Abstract
After transcription initiation, RNA polymerase (Pol) II escapes from the promoter and recruits elongation factors. The molecular basis for the initiation-elongation factor exchange during this transition remains poorly understood. Here, we used chromatin immunoprecipitation (ChIP) to elucidate the initiation-elongation transition of Pol II in the budding yeast Saccharomyces cerevisiae. We show that the early Pol II elongation factor Spt5 contributes to stable recruitment of the mRNA capping enzymes Cet1, Ceg1, and Abd1. Genome-wide occupancy for Cet1 and Ceg1 is restricted to the transcription start site (TSS), whereas occupancy for Abd1 peaks at ∼110 nucleotides downstream, and occupancy for the cap-binding complex (CBC) rises subsequently. Abd1 and CBC are important for recruitment of the kinases Ctk1 and Bur1, which promote elongation and capping enzyme release. These results suggest that cap completion stimulates productive Pol II elongation.
INTRODUCTION
Transcription of protein-coding genes by RNA polymerase (Pol) II begins with assembly of an initiation complex at the promoter. As the RNA grows, initiation factors dissociate from Pol II, and elongation factors are recruited (1–3). The initiation-elongation transition begins when the nascent RNA grows to about 12 nucleotides (nt), which releases initiation factor TFIIB (4, 5). Release of initiation factors frees the Pol II clamp domain, which binds the conserved elongation factor Spt5 (6, 7). Here, we refer to the exchange of initiation by elongation factors as the initiation-elongation transition. The transition starts with the dissociation of initiation factors and ends with completed recruitment of elongation factors.
During initiation, the C-terminal repeat domain (CTD) of Pol II gets phosphorylated at the serine 5 (S5) and S7 residues (8–10). CTD S5 phosphorylation is important for recruitment of RNA 5′ capping enzymes (11–13). Capping starts with removal of the terminal γ-phosphate from the 5′ triphosphate end of the nascent RNA, which is catalyzed by the RNA triphosphatase (14–16). The truncated RNA 5′ end is then linked to an inverted guanylyl group by the guanylyltransferase. Finally, the cap methyltransferase methylates position N7 of the newly added terminal guanine. In Saccharomyces cerevisiae the three catalytic activities are encoded by Cet1, Ceg1, and Abd1, respectively. In metazoans the first two steps of the capping reaction are catalyzed by a single capping enzyme consisting of an N-terminal triphosphatase and a C-terminal guanylyltransferase domain. The resulting 7-methyl-guanosine (m7G) cap protects the transcript from degradation and promotes translation initiation of the mRNA (17, 18). The complete cap associates with the cap-binding complex (CBC) that functions in pre-mRNA splicing and mRNA export (19, 20).
We have previously obtained high-resolution genome-wide occupancy profiles for Pol II initiation and elongation factors by chromatin immunoprecipitation (ChIP) in S. cerevisiae. These studies showed that initiation factor occupancy peaked around 50 bp upstream of the transcription start site (TSS) (21), consistent with recent nucleotide resolution studies combining ChIP with exonuclease digestion (ChIP-exo) (22). Exchange of Pol II initiation factors by elongation factors mainly occurred within 150 bp downstream of the TSS (21). Further downstream, CTD phosphorylation levels decreased at residues S5 and S7 and increased at residues Y1 (23) and S2 (8, 9, 21). S2 phosphorylation is achieved by the CTD kinases Bur1 (24) and Ctk1 (25) that enable efficient RNA elongation and 3′ processing (26, 27). The mechanisms to recruit these elongation-promoting kinases are poorly understood although recent work showed that Bur1 recruitment is stimulated by its C-terminal region that binds the S5-phosphorylated CTD (24).
Bur1 and Ctk1 share homology with mammalian Cdk9, the kinase of the P-TEFb (positive transcription elongation factor b) complex that triggers the transition into productive elongation (28–30). For a long time it was believed that Cdk9 was the major contributor to S2 phosphorylation in higher eukaryotes, but recent evidence suggested that the Drosophila and human Cdk12 proteins also contribute to S2 phosphorylation and that Bur1 and Ctk1 might be the orthologs of Cdk9 and Cdk12, respectively (31). P-TEFb can release Pol II from promoter-proximal pause sites by phosphorylating the CTD, DSIF (human Spt4-Spt5), and NELF (negative elongation factor), leading to dissociation of NELF and transformation of DSIF into a positive elongation factor (32, 33). While most of the key players controlling the initiation-elongation transition are conserved between S. cerevisiae and higher eukaryotes, yeast and Caenorhabditis elegans apparently lack an NELF ortholog and promoter-proximal pausing.
Human Cdk9 physically interacts with the CBC (34), and Cdk9 from the fission yeast Schizosaccharomyces pombe interacts with the cap methyltransferase (35, 36). Further, Spt5 interacts with human, S. cerevisiae, and S. pombe capping enzymes (37–40). These interactions have led to a capping checkpoint model that suggests that Pol II pauses near the TSS to allow for cotranscriptional capping before entering productive elongation (2, 38, 41, 42).
The relevance of these interactions for the recruitment of the capping machinery and early elongation factors to transcribed genes has not been studied genome-wide in vivo. To address this, we extended our combined ChIP-microarray technology (ChIP-chip) occupancy profiling to capping enzymes and the cap-binding complex (CBC). We found that the first two capping enzymes, Cet1 and Ceg1, are recruited near the TSS, whereas the third enzyme, the methyltransferase Abd1, is recruited about 110 bp downstream of the TSS, and CBC is recruited further downstream. We report that capping enzyme recruitment requires the C-terminal region (CTR) of Spt5 and that Bur1 helps to release capping enzymes from Spt5 once capping is completed. Moreover, we show that Abd1 and CBC are involved in the recruitment of Ctk1 and Bur1 in vivo, thus maintaining proper levels of S2-phosphorylated Pol II and elongation factors.
Recently, a study appeared that showed that gene recruitment of Bur2 and Ctk2, the cyclin partners of Bur1 and Ctk1, respectively, required the CBC and that the kinase complexes interacted with the CBC (43). This study demonstrated an effect of CBC deletion on S2 phosphorylation. Our data are consistent with these findings and also provide additional insights that lead to a model for factor exchange during the initiation-elongation transition of Pol II that supports the capping checkpoint model and has implications for understanding the promoter-proximal transition in higher cells.
MATERIALS AND METHODS
Yeast strains and epitope tagging.
All S. cerevisiae strains used in this study are listed in Table 1. Deletion of the 15 C-terminal hexapeptide repeats (CTR; amino acids 931 to 1063) of Spt5 was done by homologous recombination with the KanMX6 cassette, amplified from the pFA6a-KanMX6 vector. For ChIP of C-terminally tandem affinity purification (TAP)-tagged versions of target proteins (TAP strains), TAP strains were either bought (Open Biosystems) or generated by integration of the TAP tag into the genome C-terminal of the respective genes by homologous recombination. For nuclear depletion of Abd1 using the anchor-away (AA) method (44), an FKBP12-rapamycin-binding (FRB) tag was introduced at the C terminus of Abd1. Anchor-away strains were generated as described previously (44) using PCR products amplified from plasmids pFA6a-FRB-KanMX6 and pFA6a-FRB-GFP-KanMX6 (where GFP is green fluorescent protein) (P30578/P30580; Euroscarf). For spot dilutions, cells were grown in yeast extract-peptone-dextrose (YPD) medium at 30°C to stationary phase and diluted to an optical density at 600 nm (OD600) of ∼1.0. Equal amounts of cells were spotted on YPD plates with or without 1 μg/ml (final concentration [f.c.]) rapamycin (LC Laboratories) in 10-fold serial dilutions. Plates were incubated at 30°C and inspected daily. For microscopy, cells were grown in YPD medium with 40 mg/liter adenine hemisulfate at 30°C to pre-log phase. Cultures were split and incubated with equal volumes of either rapamycin (1 μg/ml f.c. in dimethyl sulfoxide [DMSO]) or DMSO at 30°C for another 60 min. Cells were resolved in water and inspected under a microscope (Leica DM2500 microscope with an EL6000 light source). A DFC365FX camera and LAS AF 6000 Modular Systems, version 2.6.0.7266, software (Leica) were used for image analysis.
Table 1.
Yeast strains used in this study
| Strain namea | Genotype | Source |
|---|---|---|
| WT | BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| Cet1-TAP | BY4741 CET1::TAP::HIS3MX6 | Open Biosystems |
| Ceg1-TAP | RS453 MATa CEG1::TAP::TRP1-KL | K. Sträßer |
| Abd1-TAP | BY4741 ABD1::TAP::HIS3MX6 | This study |
| Cet1-TAP Spt5 ΔCTR | BY4741 CET1::TAP::HIS3MX6 SPT5Δ931–1063::KANMX6 | This study |
| Ceg1-TAP Spt5 ΔCTR | RS453 CEG1::TAP::TRP1-KL SPT5Δ931–1063::KANMX6 | This study |
| Abd1-TAP Spt5 ΔCTR | BY4741 ABD1::TAP::HIS3MX6 SPT5Δ931–1063::KANMX6 | This study |
| bur2Δ strain | BY4741 bur2::KANMX6 | Open Biosystems |
| bur2Δ Cet1-TAP | BY4741 bur2::KANMX6; Cet1::TAP::HIS3MX6 | This study |
| bur2Δ Abd1-TAP | BY4741 bur2::KANMX6; Abd1::TAP::HIS3MX6 | This study |
| CBP20-TAP | BY4741 CBP20::TAP::HIS3MX6 | Open Biosystems |
| CBP80-TAP | BY4741 CBP80::TAP::HIS3MX6 | Open Biosystems |
| cbp20Δ strain | BY4741 cbp20::KANMX6 | Invitrogen |
| cbp20Δ Bur1-TAP | BY4741 cbp20::KANMX6 BUR1::TAP::HIS3MX6 | This study |
| cbp20Δ Ctk1-TAP | BY4741 cbp20::KANMX6; CTK1::TAP::HIS3MX6 | This study |
| cbp20Δ Elf1-TAP | BY4741 cbp20::KANMX6 ELF1::TAP::HIS3MX6 | This study |
| cbp20Δ Spn1-TAP | BY4741 cbp20::KANMX6 SPN1::TAP::HIS3MX6 | This study |
| cbp20Δ Spt4-TAP | BY4741 cbp20::KANMX6 SPT4::TAP::HIS3MX6 | This study |
| cbp20Δ Spt5-TAP | BY4741 cbp20::KANMX6 SPT5::TAP::HIS3MX6 | This study |
| cbp20Δ Spt6-TAP | BY4741 cbp20::KANMX6 SPT6::TAP::HIS3MX6 | This study |
| cbp20Δ Spt16-TAP | BY4741 cbp20::KANMX6 SPT16::TAP::HIS3MX6 | This study |
| Anchor-away WT (AA) | HHY168 MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL psi+ tor1-1 fpr1::NAT RPL13A-2×FKBP12::TRP1 | Euroscarf |
| AA Bur1-TAP | HHY168 BUR1::TAP::HIS3MX6 | This study |
| AA Ctk1-TAP | HHY168 CTK1::TAP::HIS3MX6 | This study |
| AA Abd1-FRB | HHY168 ABD1::FRB::KANMX6 | This study |
| AA Abd1-FRB-GFP | HHY168 ABD1::FRB::GFP::KANMX6 | This study |
| AA Abd1-FRB Bur1-TAP | HHY168 ABD1::FRB::KANMX6 BUR1::TAP::HIS3MX6 | This study |
| AA Abd1-FRB CBP20-TAP | HHY168 ABD1::FRB::KANMX6 CBP20::TAP::HIS3MX6 | This study |
| AA Abd1-FRB Ctk1-TAP | HHY168 ABD1::FRB::KANMX6 CTK1::TAP::HIS3MX6 | This study |
WT, wild type.
Chromatin immunoprecipitation.
For standard ChIP experiments, yeast cultures were grown in 40 ml of YPD medium at 30°C to mid-log phase (OD600 of ∼0.8) and treated with 1% formaldehyde (F1635; Sigma) for 20 min at 20°C, and cross-linking was quenched with 5 ml of 3 M glycine for 10 min. For Abd1 anchor-away ChIP experiments, yeast cultures were grown in 80 ml of YPD medium to an OD600 of ∼0.6, split, and incubated with equal volumes of either rapamycin (1 μg/ml f.c. in DMSO) or DMSO at 30°C for another 60 min before formaldehyde cross-linking. Subsequent steps were performed at 4°C with precooled buffers containing protease inhibitors (1 mM leupeptin, 2 mM pepstatin A, 100 mM phenylmethylsulfonyl fluoride, 280 mM benzamidine). Cells were collected by centrifugation, washed twice with 1× Tris-buffered saline (TBS; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl) and twice with FA lysis buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS). Cell pellets were flash-frozen in liquid nitrogen and stored at −80°C. Pellets were thawed, resuspended in 1 ml of FA lysis buffer, and disrupted by bead beating (Retsch) in the presence of 1 ml of silica-zirconia beads for 30 min at 4°C. Lysis efficiency was typically >80%. Chromatin was solubilized and fragmented via sonication with a Bioruptor UCD-200 (Diagenode, Inc.). A total of 700 μl of sample was immunoprecipitated with 20 μl of IgG-Sepharose 6 Fast Flow beads (GE Healthcare) at 4°C for 1 h. The IgG beads were directed against the protein A content of the C-terminal TAP tag. For ChIP of RNA polymerase II, a monoclonal antibody directed against the Rpb3 subunit (1Y26; NeoClone) or a polyclonal antibody directed against the Ser-2 phosphorylated CTD (Bethyl Laboratories) was used. Chromatin (700 μl) was immunoprecipitated with either 5 μl of 1Y26 or 2 μl of anti-Ser-2 CTD antibody for 18 h at 4°C, followed by the addition of 25 μl of protein G and protein A-Sepharose 4 Fast Flow (GE Healthcare) and incubation for another 1.5 h at 4°C. Immunoprecipitated chromatin was washed three times with FA lysis buffer, twice with FA lysis buffer containing 500 mM NaCl, twice with ChIP wash buffer (10 mM Tris-HCl, pH 8.0, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% Na deoxycholate), and once with TE buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA). Immunoprecipitated chromatin was eluted for 10 min at 65°C with ChIP elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% SDS). Eluted chromatin was digested with proteinase K (Sigma) at 37°C for 2 h, and the reversal of cross-links was performed at 65°C overnight. DNA was purified with a QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. For ChIP-chip experiments, the above protocol was modified. Yeast cultures were grown in 600 ml of YPD medium, and cell disruption by bead beating was performed for 2 h. Chromatin was immunoprecipitated for 4 h and, after washing (see above), incubated for 60 min at 65°C.
qPCR.
Input and immunoprecipitated samples were assayed by quantitative real-time PCR (qPCR) to assess the extent of protein occupancy at different genomic regions. Primer pairs directed against different regions of the ADH1, PMA1, ACT1, and ILV5 genes as well as against an untranscribed control region of chromosome V were used to determine PCR efficiencies. All PCR efficiencies ranged between 95 and 100%. PCR mixtures contained 1 μl of DNA template, 2 μl of 10 μM primer pairs, and 12.5 μl of iTaq SYBR green Supermix (Bio-Rad). qPCR was performed on a CFX96 real-time system (Bio-Rad Laboratories, Inc.) using a 3-min denaturing step at 95°C, followed by 49 cycles of 30 s at 95°C, 30 s at 61°C, and 15 s at 72°C. Threshold cycle (CT) values were determined by application of the corresponding Bio-Rad CFX Manager software, version 1.1, using the CT determination mode “regression.” Fold enrichment of any given region over an open reading frame (ORF)-free untranscribed region on chromosome V was determined as described previously (45). Sequence information of primer pairs used in this study is available upon request.
DNA labeling and microarray handling.
DNA samples were amplified and reamplified with a GenomePlex Complete Whole Genome Amplification 2 (WGA2) kit using the Farnham laboratory WGA protocol for ChIP-chip (http://www.genomecenter.ucdavis.edu/farnham/protocol.html). The reamplification was performed in the presence of 0.4 mM dUTP (U1191; Promega) to allow later enzymatic fragmentation. The enzymatic fragmentation, labeling, hybridization, and array scanning were done according to the manufacturer's instructions (Affymetrix chromatin immunoprecipitation assay protocol, product number [P/N] 702238) (46). Enzymatic fragmentation and terminal labeling were performed by application of a GeneChip WT double-stranded DNA terminal labeling kit (P/N 900812; Affymetrix). Briefly, reamplified DNA was fragmented in the presence of 1.5 μl of uracil-DNA-glycosylase (10 U/μl) and 2.25 μl of APE1 (100 U/μl) at 30°C for 1 h 15 min. The fragmented DNA was then labeled at the 3′ end by adding 2 μl and 1 μl of terminal nucleotidyl transferase (TdT; 30 U/μl) and GeneChip DNA labeling reagent (5 mM), respectively. A total of 5.5 μg of fragmented and labeled DNA was hybridized to a high-density custom-made Affymetrix tiling array (P/N 520055) at 45°C for 16 h with constant rotational mixing at 60 rpm in a GeneChip Hybridization Oven 640 (Affymetrix, Santa Clara, CA). Washing and staining of the tiling arrays were performed using the FS450_0001 script of the Affymetrix GeneChip Fluidics Station 450. The arrays were scanned using an Affymetrix GeneChip Scanner 3000 7G.
ChIP-chip occupancy profiling.
ChIP-chip experiments were performed and analyzed as described previously (21), with some modifications. Briefly, two independent biological replicates were analyzed for each factor. First, we performed quantile normalization between replicate measurements and averaged the signal for each probe over the replicate intensities. For the capping enzymes, ChIP enrichments were obtained by dividing ChIP intensities by input intensities (log2 IP/input). For CBP20, ChIP data were normalized using both mock IP and input measurements. Normalized signal was converted to occupancy values between 0 and 100% by setting the genome-wide 99.8% quantile to 100% occupancy and the 10% quantile to 0% occupancy. The normalized ChIP-chip signal at each nucleotide was calculated as the median of all probes overlapping this position. Profiles were smoothed using running-median smoothing with a window half-size of 75 bp. To average profiles, genes were filtered. Of 4,366 yeast genes with annotated TSS and polyadenylation (pA) sites (47), we examined the 50% most highly expressed genes (48) that were at least 200 bp away from neighboring genes (1,140 genes; total set). Furthermore, the total set was subdivided into three length classes, short (512 to 937 bp), medium (938 to 1,537 bp), and long (1,538 to 2,895 bp), comprising 266, 339, and 299 filtered genes, respectively. Profiles within these classes were aligned at their TSS and pA sites, scaled to median length, and averaged using a 5% trimmed mean at each genomic position. To avoid scaling of profiles, profiles within gene classes were cut around the TSS (250 bp upstream to 650 bp downstream; only genes of >680 bp were considered) and averaged using a 5% trimmed mean at each genomic position. To calculate peak positions, profiles were cut around the TSS (150 bp upstream to 350 bp downstream) and smoothed with cubic splines (R package: stats, function: smooth. spline, parameter: spar = 0.9). The maximum value of the smoothed curve was selected as peak position. For calculation of gene-wise peak distances between Cet1-Ceg1 and Abd1, genes with detectable peaks were selected.
Correlation analysis and network construction.
Analyses were done using 4,366 genes with available TSS and pA annotations. Pairwise Pearson correlations over factor occupancy profiles were calculated between concatenated gene profiles, ranging each from the TSS minus 250 bp to the pA site plus 250 bp, and were provided as a similarity metric. The correlation-based network was calculated using the GraphViz's Neato algorithm (49) employing an edge-weighted, spring-embedded layout procedure, attempting to minimize a global energy function, which is equivalent to statistical multidimensional scaling.
Microarray data accession number.
Raw and normalized data have been deposited in Array Express under accession number E-MTAB-1552.
RESULTS
Involvement of the Spt5 CTR in recruitment of capping enzymes.
Spt5 may help to recruit capping enzymes to transcribed genes since it copurifies with Cet1, Ceg1, and Abd1 (39). In vitro, Spt5 binds and activates the human capping enzyme (37), and its C-terminal repeat region (CTR) binds S. pombe triphosphatase and guanylyltransferase (38, 40). To test whether Spt5 is involved in recruiting capping enzymes in vivo, we carried out ChIP analysis in S. cerevisiae strains expressing only Spt5 lacking the CTR (50, 51). We detected a decrease in the occupancy of Cet1, Ceg1, and Abd1 to about 65%, 50%, and 35% of wild-type levels, respectively, at the 5′ region of the ADH1 gene, whereas Pol II occupancy was relatively unaffected (Fig. 1A). Similar results were obtained for ACT1 (see Fig. S1B in the supplemental material). Total protein levels of Cet1, Ceg1, and Abd1 remained unchanged in the CTR-deleted cells (see Fig. S1A). These results show that replacing Spt5 with a mutant lacking the CTR reduces occupancy of capping enzymes over the ADH1 and ACT1 genes. Since S5 phosphorylation of the CTD by the initiation factor TFIIH kinase Kin28 is also required for capping enzyme recruitment (11–13), these results suggest that the Spt5 CTR and the Pol II CTD both contribute to ensure full recruitment of capping enzymes to the 5′ region of genes in vivo.
Fig 1.
The Spt5 CTR is involved in recruitment of capping enzymes. ChIP-qPCR analysis was performed to monitor capping enzyme and Pol II recruitment at three different gene regions of ADH1: TSS (5′), coding (open reading frame [ORF]), and terminator region (3′). Occupancies were calculated as fold enrichments over an ORF-free untranscribed region on chromosome V and are indicated on the y axes (see Materials and Methods). Error bars show standard deviations from three independent experiments of biological replicates, and the asterisk indicates factor occupancies that are significantly different (P < 0.05) between the wild-type and mutant conditions using Student's t test. (A) Cet1, Ceg1, Abd1, and Pol II occupancies for the wild type compared to Spt5ΔCTR cells are shown. (B) Abd1 and Pol II occupancies for the wild type compared to bur2Δ cells are shown. Abd1 fold enrichments relative to Pol II (Abd1/Pol II) were calculated by dividing Abd1 occupancies by Pol II occupancies. (C) Occupancies as described in panel B for Cet1. (D) Cet1/Pol II and Abd1/Pol II ChIP signals for the ORF region of ADH1 relative to the 5′ end.
The Bur1 kinase complex promotes release of capping enzymes.
We demonstrated that deletion of the CTR of Spt5 reduced Abd1 occupancy at the 5′ regions of ADH1 and ACT1 but not further downstream (Fig. 1A; see also Fig. S1B in the supplemental material), suggesting that Abd1 is released from Spt5 during the initiation-elongation transition. Since the elongation-promoting kinase complex Bur1-Bur2 phosphorylates the Spt5 CTR during this transition (50, 52), Bur1 may help to release capping enzymes from Spt5 after cap completion. To investigate this, we carried out ChIP analysis of Cet1 and Abd1 in a strain lacking Bur2, the cyclin subunit of the Bur1 complex (Fig. 1B and C). In the bur2Δ mutant, phosphorylation of Spt5 was reduced (52). Cet1 and Abd1 occupancies were normalized against Pol II occupancy since the Bur2 deletion had a negative effect on Pol II recruitment (Fig. 1B). We found that deletion of Bur2 led to a significant increase (>2-fold) of Abd1/Pol II ratios at the ORF and 3′ regions of ADH1, ACT1, and ILV5 (Fig. 1B; see also Fig. S1C). For Cet1 we observed a similar increase in Cet1/Pol II ratios, suggesting that residual amounts of Cet1 molecules remain associated with the transcription complex until the 3′ ends of the genes when Bur2 is deleted (Fig. 1C; see also Fig. S1D). At the ADH1 gene the Cet1/Pol II ratio was also increased at the 5′ region (Fig. 1C). We do not know the reason for this increase, but to check whether the Cet1/Pol II increase at the ORF region is an indirect effect due to higher levels at the 5′ region, we compared the ORF/5′ ChIP ratio between wild-type and bur2Δ experiments. In the wild type, the Cet1/Pol II ratio decreased from to 100% (5′) to 3.9% (ORF), while in bur2Δ it decreased to 9.5% (ORF) (Fig. 1D). Likewise, the Abd1/Pol II ratio decreased from 100% (5′) to 18.8% (ORF) in the wild type and to in 48.8% (ORF) in bur2Δ cells. Similar effects were seen at other tested genes (see Fig. S1E). Overall these results support a model in which the Bur1-Bur2 kinase complex helps to release capping enzymes from Spt5, perhaps by phosphorylating the CTR (50, 52).
Cet1-Ceg1 recruitment is restricted to the TSS.
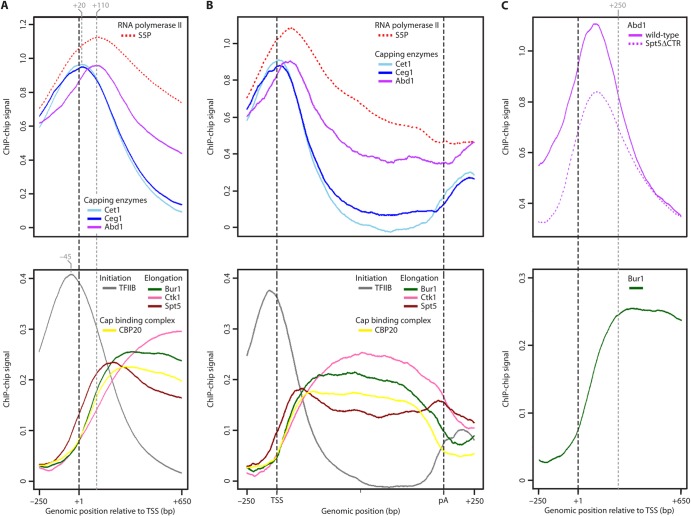
To determine the regions of capping enzyme recruitment, we collected genome-wide occupancy profiles for capping enzymes by ChIP-chip in exponentially growing S. cerevisiae cultures. ChIP-chip and data analysis were performed as described previously (21). Averaging of ChIP profiles after alignment of genes at their TSSs (47) revealed a sharp peak for Ceg1 (Fig. 2A and B, upper panels) ∼20 bp downstream of the TSS. ChIP-chip profiles for total Pol II (Rpb3 subunit) are shown as a reference (see Fig. S2C in the supplemental material). The peak location and shape for Ceg1 were virtually identical to those for Cet1 (21) (Fig. 2A and B, upper panels). Both peaks were independent of gene length, gene type, or expression level (see Fig. S2A, B, and E). The highly similar profiles for Cet1 and Ceg1 (R = 0.93) are consistent with the formation of a stable Cet1-Ceg1 heterodimer (53) and confirm the high resolution of our occupancy profiling. Cet1-Ceg1 occupancy increases and decreases sharply, with a peak width similar to that obtained for the initiation factor TFIIB (21) that binds a defined promoter region (Fig. 2A and B, lower panels). Thus, the recruitment and activity of the Cet1-Ceg1 heterodimer are apparently restricted to a narrow window near the TSS. Although ChIP data reflect only the physical presence of proteins and although cross-linking can be indirect via other proteins, these results are consistent with the model that the first two capping reactions occur on the polymerase surface when the nascent RNA 5′ end emerges from the Pol II RNA exit channel.
Fig 2.
Genome-wide occupancy profiling of the capping machinery. (A) Gene-averaged ChIP-chip profiles for capping enzymes and Pol II phosphorylated at serine 5 (S5P) residues of the CTD (upper panel) and for initiation factor TFIIB, CBC subunit CBP20, and elongation factors Bur1, Ctk1, and Spt5 (lower panel). Profiles in the lower panel were described previously (21) except that for CBP20. Occupancy profiles taken from the quality-filtered total gene set (1,140 genes) were cut around the TSS (250 bp upstream to 650 bp downstream; only genes of >680 bp were considered) and averaged using a 5% trimmed mean at each genomic position. Profiles for gene length classes are similar (see Fig. S2A in the supplemental material). ChIP-chip signal intensity is expressed as log2 IP/input. For details refer to Materials and Methods. Dashed gray lines mark the peak positions of the averaged ChIP-chip profiles. (B) Gene-averaged ChIP-chip profiles as described for panel A but for medium-length genes (1,238 ± 300 bp; n = 339). Genes were aligned at their TSS and pA sites (47), scaled to median length, and averaged using a 5% trimmed mean at each genomic position. Profiles of other gene length classes are similar (see Fig. S2B). For details refer to Materials and Methods. (C) Gene-averaged ChIP-chip profiles as described in panel A for Abd1 in wild-type compared to Spt5ΔCTR cells (upper panel) and for Bur1 (lower panel). The dashed gray line marks the position where full recruitment of Bur1 is reached ∼250 bp downstream of the TSS.
Abd1 recruitment occurs downstream of Cet1-Ceg1.
ChIP-chip profiling of the third capping enzyme, the methyltransferase Abd1, revealed a peak ∼110 bp downstream of the TSS, about 90 bp further downstream from the peak for Cet1-Ceg1 (Fig. 2A and B, upper panels). This separation of occupancy peaks for Cet1-Ceg1 and Abd1 is highly significant and independent of gene length, gene type, or expression level (Fig. 3; see also Fig. S2A and B in the supplemental material). A separation of occupancy peaks was also visible for the Cet1-Ceg1 dimer and the initiation factor TFIIB, which is located ∼70 bp upstream of Cet1-Ceg1 (Fig. 2A, compare upper and lower panels), a location that was confirmed by nucleotide resolution ChIP-exo data (22). To further demonstrate the resolution of our ChIP-chip data, we provide tracks of example genes (see Fig. S2D). The Abd1 profile is very similar to the profile for S5-phosphorylated Pol II (R = 0.75), and the peaks of both profiles are at the same positions (Fig. 2A and B, upper panels). This is consistent with Abd1 binding to the S5-phosphorylated CTD in vitro (11). These results suggest that cotranscriptional pre-mRNA capping occurs in two steps, triphosphatase/guanylyltransferase action and methylation, and that completion of the cap by methylation is likely a spatially separated event that occurs around 110 bp downstream of the TSS.
Fig 3.

Capping enzyme profile peak distances are independent of expression level and gene length. (A) Gene-averaged profiles for Ceg1 and Abd1 for genes in three expression level classes (for Cet1, see Fig. S2E in the supplemental material). The quality-filtered set of medium length genes (Fig. 2B) was partitioned into three groups: low (25% to 50% quantile), medium (50% to 75% quantile), and high (>75% quantile) expression levels (see Materials and Methods). (B) Box plots showing the gene-wise variation of peak distances between Cet1 and Abd1 for short (725 ± 213 bp; n = 266), medium (1,238 ± 300 bp; n = 339), and long (2,217 ± 679 bp; n = 299) genes. Mean distances are indicated by filled black circles. The distributions of peak distances between Ceg1 and Abd1 are similar (data not shown). For details see Materials and Methods.
Spt5 is involved in Abd1 recruitment genome wide.
We demonstrated that full Abd1 recruitment to the 5′ region of ADH1 and ACT1 requires the Spt5 CTR (Fig. 1A; see also Fig. S1B in the supplemental material). To further investigate whether deletion of the Spt5 CTR has an effect on Abd1 occupancy genome-wide, we carried out ChIP-chip profiling of Abd1 in cells expressing only Spt5 lacking the CTR. We performed metagene analysis and detected a decrease in Abd1 occupancy across the 5′ region of the genes (Fig. 2C, upper panel). Clustering analysis revealed that the effect is of a general nature and is observed at most genes (data not shown). The main differences in Abd1 occupancy between the wild-type and Spt5ΔCTR backgrounds were restricted to a region from upstream of the TSS to ∼250 bp downstream (Fig. 2C, upper panel), consistent with our ChIP data at ADH1 and ACT1 (Fig. 1A; see also Fig. S1B). Bur1 occupancy is low around the TSS and increases further downstream, with peak occupancy also reached ∼250 bp downstream of the TSS (Fig. 2C, lower panel). This is consistent with the model that the Bur1-Bur2 kinase complex promotes release of capping enzymes from Spt5 (Fig. 1B to D; see also Fig. S1C to E). Taken together, our ChIP-chip results indicate that the requirement of Spt5 for full Abd1 recruitment occurs globally and is restricted to the 5′ regions of the genes, where Bur1 occupancy is low.
Recruitment of the cap-binding complex.
The ChIP-chip data of the capping enzymes suggested that the cap structure can generally be completed only around 110 bp downstream of the TSS, where peak occupancies of Abd1 were observed. If this is true, then recruitment of the cap-binding complex (CBC), which binds the complete cap and interacts directly with the N7 methyl group in vitro (54, 55), should occur only from this point on downstream. Indeed, ChIP-chip profiling revealed that the two CBC subunits CBP20 and CBP80 are recruited around 110 bp downstream of the TSS and show full occupancy from 200 to 300 bp downstream onwards (Fig. 2A and B, lower panels; see also Fig. S2A, B, and F in the supplemental material). CBP20 showed higher signals than CBP80 (see Fig. S2F), probably since it contains the cap-binding site. CBC occupancy drops sharply just upstream of the polyadenylation (pA) site. This is consistent with release of the CBC-containing mRNA from DNA after 3′ RNA cleavage that occurs upon polymerase passage over the pA site. These results indicate that direct binding to the complete m7G cap dominates CBC recruitment to active genes in vivo.
Involvement of CBC in recruitment of kinases Bur1 and Ctk1.
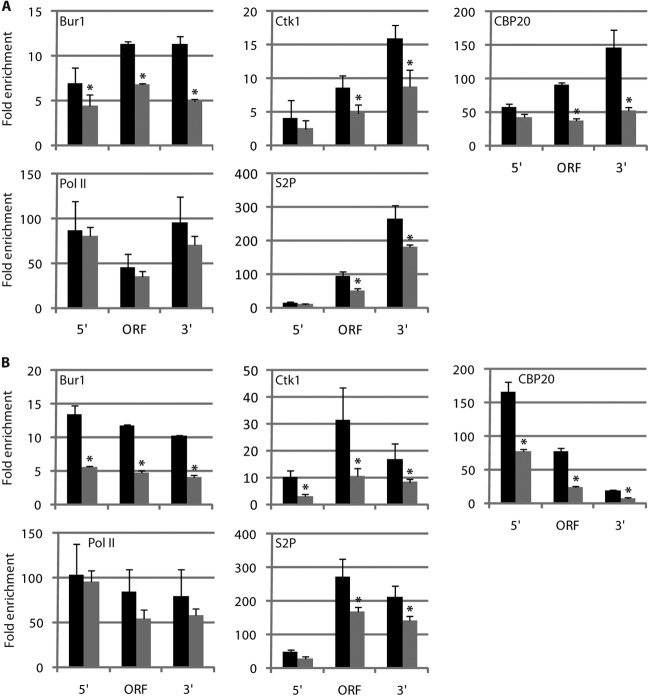
The CBC subunit occupancy profiles resembled those of the CTD kinases Bur1 and Ctk1, which are also released before the pA site (21) (Fig. 2A and B, lower panels). Since binding of these kinases is generally completed 200 to 300 bp downstream of the TSS, we wondered whether cap completion and CBC binding trigger Bur1 and Ctk1 recruitment in vivo. We carried out ChIP analysis of Bur1 and Ctk1 in knockout strains lacking CBP20 (Fig. 4), in which the CBC-cap complex is disrupted (56). In cbp20Δ strains, Bur1 occupancy was decreased up to 50% of wild-type levels throughout the ADH1 and PMA1 genes (Fig. 4) although Bur1 protein levels increased for unknown reasons (see Fig. S4A in the supplemental material). The reduction in kinase occupancy was not due to a difference in Pol II occupancy (Fig. 4), consistent with the finding that the CBC does not affect Pol II processivity (56). In contrast, CBP20 deletion did not affect Ctk1 recruitment to the 5′ region of ADH1 and PMA1 but reduced Ctk1 occupancy in the central and the 3′ regions of the genes (Fig. 4) although Ctk1 protein levels were increased for an unknown reason (see Fig. S4A). Taken together, these results demonstrate that the CBC is required for normal recruitment of the CTD kinase Bur1 to active genes and for maintaining high occupancy of Ctk1 further downstream.
Fig 4.
CBP20 is involved in recruitment of kinases Bur1 and Ctk1. ChIP-qPCR analysis was performed to monitor changes in elongation factor and Pol II recruitment upon CBP20 deletion. Changes in levels of Ser-2 phosphorylated Pol II CTD (S2P) were also analyzed. Three gene regions of ADH1 (A) (Fig. 1) and PMA1 (B) (see Fig. S4B in the supplemental material) were investigated. Occupancies for wild-type (black bars) compared to cbp20Δ (gray bars) cells are shown. Occupancies were calculated as fold enrichments over an ORF-free untranscribed region on chromosome V and are indicated on the y axes (see Materials and Methods). Error bars show standard deviations from at least three independent experiments of biological replicates, and the asterisk indicates factor occupancies that are significantly different (P < 0.05) between the wild-type and mutant conditions using Student's t test.
Role of CBC in CTD S2 phosphorylation and elongation factor recruitment.
Since Bur1 and Ctk1 phosphorylate S2 residues of the Pol II CTD (24, 25), we suspected that CBP20 deletion would affect levels of S2-phosphorylated Pol II and elongation factor recruitment. To test this, we performed ChIP experiments using specific antibodies that recognize the S2-phosphorylated CTD. As expected, deletion of CBP20 led to a 2-fold decrease in S2 phosphorylation levels in the central and 3′ regions of the ADH1 and PMA1 genes (Fig. 4). Also, CBP20 deletion affected downstream occupancy with the Pol II elongation factors Elf1, Spn1, and Spt6 (Fig. 4; see also Fig. S4B in the supplemental material), the last of which binds the S2-phosphorylated CTD in vitro (57). Moreover, genome-wide occupancy of Elf1 and Spn1 correlates with S2 phosphorylation levels (21). Elongation factor Spt4 was reduced at the 3′ region of ADH1 but not PMA1, and elongation factors Spt5 and Spt16 were unaffected (see Fig. S4B to D; also data not shown). Thus, proper Bur1 and Ctk1 recruitment by CBC is crucial for maintaining high levels of S2-phosphorylated Pol II and elongation factors.
Abd1 contributes to Bur1 and Ctk1 recruitment.
To further investigate a role for capping in recruitment of the CTD kinases Bur1 and Ctk1 in vivo, we conditionally depleted the essential Abd1 methyltransferase from the nucleus using the anchor-away (AA) method (44). Abd1 was tagged with the FKBP12-rapamycin binding domain (FRB) and depleted from the nucleus upon rapamycin treatment by its anchoring to FKBP12 fused to the ribosomal protein RPL13A. Strains expressing Abd1-FRB from the endogenous ABD1 promoter grew normally, but rapamycin addition induced a growth defect (see Fig. S5A in the supplemental material). To confirm nuclear depletion of Abd1, we tagged the Abd1-FRB fusion protein with GFP and monitored fluorescence upon rapamycin treatment. Abd1-FRB-GFP was exclusively located in nuclei, and rapamycin treatment led to cytoplasmic fluorescence (see Fig. S5B), as expected.
Nuclear depletion of Abd1 decreased Bur1 occupancy at the ADH1 and PMA1 genes (Fig. 5), as observed upon CBP20 deletion. This suggested that a lower level of cap completion could have led to reduced CBC binding and thus reduced Bur1 recruitment. Consistent with this, CBP20 occupancy was reduced to about 40% throughout PMA1 and in the central and 3′ regions of ADH1 upon nuclear depletion of Abd1 (Fig. 5). Moreover, Abd1 depletion reduced Ctk1 recruitment to the central and 3′ regions of ADH1 and PMA1 by ∼50% (Fig. 5). In addition, Ctk1 occupancy was also decreased at the 5′ region of PMA1 upon Abd1 depletion but was unaffected by CBP20 deletion (compare Fig. 4B and 5B). Changes in ChIP signals did not originate either from differences in Pol II occupancy (Fig. 5) or from rapamycin treatment (see Fig. S6A and B in the supplemental material). Total protein levels of Bur1 and Ctk1 remained unchanged upon Abd1 depletion (see Fig. S6C). Taken together, these results indicate that the cap methyltransferase Abd1 is important for recruitment of CBC and for maintaining normal levels of the CTD kinases Bur1 and Ctk1.
Fig 5.
Abd1 contributes to recruitment of the CBC, Bur1, and Ctk1. ChIP-qPCR analysis was performed to monitor changes in Bur1, Ctk1, CBP20, and Pol II recruitment upon Abd1 nuclear depletion using the anchor-away technique. Changes in levels of Ser-2-phosphorylated Pol II CTD (S2P) were also analyzed. Three gene regions of ADH1 (A) (Fig. 1) and PMA1 (B) (see Fig. S4B in the supplemental material) were investigated. Occupancies in the Abd1 anchor-away strain that was left untreated (black bars) or treated with rapamycin for 60 min (gray bars) are indicated. Occupancies were calculated as fold enrichments over an ORF-free untranscribed region on chromosome V and are indicated on the y axes (see Materials and Methods). Error bars show standard deviations from at least three independent experiments of biological replicates, and the asterisk indicates factor occupancies that are significantly different (P < 0.05) between the treated and untreated conditions using a paired-sample Student's t test.
A global view of initiation-elongation factor exchange.
We carried out a correlation analysis with genome-wide occupancy profiles reported here and elsewhere (21, 23, 51). Pearson correlation coefficients between profiles were provided as a similarity metric to calculate a correlation-based two-dimensional network using the GraphViz's Neato algorithm (49). In the resulting network, elongation factors such as Spt5 and CBC cluster with Pol II, indicating that the elongating form of Pol II dominates its genome-wide occupancy profile (Fig. 6; see also Fig. S3 in the supplemental material). Initiation and termination factors form distinct clusters, reflecting different phases of the transcription cycle. Profiles for the capping enzymes lie between clusters for initiation and elongation factors. This unbiased analysis highlights the central role of pre-mRNA capping in the initiation-elongation transition. Profiles of the Pol II isoforms phosphorylated at S5 and S7 fall between the initiation and elongation clusters, consistent with the role of these modifications in the transition (11–13, 36, 58).
Fig 6.
Correlation-based network of occupancy profiles reflects different phases of the transcription cycle. A correlation analysis of genome-wide ChIP-chip profiles reported here and elsewhere (21, 23, 51) was performed. Pairwise Pearson correlation coefficients were calculated between concatenated gene profiles ranging each from the TSS minus 250 bp to the pA site plus 250 bp (see also Table S1 and Fig. S3 in the supplemental material) and were provided as a similarity metric to calculate a two-dimensional network using the GraphViz's Neato algorithm (see Materials and Methods). Solid lines represent known direct interactions between factors. Dashed lines represent known direct interactions between factors and the phosphorylated CTD of Pol II. Essential and nonessential factors are represented as boxes and circles, respectively. Pol II phosphoisoforms are represented as hexagons.
DISCUSSION
We previously observed that genome-wide ChIP occupancy profiles for yeast transcription initiation and elongation factors are not overlapping, consistent with the exchange of factors during a general initiation-elongation transition (21). Here, we show that this transition and factor exchange involve stepwise recruitment of proteins that form and recognize the RNA 5′ cap structure. We first analyzed the occupancy of cap-forming and cap-binding proteins on active Pol II genes and then investigated how formation of a complete cap triggers the recruitment of the elongation-promoting kinases Bur1 and Ctk1.
In the first part of this work, we found that the RNA triphosphatase Cet1 and the guanylyltransferase Ceg1 have virtually identical occupancy profiles, with sharp peaks ∼20 bp downstream of the TSSs. This is consistent with the existence of a stable Cet1-Ceg1 heterodimer in fungi (53) and the presence of both activities within a single enzyme in metazoa (59). Occupancy for the cap methyltransferase Abd1 was clearly distinct and peaked ∼110 bp downstream of the TSS. This is consistent with earlier studies also showing a significant lag from guanylylation to methylation using a functionally coupled human in vitro transcription/capping system (60, 61). Thus, whereas Cet1 and Ceg1 are obviously recruited in a single step and are present only near the TSS, Abd1 is recruited further downstream and can remain until the 3′ end of a gene, consistent with ChIP data at individual genes (13). Abd1 recruitment apparently leads to cap completion immediately downstream because the cap-binding complex reaches full occupancy levels about 200 to 300 bp downstream of the TSS, supporting the hypothesis that the methylated cap recruits the CBC to nascent mRNA in yeast (56). We further demonstrated that full capping enzyme recruitment requires the CTR of Spt5. Since our genome-wide ChIP-chip data revealed that the main peak for Spt5 occupancy lies downstream of the capping enzyme peaks, we suggest that the capping factors are loaded onto the Pol II machinery first via binding to the S5-phosphorylated CTD (11–13) and that this association is subsequently stabilized by Spt5.
In the second part of our work we found that cap completion is coupled to the recruitment of the kinases Bur1 and Ctk1. These yeast enzymes share homology with mammalian Cdk9, the kinase subunit of the P-TEFb complex that triggers the transition into productive transcription elongation (28–30). Normal Bur1 and Ctk1 recruitment to active genes, and thus proper S2 phosphorylation levels of Pol II, required Abd1 and CBC. The CBC and its effect on CTD S2 phosphorylation were also important for maintaining high levels of elongation factors. Recruitment of Bur1 additionally depends on binding the S5-phosphorylated CTD (24), suggesting that the CBC and the CTD cooperate for Bur1 recruitment. Since Bur1 also phosphorylates the Spt5 CTR (50, 52) and since we show here that the Spt5 CTR contributes to capping enzyme recruitment, Bur1 may help to release capping enzymes from Spt5 after cap completion. This is consistent with our finding that Abd1 occupancy downstream of Bur1 recruitment was unaffected by CTR deletion (Fig. 1A and 2C). In addition, we demonstrated that Bur2 deletion leads to increased Cet1 and Abd1 occupancies along the gene bodies of ADH1, ACT1, and ILV5, further supporting our model.
Our findings agree with published results that showed physical interactions between the capping machinery and early elongation factors (34, 35, 37–40, 43). In addition, a recent study showed that gene recruitment of Bur2 and Ctk2, the cyclin partners of Bur1 and Ctk1, respectively, required the CBC (43). Furthermore, this study demonstrated an effect of CBC deletion on S2 phosphorylation. Our data are consistent with these findings but also provide additional insights that lead to a more complete model for factor exchange during the initiation-elongation transition of Pol II. All available results support a previously proposed capping-dependent checkpoint during early elongation (2, 38, 41, 42).
From these studies emerges the following model for the initiation-elongation transition (Fig. 7). During initiation, the growing RNA transcript triggers initiation factor dissociation, which liberates the Pol II clamp domain for binding Spt5 (6, 7). The S5-phosphorylated CTD stimulates recruitment of capping enzymes to Pol II (11–13), and this association is subsequently stabilized by the CTR of Spt5. When the nascent RNA appears on the Pol II surface, it receives an inverse GMP moiety by the action of the Cet1-Ceg1 heterodimer. Rapid dissociation of Cet1-Ceg1 gives way to the cap methyltransferase Abd1, which is recruited downstream and can remain with partially S5-phosphorylated elongating Pol II. Abd1 and the S5-phosphorylated CTD (24) initially recruit Ctk1 and Bur1, respectively. The cap is completed by methylation and binds CBC, which ensures high occupancy for both Ctk1 and Bur1 when levels of Abd1 and S5 phosphorylation drop downstream. Nascent RNAs with unmethylated cap structures are removed by the Rai1-Rat1 decay pathway (62). Bur1 phosphorylates promoter-proximal CTD S2 residues and the Spt5 CTR, promoting release of capping enzymes. This facilitates recruitment of the Paf complex (24, 50, 52, 63), which establishes histone modifications linked to active transcription (64). Ctk1 phosphorylates S2 residues of the CTD further downstream (24, 25) and stimulates Pol II association with elongation, chromatin-modifying, and RNA-processing factors (57, 65, 66), resulting in a productive elongation complex. Because recent evidence indicates that Bur1 is the P-TEFb (Cdk9) homologue (31), the same events likely occur during the initiation-elongation transition in higher cells.
Fig 7.

Model for the Pol II initiation-elongation transition. Pol II and its CTD (black) transcribe DNA (horizontal black line) from left to right to produce capped mRNA (gray). PIC, preinitiation complex; EC, elongation complex; IIA/B/D/E/F/H, initiation factors; S2/5P, phosphorylated serine 2 and 5 residues; m7, N7 methyl group of the cap; Paf1C, Paf1 complex. For details, see the text.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Sun, A. Schreieck, and S. Etzold for help, K. Sträßer for providing the Ceg1-TAP strain, and J. Söding, A. Tresch, J. Gagneur, H. Feldmann, and A. Mayer for discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB646, SFB960, TR5, CIPSM, NIM, GRK1721, and QBM), the Center for Nanoscience, the Jung-Stiftung, and the Vallee Foundation.
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00361-13.
REFERENCES
- 1.Orphanides G, Reinberg D. 2000. RNA polymerase II elongation through chromatin. Nature 407:471–475 [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G, Reinberg D. 2002. A unified theory of gene expression. Cell 108:439–451 [DOI] [PubMed] [Google Scholar]
- 3.Pokholok D, Hannett N, Young R. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- 4.Pal M, Ponticelli AS, Luse DS. 2005. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19:101–110 [DOI] [PubMed] [Google Scholar]
- 5.Sainsbury S, Niesser J, Cramer P. 2013. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493:437–440 [DOI] [PubMed] [Google Scholar]
- 6.Grohmann D, Nagy J, Chakraborty A, Klose D, Fielden D, Ebright RH, Michaelis J, Werner F. 2011. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell 43:263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P. 2011. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 30:1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perales R, Bentley D. 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36:178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsin J-P, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26:2119–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Program AE, Shuman S, Bentley DL. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komarnitsky P, Cho E, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder SC, Schwer B, Shuman S, Bentley D. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shatkin A. 1976. Capping of eucaryotic mRNAs. Cell 9:645–653 [DOI] [PubMed] [Google Scholar]
- 15.Cowling VH. 2010. Regulation of mRNA cap methylation. Biochem. J. 425:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh A, Lima CD. 2010. Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA 1:152–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwer B, Mao X, Shuman S. 1998. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Res. 26:2050–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonenberg N, Hinnebusch A. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwer B, Shuman S. 1996. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA 2:574–583 [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis J, Izaurralde E. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247:461–469 [DOI] [PubMed] [Google Scholar]
- 21.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272–1278 [DOI] [PubMed] [Google Scholar]
- 22.Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. 2012. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336:1723–1725 [DOI] [PubMed] [Google Scholar]
- 24.Qiu H, Hu C, Hinnebusch AG. 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keogh M, Podolny V, Buratowski S. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahn SH, Kim M, Buratowski S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67–76 [DOI] [PubMed] [Google Scholar]
- 28.Marshall N, Price D. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335–12338 [DOI] [PubMed] [Google Scholar]
- 29.Cheng B, Price DH. 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282:21901–21912 [DOI] [PubMed] [Google Scholar]
- 30.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez J-R, Webb WW, Lis JT. 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 28:1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24:2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51 [DOI] [PubMed] [Google Scholar]
- 33.Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13:720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenasi T, Peterlin BM, Barboric M. 2011. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb). J. Biol. Chem. 286:22758–22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guiguen A, Soutourina J, Dewez M, Tafforeau L, Dieu M, Raes M, Vandenhaute J, Werner M, Hermand D. 2007. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 26:1552–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St Amour CV, Sansó M, Bösken CA, Lee KM, Larochelle S, Zhang C, Shokat KM, Geyer M, Fisher RP. 2012. Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) Rpb1 carboxyl-terminal domain substrate recognition. Mol. Cell. Biol. 32:2372–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen Y, Shatkin AJ. 1999. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13:1774–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei Y, Shuman S. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639–19648 [DOI] [PubMed] [Google Scholar]
- 39.Lindstrom D, Squazzo S, Muster N, Burckin T, Wachter K, Emigh C, McCleery J, Yates J, Hartzog G. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider S, Pei Y, Shuman S, Schwer B. 2010. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell. Biol. 30:2353–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei Y, Schwer B, Shuman S. 2003. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 278:7180–7188 [DOI] [PubMed] [Google Scholar]
- 42.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 101:7572–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hossain MA, Chung C, Pradhan SK, Johnson TL. 2013. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol. Cell. Biol. 33:785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 31:925–932 [DOI] [PubMed] [Google Scholar]
- 45.Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K. 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. Chapter 21:Unit 21.3. 10.1002/0471143030.cb1707s23 [DOI] [PubMed] [Google Scholar]
- 46.Affymetrix 2011. User guide: Affymetrix chromatin immunoprecipitation assay protocol. Affymetrix, Inc., Santa Clara, CA [Google Scholar]
- 47.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dengl S, Mayer A, Sun M, Cramer P. 2009. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J. Mol. Biol. 389:211–225 [DOI] [PubMed] [Google Scholar]
- 49.Gansner E, North S. 1998. Improved force-directed layouts. Graph Drawing 1547:364–373 [Google Scholar]
- 50.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29:4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P. 2012. The Spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol. Cell. Biol. 32:1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou K, Kuo WHW, Fillingham J, Greenblatt JF. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U. S. A. 106:6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu M, Rajashankar K, Lima C. 2010. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 18:216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. 2002. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 9:912–917 [DOI] [PubMed] [Google Scholar]
- 55.Mazza C, Segref A, Mattaj IW, Cusack S. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21:5548–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong C-M, Qiu H, Hu C, Dong J, Hinnebusch AG. 2007. Yeast cap binding complex impedes recruitment of cleavage factor IA to weak termination sites. Mol. Cell. Biol. 27:6520–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun M, Larivière L, Dengl S, Mayer A, Cramer P. 2010. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J. Biol. Chem. 285:41597–41603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czudnochowski N, Bösken CA, Geyer M. 2012. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 3:842. 10.1038/ncomms1846 [DOI] [PubMed] [Google Scholar]
- 59.Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 273:9577–9585 [DOI] [PubMed] [Google Scholar]
- 60.Chiu Y-L, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10:585–597 [DOI] [PubMed] [Google Scholar]
- 61.Moteki S, Price D. 2002. Functional coupling of capping and transcription of mRNA. Mol. Cell 10:599–609 [DOI] [PubMed] [Google Scholar]
- 62.Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu H, Hu C, Gaur NA, Hinnebusch AG. 2012. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 31:3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799:379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430:223–226 [DOI] [PubMed] [Google Scholar]
- 66.Kizer K, Phatnani H, Shibata Y, Kizer KO, Phatnani HP, Hall H, Greenleaf AL, Strahl BD. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25:3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.