Abstract
Human papillomavirus type 16 (HPV-16) associated oropharyngeal cancers are on a significant increase and better therapeutic strategies are needed. The HPV-16 oncogenes E6 and E7 are expressed in HPV-associated cancers and are able to transform human tonsillar epithelial cells (HTECs). We used cell-SELEX (Systematic Evolution of Ligands by Exponential Enrichment) to select for RNA aptamers that entered into HPV-16 E6/E7-HTECs. After 12 rounds of cell-SELEX, a pool of aptamers was obtained that had significantly greater internalization capacity (~5-fold) into E6/E7-HTECs as compared to primary HTECs or fibroblasts. Analysis of individual aptamers from the pool indicated variable internalization into E6/E7-HTECs (1 to 8-fold as compared to a negative control). Most of the individual aptamers internalized into E6/E7 and primary HTECs with similar efficiency, while one aptamer exhibited ~3-fold better internalization into E6/E7-HTECs. Aptamers that internalize into cells may be useful for delivering therapeutic agents to HPV-16 associated malignancies.
Keywords: Papillomavirus, SELEX, HPV, Aptamers, Oropharyngeal, Head Neck
INTRODUCTION
Ninety-nine percent of cervical cancer, more than 50% of anal, vaginal, vulvar, and penile cancers, and a subset of head and neck cancers are caused by high-risk human papillomaviruses (HPVs) (Moody and Laimins, 2010). While there are several types of HPVs associated with cancer, HPV-16 accounts for the majority of cervical cancers and nearly all HPV-associated head and neck cancers (HNCs). Currently available vaccines can do nothing for the millions of people that are already infected with HPV and compliancy rates for vaccination have fallen well short of expectations (Dillard and Spear, 2010). HPV-associated HNCs, particularly oropharyngeal (tonsillar) cancers, are on the rise in both men and women (Chaturvedi et al., 2011; Marur et al., 2010). Few new treatment strategies for HPV positive or negative HNCs have emerged in recent decades and there is a pressing need for novel therapies. HPV-positive HNCs are considered to have a better prognosis than HPV-negative HNCs, in general, but a significant subset do not respond to therapy and metastasize (Huang et al., 2013). Genetic and microarray expression studies of HPV positive and HPV negative HNCs indicate that they are clearly two separate entities, with HPV-positive HNCs having many fewer mutations than HPV-negative HNCs, which goes along with their viral etiology (Agrawal et al., 2011; Pyeon et al., 2007; Stransky et al., 2011). Two viral-specific oncogenes called E6 and E7 are expressed in HPV-associated cancers (Klingelhutz and Roman, 2012). We and others have demonstrated that HPV-16 E6 and E7 together can immortalize human and mouse head and neck epithelial cells (Al Moustafa et al., 2004; Hoover et al., 2007; Spanos et al., 2008). The E6 and E7 proteins inactivate the p53 and pRb tumor suppressor proteins, respectively, and also activate telomerase (Dyson et al., 1989; Klingelhutz et al., 1996; Scheffner et al., 1990). HPV-16 E6 and E7 interact with numerous cellular proteins and alter a vast array of pathways involved in transformation (Klingelhutz and Roman, 2012). Expression of E6 and E7 are essential for transformation and continuous proliferation. Inhibition of expression or knockdown by siRNA causes HPV transformed cells to rapidly senesce and/or apoptose (Butz et al., 2003; Jiang and Milner, 2002; Tang et al., 2006).
RNA aptamers are synthetic RNAs that can specifically bind to targets of interest (Thiel and Giangrande, 2009). They are derived from combinatorial sequence libraries by a process called SELEX (Systematic Evolution of Ligands by EXponential Enrichment)(Blank et al., 2001). This methodology has been modified in such a way that it can be cell based (cell-SELEX)(Cerchia et al., 2009b; Shamah et al., 2008). Multiple rounds of selection and amplification allow enrichment of RNA aptamers that can bind selectively to certain cell types and internalize. Contrary to selection conducted by in vitro expressed proteins, cell-SELEX enables the isolation of RNA aptamers that bind to receptors in their native state and selects specifically for those that can be internalized into cells. The aptamers are synthesized with 2′-fluoropyrimidines to make them resistant to nuclease-mediated degradation (Cerchia et al., 2009b). Aptamers can be easily synthesized in large quantities and the smaller size, as compared to antibodies, facilitates delivery by allowing better tissue penetration. Aptamers have been utilized to target different proteins including transcription factors and cell surface receptors (Cerchia and de Franciscis, 2010; Giangrande et al., 2007; Mi et al., 2008; Shangguan et al., 2008; Shangguan et al., 2006; Wan et al., 2010). Conjugation of fluorescent or radioactive tags to protein-specific aptamers has been shown to be a means to tag/detect cancer cells that express that protein (Cerchia and de Franciscis, 2010). In addition, immobilized protein-specific aptamers (e.g. aptamers against EGFR) have been utilized to specifically capture cancer cells from the blood, a method that could be used as a possible means for early cancer detection (Pu et al., 2010). Selecting aptamers that specifically bind to transformed cells also offers the possibility of identifying proteins that are upregulated in cancer cells (Shangguan et al., 2007a). A number of publications have demonstrated that protein or cell-specific aptamers can be utilized to inhibit protein function (McNamara et al., 2008; Mi et al., 2008). In addition to direct interaction of aptamers with cellular proteins, aptamers can be used to deliver siRNA into cells. For example, aptamer-siRNA chimeras have been developed that mediate binding to PSMA, a cell-surface receptor expressed on prostate cancer cells, followed by internalization and specific delivery of growth inhibiting siRNA to PSMA expressing prostate cells in vitro and in vivo (Dassie et al., 2009; Lupold et al., 2002; McNamara et al., 2006). A similar aptamer-siRNA methodology has been utilized to target the HIV gp120 protein to deliver siRNA to inhibit tat/rev and suppress HIV replication (Zhou et al., 2008). It has also been proposed that cell-specific aptamers could be used in conjunction with nanoparticles to increase nanoparticle specificity and internalization (Cerchia and de Franciscis, 2010; Dua et al., 2011; Yang et al., 2012).
In this study, we have utilized cell-SELEX to identify RNA aptamers that efficiently internalize into HPV-16 E6/E7 transformed human tonsillar epithelial cells. These aptamers may provide a mechanism to deliver therapeutic siRNA or other cytotoxic agents to HPV-16 associated cancers.
MATERIALS AND METHODS
Cell Lines
Primary human tonsillar epithelial cells (HTECs) were isolated from discarded tonsillar tissue obtained from the University of Iowa Tissue Procurement Facility under IRB approval using methods that have been previously described (Hoover et al., 2007). After establishment, cells were grown in KSFM (Invitrogen) and passaged using 1:4 as described (Darbro et al., 2005). HTECs were transduced at early passage with a replication defective retrovirus 16E6/E7-LXSN expressing HPV-16 E6/E7 and a neomycin resistance marker and selected in G418 as described (James et al., 2006). 16E6/E7 cells were used at early to mid-passage (P11-P17) for all experiments.
Cell-SELEX
Generation of aptamers and cell-SELEX was performed according to methods described previously (Cerchia et al., 2009b; Thiel et al., 2012a). DNA oligonucleotides were obtained from Integrated DNA Technology (IDT, Coralville, IA) that contained the following sequence: 5′-TCGGGCGAGTCGTCTG-N30-CCGCATCGTCCTCCC-3′. This generation of aptamers is referred to as Sel2 (Thiel et al., 2012a). PCR primers for amplification and addition of a T7 polymerase site were as follows: 5′primer-TAATACGACTCACTATAGGGAGGACGATGCGG; 3′primer-TCGGGCGAGTCGTCTG. The RNA aptamer library was generated using a mutant Y639F T7 polymerase (Huang et al., 1997) with 2′-fluoro modified CTP and UTP (TriLink Biotechnologies) to make the aptamers nuclease-resistant. The aptamers were purified by running on denaturing 10% acrylamide gels. Bands were visualized by UV shadowing, excised, and eluted with Tris-EDTA followed by filtering with a Centrex spin filter and concentrating with an Amicon Ultracel as described. Aptamers were folded in the presences of tRNA and calcium by heating to 95°C for 5 minutes followed by incubation by incubation at 37°C for 20 minutes. The purified and folded aptamers were then used in a series of pre-clearing and internalization steps followed by reverse transcription and PCR to isolate those aptamers that internalized differentially into 16E6/E7-HTECs as compared to primary HTECs. In each round of cell-internalization SELEX, RNA aptamer pools were first folded in the presence of calcium in serum-free KSFM and supplemented with yeast tRNA (Invitrogen) and then pre-cleared with primary HTECs (at passage 3 to 5) that had been pre-incubated with tRNA as a pre-clearing step for 20 minutes at 37°C. After pre-clearing, the supernatant was transferred to 16E6/E7-HTECs (always at passage 16) and incubated for varying lengths of time to allow internalization starting with 75 minutes in the first rounds and decreasing to 20 minutes in later rounds to increase stringency. After incubation, to remove unbound and surface-bound aptamers, the target 16E6/E7 cells were washed with ice-cold PBS, followed by two high salt washes (0.5 M NaCl), first quickly and then for 5 minutes, followed by another PBS wash (all on ice). Internalized RNA aptamers were then recovered using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The RNA was reverse transcribed into DNA using SuperScript III (Invitrogen) and amplified by PCR using AmpliTaq and Sel2 primers (as above) to amplify and add back the T7 polymerase promoter. The PCR products were purified using a Qiagen Miniprep Column and then in vitro transcribed to generate an enriched pool of RNA aptamers for subsequent rounds of cell-SELEX. PCR samples were also removed from each round for later sequencing. A total of 12 pre-clearing and selection rounds were performed.
Sequencing and Structure Analysis
Pools of aptamers were sequenced using 454-deep sequencing (University of Iowa DNA Facility), processed, and analyzed as described (Thiel et al., 2012a; Thiel et al., 2012b). A script was written using the Perl programming language to identify unique sequences and count the number of times each sequence appeared in the dataset. The percent enrichment was calculated as %Enrichment = 1-(Unique/Total)*100 as described previously (Thiel et al., 2012b; Thiel et al., 2011). All unique sequences were aligned using ClustalX2 (Larkin et al., 2007). If clear families of related sequences were present, the sequence that was represented most in the family was chosen for follow-up studies. Theoretical structures of RNA were evaluated using M-Fold (Zuker, 2003). The structure with the minimum free energy was determined for aptamers that increased in number with more selection.
Internalization Assays
RT-qPCR
To assess internalization, primary HTECs (passage 3-5), 16E6/E7-HTECs (passage 15-17), or fibroblasts (passage 8) were plated in 6-well dishes at 2 ×105 per well. Internalization was performed and assessed as described previously (Thiel et al., 2012a). Briefly, 24 hours after plating, the cells were washed and incubated first with tRNA followed by incubation with 100 nM folded aptamer in tRNA mix (see above). The folded aptamer was incubated for 20 minutes at 37°C followed by washes in ice-cold PBS followed by high salt wash (0.5 M). After washing, RNA was collected and processed as above with the addition that the Trizol samples were spiked with an internal 2′-fluoropyrmidine aptamer control called Sel1. Sel1 refers to a generation of aptamers that utilizes a different set of primers than Sel2 for amplification (McNamara et al., 2008). In our assays, we utilized a Sel1 spike aptamer called M12-23 (McNamara et al., 2008). The Sel1 allows for rigid control of cDNA synthesis, pipetting, and PCR amplification during the purification and quantitation process. Purified RNA was split into two aliquots. One was treated with RNase to remove cellular RNA (Aptamer RNA is resistant to RNase). The other was used for quantification of GAPDH (as another control for cell number). Samples were reverse transcribed as above and aptamer amount (as well as GAPDH for the sample that was not treated with RNase) was assessed by quantitative PCR using appropriate primers as described (Thiel et al., 2012a). Results are expressed as a Sel2/Sel1 ratio, normalized to GAPDH levels (obtained from untreated samples). For experiments with individual aptamers, we also normalized to a negative control aptamer that had the same constant regions as the tested aptamers. The negative control that we chose to use was an aptamer that was present in the original aptamer library but was not found after later rounds of cell-SELEX.
Fluorescent-Labeled Aptamers
For the experiments to assess internalization by FACS, cells were incubated with the folded labeled aptamer in the presence of tRNA for 45 minutes at 37° C then washed with high-salt as above, trypsinized, and stained with Hoechst (to differentiate live and dead cells). Cells were then analyzed by FACS for Cy3 and Hoechst uptake using a FACSDiva at the University of Iowa Flow Cytometry Facility. The percent shift represents the number of cells that are positive for Cy3 (i.e. aptamer positive) as compared to background levels.
For the assays to assess internalization at different temperatures, the fluorescently labeled aptamers were folded and incubated in the presence of tRNA with cells grown on coverslips. During the 90 minute incubation, the cells were either kept at 37°C in the incubator or on ice. After incubation, the cells were washed with cold PBS and high-salt as above before microscopic analysis using a 40× oil immersion objective on an Olympus 1×71 inverted microscope with a cooled charge-coupled device camera and filters for Texas Red. Intracellular localization was confirmed with nuclear DAPI stain and overlapping with Image J software.
RESULTS
Aptamer Selection Strategy
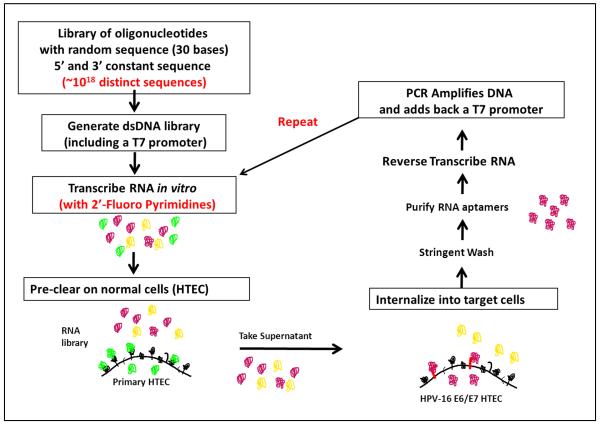
For these studies, we used HPV-16 E6/E7 transformed HTECs that were generated as previously described (Spanos et al., 2008). The HPV-16 E6/E7 transformed HTECs represent an early stage of HPV-associated transformation and should exhibit alterations that are specific for HPV transformation and not individualized alterations that have been selected for in specific cancer cell lines. Previous microarray expression studies from our lab and others have demonstrated that HPV-16 E6/E7 expression causes changes in expression of a large number of cellular genes, including many that code for cell-surface proteins (and hence would be good targets for aptamers) (Duffy et al., 2003; Pyeon et al., 2007; Wan et al., 2008). To identify aptamers that specifically bind to and internalize into HPV-16 transformed head and neck epithelial cells, we utilized cell-SELEX with a complex RNA aptamer library with a 30 nucleotide variable sequence and an estimated starting complexity of greater than 1018. The RNA aptamers were synthesized using 2′-fluoropyrimidine to provide stability and inhibit degradation by RNAses. Each round of cell-SELEX involved a pre-clearing step with primary HTECs for removal of aptamers that bind to non-transformed cells (negative selection) and a positive selection step against the target using HPV-16 E6/E7-HTECs (Figure 1). With each round, we progressively increased the selective pressure by decreasing concentration and increasing incubation times in the primary HTECs and shortening binding/internalization time in the target HPV-16 E6/E7 transformed cells (Table I). A total of 12 rounds of negative and positive selection were performed. With each round, cell extracts were prepared and RNA isolated. After removal of cellular RNA, extracted aptamers were precipitated, reverse transcribed, and PCR amplified with an added T7 promoter. New 2′-fluoropyrimidine modified RNA was then generated for the next round of selection.
Figure 1. Strategy for cell-SELEX (Systematic Evolution of Ligands by Exponential Enrichment).
A pool of oligonucleotides with a 30 base random internal sequence were converted to double-stranded DNA and then into 2′ fluoropyrimidine modified RNA aptamers using a T7 polymerase. The pool of aptamers was first pre-cleared using primary human tonsillar epithelial cells (HTECs). Aptamers that did not bind and internalize were incubated with HPV-16 E6/E7 transformed HTECs and allowed to internalize. The cells were stringently washed with high salt, followed by lysis and purification of the RNA. After RNAse treatment (to remove the cellular RNA but not the resistant aptamers), the RNA was reverse transcribed and PCR-amplified. The amplified DNA was then transcribed into new aptamers and used in subsequent rounds of selection.
Table I. Parameters of cell-SELEX selection.
| *Rd1 | Rd2 | Rd3 | Rd4 | Rd5 | Rd6 | Rd7 | Rd8 | Rd9 | Rd10 | Rd11 | Rd12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Concentration (beginning) |
2 nM | 2 nM | 2 nM | 1 nM | 1 nM | 1 nM | 0.5 nM |
0.5 nM |
0.5 nM |
0.5 nM |
0.5 nM |
0.5 nM |
|
Pre-clearing time on HTECs and number of sequential plates |
20 min × 2 plates |
20 min 1 plate |
40 min 1 plate |
40 min × 3 plate s |
15 min × 3 plate s |
15 min × 3 plate s |
15 min × 3 plate s |
20 min × 4 plate s |
20 min × 4 plate s |
20 min × 4 plate s |
20 min × 4 plate s |
20 min × 4 plate s |
|
Internalization time on 16E6/E7 HTECs |
75 min | 75 min |
75 min |
75 min |
75 min |
75 min |
30 min |
30 min |
30 min |
20 min |
20 min |
20 min |
Rd=Round of cell-SELEX
Convergence of Aptamer Sequences that Differentially Internalize into E6/E7-HTECs
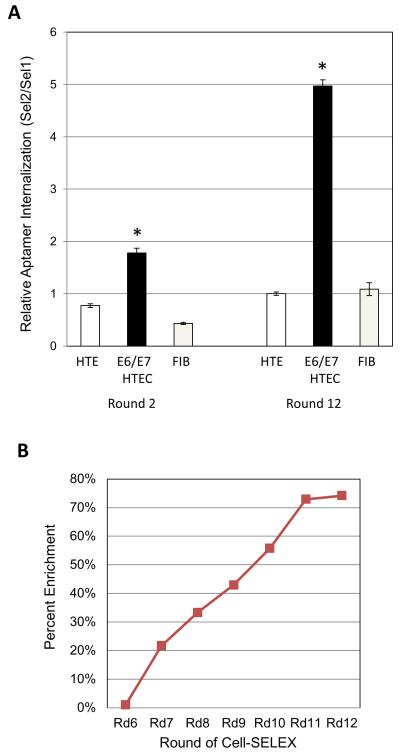
Aptamer pools from selected rounds (early round 2 and late round 12) were then tested for their differential ability to internalize into E6/E7-HTECs as compared to primary HTECs. The pool of aptamers in round 12 were much more efficient at internalizing into E6/E7-HTECs and less efficient at internalizing into primary HTECs than the earlier round aptamers (Figure 2A). Thus, we were able to enrich for aptamers that bind/internalize better into 16E6/E7 transformed cells as compared to primary normal cells. The difference was approximately 5-fold (compare round 12 to round 2). In addition, the aptamer pools from the later rounds were poor at internalizing into human fibroblasts, indicating cell-type specificity (Figure 2A).
Figure 2. Internalization of selected RNA aptamer pools and convergence of aptamer sequences.
A. Aptamer pools from rounds 2 and 12 were tested on primary HTECs (HTEC), 16E6/E7 HTECs (E6/E7 HTEC), and primary fibroblasts. The pools were incubated with the cells to allow for binding and internalization, followed by stringent washing, isolation of RNA, and quantification as described in the Materials and Methods. The quantity is displayed as the ratio of the aptamers (Sel2) over an internal spiked control (added after Trizol extraction) called Sel1 all normalized to GAPDH (measured from a separated non-RNAse treated fraction) and made relative to primary HTECs (*=p<0.05, Student’s t-test). B. An analysis of sequences from 454 sequencing was performed as described in the Materials and Methods to determine library complexity with increasing rounds of selection. A higher percent indicates enrichment and less complexity.
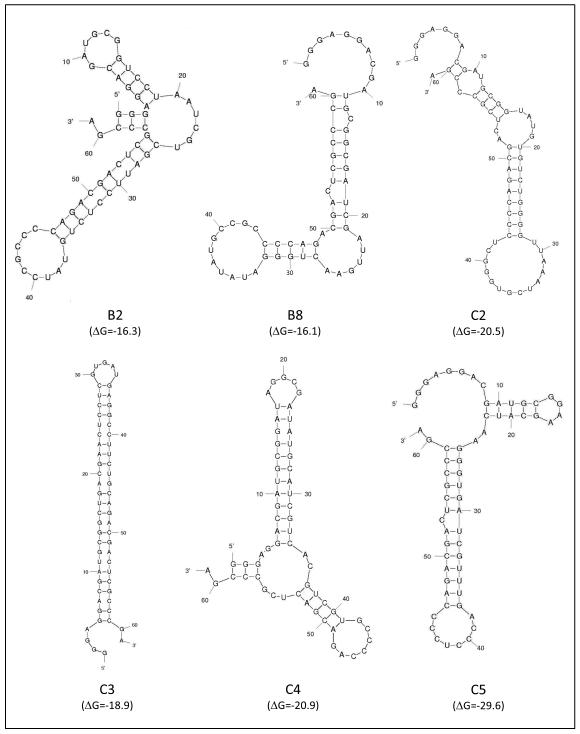
The above studies demonstrate that the cell-SELEX methodology resulted in the selection of a pool of aptamers that could differentially bind and internalize into E6/E7-HTECs. It would therefore be expected that the later aptamer rounds would be enriched for aptamers of certain sequences or structures that provided for better and more specific internalization into 16E6/E7 transformed cells. To identify individual aptamers, we performed 454 sequencing on reverse transcribed aptamer pools from the different rounds. Comparison of sequences that were obtained allowed a determination of whether enrichment had occurred. Percent enrichment was calculated as 1-(Unique Sequences/Total Sequences)X100 where fewer unique sequences represents less complexity and more enrichment (Thiel et al., 2012b; Thiel et al., 2011). We found that the percent enrichment did not increase substantially after 5 rounds of selection (data not shown) but increased with subsequent rounds (rounds 6-12) of SELEX, indicating that aptamer pool complexity decreased with increasing selection (Figure 2B and Table II). Still, a large number of different sequences were present in the population. We chose to follow up on six aptamers that were the most common and that showed increases in number with increasing rounds of selection. If families of aptamers with clearly related sequences were present, we chose the most highly represented aptamer for follow-up studies. Theoretical structures of least energy were generated by M-fold for the six aptamers. These structures along with the sequence of the variable region for each aptamer are shown in figure 3 and Table III. While some of the aptamers have similar theoretical structures, there is some diversity, suggesting that the selected aptamers may have different mechanisms of binding and/or entry into the E6/E7 transformed cells. It should be noted that these structures are theoretical; other folding patterns are possible.
Table II. Common aptamers found by 454 sequencing that increase in later rounds of cell-SELEX.
| Aptamer | Rd6 | Rd7 | Rd8 | Rd9 | Rd10 | Rd11 | Rd12 |
|---|---|---|---|---|---|---|---|
| B2 | 3* | 88 | 382 | 555 | 823 | 950 | 567 |
| B8 | 1 | 79 | 119 | 232 | 259 | 486 | 534 |
| C2 | 1 | 51 | 77 | 201 | 344 | 946 | 967 |
| C3 | 1 | 3 | 11 | 36 | 249 | 1028 | 927 |
| C4 | 1 | 80 | 139 | 203 | 266 | 340 | 407 |
| C5 | 1 | 1 | 51 | 85 | 193 | 397 | 437 |
Represents the number of times that an individual aptamer was identified by deep sequencing of the pool of aptamers from each round of cell-SELEX.
Figure 3. Theoretical structures of selected aptamers.
The structure with the minimum free energy was determined for aptamers that increased in number with more selection. Sequences of the variable region of the aptamers are shown. Theoretical structures of RNA were evaluated using M-Fold. DeltaG (ΔG) values are noted below the structure.
Table III. Aptamer sequences.
| Aptamer | 5′Constant | Variable Sequence | 3′ Constant |
|---|---|---|---|
| B2 | GGGAGGACGAUGCGG | TCCTAATCGTCGATTCCTCTGTATCCGCCC | CAGACGACUCGCCCGA |
| B8 | GGGAGGACGAUGCGG | CGATCGATTGAACTGGGATATATGCCGCCC | CAGACGACUCGCCCGA |
| C2 | GGGAGGACGAUGCGG | TATGTGTCTGGGGTTAAATCGTGGGCTCCC | CAGACGACUCGCCCGA |
| C3 | GGGAGGACGAUGCGG | CTGACGAACTCCTCGTGATGAGGCCTTCTG | CAGACGACUCGCCCGA |
| C4 | GGGAGGACGAUGCGG | ATAGGCGATATGCATCGTCACGTCGTGCCC | CAGACGACUCGCCCGA |
| C5 | GGGAGGACGAUGCGG | AAGCATCAAGGGTGATCGTTTGACCCTCCC | CAGACGACUCGCCCGA |
| Neg. Con. |
GGGAGGACGAUGCGG | GTAATAAACACGACAACGCTTTATTGCCCC | CAGACGACUCGCCCGA |
Internalization of Individual Aptamers into E6/E6 Transformed Cells
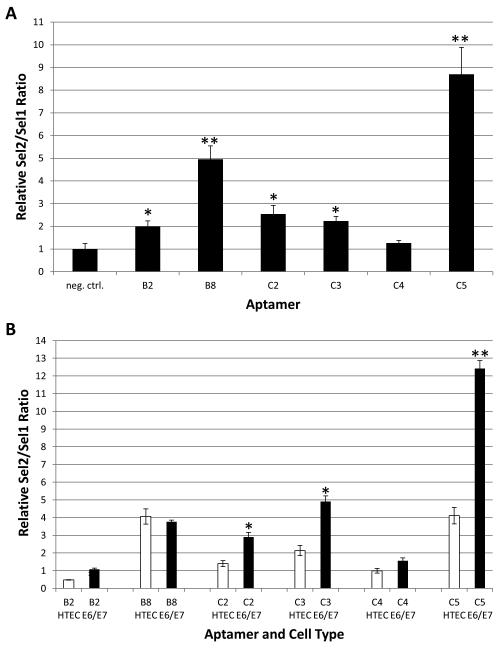
To assess internalization, the individual aptamers were incubated with E6/E7-HTECs followed by stringent washing, RNA extraction, and RT-qPCR. Although variability in internalization was observed, most of the individual aptamers, except C4, internalized better into E6/E7-HTECs as compared to internalization by a negative control aptamer (Figure 4A). Two of the aptamers, B8 and C5, internalized approximately 5 and 8-fold, respectively, better than a negative control aptamer. The heterogeneity of internalization between the individual aptamers indicates some measure of specificity. Unexpectedly, most of the tested aptamers exhibited similar internalization into E6/E7-HTECs and primary HTECs, with ~2-fold or less difference (Figure 4B). One aptamer, C5, which internalized with the highest efficiency, also differentially internalized ~3.0 fold better into E6/E7-HTECs as compared to primary HTECs. This aptamer was chosen for further studies.
Figure 4. Internalization of individual aptamers into cells.
A. The individual aptamers were incubated with the 16E6/E7 HTECs to allow for binding and internalization, followed by stringent washing, isolation of RNA, and quantification as described in the Materials and Methods. The quantity is displayed as the ratio of the aptamers (Sel2) over an internal spiked control (added after Trizol extraction) called Sel1 all normalized to GAPDH (measured from a separated non-RNAse treated fraction) and compared to internalization of a negative control aptamer (*=p<.05; **=p<.01, Student’s t-test). B. Internalization of individual aptamers into 16E6/E7 HTECs (E6/E7) as compared to internalization into primary HTECs (HTEC). Experiments and quantification were performed as in A (*=p<.05;**p<.01, Student’s t-test comparing internalization into E6E7 with internalization into HTEC).
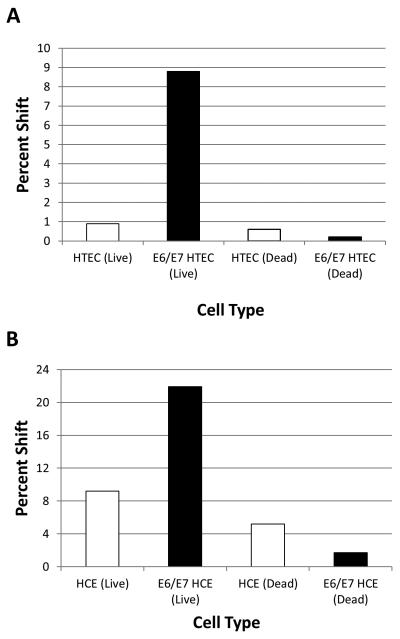
As another measure of internalization, the C5 aptamer was fluorescently labeled with Cy3. We then assessed internalization by incubating with E6/E7-HTECs or primary HTECs, followed by stringent washes, trypsinization, and FACS analysis. In this assay, the labeled C5 aptamer was found to internalize better into E6/E7-HTECs better than it did into primary HTECs (~9% shift compared to 1%)(Figure 5A). Using Hoechst stain, we were also able to distinguish between live and dead cells, and we found that most internalization was in live cells with minimal background in dead cells. To determine if the aptamer internalized into other HPV-16 transformed cells, we incubated the labeled C5 aptamer with primary cervical and 16E6/E7 immortalized human cervical epithelial (HCE) cells. The aptamer internalized into primary HCE cells, but it was better at internalizing into 16E6/E7-HCE cells (22% shift as compared to 9%)(Figure 5B), suggesting that E6/E7 expressing cells were more efficient at binding and internalizing the C5 aptamer.
Figure 5. Internalization of fluorescently tagged C5 aptamer into 16E6/E7 transformed cells.
The aptamer was labeled with Cy3 and purified and incubated with cells as described in the Materials and Methods. After washing and trypsinization the cells were analyzed by FACS. The quantity shown represents the percentage shift in the number of cells as compared to background autofluorescence. A. Internalization into 16E6/E7-HTEC or primary HTEC; B. Internalization into 16E6/E7 human cervical epithelial (E6/E7 HCE) or primary cervical epithelial (HCE) cells.
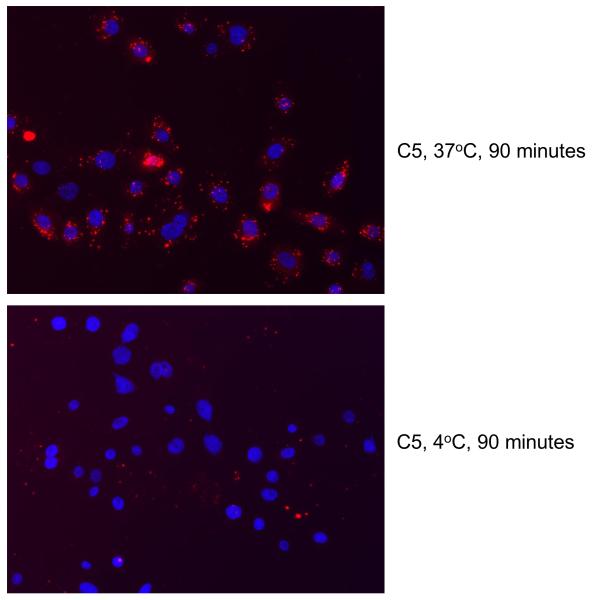
To assess cellular localization after internalization, we incubated E6/E7-HTECs with labeled C5 aptamer and examined the cells using fluorescence microscopy. The aptamer localized to punctate spots in the cytoplasm (Figure 6). Internalization was found to be dependent on incubation at 37°C, indicating a mechanism of active uptake (Figure 6).
Figure 6. Localization of C5 aptamer and inhibition of internalization by cold.
Cy3 labeled C5 aptamer was incubated with E6/E7-HTECs as described in the Materials and Methods at either 37°C or 4°C. The cells were visualized using fluorescence microscopy as described in the Materials and Methods.
DISCUSSION
In this study, we utilized cell-SELEX to identify RNA aptamers that can internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Our selection strategy resulted in a pool of aptamers that were better at internalizing into E6/E7-HTECs than primary HTECs or fibroblasts. The individual aptamers exhibited different levels of internalization ability and one aptamer, C5, exhibited increased 3-fold increased internalization for E6/E7-HTECs as compared to primary HTECs.
Cell-SELEX has provided a means for identifying aptamers that differentially bind and internalize better into cancer cells as compared to non-transformed or non-metastasizing cells (Cerchia et al., 2009a; Chen et al., 2008; Mi et al., 2010; Sefah et al., 2010; Shangguan et al., 2006; Shangguan et al., 2007b; Tang et al., 2007; Thiel et al., 2012a). To our knowledge, aptamers that specifically internalize into HPV transformed cells have not been reported. However, in a recent study, cell-SELEX was used to identify aptamers that bound and internalized into revertants of Hela (HPV-18 positive) cells that were no longer tumorigenic (Graham and Zarbl, 2012). In other words, those aptamers that were identified were better at internalizing into non-tumorigenic cells than tumorigenic cells. In our studies, we were interested in isolating aptamers that internalized specifically into HPV transformed tonsillar epithelial cells. To do this, we used a pre-clearing step in which we first incubated the aptamer library with primary normal HTECs. This pre-clearing should have removed a population of aptamers that can bind and enter into primary cells. The aptamers that did not internalize were then used for internalization into E6/E7-HTECs to select for those that were better at internalizing into HPV-16 transformed cells. These latter aptamers were then isolated and re-amplified for subsequent rounds of pre-clearing and selection. After 12 rounds, the pool of aptamers that remained was better able to internalize into E6/E7-HTECs as compared to primary HTECs, indicating that selection and convergence had been successful. However, while most of the individual aptamers that we selected from round 12 internalized into E6/E7-HTECs, our results would suggest that most of them did not differentially internalize to any large degree. In other words, they bound and internalized into both HTECs and E6/E7-HTECs with similar efficiency. The reason for this result is not entirely clear. One possibility is that the cell-SELEX strategy we utilized was not stringent enough in the pre-clearing step and only a subpopulation of aptamers in the pool can differentially internalize. If that is the case, we would simply need to increase the number of rounds of cell-SELEX and/or test more individual aptamers for differential internalization. In addition, E6/E7-HTECs and primary HTECs are likely to share many of the same aptamer targets since they are from the same origin and it may be difficult to find aptamers that can differentiate between the two cell types. Using HPV transformed cells that represent a more malignant stage in cell-SELEX might have resulted in more cell-specific aptamers. Another possibility is that specificity and internalization could arise through interactions that involve combinations of aptamers. A mechanism such as this is certainly not unprecedented in the virus world, in which receptors and co-receptors are needed for efficiency and specificity. This hypothesis could be tested by using various combinations of different aptamers to determine if specificity or efficiency is increased. While the individual aptamers that we identified did not have high specificity, characterization of aptamer targets through mass spectrometry techniques (Li et al., 2009; Mallikaratchy et al., 2007; Mi et al., 2010) or other methods might allow the development of more stringent strategies to increase specificity (e.g. through further selection against a known protein target). Determination of the mechanism of uptake could also lead to better insight into what is necessary for internalization and specificity which, in turn, could lead to identification of aptamers with higher specificity.
The single aptamer, C5, was found to consistently internalize better into 16E6/E7 cells as compared to primary cells. As mentioned, the level of differential internalization was not remarkable (~3-fold) and depended on the type of assay that was used. For RT-qPCR, the difference was approximately 3-fold, whereas using fluorescently labeled aptamer, the difference was represented by a small shift in fluorescence intensity in the pool of cells. The reasons for these differences in results are not entirely clear, but it has been demonstrated that assays to measure aptamer internalization often yield variable results and can depend upon the specific aptamer that is being studied (Hernandez et al., 2013). Modifications to the aptamer (e.g. through the fluorescent tag) could have also resulted in a change in structure and the ability to bind and internalize. Further studies using different aptamer concentrations, different incubation times, and different tagging methods will be essential to determine what methods are best for assessing internalization capacity. Whether C5 is an aptamer that would be useful for cell-specific targeting of HPV-16 transformed cells remains to be determined. Higher specificity might be necessary, particularly if an aptamer (or its target) is to be used as a biomarker.
As mentioned, previous studies have shown that RNA aptamers can be used for delivery of siRNA to cells, both in vitro and in vivo. One possibility would be to generate aptamer-siRNA chimeras that target HPV E6 and/or E7 expression, as has been done with PSMA-specific chimeras for prostate cancer (Dassie et al., 2009; McNamara et al., 2006). These chimeras would enter cells and specifically target viral genes. Such a strategy may allow more efficient and specific delivery of siRNAs to HPV-associated malignancies and could be used in combination with chemotherapeutic agents for treatment of HPV-positive cancers. In the case of HPV transformation, aptamer internalization would not necessarily have to be cell-specific since siRNA against HPV genes would be specific for HPV-transformed cells. Another possible use of cell-internalizing aptamers would be to use them to carry in aptamers that target the E6 and/or E7 proteins specifically. Interestingly, RNA aptamers that associate with 16E7 have been identified (Nicol et al., 2013; Toscano-Garibay et al., 2011). Theoretically, the internalizing aptamer and the E7 aptamer could be linked to create a chimera that could both internalize and inhibit E7.
In summary, the cell-SELEX strategy we utilized allowed us to identify aptamers that internalize into HPV transformed tonsillar epithelial cells. These aptamers may be of use for delivery of HPV-specific therapeutics to HPV-transformed head and neck epithelial cells. Further studies will be necessary to increase specificity and to characterize the mechanism of entry. In addition, it will be important to test the identified aptamers for the ability to internalize into HPV-positive cancer cell lines.
Highlights.
Experience with cell-SELEX is to isolate internalizing RNA aptamers is described
Aptamers that internalize into HPV-16 transformed tonsillar epithelial cells are identified
Identifying aptamers with high specificity for transformed cells is problematic
Aptamers may be useful for delivering therapeutic agents to cells
ACKNOWLEDGEMENTS
We thank Xiuying Liu for technical support and John Lee, James McNamara, Frank Hernandez, and Luiza Hernandez for technical advice. The Holden Cancer Center Tissue Procurement Core provided tonsil specimens. Sequencing and flow cytometry was performed at the University of Iowa DNA Core Facility and the Flow Cytometry Facility, respectively. This work was supported by NIH R21DEO19953 awarded to AJK and PHG and a Department of Microbiology Developmental Award (AJK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Moustafa AE, Foulkes WD, Benlimame N, Wong A, Yen L, Bergeron J, Batist G, Alpert L, Alaoui-Jamali MA. E6/E7 proteins of HPV type 16 and ErbB-2 cooperate to induce neoplastic transformation of primary normal oral epithelial cells. Oncogene. 2004;23:350–358. doi: 10.1038/sj.onc.1207148. [DOI] [PubMed] [Google Scholar]
- Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J Biol Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22:5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- Cerchia L, de Franciscis V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010;28:517–525. doi: 10.1016/j.tibtech.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Cerchia L, Esposito CL, Jacobs AH, Tavitian B, de Franciscis V. Differential SELEX in human glioma cell lines. PloS one. 2009a;4:e7971. doi: 10.1371/journal.pone.0007971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. Cell-specific aptamers for targeted therapies. Methods Mol Biol. 2009b;535:59–78. doi: 10.1007/978-1-59745-557-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbro BW, Schneider GB, Klingelhutz AJ. Co-regulation of p16INK4a and migratory genes in culture conditions that lead to premature senescence in human keratinocytes. Journal of Investigative Dermatology. 2005;125:499–509. doi: 10.1111/j.0022-202X.2005.23844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, 2nd, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, Spear ME. Knowledge of human papillomavirus and perceived barriers to vaccination in a sample of US female college students. J Am Coll Health. 2010;59:186–190. doi: 10.1080/07448481.2010.493189. [DOI] [PubMed] [Google Scholar]
- Dua P, Kim S, Lee DK. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54:215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Duffy CL, Phillips SL, Klingelhutz AJ. Microarray analysis identifies differentiation associated genes regulated by HPV-16 E6. Virology. 2003;314:196–205. doi: 10.1016/s0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The Human Papilloma Virus-16 E7-Oncoprotein Is Able to Bind to the Retinoblastoma Gene-Product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Giangrande PH, Zhang J, Tanner A, Eckhart AD, Rempel RE, Andrechek ER, Layzer JM, Keys JR, Hagen PO, Nevins JR, Koch WJ, Sullenger BA. Distinct roles of E2F proteins in vascular smooth muscle cell proliferation and intimal hyperplasia. Proc Natl Acad Sci U S A. 2007;104:12988–12993. doi: 10.1073/pnas.0704754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JC, Zarbl H. Use of Cell-SELEX to Generate DNA Aptamers as Molecular Probes of HPV-Associated Cervical Cancer Cells. PloS one. 2012;7:e36103. doi: 10.1371/journal.pone.0036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LI, Stockdale-Flenker K, Hernandez FJ, Klingelhutz AJ, McNamara JO, Giangrande PH. Methods for evaluating cell-specific, cell-internalizing RNA aptamers. Pharmaceuticals. 2013;6:295–319. doi: 10.3390/ph6030295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover AC, Spanos WC, Harris GF, Anderson ME, Klingelhutz AJ, Lee JH. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch Otolaryngol Head Neck Surg. 2007;133:495–502. doi: 10.1001/archotol.133.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SH, Perez-Ordonez B, Weinreb I, Hope A, Massey C, Waldron JN, Kim J, Bayley AJ, Cummings B, John Cho BC, Ringash J, Dawson LA, Siu LL, Chen E, Irish J, Gullane P, Hui A, Liu FF, Shen X, Xu W, O’Sullivan B. Natural course of distant metastases following radiotherapy or chemoradiotherapy in HPV-related oropharyngeal cancer. Oral Oncol. 2013;49:79–85. doi: 10.1016/j.oraloncology.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Huang Y, Eckstein F, Padilla R, Sousa R. Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry. 1997;36:8231–8242. doi: 10.1021/bi962674l. [DOI] [PubMed] [Google Scholar]
- James MA, Lee JH, Klingelhutz AJ. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int J Cancer. 2006;119:1878–1885. doi: 10.1002/ijc.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Roman A. Cellular transformation by human papillomaviruses: lessons learned by comparing high- and low-risk viruses. Virology. 2012;424:77–98. doi: 10.1016/j.virol.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, Meng Y, Li X, Zhao Q, Shen B, Shao N. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J, Zhang X, Rabbani ZN, Liu Y, Reddy SK, Su Z, Salahuddin FK, Viles K, Giangrande PH, Dewhirst MW, Sullenger BA, Kontos CD, Clary BM. RNA aptamer-targeted inhibition of NF-kappaB suppresses non-small cell lung cancer resistance to doxorubicin. Mol Ther. 2008;16:66–73. doi: 10.1038/sj.mt.6300320. [DOI] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Nicol C, Cesur O, Forrest S, Belyaeva TA, Bunka DH, Blair GE, Stonehouse NJ. An RNA Aptamer Provides a Novel Approach for the Induction of Apoptosis by Targeting the HPV16 E7 Oncoprotein. PLoS One. 2013;8:e64781. doi: 10.1371/journal.pone.0064781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Zhu Z, Liu H, Zhang J, Liu J, Tan W. Using aptamers to visualize and capture cancer cells. Anal Bioanal Chem. 2010;397:3225–3233. doi: 10.1007/s00216-010-3715-7. [DOI] [PubMed] [Google Scholar]
- Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan W. DNA aptamers as molecular probes for colorectal cancer study. PLoS One. 2010;5:e14269. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc Chem Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem. 2007a;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem. 2007b;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- Spanos WC, Geiger J, Anderson ME, Harris GF, Bossler AD, Smith RB, Klingelhutz AJ, Lee JH. Deletion of the PDZ motif of HPV16 E6 preventing immortalization and anchorage-independent growth in human tonsil epithelial cells. Head Neck. 2008;30:139–147. doi: 10.1002/hed.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Tao M, McCoy JP, Jr., Zheng ZM. Short-term induction and long-term suppression of HPV16 oncogene silencing by RNA interference in cervical cancer cells. Oncogene. 2006;25:2094–2104. doi: 10.1038/sj.onc.1209244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, 2nd, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012a;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, Behlke MA, Miller FJ, Jr., Giangrande PH. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One. 2012b;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, Bair T, Wyatt Thiel K, Dassie JP, Rockey WM, Howell CA, Liu XY, Dupuy AJ, Huang L, Owczarzy R, Behlke MA, McNamara JO, Giangrande PH. Nucleotide Bias Observed with a Short SELEX RNA Aptamer Library. Nucleic Acid Ther. 2011;21:253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano-Garibay JD, Benitez-Hess ML, Alvarez-Salas LM. Isolation and characterization of an RNA aptamer for the HPV-16 E7 oncoprotein. Archives of medical research. 2011;42:88–96. doi: 10.1016/j.arcmed.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Wan F, Miao X, Quraishi I, Kennedy V, Creek KE, Pirisi L. Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int J Cancer. 2008;123:32–40. doi: 10.1002/ijc.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Kim YT, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Surface-immobilized aptamers for cancer cell isolation and microscopic cytology. Cancer Res. 2010;70:9371–9380. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- Yang J, Xie SX, Huang Y, Ling M, Liu J, Ran Y, Wang Y, Thrasher JB, Berkland C, Li B. Prostate-targeted biodegradable nanoparticles loaded with androgen receptor silencing constructs eradicate xenograft tumors in mice. Nanomedicine. 2012;7:1297–1309. doi: 10.2217/nnm.12.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel Dual Inhibitory Function Aptamer-siRNA Delivery System for HIV-1 Therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]