Abstract
Type I interferons (IFNs) stimulate transcription through a latent heterotrimeric transcription factor composed of tyrosine-phosphorylated STAT1 and STAT2 and the DNA binding partner IRF9, with STAT2 contributing a critical transactivation domain. Human RVB1 and RVB2, which are highly conserved AAA+ ATP binding proteins contained in chromatin-remodeling complexes such as Ino80, SNF2-related CBP activator protein (SRCAP), and Tip60/NuA4, interacted with the transactivation domain of STAT2 in the nuclei of IFN-stimulated cells. RNA interference (RNAi) experiments demonstrated that RVB proteins were required for robust activation of IFN-α-stimulated genes (ISGs). The requirement for RVB proteins was specific to IFN-α/STAT2 signaling; transcription of tumor necrosis factor alpha (TNF-α)- and IFN-γ-driven genes was not affected by RVB1 depletion. Using RNAi-based depletion, we assessed the involvement of catalytic subunits of the RVB-containing Tip60, BRD8, Ino80, SRCAP, and URI complexes. No component other than RVB1/2 was uniquely required for ISG induction, suggesting that RVB1/2 functions as part of an as yet unidentified complex. Chromatin immunoprecipitation assays indicated that RVB1/2 was required for recruitment of RNA polymerase II (Pol II) to ISG promoters but was dispensable for STAT2 recruitment to chromatin. We hypothesize that an RVB1/2 chromatin-remodeling complex is required for efficient Pol II recruitment and initiation at ISG promoters and is recruited through interaction with the STAT2 transactivation domain.
INTRODUCTION
Interferons (IFNs) are a family of pleiotropic cytokines primarily known for their ability to establish a potent antiviral state via modulation of the Janus kinase-signal transducer and activator of transcription (JAK-STAT) signal transduction pathway. Alternatively, aberrant production of IFNs correlates with systemic autoimmunity, underscoring the importance of the stringent regulation of IFN responses. Type I IFNs activate the heterotrimeric transcription factor ISGF3, composed of tyrosine-phosphorylated STAT1 and STAT2 and an auxiliary DNA binding protein, IRF9. Following IFN stimulation, phosphorylated ISGF3 translocates into the nucleus, binds the interferon-stimulated response elements (ISREs) in the promoters of IFN-α/β-stimulated genes (ISGs), and rapidly and robustly induces the transcription of a large family of previously silent genes. ISGs subsequently influence multiple cellular pathways involved in innate immunity in order to protect against viral and bacterial replication, including modulation of cell stress responses, apoptosis, proliferation, translation, and innate and adaptive immune signaling (1). In the context of ISGF3, STAT2 contains an essential transactivation domain (TAD) that provides most of the transcriptional function to the transcription factor complex, whereas STAT1 and IRF9 confer DNA sequence specificity (2). Induction of IFN target genes provides a robust system for studying the molecular mechanisms underlying transcriptional regulation. However, little is known concerning the molecular mechanisms underpinning the action of the STAT2 transactivation domain.
The transcription of target genes by RNA polymerase II (Pol II) is preceded by an ordered, multistep cascade of events that includes histone modification and chromatin remodeling, often initiated following binding of a site-specific transcription factor. This process is thought to facilitate signal-dependent access of Pol II to regulatory sequences. ATP-dependent chromatin-remodeling complexes that use the energy of ATP hydrolysis to mobilize nucleosomes and factors that influence chromatin compaction by covalently modifying histone tails are believed to play an essential role in the molecular switch that regulates the rapid induction of genes in the presence of cytokines (3). It is hypothesized that the C-terminal TAD of STAT2 serves as a platform for coactivator assembly. STAT2 has been shown to interact with the histone acetyltransferase (HAT) p300, and STAT2-mediated transcription depends on a GCN5-containing complex that lacks TATA box binding protein (TBP), suggesting that IFN-stimulated transcription can proceed in the event of virus-targeted degradation of TBP (4, 5). The recruitment of HATs is expected to activate genes through histone acetylation and nucleosome displacement, similar to the process required to activate the IFN-β locus in response to virus infection (6). Additionally, a subset of ISGs depends on the Brahma-related gene 1 (BRG1) subunit of the mammalian SWI-SNF chromatin-remodeling complex, which acts as a molecular motor to alter chromatin accessibility at ISG promoters (7–9). Recent studies have revealed further complexity by demonstrating the activating role of histone deacetylases (HDACs), rather than the more canonical repressive one, in the context of IFN-stimulated transcription, via unelucidated mechanisms (10, 11).
In an effort to develop a better understanding of how the specificity of type I IFN-stimulated transcription is dictated, we utilized an affinity purification strategy to isolate novel STAT2-interacting proteins from nuclear extracts of IFN-α-treated cells. Here we report the identification of RVB1 and RVB2 (also known as Tip49 and Tip48, pontin52 and reptin52, or Tih1p and Tih2p), two putative ATPases that are highly conserved in eukaryotes, as STAT2 interactors and coactivators. RVB1 and RVB2 interacted with the STAT2 transactivation domain and were essential for robust induction of ISGs. Metazoan RVBs have been identified before as components of both ATPase-type chromatin-remodeling complexes (Ino80 and SNF2-related CBP activator protein [SRCAP]) and the lysine acetyltransferase human complex Tip60 (12); however, their exact molecular function is incompletely understood. We provide evidence that human RVBs are required specifically for successful initiation of type I IFN-stimulated transcription, as part of an as yet unknown complex.
MATERIALS AND METHODS
Cell culture, transfection with siRNAs, plasmids, and cytokine treatments.
HeLa, HEK293, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium containing 7% calf serum. Cells were transfected by using the calcium phosphate method, and cellular extracts were collected for luminescent luciferase and β-galactosidase assays, as previously described (4). Luciferase assays for ISG54-luc were performed in triplicate, and results were normalized to cotransfected β-galactosidase activity. For transfection with small interfering RNAs (siRNAs), HEK293 cells were grown in 6-well plates and transfected at 24-h intervals with 20 nM annealed siRNA duplexes by the calcium phosphate method. Five hours after the first transfection, fresh medium was added. Forty-eight hours after the first transfection, cells were split 1:3. Ninety-six hours after the first transfection, cells were harvested and processed for Western blotting and reverse transcription-quantitative PCR (RT-qPCR). Flag-RVB1 and HA-RVB2 plasmids were a kind gift from Anindya Dutta (University of Virginia). Cells were treated with IFN-α2a (Hoffman-La Roche) at 1,000 units/ml, IFN-γ (Amgen Biologicals) at 5 ng/ml, tumor necrosis factor alpha (TNF-α) (NIH) at 10 ng/ml, and doxycycline (Sigma-Aldrich) at 1 μg/ml. Anacardic acid was obtained from Calbiochem.
Western blotting and immunoprecipitation.
For Western blotting, cells were lysed in ELB buffer (50 mM HEPES [pH 7.0], 250 mM NaCl, 5 mM EDTA, 0.1% [vol/vol] NP-40, 1 mM dithiothreitol, and a protease inhibitor mixture [Sigma-Aldrich]). Nuclear extracts were prepared using the protocol of Dignam et al. (13). All membranes were scanned using a LiCor Odyssey system, except for that in Fig. 5C, which was generated by film-based enhanced chemiluminescence (ECL). All quantifications were performed with LiCor Odyssey software.
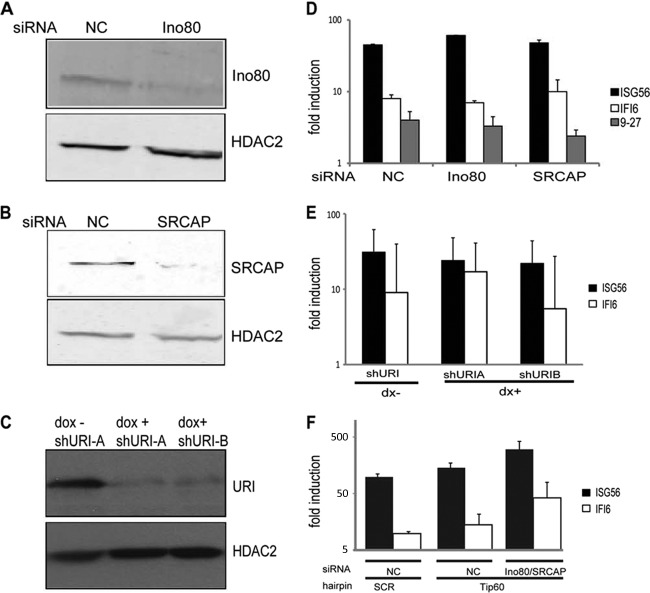
Fig 5.
Ino80, SRCAP, and URI are not essential for type I IFN-stimulated transcription. (A and B) Nuclear proteins prepared from HEK293 cells transfected with the indicated siRNAs were immunoblotted for Ino80 (A) or SRCAP (B) and HDAC2 (loading control). (C) HEK293 cells carrying two independent inducible hairpins against URI were treated with doxycycline for 72 h or left untreated. The lysates were blotted for URI and HDAC2. (D) Cells were transfected with the indicated siRNA duplexes and treated with IFN-α2a for 8 h before isolation of RNA. Expression of ISG56, IFI6, 9-27, and actin mRNAs was quantified by real-time PCR and is presented as the fold induction over uninduced levels. The averages for three experiments are shown. (E) The HEK293 cells described for panel C were incubated with IFN-α2a for 8 h before RNA isolation. Expression of ISG56, IFI6, and actin was quantified by real-time PCR and is presented as the fold induction over uninduced levels before and after URI depletion. The averages for three experiments are shown. (F) HEK293 cells were infected with the indicated hairpin, transfected with the indicated siRNA duplexes, and treated with IFN-α2a for 8 h before isolation of RNA. Expression of ISG56, IFI6, and actin mRNAs was quantified by real-time PCR and is presented as the fold induction over uninduced levels. The averages for three experiments are shown.
RT-qPCR.
RNA was isolated and converted to cDNA by use of Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) according to the manufacturer's instructions. Relative abundances of specific mRNA sequences were determined by real-time fluorescent PCR with SYBR green (Molecular Probes) by comparison with a standard curve generated by serial dilution of a cDNA sample containing abundant target sequences and normalized to the expression of β-actin.
Quantitative ChIP.
Ten million cells were treated with 1% formaldehyde for 10 min at room temperature and lysed in SDS-containing buffer. Cell extracts were sonicated to sheer DNA to ≈500 bp and then immunoprecipitated with antibodies; the recovered chromatin was digested with proteinase K, treated at 65°C, and purified on ion-exchange spin columns. The presence of specific immunoprecipitated DNA sequences was quantified by real-time PCR compared with input genomic DNA. For STAT2 and total RNA Pol II chromatin immunoprecipitation (ChIP) assays (see Fig. 6A and B), data were normalized by subtracting the IgG control % input. For Ser5-phosphorylated Pol II (S5P-PolII) assays and acetylated H4 (AcH4) and H3 assays (see Fig. 6C and D), the IgG control was 0.03 to 0.05% input.
Fig 6.
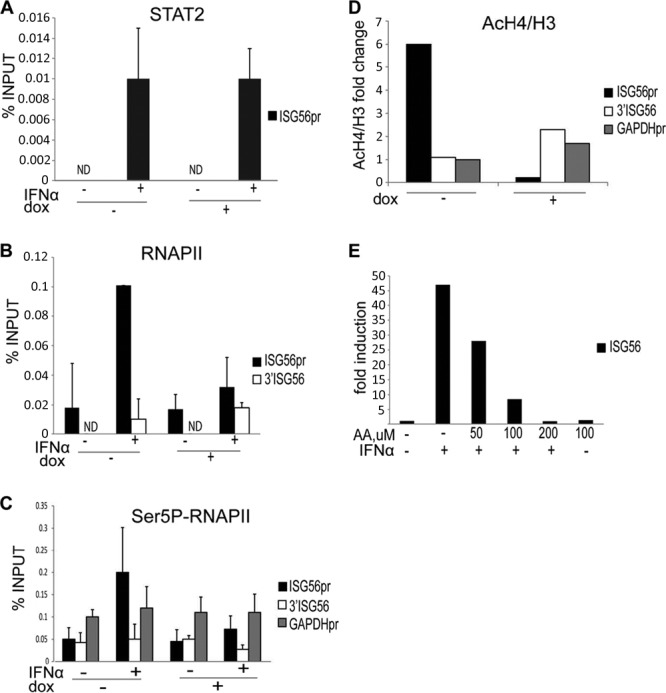
RVB1 is dispensable for STAT2 but required for RNA polymerase II recruitment to the ISG56 promoter. (A) HEK293 cells carrying an inducible hairpin against RVB1 were treated with doxycycline for 72 h or left untreated. Cells were then incubated with IFN-α2a for 45 min or left untreated, as indicated, and processed for ChIP assays. STAT2 antibody-precipitated ChIP DNA and input material were used to quantify the ISG56 promoter. (B) Same as in panel A, but a total Pol II antibody was used for ChIP assays to examine both the ISG56 promoter and the intragenic region downstream of ISG56. (C) Same as in panel A, but an antibody against the phosphorylated serine 5 Pol II CTD was used for ChIP assays. The GAPDH promoter was quantified as a positive control. Data are presented as percentages of input material. Data from one of three independent experiments are shown. Error bars show data ranges. (D) Same as in panel A, but antibodies against acetylated H4 and H3 were used for ChIP assays. AcH4/H3 ratios are presented as fold changes of IFN-treated over untreated samples before and after RVB1 depletion. Data from one of two independent experiments are shown. (E) HEK293 cells were treated with the indicated amount of anacardic acid (AA) and/or IFN for 8 h before isolation of RNA. Expression of ISG56 and actin mRNAs was quantified by real-time PCR, and normalized data are presented as the fold induction over uninduced levels. Data from one of three independent experiments are shown.
Antibodies.
Rabbit anti-RVB1, -Ino80, and -SRCAP antibodies were a kind gift from Anindya Dutta (University of Virginia). Tip60 antibody was a gift from Bruno Amati (Italian Institute of Technology). URI antibody was a gift from Susan Logan (NYU School of Medicine). Anti-RVB2 antibody was a gift from Yixian Zheng (Carnegie Institution of Washington). Other antibodies used were as follows: M2 anti-Flag (Sigma-Aldrich), antihemagglutinin (anti-HA) (12CA5), anti-STAT2 (sc-476; Santa Cruz), mouse anti-RVB1 (ab51500; Abcam), anti-pSTAT2, -pSTAT1, and -IRF9 (Zymed, San Francisco, CA), anti-HDAC2, clone 3F3 (05-814; Millipore), anti-Pol II (N-20) (sc-899; Santa Cruz), anti-Ser5P-PolII[4H8] (ab5408; Abcam), anti-acetyl-histone H4 (06-866; Millipore), anti-histone H3 (ab1791; Abcam), anti-E2F4 (sc-1082; Santa Cruz), and anti-α-tubulin (T9026; Sigma-Aldrich).
RNA interference (RNAi).
The siRNA sequences used in this study were as follows: NC, TTCTCCGAACGTGTCACGTTT; RVB1-A, TAAAGGAGACCAAGGAAGT; RVB1-B, TCTTCTCTCTCTCTCTCTA; INO80, TGACCTGCGTCTACACTTA; SRCAP, GGAAACGATTGAAGTTGAA; and TIP60, TGATCGAGTTCAGCTATGA (all purchased from Shanghai GenePharma). An siGENOME SMARTpool against URI was purchased from Dharmacon.
For lentiviral short hairpin RNA (shRNA) vectors, previously described hairpin sequences for RVB1 and RVB2 were cloned into the pGIPZ vector from Open Biosystems (14, 15). Another shRNA against RVB1, identified by the A. Dutta group (16), was cloned into the doxycycline-inducible pTRIPZ vector from Open Biosystems. Lentiviruses were generated by cotransfecting plasmids pMD2.G (vesicular stomatitis virus glycoprotein [VSV-G]) and psPax2 (packaging vector) and shRNA plasmids into HEK293T cells by using the calcium phosphate precipitation method. pTRIPZ shRNAs against URI (RHS4696-99705821 V2THS_33824 and RHS4696-99709379 V2THS_23229) were obtained from Open Biosystems.
pLKO.1 hairpins against Tip60 (TRCN0000020314) and BRD8 (TRCN0000019380 and TRCN0000019381) were from Sigma-Aldrich and were provided by the NYU School of Medicine shRNA Core Facility.
STAT2 complex purification and mass spectrometry.
HeLa S3 cells retrovirally transduced with pLPC-N-Flag-2HA-WT-STAT2 were grown at 5 × 105 cells/ml in spinner flasks in Joklik's minimum essential medium (MEM) supplemented with 5% calf serum and antibiotics. Five-liter cultures were treated overnight with IFN-γ at 1 ng/ml and then were spun down, resuspended in 50 ml of medium, and treated further with IFN-α2a for 45 min. Subsequently, nuclear extracts were isolated using the protocol of Dignam et al., and Flag-STAT2 was purified using M2 anti-Flag affinity gel (Sigma-Aldrich) and eluted with the Flag peptide followed by further purification by use of anti-STAT2 affinity columns. Beads were washed with the following washing buffer: 20 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 0.1% Tween, 10 mM 2-mercaptoethanol, and 0.25 mM phenylmethylsulfonyl fluoride (PMSF). Nonreducing Laemmli buffer (Boston BioProducts) was used for elution at room temperature, followed by fractionation in an SDS-PAGE gel, fixation, and staining with Coomassie blue. Five gel regions corresponding to a broad molecular size range were excised from the lane containing STAT2-interacting polypeptides and submitted for sequencing to the Taplin Mass Spectrometry Facility at Harvard University. A complete list of identified factors is available in the supplemental material.
RESULTS
Gene expression analysis of HeLa cells carrying tagged STAT2.
The human STAT2 open reading frame was cloned into the retroviral vector pLPC-N-Flag-2HA and transduced into HeLa cells to generate stable cell lines expressing STAT2 containing both Flag and HA epitopes at its amino terminus. Additionally, we created a similar cell line with truncated STAT2 lacking the transactivation domain (TAD) and known to be transcriptionally incompetent (17). Whole-cell extracts from these cell lines were immunoblotted for total and phosphorylated STAT2. Comparison of the slower-migrating band corresponding to Flag-tagged STAT2 to the band for the slightly smaller wild-type STAT2 for the cells carrying the empty vector demonstrated that in IFN-α-treated cells, both total and phosphorylated tagged STAT2 was overproduced 6- to 7-fold (Fig. 1A). Moreover, Flag-WTSTAT2-expressing cells exhibited an approximately 20% increased induction of ISG56 mRNA, indicating that cells with overexpressed STAT2 display normal but somewhat enhanced induction of a typical ISG (Fig. 1B). Interestingly, the expression of truncated STAT2 severely blunted ISG56 induction (Fig. 1B), suggesting that STAT2ΔTAD acts as a dominant negative inhibitor.
Fig 1.
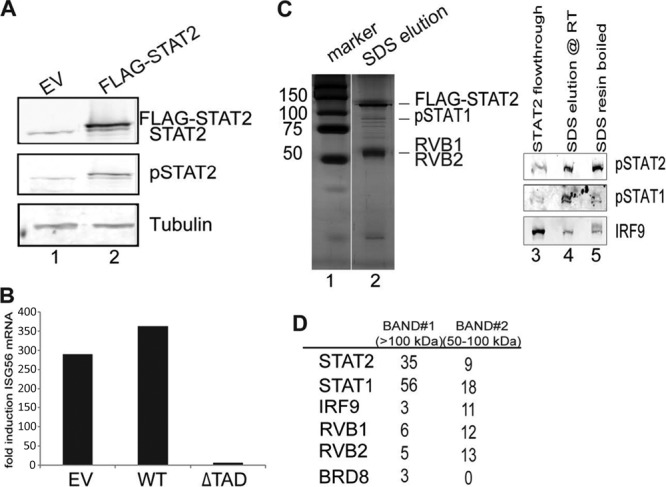
Purification and mass spectrometric analysis of polypeptides associated with STAT2. (A) Whole-cell lysates prepared from IFN-α2a-treated (45 min) cells expressing either empty vector (EV) or wild-type STAT2 were immunoblotted for STAT2, pY690-STAT2, and tubulin (loading control). (B) Cells carrying EV, wild-type STAT2, or STAT2 lacking its TAD (ΔTAD) were treated with IFN-α2a for 8 h before isolation of RNA. Expression of ISG56 relative to that of actin was quantified by real-time PCR and is presented as the fold induction over the uninduced level. (C) Nuclear extracts from HeLa cells expressing Flag-STAT2 were sequentially immunoprecipitated with Flag and STAT2 antibody affinity resins. Lanes 1 and 2, STAT2-associated polypeptides were detected by silver staining; lanes 3 to 5, ISGF3 components were detected by Western blotting. The eluate was analyzed by mass spectrometry. (D) Numbers of unique peptides of selected proteins from the indicated size ranges obtained by mass spectrometry.
Purification and mass spectrometric analysis of polypeptides associated with STAT2.
To define human STAT2-interacting proteins, HeLa S3 cells expressing Flag-STAT2 were grown in suspension cultures, and nuclear extracts of IFN-α-treated cells were subjected to sequential purification by anti-Flag and anti-STAT2 immunopurification as described in Materials and Methods. Multiple peptides were found to copurify with tagged STAT2 (Fig. 1C, lanes 1 and 2). The pattern of STAT2-interacting peptides differed from that for bead-only- and anti-E2F4 antibody-purified polypeptides (data not shown), indicating a degree of specificity. All the components of ISGF3 were also detected by immunoblotting of the final eluate (Fig. 1C, lanes 3 to 5). After staining with Coomassie blue, five bands were excised according to their molecular size, and the identities of STAT2-interacting polypeptides were determined by mass spectrometry (see the supplemental material). After eliminating factors known to nonspecifically interact with agarose beads, such as myosin and DEAD box proteins (18), and those with no known connection to transcription-related processes, we narrowed the list of candidates to the ones shown in Fig. 1D. All the known components of ISGF3 were identified, indicating that the purification of STAT2 was successful. Three additional candidate factors were also identified, namely, RVB1, RVB2, and BRD8 (TRCp120), which are known components of the human Tip60 complex (19). However, Tip60 itself was not recovered in the STAT2 complex. The TRCp120 protein (thyroid receptor-coactivating protein p120) was originally identified in a yeast two-hybrid screen as a thyroid hormone receptor-interacting protein that is capable of stimulating thyroid hormone receptor-dependent transcription (20). RVB1 and RVB2 are putative ATP-dependent helicases that have been linked extensively to transcriptional regulation (12).
STAT2 interacts with RVB1 and RVB2.
To confirm a physical interaction between RVBs and STAT2, we performed immunoprecipitation experiments with the HeLa cell lines described above and used for Fig. 1A. Using anti-Flag antibodies, endogenous RVB1 and RVB2 were coimmunoprecipitated with Flag-STAT2 from nuclear extracts of IFN-stimulated cells but not with STAT2 lacking the TAD (Fig. 2A), indicating that the interaction depended on the STAT2 transactivation domain. Thus, we concluded that RVB proteins are contained in a STAT2 complex in IFN-treated cells. We further tested whether the STAT2-RVB interaction is IFN dependent. We immunoprecipitated Flag-STAT2 from the cytoplasmic fraction of HeLa cells before and after treatment with IFN-α2a. As shown in Fig. 2B, endogenous RVB1 and RVB2 coprecipitated with STAT2 regardless of IFN treatment. As seen before, Flag-STAT2 interacted with RVB proteins in the nuclear fraction of IFN-treated cells. Detection of HDAC2 was used to verify a good separation between the nucleus and the cytoplasm. This result suggests that RVB proteins interact constitutively with STAT2 such that the STAT2-RVB interaction is not IFN dependent.
Fig 2.
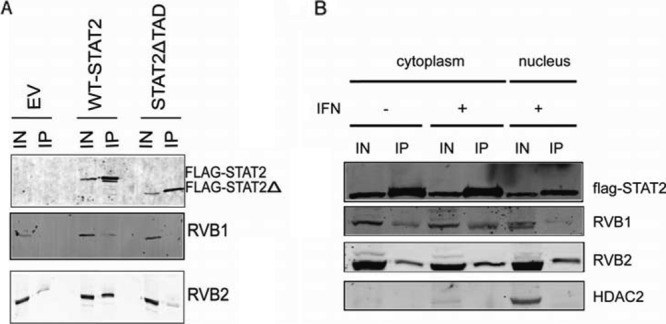
Flag-STAT2 interacts with RVB1 and RVB2 in vivo. (A) Nuclear extracts from IFN-α2a-treated HeLa cells stably expressing either wild-type or truncated Flag-STAT2ΔTAD were used to immunoprecipitate (IP) Flag-STAT2 and subjected to Western blotting using antibodies against Flag, RVB1, and RVB2. Input lanes (IN) represent ∼1% of the immunoprecipitated material. In a separate IP, Flag-STAT2ΔTAD was shown to interact with pSTAT1 and IRF9 under these conditions (data not shown). The individual panels shown were obtained from independent blots. (B) Cytoplasmic and nuclear extracts from untreated and IFN-α2a-treated HeLa cells stably expressing wild-type Flag-STAT2 were used to immunoprecipitate Flag-STAT2 and subjected to Western blotting using antibodies against Flag, RVB1, RVB2, and HDAC2. Each panel was obtained from an independent blot.
Loss of RVB1 attenuates transcription of ISGs.
Based on the interaction data, we asked whether RVB1 deficiency would affect the induction of ISGs. We also wanted to test the role of Tip60, because this complex contains RVB proteins and RVB1 is required for the acetyltransferase activity of the Tip60 complex (16, 21–23). In addition, we identified one of the human Tip60 complex components, BRD8, as a STAT2-interacting protein, further implicating a potential role for this complex, although the Tip60 catalytic component itself was not detected (Fig. 1D). Two siRNA duplexes targeting different parts of RVB1 were transfected into HEK293 cells, resulting in reductions of RVB1 protein expression of approximately 60 to 80%, whereas duplexes against Tip60 reduced its abundance ∼50% (Fig. 3A). Additionally, Tip60 appeared to be destabilized by RVB1 depletion, although Tip60 knockdown did not perturb the levels of RVB1. Levels of endogenous RVB1 and Tip60 protein expression were unperturbed by IFN treatment. The induction of two representative ISGs, ISG56 and IFI6, in response to IFN stimulation was attenuated in RVB1-depleted cells (Fig. 3B). It should be noted that we did not observe any effect on basal expression of ISGs, suggesting that RVB1 is selectively required for induced transcription. In contrast, depletion of Tip60 did not impair ISG induction. To further evaluate the lack of involvement of Tip60, we depleted its expression with a lentiviral shRNA targeting construct. Again, depletion of Tip60 did not affect ISG induction (Fig. 3D and E).
Fig 3.
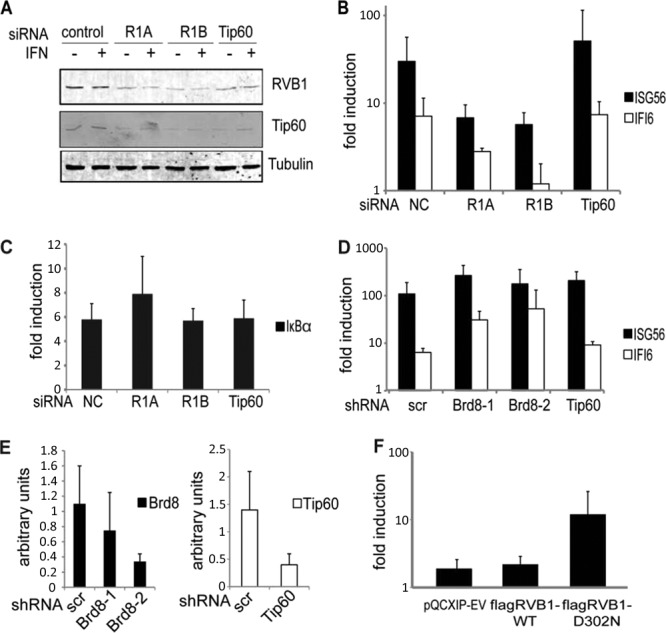
Depletion of RVB1 attenuates type I IFN-stimulated transcription but not TNF-α-stimulated transcription. (A) Whole-cell lysates prepared from HEK293 cells transfected with the indicated siRNAs were immunoblotted for RVB1, Tip60, and tubulin. (B) Cells were transfected with the indicated siRNA duplexes and treated with IFN-α2a for 8 h before isolation of RNA. Expression of ISG56, IFI6, and actin mRNAs was quantified by real-time PCR, and normalized data are presented as the fold induction over uninduced levels. (C) Same as in panel B, but HEK293 cells were treated with TNF-α for 4 h and IκBα mRNA was quantified by real-time PCR. (D) Lentiviruses carrying hairpins against BRD8 and Tip60 were transduced into HEK293 cells. Expression of ISG56, IFI6, and actin mRNAs was quantified by real-time PCR after 8 h of treatment with IFN-α2a, and normalized data are presented as the fold induction over uninduced levels. (E) Real-time PCR quantification of BRD8 and Tip60 knockdown relative to actin levels. (F) HEK293 cells transfected with ISG54-luciferase, the cytomegalovirus (CMV) LacZ gene, and the indicated constructs were treated with IFN-γ overnight and with IFN-α for 7 h. Cell extracts were analyzed for luciferase activity, and data were normalized to β-galactosidase activity and reported as the fold induction over uninduced levels. Data shown are means with standard deviations (SD) obtained from two independent experiments, each performed in triplicate.
To determine the specificity of the requirement for RVB1 for transcriptional responses to IFN, we evaluated the induction of the TNF-α-induced gene IκBα in cells in which RVB1 and Tip60 were depleted using the same siRNA duplexes. As shown in Fig. 3C, depletion of neither RVB1 nor Tip60 had any discernible effect on IκBα expression. TNF-α activates a distinct pathway, resulting in activation and nuclear translocation of the p50/p65 NF-κB heterodimer, rather than using STAT proteins (24). Taken together, these data suggest that RVB1 is specifically employed by STAT2 for induction of target genes but not by NF-κB, while Tip60 is not essential for either IFN- or TNF-stimulated transcription.
We also used two hairpins against BRD8, another component of the Tip60 complex (Fig. 3E), to test the role of BRD8, which was identified as a STAT2-copurifying protein (Fig. 1D). As shown in Fig. 3D, BRD8 depletion did not impair the induction of ISG56 or IFI6 in response to IFN. In contrast, BRD8 depletion appeared to modestly enhance induction of IFN-stimulated transcripts, while Tip60 depletion showed a similar, though smaller, enhancement. From these data, we concluded that neither Tip60 nor BRD8 is a required positive regulator for type I IFN-driven transcription, although these factors may serve as negative modulators of ISG induction.
Since RVB proteins appear to act as DNA helicases in some but not all contexts (25, 26), we were interested in testing whether the helicase activity of RVB1 was required during IFN-stimulated transcription. A conserved aspartic acid residue in the Walker B motif in RVB1 was mutated to asparagine (RVB1-D302N). The equivalent mutation was previously shown to inactivate the RVB1 protein in vivo (25) and to inhibit the ATPase activity of bacterial RVB (27). The effects of wild-type and mutated RVB1 on IFN-stimulated transcription were tested by using ISG promoter-transient reporter assays (2). Cells were transfected with ISG54-luciferase with or without wild-type or mutant RVB1. We found that IFN-stimulated promoter activity was not affected by overexpression of wild-type RVB1, whereas the presence of mutated RVB1 augmented reporter activity (Fig. 3F). Thus, we conclude that the helicase activity of RVB1 is probably not required for its ability to cooperate with STAT2 for ISG transcription. The increased expression of the IFN-stimulated reporter in the presence of mutated RVB1 is reminiscent of the effects observed following Tip60 and BRD8 depletion (Fig. 3D). We speculate that ectopic expression of mutated RVB1 could have a dominant negative effect on processes requiring helicase activity, such as the integrity of the Tip60 complex, and thus might mimic the effects of Tip60 reduction. As noted previously, Tip60 abundance is dependent on RVB expression (though not vice versa), since Tip60 levels were reduced in concert with RVB depletion (Fig. 3A).
To further assess the role of RVB1 and reduce the possibility of off-target effects of RNAi-mediated depletion, we tested depletion by use of lentiviral shRNA constructs. Short hairpin sequences against RVB1 and RVB2 (15, 16) were cloned either into a doxycycline-inducible short hairpin vector, pTRIPZ, or into a constitutively expressed construct, pGIPZ (OpenBiosystems). As shown in Fig. 4A, the inducible hairpin very efficiently silenced RVB1 expression in response to doxycycline treatment. Similar to siRNA duplexes, this hairpin impaired the induction of several ISGs, including both Brg1-dependent (9-27) and Brg1-independent (ISG15) genes, indicating that BAF and RVB dependence do not correlate (Fig. 4C). To further examine the specificity of RVB1's role, we tested the induction of IFN-γ-stimulated transcripts. IFN-γ activates primarily STAT1 homodimers and does not depend on STAT2 (1). As demonstrated in Fig. 4D, the induction of two IFN-γ-stimulated transcripts, IRF1 and IP10, was unaffected by RVB1 knockdown. This result provides additional validation to the hypothesis that the RVB1 requirement is unique to the type I IFN-STAT2 signaling pathway. Additional confirmation of the requirement for RVB1 and RVB2 is presented in Fig. 4. Two independent hairpins efficiently depleted RVB1 expression, and an RVB2 hairpin partially depleted RVB2 protein levels (Fig. 4B). As reported previously (15), RVB2 knockdown codepleted the RVB1 protein (Fig. 4B), indicating that these two related factors depend on their association for stability. Depletion of RVB protein levels by use of these hairpins significantly impaired ISG54, ISG56, and IFI6 gene expression (Fig. 4E). Although impaired gene expression following RVB2 knockdown may be due, at least in part, to loss of RVB1, these data demonstrate that RVB1 and RVB2 are interdependent and are required for robust IFN-α-stimulated transcription.
Fig 4.
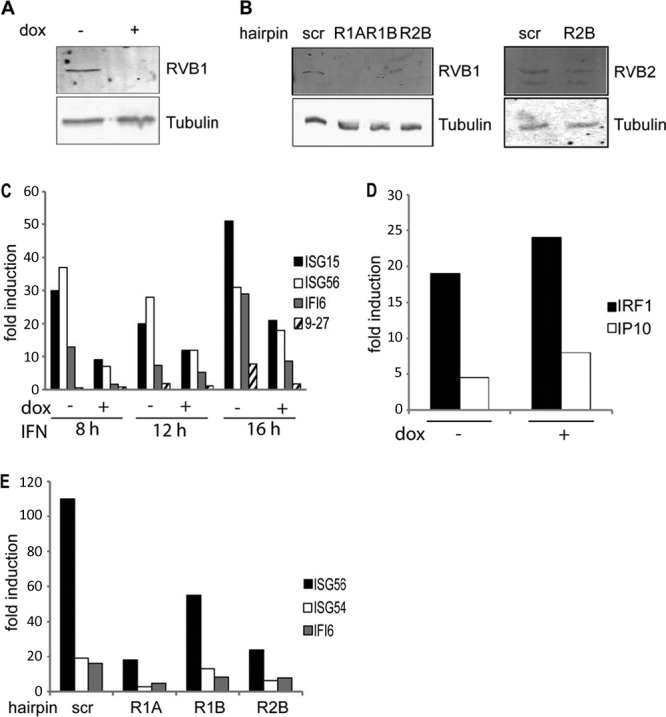
shRNAs against RVB1 and RVB2 reduce type I IFN- but not type II IFN-stimulated gene expression. (A) HEK293 cells carrying an inducible hairpin against RVB1 were treated with doxycycline (dox) for 72 h or left untreated. The lysates were blotted for RVB1 and tubulin. (B) Lentiviruses carrying two independent hairpins against RVB1 and one against RVB2 were transduced into HEK293 cells. Whole-cell extracts were blotted for RVB1, RVB2, and tubulin. scr, scrambled hairpin. (C) The HEK293 cells described for panel A were incubated with type I IFN for the indicated numbers of hours before RNA isolation. Expression of ISG15, ISG56, IFI6, 9-27, and actin was quantified by real-time PCR, and normalized data are presented as the fold induction over uninduced levels before and after RVB1 depletion. Data representative of three independent experiments are shown. (D) Same as in panel C, but cells were treated with IFN-γ for 8 h before RNA isolation. IRF1, IP10, and actin were quantified by real-time PCR, and normalized data are presented as the fold induction over uninduced levels. Data from one of two independent experiments are shown. (E) The cells described for panel B were treated with IFN-α2a for 8 h before RNA isolation. Expression of ISG54, ISG56, IFI6, and actin was quantified by real-time PCR, and normalized data are presented as the fold induction over uninduced levels.
Knockdown of Ino80, SRCAP, or URI does not affect ISG induction.
RVB1 and RVB2 have been identified as components of a number of distinct multiprotein complexes that could be considered candidates to mediate the requirement for RVB proteins by STAT2. In an attempt to discriminate among these complexes, catalytic subunits of known RVB-containing complexes were depleted. The human chromatin-remodeling complexes Ino80 and SRCAP contain RVB proteins, and at least in the case of Ino80, these proteins are required for function. For instance, studies in Saccharomyces cerevisiae have shown that RVB1 and RVB2 recruit Arp5 into the Ino80 complex and are required for the catalytic activity of the Ino80 complex (26). Human Ino80 is also associated with RVB proteins (28). Using siRNA duplexes, we reduced the levels of Ino80 by 85% (Fig. 5A). However, this reduction did not impair ISG induction (Fig. 5D).
RVB1 and RVB2 are also found in another complex, Swr1, that is involved in depositing the H2A.Z histone variant into nucleosomes, thus creating a distinct chromatin state (23, 29–31). Therefore, we tested whether the SRCAP complex, a mammalian homolog of the Swr1 complex containing RVB1, is involved in ISG induction. Again, siRNA duplexes were deployed against SRCAP and efficiently decreased the amount of SRCAP protein, by >70% (Fig. 5B). However, the depletion of SRCAP did not affect the induction of ISGs in response to IFN stimulation (Fig. 5D).
Following the same reasoning, we also tested whether URI has a role in ISG induction. The URI complex contains RVB proteins (32) and is known to negatively modulate VP16- and androgen receptor (AR)-driven transcription (33, 34). Two doxycycline-inducible hairpins against URI efficiently decreased URI protein levels (Fig. 5C), but the depletion did not decrease the extent of ISG induction in response to IFN stimulation (Fig. 5D). Therefore, none of the known catalytic subunits of previously identified RVB complexes is essential for gene expression in response to IFN stimulation.
Taken together, these results suggest that a novel RVB-containing complex is essential for full ISG induction in response to IFN treatment. Alternatively, it is possible that one or more of the Ino80/SRCAP/URI complexes that contain RVB proteins function in a mutually redundant fashion for IFN-stimulated transcription. To test the hypothesis that Tip60, Ino80, and SRCAP act redundantly in this regard, we depleted these factors simultaneously by using a lentiviral hairpin against Tip60 (Fig. 3D and E) and siRNA duplexes targeting Ino80 and SRCAP (Fig. 5F). As previously shown, Tip60 depletion alone did not inhibit ISG induction and instead resulted in enhanced induction of ISG expression. Knockdown of Ino80 and SRCAP in Tip60-depleted cells had no negative impact on transcription of IFN-stimulated genes. From these data, we conclude that none of the Tip60/Ino80/SRCAP complexes is required during IFN-stimulated transcription and that they do not function redundantly in this context.
RVB1 is not required for ISGF3 activation and assembly on chromatin.
Given the lack of interaction between RVB1 and STAT2ΔTAD (Fig. 2A), it is likely that RVB1 is recruited to chromatin following ISGF3 nuclear translocation and assembly on ISG promoters. However, because RVB proteins are components of chromatin-remodeling complexes, we examined whether RVB1 was required for ISGF3 recruitment to chromatin. To address this, we compared the recruitment of STAT2 to ISG promoters with and without RVB1 expression by using our doxycycline-inducible hairpin against RVB1 (Fig. 4A). By using ChIP assays, we precipitated STAT2-chromatin complexes from nuclei of IFN-stimulated cells and assayed the recovery of ISG promoter sequences. As expected, the presence of STAT2 at the ISG56 promoter was increased 4- to 5-fold following IFN treatment (Fig. 6A). However, depletion of RVB1 did not prevent STAT2 from binding to the target promoter, indicating that the requirement for RVB1 for ISG induction occurs after ISGF3 assembly on chromatin. These results demonstrate that assembly of STAT2 on chromatin in vivo, presumably as a component of the ISGF3 complex, does not require RVB1, despite the impaired transcriptional activation of target genes.
RVB1 is required for successful initiation of ISG transcription.
The apparent lack of defect in ISGF3 recruitment prompted us to test whether the failure to adequately activate ISGs in the absence of RVB1 was due to impaired RNA Pol II association with ISG promoters. Selective recruitment of RNA Pol II is a crucial step in transcriptional initiation. As previously reported (10), we detected an IFN-dependent increase in recruitment of total RNA Pol II at the ISG56 promoter but not at an intragenic region downstream of the gene. However, in the absence of RVB1, there was a reduction of Pol II density at the ISG promoter in IFN-treated cells (Fig. 6B).
To further validate and extend this finding, we evaluated the presence of initiated RNA Pol II by using an antibody that recognizes a Ser5-phosphorylated (S5P) form of the enzyme (Fig. 6C). The C-terminal domain (CTD) of the largest subunit of Pol II contains a series of heptapeptide repeats (YS2PTS5PS) that can be phosphorylated differentially during the transcription cycle. The S5P CTD of RNA Pol II is enriched at the 5′ end of transcribed genes and is believed to decorate poised, transcriptionally engaged Pol II, whereas phosphorylation of Ser2 (S2P) is associated with the transition to productive elongation (35). As expected, IFN treatment resulted in enhanced detection of initiated RNA Pol II at the ISG56 promoter. On the other hand, we observed an impaired recruitment of the initiated polymerase at the ISG56 promoter when RVB1 was depleted, while its presence at the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene promoter remained unperturbed regardless of IFN treatment or RVB1 status. The recovery of the intragenic DNA fragment downstream from ISG56 remained near background levels under all conditions. A similar pattern was observed for two other ISG promoters: ISG54 and IFI6 (data not shown). Therefore, RVB1 appears to act upstream of RNA Pol II recruitment and initiation in response to IFN stimulation, mediated by the action of STAT2. Interestingly, we did not detect appreciable amounts of recruited or initiated polymerase at the ISG56 promoter under nonstimulated conditions that could represent a stalled enzyme, suggesting that gene expression is regulated by selective RNA Pol II recruitment following ISGF3 assembly on chromatin rather than by release of preengaged molecules. This is in contrast to the elongation-dependent regulation observed for many inducible genes in lipopolysaccharide (LPS)-treated macrophages (36, 37) but consistent with the IFN dependence of STAT2 activation, nuclear translocation, and chromatin association.
Induction of gene expression is often associated with histone acetylation following recruitment of DNA-bound transcription factors. To examine the role of RVB1 in this process, we examined acetylation of histone H4 at the ISG56 promoter (Fig. 6D). IFN stimulated a 6-fold increase in H4 acetylation at the promoter but not the 3′ intragenic region of ISG56. However, this increased acetylation was largely abolished by depletion of RVB1 following expression of the doxycycline-inducible hairpin. In contrast, acetylation of histones at the GAPDH promoter was largely unaffected regardless of IFN treatment or the absence of RVB1 (Fig. 6D). Pharmacologic inhibition of histone acetyltransferase activity by use of anacardic acid effectively blocked ISG56 induction (Fig. 6E). Altogether, these results suggest that an RVB1-containing complex promotes histone acetylation and RNA Pol II recruitment in response to IFN stimulation in order to activate the promoters of type I IFN-stimulated genes.
DISCUSSION
A swift and regulated cellular response is crucial for host defense and is coordinated by an assortment of transcription factors that rapidly alter the expression of a battery of relevant genes depending on the cell type and the nature of the invading pathogen. Viral infection first activates the transcription factors IRF3 and -7 and NF-κB, which bind recognition elements in IFN promoters and induce their transcription. Induced type I IFNs bind their specific receptors and drive the formation of the ISGF3 complex following tyrosine phosphorylation of STAT1 and STAT2, triggering transcription of genes with multiple antiviral functions. In this study, we identified RVB proteins that are recruited to chromatin by DNA-bound STAT2 as essential components that mediate RNA Pol II recruitment and histone acetylation at ISG promoters in order to induce transcription in response to IFN.
RVB1 was originally identified as part of a complex with TATA box binding protein (TBP) and was named TBP-interacting protein 49 (Tip49) (38). Since then, RVB proteins have been studied in multiple organisms and have been identified in several multisubunit complexes involved in both ATP-dependent remodeling and histone modification. The human Ino80 complex has a nucleosome-stimulated ATPase activity and ATP-dependent remodeling activity in vitro (28). In human cells, Ino80 is also required for transcription of Ying-Yang1 (YY1)-activated genes (39). The SRCAP complex contains RVB proteins and modifies chromatin via incorporation of H2A.Z into nucleosomes by a dimer exchange reaction (29). One of the few functions directly attributed to RVB proteins is their role in the assembly and acetyltransferase activity of the human Tip60 complex (16). Tip60 and RVB1/2 are also recruited by c-myc to its target promoters, and Tip60 is required for AR- and p53-mediated transcription (40, 41). Yet another RVB-containing complex, URI, negatively regulates RNA Pol II during activated transcription driven by VP16 and hepatitis B virus X protein (34), and it was recently shown to negatively regulate AR-driven transcription (33). Although these complexes contain RVB proteins, the role of these proteins in regulating many of these processes remains unexplored.
Biochemical purification coupled with mass spectrometry has become a powerful tool for identifying the protein networks of diverse cellular pathways, including cytokine signaling and chromatin-related complexes. In this study, RVB proteins were identified as STAT2-interacting partners in a proteomic screen. RVB proteins are reported to interact with several other transcription factors, such as c-myc and β-catenin, suggesting that they can serve as cofactors for diverse transcription factors and providing precedence for a functionally relevant STAT2-RVB interaction. Complexes containing RVB proteins support transcription in a variety of settings. Tip60 was shown to act as a coactivator for UV-induced p53-dependent transcription of PUMA in MCF7 cells, while BRD8 was shown to be a nuclear coactivator for thyroid hormone receptor and for peroxisome proliferator-activated receptor gamma (PPARgamma) and 9-cis-retinoic acid receptor (RXR) heterodimers in transient-transfection assays (20, 41, 42). The Ino80 complex is reported to be a coactivator for YY1-directed transcription of an endoplasmic reticulum (ER) stress-inducible gene in HeLa cells (39).
Our validation studies mapped the interaction between RVB1/2 and STAT2 to the transactivation domain of STAT2, implicating RVB proteins in transcriptional regulation. This notion was supported by our gene expression experiments showing that RVB1 or RVB2 silencing specifically impaired responses to IFN-α but not to other inducers, such as TNF-α or IFN-γ. Moreover, RVB was required for expression of the IFN-induced expression of genes that require BRG1 as well as those that do not require the presence of this chromatin remodeler. Therefore, RVB proteins appear to be generally required for IFN-stimulated gene expression that is dependent on STAT2.
In our search for the identity of the RVB complex involved in IFN-α signaling, we ruled out several known complexes based on the lack of a specific requirement for their catalytic subunits (Fig. 3B and D and 5). These data suggest that the function of RVB proteins during IFN-stimulated transcription may not be the modification of DNA-nucleosome interactions previously characterized for SRCAP and Ino80. It is possible that IFN-α treatment causes some fraction of RVBs to assemble into a unique, previously unknown complex that may have a chromatin-remodeling or histone acetyltransferase activity.
Our results further demonstrate that the RVB1 requirement is likely downstream of ISGF3 assembly on chromatin, since it was dispensable for STAT2 recruitment (Fig. 6A), whereas recruitment of initiated RNA Pol II was dependent on RVB1 (Fig. 6B and C). To our knowledge, this is the first documented example of a direct role for RVB1 in transcriptional initiation. Interestingly, we were unable to detect endogenous or transiently overexpressed RVB1 at ISG promoters by ChIP assays using two different antisera. This result may indicate that RVB proteins play an indirect role in transcriptional initiation following transient recruitment by STAT2, a notion that is not without precedence. Yeast RVB proteins have been shown to assist in the assembly of the Ino80 complex, and while promoter-bound Ino80 could be detected, RVB1 was not observed (26). Alternatively, chromatin-bound RVB1 may be inaccessible to immunoprecipitation for technical or conformational reasons.
The exact molecular function of RVB proteins in the context of ISG transcription remains undefined. The importance of ATPase/helicase motifs, which are present in RVB proteins, has been established for several cofactors involved in transcription. For instance, TFIIH is a multiprotein complex containing subunits with intrinsic helicase activity (43). TFIIH functions both as a general transcription factor and in nucleotide excision repair, and its two helicases exhibit opposing helicase polarities (44, 45). RVB1 and RVB2 have also been reported to have helicase activities with opposing polarities (46, 47), although multiple groups have been unable to confirm the helicase activity ascribed to RVB proteins (48, 49), possibly due to the absence of necessary cellular partners in the in vitro assays used. Notably, however, mutation of the ATPase motif of RVB1 inhibits the oncogenic activity of c-myc (25), implicating helicase activity in at least this RVB function. One attractive hypothesis, therefore, is that the ATPase/helicase activity of RVB proteins promotes DNA strand displacement during the formation of a transcriptional bubble. However, our analysis of ATPase/helicase mutant RVB1 (Fig. 3F) demonstrated that ATPase activity was not required for the function of RVB proteins in the context of ISG transcription. Therefore, while strand displacement may be responsible for some functions of RVB1, a helicase-dependent function is unlikely to be involved in the ability of RVB1 to cooperate with STAT2 during ISG expression.
Interestingly, it has been reported that STAT1 homodimers that mediate transcriptional responses in response to IFN-γ utilize MCM5 as a coactivator. MCM5 is an ATPase/helicase and a member of the minichromosome maintenance family of proteins involved in DNA replication. MCM5 interacts with the STAT1 TAD, and it appears to travel along chromatin with elongating RNA Pol II, presumably to facilitate DNA denaturing during transcription (50). Another RNA/DNA helicase, RNA helicase A (RHA), is employed by the STAT6 TAD during interleukin-4 (IL-4)-regulated transcription, and its helicase activity is required for gene expression. It is hypothesized that RHA facilitates entry of the transcriptional complex by interacting with chromatin-modifying enzymes (51). Combined with the findings reported here, it emerges that different STAT proteins have evolved distinct mechanisms to exploit coactivator factors with ATPase/helicase activity to facilitate induction of transcription.
In sum, these results suggest that induction of transcription of previously silent genes in response to acute stimulation with cytokines requires ATPase-dependent modification of chromatin for efficient transcriptional initiation by RNA Pol II. In contrast, actively transcribed housekeeping genes appear to be independent of this activity. In the case of IFN-stimulated transcription, an RVB-containing complex is hypothesized to play this role, being recruited in a gene-specific manner through interaction with the STAT2 transactivation domain. It will be of interest to determine the molecular composition of the RVB complex recruited by STAT2, why different STAT proteins and other inducible transcription factors (e.g., NF-κB) have evolved to be dependent on distinct ATPase/helicase coactivators, and whether helicase dependence is restricted to early pioneer rounds of transcription and is therefore dispensable for constitutively expressed genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank William Y. Tsang and Jia Meng for technical assistance, Isabelle Marié and Susan Logan for helpful discussions, and Anindya Dutta, Yixian Zheng, and Bruno Amati for gifts of reagents.
This work was supported by grants from the NIH (R01AI28900, U54AI057157, and T32CA009161).
Footnotes
Published ahead of print 22 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01562-12.
REFERENCES
- 1.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264 [DOI] [PubMed] [Google Scholar]
- 2.Bluyssen HA, Levy DE. 1997. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 272:4600–4605 [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Young RA. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77–137 [DOI] [PubMed] [Google Scholar]
- 4.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343–25349 [DOI] [PubMed] [Google Scholar]
- 5.Paulson M, Press C, Smith E, Tanese N, Levy DE. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140–147 [DOI] [PubMed] [Google Scholar]
- 6.Agalioti T, Chen G, Thanos D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381–392 [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Kang H, Liu R, Chen X, Zhao K. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22:6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. 2004. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell. Biol. 24:4476–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. 2002. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 4:774–781 [DOI] [PubMed] [Google Scholar]
- 10.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I, Levy DE. 2004. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 101:9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nusinzon I, Horvath CM. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. U. S. A. 100:14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jha S, Dutta A. 2009. RVB1/RVB2: running rings around molecular biology. Mol. Cell 34:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam JD, Martin PL, Shastry BS, Roeder RG. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101:582–598 [DOI] [PubMed] [Google Scholar]
- 14.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat. Genet. 37:1289–1295 [DOI] [PubMed] [Google Scholar]
- 15.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha S, Shibata E, Dutta A. 2008. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28:2690–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr 1996. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell. Biol. 16:288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, Lamond A. 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183:223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278:42733–42736 [DOI] [PubMed] [Google Scholar]
- 20.Monden T, Wondisford FE, Hollenberg AN. 1997. Isolation and characterization of a novel ligand-dependent thyroid hormone receptor-coactivating protein. J. Biol. Chem. 272:29834–29841 [DOI] [PubMed] [Google Scholar]
- 21.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–473 [DOI] [PubMed] [Google Scholar]
- 23.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665–13670 [DOI] [PubMed] [Google Scholar]
- 24.Barnes PJ, Karin M. 1997. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336:1066–1071 [DOI] [PubMed] [Google Scholar]
- 25.Wood MA, McMahon SB, Cole MD. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321–330 [DOI] [PubMed] [Google Scholar]
- 26.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16:465–477 [DOI] [PubMed] [Google Scholar]
- 27.Mezard C, Davies AA, Stasiak A, West SC. 1997. Biochemical properties of RuvBD113N: a mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J. Mol. Biol. 271:704–717 [DOI] [PubMed] [Google Scholar]
- 28.Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. 2005. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280:41207–41212 [DOI] [PubMed] [Google Scholar]
- 29.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343–348 [DOI] [PubMed] [Google Scholar]
- 30.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Roberts DN, Cairns BR. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, Vigneron M, Peter M, Krek W. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302:1208–1212 [DOI] [PubMed] [Google Scholar]
- 33.Mita P, Savas JN, Djouder N, Yates JR, 3rd, Ha S, Ruoff R, Schafler ED, Nwachukwu JC, Tanese N, Cowan NJ, Zavadil J, Garabedian MJ, Logan SK. 2011. Regulation of androgen receptor-mediated transcription by RPB5 binding protein URI/RMP. Mol. Cell. Biol. 31:3639–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N, Murakami S. 1998. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18:7546–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phatnani HP, Greenleaf AL. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922–2936 [DOI] [PubMed] [Google Scholar]
- 36.Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. 2009. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. U. S. A. 106:18207–18212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanemaki M, Makino Y, Yoshida T, Kishimoto T, Koga A, Yamamoto K, Yamamoto M, Moncollin V, Egly JM, Muramatsu M, Tamura T. 1997. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235:64–68 [DOI] [PubMed] [Google Scholar]
- 39.Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. 2007. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14:872–874 [DOI] [PubMed] [Google Scholar]
- 40.Gaughan L, Brady ME, Cook S, Neal DE, Robson CN. 2001. Tip60 is a co-activator specific for class I nuclear hormone receptors. J. Biol. Chem. 276:46841–46848 [DOI] [PubMed] [Google Scholar]
- 41.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24:841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monden T, Kishi M, Hosoya T, Satoh T, Wondisford FE, Hollenberg AN, Yamada M, Mori M. 1999. p120 acts as a specific coactivator for 9-cis-retinoic acid receptor (RXR) on peroxisome proliferator-activated receptor-gamma/RXR heterodimers. Mol. Endocrinol. 13:1695–1703 [DOI] [PubMed] [Google Scholar]
- 43.Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S. 1993. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365:852–855 [DOI] [PubMed] [Google Scholar]
- 44.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D. 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368:769–772 [DOI] [PubMed] [Google Scholar]
- 45.Sung P, Guzder SN, Prakash L, Prakash S. 1996. Reconstitution of TFIIH and requirement of its DNA helicase subunits, Rad3 and Rad25, in the incision step of nucleotide excision repair. J. Biol. Chem. 271:10821–10826 [DOI] [PubMed] [Google Scholar]
- 46.Makino Y, Kanemaki M, Kurokawa Y, Koji T, Tamura T. 1999. A rat RuvB-like protein, TIP49a, is a germ cell-enriched novel DNA helicase. J. Biol. Chem. 274:15329–15335 [DOI] [PubMed] [Google Scholar]
- 47.Kanemaki M, Kurokawa Y, Matsu-ura T, Makino TY, Masani A, Okazaki K, Morishita T, Tamura TA. 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274:22437–22444 [DOI] [PubMed] [Google Scholar]
- 48.Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. 1998. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 273:27786–27793 [DOI] [PubMed] [Google Scholar]
- 49.Matias PM, Gorynia S, Donner P, Carrondo MA. 2006. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 281:38918–38929 [DOI] [PubMed] [Google Scholar]
- 50.Snyder M, He W, Zhang JJ. 2005. The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 102:14539–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valineva T, Yang J, Silvennoinen O. 2006. Characterization of RNA helicase A as component of STAT6-dependent enhanceosome. Nucleic Acids Res. 34:3938–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.