numbers were estimated based on optical density and were confirmed retrospectively by viable counts on chocolate horse blood agar plate), 500 ng/ml lipopolysaccharide (LPS) (from Escherichia coli 0111:B4; Sigma), 100 ng/ml flagellin (Adipogen; Caltag Medsystems), 100 ng/ml pamCSK4 (Imgenex), or 10 μg/ml R848 (Enzo Life Sciences). In some experiments, cells were treated with 10 μM cytochalasin D (Sigma), 1 μM latrunculin A (Cayman Chemical), 80 μM Dynasore (Sigma), 10 μM 1-(2-(4-morpholinyl)ethyl)-2-(3-nitrobenzoylamino)benzimidazole (Sigma), 20 μM Bay 11-7082 (Sigma), 10 μM SB203580, or 10 μM SP600125 (both from Calbiochem) for 1 h before cell stimulation. Supernatants were harvested after 8 h, frozen, and stored at −80°C before analysis for cytokine concentration determinations.
Microarray sample preparation, hybridization, and data analysis.
Total RNA from three independent experiments was extracted from uninfected or infected cells after 8 h of incubation using an RNeasy Minikit (Qiagen) according to the manufacturer's instructions. Dye-labeled amplified RNA (aRNA) for microarray analysis was generated according to the Amino Allyl MessageAmp II aRNA amplification manual (Ambion), using 0.5 μg of total input RNA per sample. Fluorescent tags used were NHS-ester derivatives of Cy5 and Cy3 (Cy-Dye; GE Lifesciences). Hybridization and wash solutions were as follows: 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS (buffer 1); 2× SSC, 0.1% SDS (buffer 2); 1× SSC (buffer 3); 0.5× SSC (buffer 4); and 0.1× SSC with 0.001% Triton X-100 (buffer 5). Labeled aRNA, in a maximum volume of 4 μl containing 40 pmol of dye equivalents, was mixed with 1 μg (1 μl) human Cot-I DNA (Sigma) and 100 μl 1× Schott hybridization buffer (Schott) and hybridized to microarrays containing 30,000 human gene-specific probes (Human 30K OciChip [A, B, and C]; Ocimum Biosolutions) and printed onto Schott Nexterion A+ APS-coated glass slides by the Post-genomics Technology Facility (University of Nottingham, Nottingham, United Kingdom). Hybridizations were performed on a HS 4800 hybridization station (Tecan) for 16 h at 50°C with moderate agitation. Washing cycles were as follows: buffer 1 at 50°C for 30s, buffer 2 at 30°C for 1 min, buffer 3 at 30°C for 1 min, buffer 4 at 30°C for 1 min, and buffer 5 at 23°C for 1 min. Slides were dried on the HS 4800 hybridization station, using dry nitrogen gas for 2.5 min. Hybridized slides were scanned using an Axon Genepix AL 4200 scanner (Molecular Devices). Resultant data files were processed using BASE software (BioArray Software Environment) (15) and further analyzed using a TIGR Multiple Experiment Viewer (TMEV) and J-Express Pro (Molmine AS) (16).
Dual-color microarray data were filtered and ratio normalized using the global Loess method (17), and the entire data set was scale normalized using J-Express Pro. Only probes with ≤1 missing value were kept for analysis. Log2 (ratio) ≥ 1 or ≤ −1 was used for filtering gene expression profiles. The significance of microarray analysis (SAM) (18) with a highly stringent 0% false-discovery rate (FDR) was used to identify differentially expressed genes. Differentially expressed genes were then investigated for network and gene functional interrelations by the use of Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems, Qiagen).
Determination of cytokine concentrations.
Supernatants were defrosted on ice and cytokine concentrations measured using a FlowCytomix multiple-analyte detection system (eBioscience) according to the manufacturer's instructions.
Gene silencing.
ON-TARGETplus small interfering RNA (siRNA) targeting TLR2, TLR4, MyD88, TRIF, and IRAK4 and ON-TARGETplus nontargeting pool control siRNA (CT) were from Dharmacon (Thermo Scientific). Confluent cells in 24-well plates were transfected with 50 nM siRNA using DharmaFECT 1 transfection reagent (Thermo Scientific). After 24 or 48 h, efficiency of inhibition was determined by quantitative PCR (QPCR) or by Western blotting.
QPCR.
Meningothelial cells were washed twice with serum-free DMEM. Total RNA was extracted using an RNeasy minikit (Qiagen, United Kingdom), according to the manufacturer's instructions. Contaminating DNA was removed using RNase-free Turbo-DNase I (Ambion, Applied Biosystem, Warrington, United Kingdom), and RNA was cleaned and concentrated using an RNeasy MinElute Cleanup kit (Qiagen). cDNA was synthesized using a QuantiTect reverse transcription kit (Qiagen) and quantified using an AB7500 real-time PCR system and Power SYBR green master mix (both from Applied Biosystems) according to the manufacturer's instructions. The primers used are listed in Table S1 in the supplemental material. Reactions started with a 2-min incubation at 50°C and then 10 min at 95°C followed by 40 cycles of denaturation at 95°C for 15 s followed by annealing/extension at 60°C for 1 min. Concentration values for targeted genes were normalized with RNA for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Western blot analysis.
Meningothelial cells were washed twice with serum-free DMEM, and proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer. Equal amounts of protein were loaded on 10% SDS polyacrylamide gels under denaturing conditions, and proteins were transferred to polyvinylidene difluoride membranes (GE Healthcare) using a TransblotSD semidry transfer cell (Bio-Rad) for 30 min at 15 V. Membranes were blocked for 1 h using 5% bovine serum albumin (BSA)–phosphate-buffered saline (PBS) at 4°C and then incubated overnight with anti-human actin, phospho-NF-κB p65 (S536), phospho-p38 mitogen-activated protein kinase (MAPK) (T180/Y182), phospho-Jun N-terminal protein kinase (JNK1) (T183/Y185), phospho-IRF3 (S396), or phospho-IRF7 (S471/472) antibodies (Cell Signaling) at a 1:1,000 dilution (except anti-phospho-JNK1, which was used at a 1:250 dilution) in PBS–5% BSA–0.05% Tween 20. After three washes in PBS–0.05% Tween 20, membranes were probed with goat anti-mouse IgG-horseradish peroxidase-conjugated antibody (Sigma) at a 1:10,000 dilution in PBS–5% BSA–0.05% Tween 20 for 1 h. Following additional washes, membranes were developed using a chemiluminescent substrate (ECL; GE Healthcare) according to the manufacturer's instructions.
Statistical analysis.
Values representing means ± standard errors of the means (SEM) are shown. Analysis of variance (ANOVA) or Student's t test was applied (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
RESULTS
Modulation of gene expression by meningothelial cells after exposure to N. meningitidis.
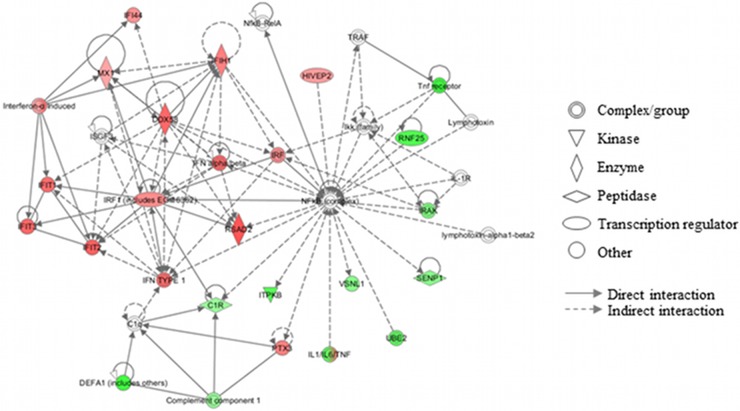
The ability of meningothelial cells to respond to the presence of N. meningitidis has been investigated in previous studies (10, 13). To date, however, a comprehensive analysis of the changes in transcription in meningothelial cells in response to interaction with Neisseria has been lacking. Using a microarray approach, we present an overview of meningothelial cell transcriptome modulation in response to infection with N. meningitidis. Meningothelial cells were stimulated with N. meningitidis MC58, and gene expression was analyzed using a 30,000-human-gene microarray. After filtering replicated microarray results to remove low-intensity signals, 9,716 gene signals remained and genes that were significantly differentially expressed were identified using the SAM algorithm. Using a 0% false-discovery rate (FDR) to minimize type 1 errors, differentially regulated genes were further filtered to 306 genes, of which 43 (14%) were significantly upregulated and 263 (86%) were significantly downregulated (Fig. 1; see also Table S2 in the supplemental material). To confirm that the cultured cells represented primary meningothelial cells derived from meningeomas and, specifically, to rule out possible contamination from fibroblasts, we first examined the data set for expression of markers characteristic of epithelial cells and of fibroblasts. It is likely that meningothelial cells represent a specialized type of epithelia. Nevertheless, we could detect high levels of expression of the epithelial cell markers desmoplakin (NM_004415); cytokeratin 17 (NM_005556); cytokeratin 14 (NM_000526); BTG3 (abundant in neuroepithelium area protein); epithelial membrane protein-1 (NM_001423), and epithlelial membrane protein-3 (NP_001416). In contrast, the fibroblast-specific genes encoding Fibroblast-specific protein 1 (NP_002952) and CD34 (a marker for undifferentiated fibroblasts; AAB24223) were not detected in our data set (data not shown). Based on these data, we are confident that the cells under study were indeed meningothelial cells derived from the primary tumors and not contaminating fibroblasts. We then used the IPA system to identify interactions between the differentially regulated genes. The top 6 networks are presented in Table 1. Three of them are associated with cellular movement or with cell-cell signaling and interaction. Network 4, associated with antimicrobial response, inflammatory response, and infectious disease, is shown in Fig. 2. It highlights the induction of type I IFN and IFN-related genes such as IFIT1, -2, and -3 or IFIH1 during meningothelial cell response.
Fig 1.
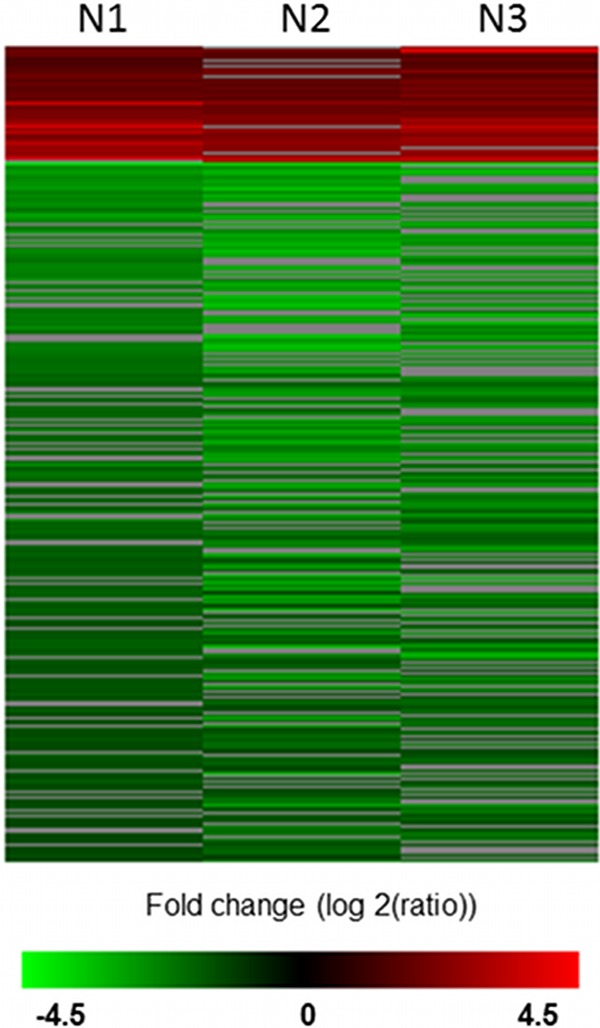
Heat map of genes differentially expressed by meningothelial cells after contact with meningococci. Data from 3 independent experiments (N1, N2, and N3) were filtered and ratio normalized using the global Loess method and scale normalized using J-Express Pro. Genes were then selected using the SAM method with a 0% FDR. Clustering showed 43 upregulated (red) and 263 downregulated (green) genes.
Table 1.
Top 6 networks identified by IPAa
| Molecules in network | Score | No. of focus molecules | Major functions |
|---|---|---|---|
| 26S proteasome, ABCC1, alpha catenin, alpha tubulin, ATPase, beta tubulin, CCT3, DERL1, ELF2, GATA2, GBAS, Gi-coupled receptor, Hdac, HHEX, HLA-DR, Hsp27, Hsp70, Hsp90, IL-8, KLK3, MAPK, MHC class I (family), NCKAP1, NDRG4, PARK2, Rsk, scavenger receptor class A, SIRPA, SSBP3, THBS2, TNF, TNFAIP3, UBE2D3, ubiquitin, VCP | 28 | 18 | Cellular movement, cardiovascular system development and function, organismal development |
| Adaptor protein 1, AMPK, ANP32A, ANXA2, APAF1, BIRC3, caspase, Cdk, cyclin A, cyclin D, cyclin E, cytochrome c, DNMT1, FASLG, FOXE1, FZR1, HBXIP, JNK, MCMBP, MIF, MTORC1, MUTYH, NADPH oxidase, OGG1, PARP14, PARP, PGD, PMAIP1, PRKAA, Rb, RMI1 (includes EG:306734), RPA, TH2 cytokine, TSH, XPC | 27 | 18 | Cancer, respiratory disease, organismal injury and abnormalities |
| 2′ 5′ oas, AIRE, CCL5, CLTCL1, CYP, EFCAB6, EIF2AK1, ferritin, HDL, hemoglobin, HSD17B2, IFN beta, IFNB1 (includes EG:15977), IL-6, IL-12 (family), immunoglobulin, ING4, alpha interferon, IRF3 dimer, JAK, LDL, MHC class I (complex), N-cor, Nr1h, Rxr, SEMA4A, SLC40A1, STAB1, STAT5a/b, STRA6, STX2, TBL1XR1, TPST1, USP18, vitamin D3-VDR-RXR | 26 | 17 | Cell-to-cell signaling and interaction, cellular movement, hematological system development and function |
| C1q, C1R, complement component 1, DDX58, DEFA1 (includes others), HIVEP2, IFI44, IFIH1, IFIT1, IFIT2, IFIT3, IFN alpha/beta, IFN type 1, IKK (family), IL-1R, IL-1/IL-6/TNF, IFN-α induced, IRAK, IRF, IRF1 (includes EG:16362), ISGF3, ITPKB, lymphotoxin, lymphotoxin-alpha1-beta2, MX1, NFκB (complex), NFκB-RelA, PTX3, RNF25, RSAD2, SENP1, TNF receptor, TRAF, UBE2, VSNL1 | 25 | 17 | Antimicrobial response, inflammatory response, infectious disease |
| ASRGL1, CCDC86, DAZ1/DAZ4, DAZAP1, DAZL, DDX50, DLG5, DLST, FADS3, FAM129A, FN3KRP, MPP2, MTR, MTRR, NDNL2, NSMCE1, NSMCE2, NSMCE4A, POLR1E, POP4 (includes EG:10775), RPP21, RPP30, RPP38, RPP40, RRN3, SLC27A3, SLC27A6, SMC6, SPARCL1, SRSF11, THAP11, TTI2 (includes EG:100334230), TUFT1, UBC, WDR3 | 24 | 16 | Developmental disorder, hereditary disorder, metabolic disease |
| Akt, c-Src, CCL20, CXCL1, CXCL10, CXCL11, CXCL13, ENSA, ESM1, EXOC4, Fgfr, HN1, HPGD, IκB kinase, IFN, gamma IFN, Ikb, IL-23, IL-17f dimer, IL17a dimer, IL17R, Integrin, IRAK4, JINK1/2, Lfa-1, MTORC2, NCAM1, NFκB (family), NfkB1-RelA, PI3K (family), Ptk, RAP2A, RIPK2, TLR9, Tlr | 22 | 15 | Cell-to-cell signaling and interaction, cellular movement, immune cell trafficking |
MHC, major histocompatibility complex; IKK, IκB kinase. Scores for each network were computed by ingenuity pathways analysis according to the fit of that network to the user-defined set of focus genes. Scores are derived from P values and indicate the likelihood of the focus genes in a network being found together due to random chance. A score of 2 indicates that there is a 1 in 100 chance that the focus genes are together in a network due to random chance. Therefore, scores of 2 or higher have at least a 99% confidence of not being generated by random chance alone.
Fig 2.
Gene network involved in “antimicrobial response, inflammatory response, and infectious disease” as identified by IPA. Upregulated and downregulated genes have been assigned red and green symbols, respectively.
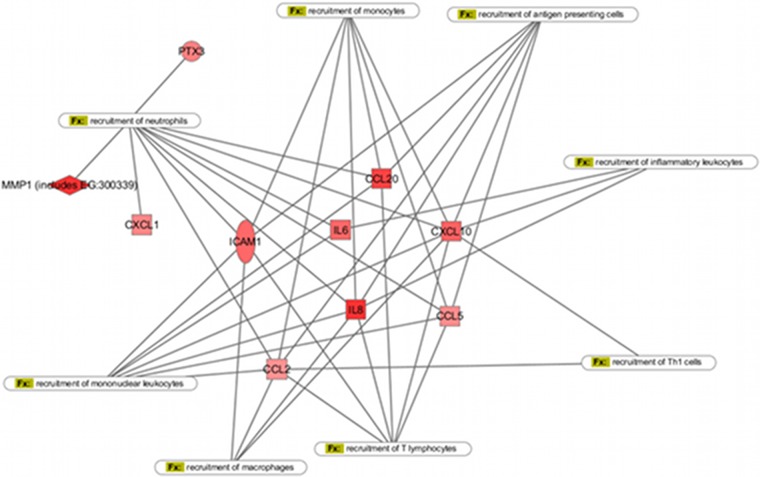
Interestingly, although a majority of genes were found to be downregulated, initiators of local and systemic inflammatory responses were upregulated. For example, transcription of a wide range of chemokines, including interleukin-8 (IL-8) (CXCL8), CXCL11, CXCL10, CXCL1, CCL20, CCL5, and CCL2, was upregulated after contact with Neisseria (see Table S2 in the supplemental material). Other proinflammatory agents whose expression is increased by interaction with meningococci include, for example, the pleiotropic cytokine IL-6 (log2[ratio], 2.53) or the soluble pattern recognition receptor PTX3 (log2[ratio], 2.15). This spectrum of molecules targets players in innate immunity, including neutrophils, monocytes, dendritic cells, and macrophages, as well as the adaptive immune system via T lymphocytes (19) (Fig. 3). Expression of colony stimulating factor 2 (CSF2) (granulocyte-macrophage colony-stimulating factor [GM-CSF]; log2[ratio], 2.63), a stimulator of granulocyte, macrophage, and eosinophil generation and differentiation, was increased. In contrast, expression of the macrophage migration inhibitory factor (MIF) was dramatically reduced in infected cells (log2[ratio], −2.97). Although expression of IL-1β mRNA was increased (log2[ratio], 1.88), the mature form of this cytokine, which requires cleavage by caspase 1 after assembly and activation of the inflammasomme multiprotein complex (20), was not detected in the cell supernatant (data not shown).
Fig 3.
Gene network associated with “immune cell recruitment” as identified by IPA. Targeted immune cell populations are indicated within the boxes.
Matrix metalloprotease 1 (MMP1) expression was upregulated (log2 [ratio], 3.80) (see Table S2 in the supplemental material). In contrast, expression of collagens VIII and XI was repressed after N. meningitidis treatment (log2 [ratios], −2.25 and −1.56, respectively).
Stimulation of meningothelial cells through TLR4.
Meningothelial cells produced a broad range of proimmune mediators in response to interaction with meningococci. The expression of TLR2 and 4 has been detected in meningothelial cells, but information on the triggering of these two major pathogen recognition receptors has been challenged recently (11).
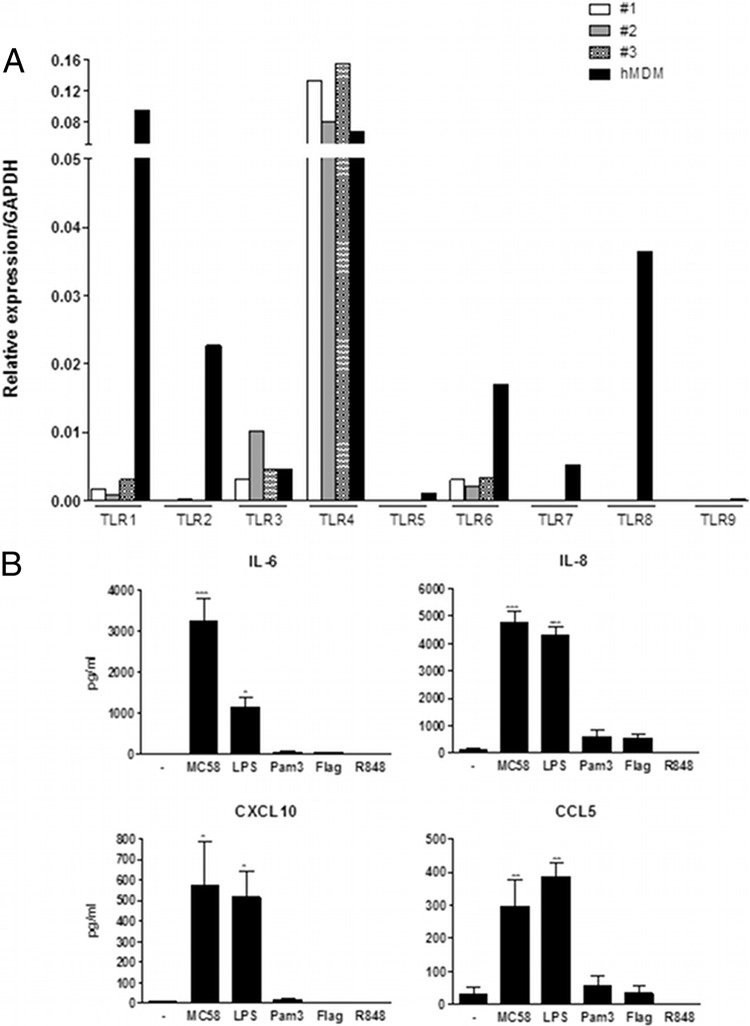
We first assessed the expression of TLR1, -2, -3, -4, -5, -6, -7, -8, and -9 in meningothelial cells. Three independently isolated meningothelial cell lines were investigated, and human monocyte-derived macrophages (hMDM) were used for comparison (Fig. 4A). High levels of TLR4 mRNA were detected in meningothelial cells. TLR1, TLR3, and TLR6 were also expressed, but at a much lower level. Expression of other TLRs was either very limited or undetectable.
Fig 4.
Meningothelial cell response to TLR stimulation. (A) Expression of TLR by meningothelial cells. Three meningothelial cell lines were analyzed and compared with hMDM for TLR expression determinations. Expression of TLR1, -2, -3, -4, -5, -6, -7, -8, and -9 was determined by QPCR. Data were standardized with reference to GAPDH expression. (B) Secretion of IL-6, IL-8, CXCL10, and CCL5 by meningothelial cells in response to contact with meningococci (strain MC58) or various TLR agonists (n ≥ 4). ANOVA was followed by Dunnett's multiple-comparison test.
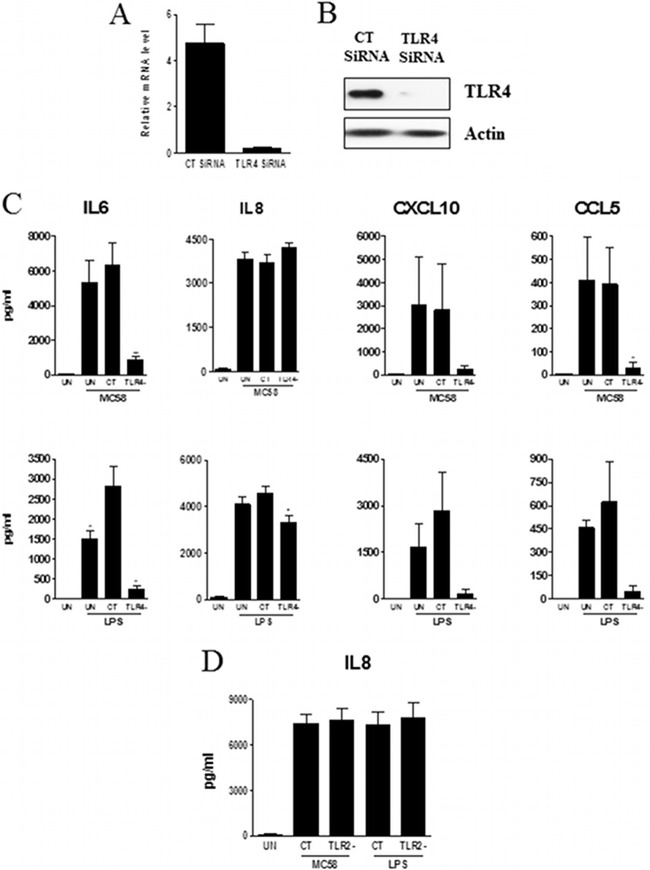
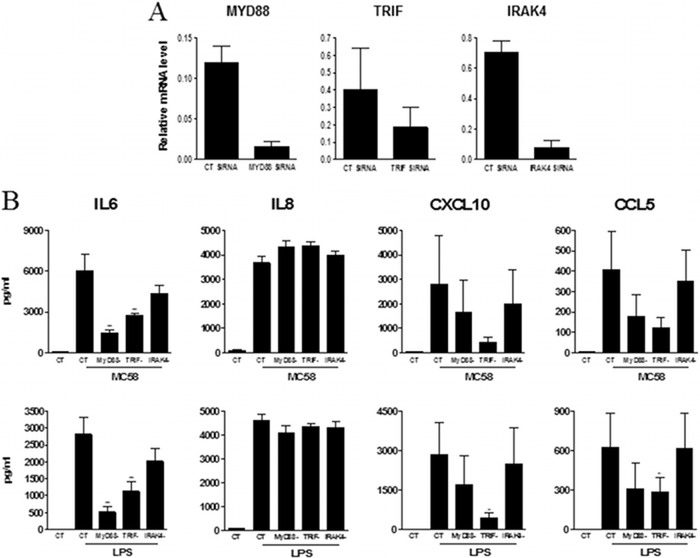
We then treated meningothelial cells with MC58 bacteria or with the following TLR agonists: LPS (TLR4 agonist), flagellin (TLR5 agonist), PamCSK4 (TLR1/2 agonist), and R848 (TLR7 and TLR8 agonist) and followed the response of meningothelial cells by measuring secretion of IL-6, IL-8, CXCL10, and CCL5. We choose these cytokines because (i) their expression is highly upregulated after stimulation, (ii) they are major players of immune response development, and (iii) their production can be easily quantified. Significant levels of these four cytokines were detected after MC58 or LPS stimulation (Fig. 4B). In contrast, and as expected from the TLR expression profiles, no significant secretion of cytokines was detected after treatment with flagellin, Pam3CSK4, or R848. Since meningothelial cells responded only to MC58 whole cells and to LPS in these experiments, we specifically silenced TLR4 expression by siRNA knockdown. TLR4 expression was effectively reduced as evidenced by QPCR (more than 90% reduction) (Fig. 5A) or by Western blotting (Fig. 5B). We also checked that the gene silencing procedure itself did not induce cell activation. Transfection alone did not trigger detectable production of the tested cytokines (not shown). After stimulation with LPS, IL-6 production was moderately boosted under the control siRNA conditions compared with the nontransfected conditions (Fig. 5C, bottom left panel). Nevertheless, levels of cytokines released by nontransfected and control siRNA-transfected cells were otherwise not significantly different (Fig. 5C).
Fig 5.
Role of TLRs in meningothelial cell activation by meningococci (strain MC58) or LPS. (A and B) TLR4 knockdown was assessed by QPCR (n = 2) (A) or by Western blotting (results from an experiment representative of 3 replicates are shown) (B). (C) Analysis of IL-6, IL-8, CXCL10, and CCL5 secretion by nontransfected (UN), control-transfected (CT), or TLR4 knockdown meningothelial cells after contact with meningococci (strain MC58) or LPS stimulation (n = 4). ANOVA was followed by Dunnett's multiple-comparison test. (D) Analysis of IL-8 secretion by TLR2 knockdown meningothelial cells treated with MC58 or LPS (n = 2).
TLR4 knockdown resulted in a significant decrease in secretion of IL-6, CXCL10, and CCL5 after stimulation either with MC58 whole cells or with LPS (Fig. 5C). The cytokine response to MC58 treatment was reduced by around 85% to 90% after TLR4 knockdown (Fig. 5C and Table 2), indicating that production of these 3 cytokines depends almost exclusively on TLR4 signaling. In contrast, TLR4 knockdown had a limited impact on IL-8 production in response either to MC58 whole-cell treatment or to LPS stimulation (Fig. 5C). Likewise, IL-8 secretion was found to be TLR2 independent (Fig. 5D).
Table 2.
Effect of TLR4, MyD88, TRIF, and IRAK4 knockdown on cytokine productiona
| Condition | % cytokine production |
|||||
|---|---|---|---|---|---|---|
| MC58 |
LPS |
|||||
| IL-6 | CXCL10 | CCL5 | IL-6 | CXCL10 | CCL5 | |
| CT | 100 | 100 | 100 | 100 | 100 | 100 |
| TLR4− | 14 ± 4 | 11 ± 3 | 3 ± 3 | 9 ± 3 | 4 ± 2 | 3 ± 3 |
| Myd88− | 24 ± 1 | 49 ± 6 | 37 ± 14 | 19 ± 5 | 47 ± 13 | 38 ± 12 |
| Trif− | 51 ± 11 | 31 ± 15 | 34 ± 15 | 43 ± 10 | 28 ± 14 | 47 ± 9 |
| IRAK4− | 75 ± 6 | 75 ± 2 | 83 ± 10 | 75 ± 11 | 84 ± 10 | 98 ± 4 |
The percentage of cytokine production after stimulation with MC58 or LPS, compared with that for CT cells, was calculated for each condition.
Role of MyD88, TRIF, and IRAK4 for TLR4 signaling.
We have shown the essential role played by TLR4 for the induction of IL-6, CXCL10, and CCL5 production by meningothelial cells in response to contact with meningococci. TLR4 signals through two different pathways dependent on the adaptor proteins MyD88 and TRIF, respectively (21). The MyD88 pathway engages the IRAK4 kinase and results notably in the activation of the transcription factor NF-κB p65, p38 MAPK, and JNK1. The TRIF pathway leads to activation of NF-κB, p38 MAPK, and JNK1 and also to activation of the IRF3 and IRF7 transcription factors. To investigate the contribution of these two pathways to meningothelial cell activation in response to meningococci, we silenced the expression of the MyD88 and TRIF adaptors and the downstream IRAK4 kinase by gene silencing (Fig. 6A) and the secretion of IL-6, CXCL10, and CCL5 was quantified. Stimulation of IL-6 secretion by contact with either MC58 whole cells or LPS was largely dependent on MyD88 signaling; secretion of this cytokine was reduced to 24% ± 1% and 19% ± 5% after stimulation with meningococci and LPS, respectively, after knockdown of MyD88 (Fig. 6B and Table 2). IL-6 production was also TRIF dependent, although to a lesser extent (51% ± 11% and 43% ± 10% left after stimulation with meningococci and LPS, respectively). IRAK4 knockdown had a more marginal effect on stimulation of IL-6 production; around 75% of the IL-6 was still produced after knockdown of this kinase in response to either stimulant (Fig. 6B and Table 2). The quantity of IL-6 was also reduced only weakly after incubation with the IRAK1/4 inhibitor 1-(2-(4-morpholinyl)ethyl)-2-(3-nitrobenzoylamino)benzimidazole (not shown), further supporting the idea of the minor role played by IRAK4 for the synthesis of this cytokine. As with IL-6 induction, induction of CXCL10 and CCL5 by contact with meningococci or LPS involved both MyD88 and TRIF. The role of TRIF in the induction of these cytokines was more significant, however, while IRAK4 knockdown had little influence on induction of these cytokines (Fig. 6B and Table 2).
Fig 6.
Contribution of MyD88, TRIFF, and IRAK4 in TLR4 signaling. (A) MyD88, TRIFF, and IRAK4 knockdown was assessed by QPCR (n = 2). (B) Secretion of IL-6, IL-8, CXCL10, and CCL5 by CT or MyD88, TRIFF, and IRAK4 knockdown meningothelial cells after contact with meningococci (strain MC58) or LPS stimulation (n = 4). ANOVA was followed by Dunnett's multiple-comparison test.
In contrast to the requirement for MyD88 and TRIF for maximal induction of IL-6, CXCL10, and CCL5, induction of IL-8 in meningothelial cells, after either contact with meningococci or treatment with LPS, was not affected by knockdown of either MyD88 or TRIF (Fig. 6B). In these experiments, IRAK4 kinase knockdown also had no effect on IL-8 induction in response to meningococci or LPS.
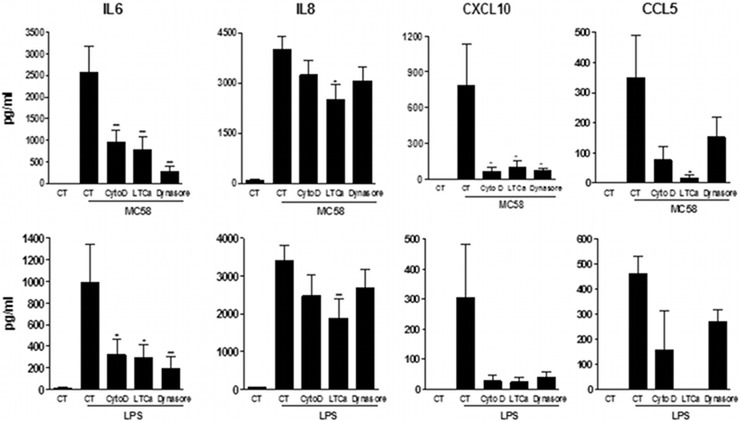
Endocytosis is needed for TLR4 signaling.
It has been reported recently that dynamin-dependent endocytosis of TLR4 after LPS triggering was a prerequisite for signaling via TRIF (22). Because of the importance of TRIF signaling observed in meningothelial cells, we investigated the role of TLR4 internalization for cytokine production. Endocytosis was blocked with the dynamin inhibitor Dynasore, with the classical inhibitor of actin polymerization cytochalasin D, or with the actin-depolarizing agent latrunculin A. Inhibition of endocytosis resulted in a dramatic drop in the induction of IL-6 in meningothelial cells exposed to either meningococci or LPS, confirming a role for internalization of TLR4 in meningothelial cells in response to stimulation (Fig. 7). IL-8 production, by contrast, was affected only slightly after treatment with latrunculin A, as is consistent with our findings that induction of IL-8 was mainly independent of the presence of TLR4. As expected, CXCL10 expression and, to a lesser extent, CCL5 expression were also affected by endocytosis inhibition.
Fig 7.
Role of endocytosis in TLR4 signaling. Meningothelial cells were treated with cytochalasin D (CytoD), latrunculin A (LTCa), or Dynasore or were given no treatment for 1 h before stimulation with MC58 or LPS. IL-6, IL-8, CXCL10, and CCL5 secretion was measured (n ≥ 3). ANOVA was followed by Dunnett's multiple-comparison test.
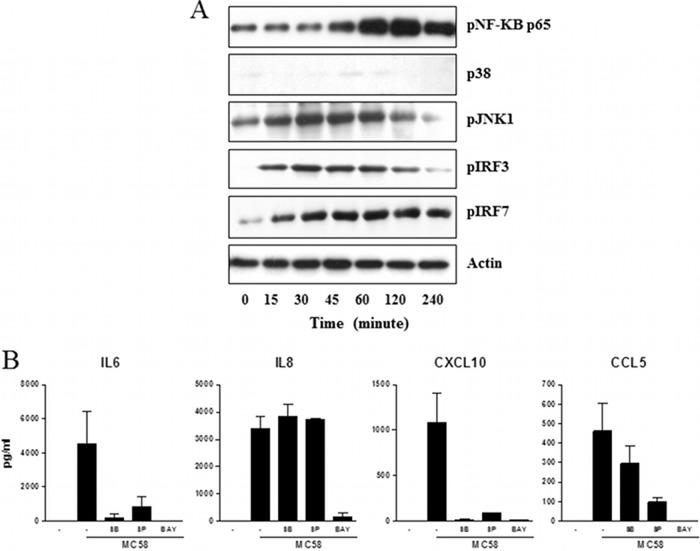
Transcription factors involved after meningothelial cell stimulation with N. meningitidis.
Since production of cytokines by meningothelial cells requires MyD88 and TRIF signaling, we investigated the phosphorylation of the transcription factors NF-κB p65, p38 MAPK, JNK, IRF3, and IRF7 after stimulation of meningothelial cells with MC58. Meningothelial cells treated with MC58 were collected at different time points for analysis of transcription factor phosphorylation. Phosphorylation of NF-κB p65 was detected 45 min after stimulation of meningothelial cells with meningococci, reaching a peak after 60 to 120 min (Fig. 8A). Phosphorylation of the other transcription factors was also detected after meningothelial cell stimulation. Finally, to further confirm the role of these transcription factors in meningothelial cell activation, we used inhibitors of NF-κB p65, p38 MAPK, and JNK1. As shown in Fig. 8B, inhibition of NF-κB p65, p38 MAPK, or JNK1 dramatically reduced the production of IL-6, CXCL10, or CCL5. Interestingly, IL-8 production was affected only after inhibition of NF-κB p65.
Fig 8.
Role of NF-κB p65, p38 MAPK, JNK1, IRF3, and IRF7 transcription factors in meningothelial cell stimulation. (A) Transcription factor phosphorylation. Meningothelial cells were stimulated with MC58. At the indicated times, proteins were collected and analyzed by Western blotting for NF-κB p65, p38 MAPK, JNK1, IRF3, and IRF7 phosphorylation. The results of 1 experiment representative of 3 are shown. (B) Role of NF-κB p65, p38 MAPK, and JNK1 transcription factors in the meningothelial cell response to meningococci. Meningothelial cells were treated with inhibitors of NF-κB p65 (Bay 11-7082; BAY), p38 MAPK (SB203580; SB), or JNK1 (SP600125; SP) for 1 h before stimulation with meningococci (strain MC58). IL-6, IL-8, CXCL10, and CCL5 secretion was then measured (n = 2).
DISCUSSION
The location of meningothelial cells makes them potentially important players in the host immune response occurring at the site of infection during microbial meningitis. Together with brain microvascular endothelial cells (BMEC), microglia, and astrocytes, meningothelial cells form part of the BBB, which constitutes a barrier protecting neuronal tissues against foreign insults. Moreover, they can respond to contact with N. meningitidis by secretion of cytokines and chemokines (10–13, 23). However, a complete picture of the meningothelial cell response to meningococcal infection is still missing and, more importantly, the molecules involved in the detection of meningococci by these cells have not been identified.
Here, we characterized the meningothelial cell response to meningococcal infection by transcriptome profiling using microarrays. Expression of a large number of genes (306) associated with various biological processes was modulated by contact with meningococci. We identified networks mainly associated with cell movement, cell-cell signaling, and interaction and antimicrobial and inflammatory response. Interestingly, whereas the majority of the altered genes were found to be downregulated, chemokine and cytokine genes involved in the development of proinflammatory response were upregulated. Increased expression of proinflammatory molecules such as IL-6, IFN-β1, CSF2, and PTX3 and of chemokines such as IL-8, CCL20, CXCL10, CXCL11, and CCL5 probably accounts for the massive influx and activation of neutrophils, monocytes, macrophages, or dendritic cells observed in cases of bacterial meningitis (8, 19). Induction of IFN-β1 gene expression is of particular interest. Although type I IFNs have been classically associated with antiviral responses, recent evidence has highlighted their contribution to bacterial immunity (24). Interestingly, microglia, the resident macrophages of the CNS, do not produce substantial levels of IFN-β1 (25). Here, meningothelial cells were able to attract Th1 lymphocytes through CCL2 or CXCL10 secretion and to potentiate their production of IFN-γ by IFN-β release. IFN-γ ultimately activates the bactericidal activity of macrophages or neutrophils. A similar mechanism has been described recently for protection against Gram-negative enteropathogens (26).
Meningothelial cells strongly upregulate the expression of MMP1 and MMP12. The precise role of these proteases in meningitis has not been explored. However, high MMP1 concentrations in the CSF have been detected in case of bacterial meningitis (27). In addition, MMP8 has been shown to be upregulated in human brain microvascular endothelial cells (HBMEC) in response to meningococcal challenge (7). Expression of this protein led to cleavage of occludin, increased permeability of the monolayer, and detachment of infected cells (7). Overexpression of MMP1, together with downregulation of collagens VIII and XI, may contribute to leukocyte chemotaxis through disruption of the extracellular matrix and blood-brain barrier. Thus, meningothelial cells not only provide chemoattractant and stimulatory signals for immune cells but also modulate extracellular matrix protein synthesis which would be expected to favor migration of immune cells through the basement membrane.
Upregulation of ICAM1, which interacts with LFA1, encourages the adhesion and diapedesis of leukocytes (28). Thus, expression of chemokines, MMPs, and adhesion molecules induced by contact of meningothelial cells with meningococci is likely to result in recruitment and activation of immune cells inside the CNS. An effective immune response to bacterial infection of the CNS is essential for pathogen elimination. However, release of compounds that include reactive oxygen species capable of damaging neural tissue, or favoring ischemia or edema, can cause severe pathology of the CNS (14).
To better understand the development of the proinflammatory response in the CNS in response to meningococcal challenge, we investigated the expression of various TLRs by meningothelial cells and showed that these cells strongly expressed TLR4. Expression of this receptor was required for expression of IL-6, CXCL10, and CCL5: the massive reduction in the production of these cytokines by TLR4-deficient cells indicates that expression of these genes is probably exclusively TLR4 dependent. In a study by Humphries et al. (11), expression of TLR2 and TLR4 was shown to be constitutive in meningothelial cells; treatment of these cells with outer membrane (OM)-derived LOS-deficient or LOS-replete meningococci did not result in upregulation of these receptors, and neither did the meningococcal OM preparations drive NF-κB activation in CHO reporter cell lines expressing CD14, TLR4, or TLR2 and an NF-κB reporter construct. It should be noted, however, that those researchers looked at expression but not at activation in meningothelial cells, while the responses of CHO, in which other components of the signaling pathway might not be present, may not accurately reproduce the signaling network in physiologically relevant meningothelial cells. TLR4 signals through two distinct pathways dependent on the presence of adaptor molecules MyD88 and TRIF, respectively (reviewed by Kawai and Akira [21]). TLR4 triggering first engages the MyD88 pathway, leading to recruitment of IRAK4, activation of NF-κB and MAPK, and the production of inflammatory cytokines. Dynamin-mediated endocytosis additionally allows the engagement of the TRIF pathway, late activation of NF-κB, MAPK, and IRF3, and the production of the inflammatory cytokines IFN, CXCL10, and CCL5. The consequences of triggering these two pathways depend on the cell type and the chemical composition of stimulating LPS. We found that both the MyD88 and TRIF adaptors were necessary for maximal IL-6 production. Production of both CXCL10 and CCL5 was dependent on TRIF expression, as was previously described for macrophages responding to LPS stimulation (29), but also on MyD88 expression. In that respect, those findings confirm recent results demonstrating synergy between the MyD88 and TRIF pathways (30, 31). Using gene silencing or pharmacological inhibitors, we then showed that IRAK4 played a limited role in IL-6, CXCL10, and CCL5 synthesis in meningothelial cells. For CXCL10 and CCL5 production, this probably reflects a dominant role for the TRIF pathway, which does not depend on IRAK4 (32). It is surprising, however, that IL-6 synthesis was not affected by these treatments, as its expression was severely reduced in IRAK4-deficient human blood mononuclear leukocytes (33) and in IRAK4 kinase-inactive knock-in mice (34). Engagement of the TRIF pathway in meningothelial cells probably accounts for the sustained IL-6 secretion, through the activation of NF-κB p65, p38 MAPK, JNK1, IRF3, and IRF7 transcription factors, even in the absence of IRAK4. The dramatic decrease of IL-6 synthesis after inhibition of endocytosis supports this hypothesis. Thus, although MyD88 and TRIF both contribute to IL-6 production, the IRAK4 contribution is rather limited, suggesting a bias toward the TRIF pathway. Interestingly, tumor necrosis factor alpha (TNF-α)—a typically MyD88/IRAK4-dependent cytokine—was not detected in meningothelial cell supernatants.
Another intriguing finding of our study concerns the regulation of IL-8 production. Although meningothelial cells produced IL-8 after exposure to meningococci or LPS, its secretion appeared to be both MyD88 and TRIF independent and did not require endocytosis of TLRs. These findings are consistent with TLR-independent induction of IL-8. TLR4-independent signaling by LPS was described previously (11, 35, 36). However, while alternative receptors for LPS have been proposed (37), none have been confirmed to date. TLR2-dependent but MyD88-independent induction of IL-8, as well as of IL-6, TNF-α, and NF-κB, has been previously described in HT-29 cells stimulated with Clostridium butyricum (38), and TLR2-dependent signaling in Caco-2 cells stimulated with Campylobacter jejuni has been shown to lead to IL-6 secretion by a MyD88-independent mechanism (39). Our data suggest that IL-8 induction by meningococci or LPS is TLR independent, but it remains a possibility that the presence of TLRs leads to induction of this cytokine by MyD88- and TRIF-independent mechanisms. Our inhibition experiments showed that NF-κB p65 was indispensable for IL-8 production. Further work will be necessary to define both the meningococcal molecules and the signaling pathways responsible for NF-κB p65-dependent induction of IL-8 production in meningothelial cells.
The signaling events observed in response to contact between meningothelial cells and meningococci could be due to contact between cells and bacterially secreted molecules, attachment of bacteria to the meningothelial cell surface, or invasion of the meningothelial cells. It is difficult, however, to distinguish between a requirement for bacterial invasion and TLR4 internalization, as both processes are inhibited by endocytosis blockade. Inhibition of endocytosis with the dynamin inhibitor Dynasore, with the classical inhibitor of actin polymerization cytochalasin D, or with the actin-depolarizing agent latrunculin A resulted in a dramatic drop in induction of IL-6, CXCL10, and CCL5 in meningothelial cells exposed to meningococci. However, since LPS signaling was also affected by endocytosis inhibition, we concluded that TLR4 internalization was necessary for the signaling, although we cannot exclude a possible role for bacterial invasion in the triggering of meningothelial cell response. IL-8 production, by contrast, was only slightly affected, and only after treatment with latrunculin. Thus, IL-8 production was mainly TLR4 and invasion independent.
In conclusion, characterization of the response of meningothelial cells to contact with meningococci emphasizes the central role these cells are likely to play in the development of the local immune response in the CNS in response to Gram-negative bacterial infection. Deciphering of the signaling pathways triggered by meningococci in these cells will help to determine potential targets for therapeutic intervention to modulate potentially damaging inflammation in the CNS during bacterial meningitis.
Supplementary Material
Footnotes
Published ahead of print 3 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00477-13.
REFERENCES
- 1.Christensen H, May M, Bowen L, Hickman M, Trotter CL. 2010. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect. Dis. 10:853–861 [DOI] [PubMed] [Google Scholar]
- 2.Akat K, Bleck CK, Lee YM, Haselmann-Weiss U, Kartenbeck J. 2008. Characterization of a novel type of adherens junction in meningiomas and the derived cell line HBL-52. Cell Tissue Res. 331:401–412 [DOI] [PubMed] [Google Scholar]
- 3.Akat K, Mennel HD, Kremer P, Gassler N, Bleck CK, Kartenbeck J. 2003. Molecular characterization of desmosomes in meningiomas and arachnoidal tissue. Acta Neuropathol. 106:337–347 [DOI] [PubMed] [Google Scholar]
- 4.Püttmann S, Senner V, Braune S, Hillmann B, Exeler R, Rickert CH, Paulus W. 2005. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab. Invest. 85:1163–1171 [DOI] [PubMed] [Google Scholar]
- 5.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala'Aldeen DA, Tuomanen EI. 2009. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J. Clin. Invest. 119:1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coureuil M, Lecuyer H, Scott MG, Boularan C, Enslen H, Soyer M, Mikaty G, Bourdoulous S, Nassif X, Marullo S. 2010. Meningococcus hijacks a beta2-adrenoceptor/beta-arrestin pathway to cross brain microvasculature endothelium. Cell 143:1149–1160 [DOI] [PubMed] [Google Scholar]
- 7.Schubert-Unkmeir A, Konrad C, Slanina H, Czapek F, Hebling S, Frosch M. 2010. Neisseria meningitidis induces brain microvascular endothelial cell detachment from the matrix and cleavage of occludin: a role for MMP-8. PLoS Pathog. 6:e1000874. 10.1371/journal.ppat.1000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Man S, Ubogu EE, Ransohoff RM. 2007. Inflammatory cell migration into the central nervous system: a few new twists on an old tale. Brain Pathol. 17:243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergman P, Johansson L, Wan H, Jones A, Gallo RL, Gudmundsson GH, Hokfelt T, Jonsson AB, Agerberth B. 2006. Induction of the antimicrobial peptide CRAMP in the blood-brain barrier and meninges after meningococcal infection. Infect. Immun. 74:6982–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christodoulides M, Makepeace BL, Partridge KA, Kaur D, Fowler MI, Weller RO, Heckels JE. 2002. Interaction of Neisseria meningitidis with human meningeal cells induces the secretion of a distinct group of chemotactic, proinflammatory, and growth-factor cytokines. Infect. Immun. 70:4035–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphries HE, Triantafilou M, Makepeace BL, Heckels JE, Triantafilou K, Christodoulides M. 2005. Activation of human meningeal cells is modulated by lipopolysaccharide (LPS) and non-LPS components of Neisseria meningitidis and is independent of Toll-like receptor (TLR)4 and TLR2 signalling. Cell. Microbiol. 7:415–430 [DOI] [PubMed] [Google Scholar]
- 12.Robinson K, Taraktsoglou M, Rowe KS, Wooldridge KG, Ala'Aldeen DA. 2004. Secreted proteins from Neisseria meningitidis mediate differential human gene expression and immune activation. Cell. Microbiol. 6:927–938 [DOI] [PubMed] [Google Scholar]
- 13.Wells DB, Tighe PJ, Wooldridge KG, Robinson K, Ala' Aldeen DA. 2001. Differential gene expression during meningeal-meningococcal interaction: evidence for self-defense and early release of cytokines and chemokines. Infect. Immun. 69:2718–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nau R, Bruck W. 2002. Neuronal injury in bacterial meningitis: mechanisms and implications for therapy. Trends Neurosci. 25:38–45 [DOI] [PubMed] [Google Scholar]
- 15.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FCP, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365–371 [DOI] [PubMed] [Google Scholar]
- 16.Dysvik B, Jonassen I. 2001. J-Express: exploring gene expression data using Java. Bioinformatics 17:369–370 [DOI] [PubMed] [Google Scholar]
- 17.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. 10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahrtz F, Piali L, Spanaus KS, Seebach J, Fontana A. 1998. Chemokines and chemotaxis of leukocytes in infectious meningitis. J. Neuroimmunol. 85:33–43 [DOI] [PubMed] [Google Scholar]
- 20.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. 2009. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384 [DOI] [PubMed] [Google Scholar]
- 22.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9:361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy SJ, Christodoulides M, Weller RO, Heckels JE. 2000. Interactions of Neisseria meningitidis with cells of the human meninges. Mol. Microbiol. 36:817–829 [DOI] [PubMed] [Google Scholar]
- 24.Decker T, Muller M, Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687 [DOI] [PubMed] [Google Scholar]
- 25.Rock RB, Gekker G, Hu S, Sheng WS, Cheeran M, Lokensgard JR, Peterson PK. 2004. Role of microglia in central nervous system infections. Clin. Microbiol. Rev. 17:942–964, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sotolongo J, Espana C, Echeverry A, Siefker D, Altman N, Zaias J, Santaolalla R, Ruiz J, Schesser K, Adkins B, Fukata M. 2011. Host innate recognition of an intestinal bacterial pathogen induces TRIF-dependent protective immunity. J. Exp. Med. 208:2705–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green JA, Thi Hong Chau T, Farrar JJ, Friedland JS, Thwaites GE. 2011. CNS infection, CSF matrix metalloproteinase concentrations, and clinical/laboratory features. Neurology 76:577–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ransohoff RM, Kivisakk P, Kidd G. 2003. Three or more routes for leukocyte migration into the central nervous system. Nat. Rev. Immunol. 3:569–581 [DOI] [PubMed] [Google Scholar]
- 29.Hirotani T, Yamamoto M, Kumagai Y, Uematsu S, Kawase I, Takeuchi O, Akira S. 2005. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-beta. Biochem. Biophys. Res. Commun. 328:383–392 [DOI] [PubMed] [Google Scholar]
- 30.Ouyang X, Negishi H, Takeda R, Fujita Y, Taniguchi T, Honda K. 2007. Cooperation between MyD88 and TRIF pathways in TLR synergy via IRF5 activation. Biochem. Biophys. Res. Commun. 354:1045–1051 [DOI] [PubMed] [Google Scholar]
- 31.Rathinam VA, Appledorn DM, Hoag KA, Amalfitano A, Mansfield LS. 2009. Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through Toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect. Immun. 77:2499–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koziczak-Holbro M, Gluck A, Tschopp C, Mathison JC, Gram H. 2008. IRAK-4 kinase activity-dependent and -independent regulation of lipopolysaccharide-inducible genes. Eur. J. Immunol. 38:788–796 [DOI] [PubMed] [Google Scholar]
- 33.Ku CL, von Bernuth H, Picard C, Zhang SY, Chang HH, Yang K, Chrabieh M, Issekutz AC, Cunningham CK, Gallin J, Holland SM, Roifman C, Ehl S, Smart J, Tang M, Barrat FJ, Levy O, McDonald D, Day-Good NK, Miller R, Takada H, Hara T, Al-Hajjar S, Al-Ghonaium A, Speert D, Sanlaville D, Li X, Geissmann F, Vivier E, Marodi L, Garty BZ, Chapel H, Rodriguez-Gallego C, Bossuyt X, Abel L, Puel A, Casanova JL. 2007. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J. Exp. Med. 204:2407–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim TW, Staschke K, Bulek K, Yao J, Peters K, Oh KH, Vandenburg Y, Xiao H, Qian W, Hamilton T, Min B, Sen G, Gilmour R, Li X. 2007. A critical role for IRAK4 kinase activity in Toll-like receptor-mediated innate immunity. J. Exp. Med. 204:1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz E, Patel DD, Hartung T, Schwartz DA. 2002. Toll-like receptor 4 (TLR4)-deficient murine macrophage cell line as an in vitro assay system to show TLR4-independent signaling of Bacteroides fragilis lipopolysaccharide. Infect. Immun. 70:4892–4896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Na HY, Mazumdar K, Moon HJ, Chang S, Seong SY. 2009. TLR4-independent and PKR-dependent interleukin 1 receptor antagonist expression upon LPS stimulation. Cell Immunol. 259:33–40 [DOI] [PubMed] [Google Scholar]
- 37.Triantafilou K, Triantafilou M, Dedrick RL. 2001. A CD14-independent LPS receptor cluster. Nat. Immunol. 2:338–345 [DOI] [PubMed] [Google Scholar]
- 38.Gao Q, Qi L, Wu T, Wang J. 2012. Clostridium butyricum activates TLR2-mediated MyD88-independent signaling pathway in HT-29 cells. Mol. Cell. Biochem. 361:31–37 [DOI] [PubMed] [Google Scholar]
- 39.Friis LM, Keelan M, Taylor DE. 2009. Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect. Immun. 77:1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.