Abstract
Microtubules (MTs) and their associated proteins (MAPs) play important roles in vesicle and organelle transport, cell motility and cell division. Perturbation of these processes by mutation typically gives rise to severe pathological conditions. In our efforts to obtain atomic information on MAP/MT interactions with the goal to understand mechanisms that might potentially assist in the development of treatments for these diseases, we have determined the 3D structure of CAP-Gly domain of mammalian dynactin by MAS NMR spectroscopy. We observe two conformations in the β2 strand encompassing residues T43-V44-A45, residues that are adjacent to the disease associated mutation, G59S. Upon binding of CAP-Gly to microtubule plus-end tracking protein EB1, the CAP-Gly shifts to a single conformer. We find extensive chemical shift perturbations in several stretches of residues of CAP-Gly upon binding to EB1, from which we define accurately the CAP-Gly/EB1 binding interface. We also observe that the loop regions may exhibit unique flexibility, especially in the GKNDG motif, which participates in the microtubule binding. This study in conjunction with our previous reports suggests that conformational plasticity is an intrinsic property of CAP-Gly likely due to its unusually high loop content and may be required for its biological functions.
Keywords: solid-state NMR, magic angle spinning, MAS, structure determination, dynactin, CAP-Gly, p150Glued domain, end-binding protein EB1, distal spinal bulbar muscular atrophy, dSBMA
Introduction
In eukaryotic cells, microtubules (MTs), together with actin and intermediate filaments, constitute the cytoskeleton system. Microtubules and their associated proteins play significant roles in a broad range of physiological functions, including cell migration, mitosis, polarization, differentiation, vesicle and organelle transport.1
Dynactin, an activator and the dominant cofactor of dynein, is a large (1.2 MDa) multisubunit complex that bridges vesicles to the MT network and is crucial to dynein motor mediated retrograde transport.2 The domain structure of dynactin is shown schematically in Figure 1. Dynactin binds dynein directly and assists dynein in its directed motion along the microtubule lattice over long distances. A central component of dynactin is the p150Glued subunit that is located in the projecting arm portion of dynactin.2 The p150Glued subunit consists of the globular head located at the very tip of the dynactin arm and the two predicted coiled coil regions. The globular head of p150Glued contains a conserved CAP-Gly (cytoskeleton-associated protein, glycine-rich) domain that binds to microtubules both in vitro3 and in vivo4. Another important binding partner of the CAP-Gly domain of the dynactin’s p150Glued subunit is an end-binding protein EB1.5; 6 EB1 is a 30-kDa homodimeric plus end tracking protein (+TIP).5; 6 +TIPS localize at and track the growing ends of microtubules, regulating their dynamics, growth and stability. While the precise regulation mechanism is unknown, direct CAP-Gly/EB1 interactions are thought to be important in this process. For instance, accumulating evidence suggests that EB1 and microtubules utilize the same binding site on the CAP-Gly domain of the CLIP-170.7 In the case of dynactin’s CAP-Gly, an additional binding interface with EB1 has been identified, which is comprised of residues A49-T54.6
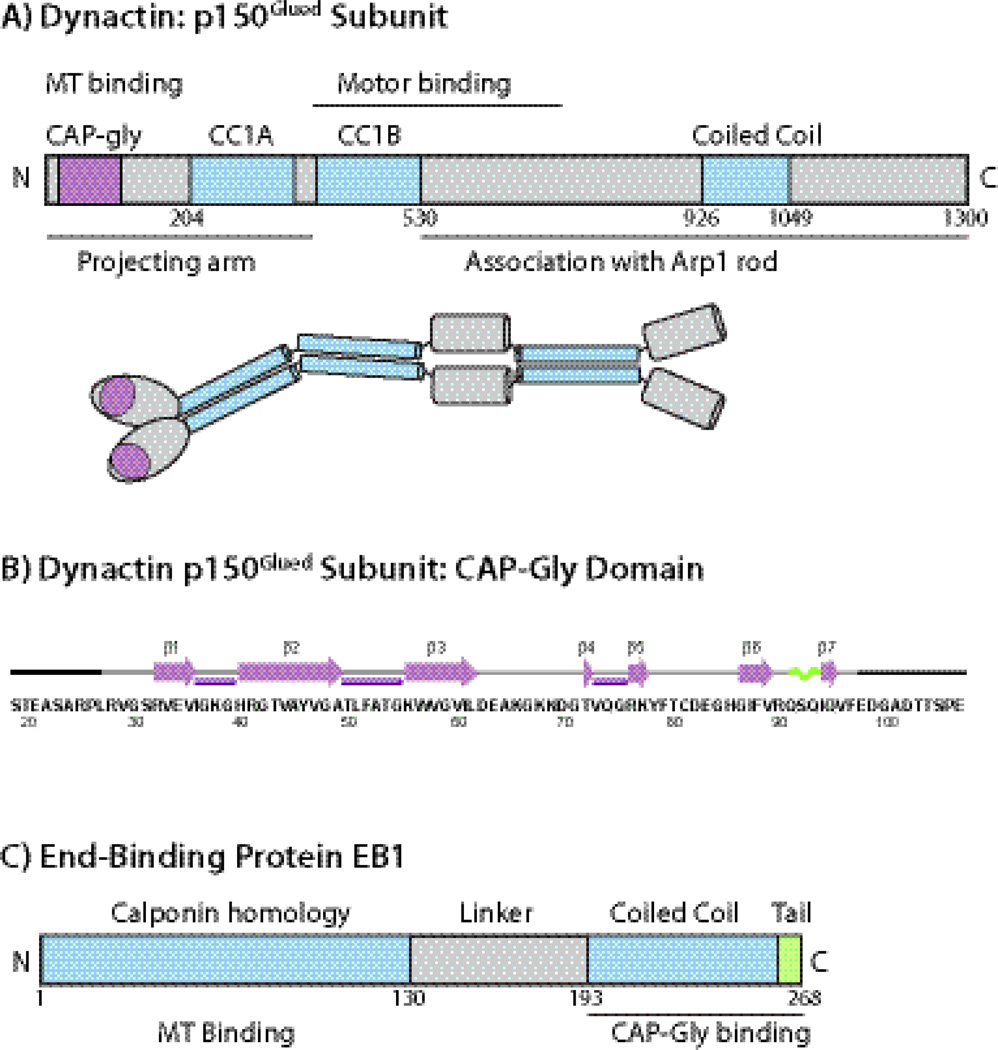
Fig. 1.
A) The domain structure and overall organization of dynactin’s p150Glued subunit. The CAP-Gly domain is located at the N-terminus of p150Glued subunit. B) Amino acid sequence and secondary structure of the CAP-Gly domain of dynactin under investigation. The secondary structures of residues L27-F97 are based on the TALOS+-derived torsion angles from solution and solid-state NMR spectroscopy and from the X-ray structures. Residues corresponding to the N-terminus (19–26) and C-terminus (98–107) do not give rise to cross peaks in the C-C correlation spectra, and were not included in the 3D structure calculation. C) The domain structure of EB1. The C-terminal dimerization domain is coiled coil with a disordered acidic tail and provides the CAP-Gly binding sites. The EB1 protein under investigation is the C-terminal domain encompassing residues 193–268.
CAP-Gly domains play important roles in many microtubule-associated proteins, which, in addition to the p150Glued subunit of dynactin,2 include α-tubulin folding factor,8 tubulin chaperone,9 cytoplasmic linker proteins (CLIPs and CLIPRs),10; 11 the familial cylindromatosis cellular suppressor CYLD,12 and the kinesin protein KIF13b.13 The CAP-Gly domains are conserved in many eukaryotes, from yeast to humans. Fig. 1 illustrates the amino acid sequence and the MAS NMR-derived14 secondary structure of mammalian dynactin CAP-Gly domain under investigation.
Mutations in this domain are associated with various neurological disorders.15; 16; 17; 18 For example, a G59S point mutation in CAP-Gly domain has been identified in patients with distal spinal bulbar muscular atrophy (dSBMA).15; 19 In vivo and in vitro studies suggest that this mutation causes misfolding of the CAP-Gly motif, and reduces the binding affinity of the mutant to microtubules.15; 16; 17 Five other mutations, G71A, G71E, G71R, T72P, and Q74P, were identified in patients diagnosed with Perry’s syndrome, a severe neurological disease whose manifestations are Parkinsonism and weight loss accompanied by depression, social withdrawal, and suicidal attempts.18 While early studies hypothesized that as the result of all these mutations binding to microtubules is disrupted,15; 16; 17 our recent results indicate that the binding affinity to microtubules is very similar or even higher for these mutants while their global fold and stability are perturbed with respect to the wild type protein, and binding to an auxiliary end-binding protein EB1 is abrogated.20 Taken together, these findings suggest that both the structure and dynamics of the CAP-Gly domain may play significant roles in the regulation of CAP-Gly’s binding to microtubules.
To further understand the mechanism of dynactin’s binding to microtubules and the mechanisms of dynein-based cargo transport, it is necessary to investigate the atomic-resolution structure of the CAP-Gly domains assembled on the microtubules. There are several high-resolution structures of CAP-Gly in complex with their interaction partners EB1 and Zn-knuckle domain of CLIP170 (ZnCLIP), determined by X-ray crystallography,6; 21; 22; 23 as well as a solution NMR structure of free CAP-Gly deposited in the Protein Data Bank (PDB file 2COY). The possible binding interfaces of CAP-Gly in complex with EB1 were determined from the analysis of the X-ray structures or from NMR chemical shift perturbations; the interaction interfaces of CAP-Gly with microtubules were also indirectly inferred from such analyses. These studies concluded that the highly conserved GKNDG motif is involved in binding to microtubules and also participates in the tubulin detyrosination-tyrosination cycles.5; 6; 9; 21; 24; 25; 26
Despite numerous biochemical and biophysical investigations summarized above, the structure of the binding interface between CAP-Gly domain and polymerized microtubules remains unknown, particularly at the atomic level, due to the large molecular size, the insolubility and the lack of long-range order of CAP-Gly/microtubule assemblies which have precluded their detailed structural and dynamics studies by X-ray crystallography and solution NMR spectroscopy so far. In contrast, magic angle spinning (MAS) NMR spectroscopy does not require solubility or long-range order, and there are no limitations on the molecular weight of the system under investigation; therefore, this approach is uniquely suited for gaining atomic-level insight into the structure and dynamics of assemblies of microtubules with their associated proteins, such as CAP-Gly. We have previously reported solution and solid-state NMR analysis of CAP-Gly in complex with polymerized microtubules and have demonstrated that the CAP-Gly/microtubule assembly gives rise to high-quality solid-state NMR DARR spectra, which reveal multiple chemical shift perturbations in CAP-Gly upon binding to the microtubules.14 These studies have established the feasibility of detailed structural and dynamics analysis of CAP-Gly/microtubule assemblies by MAS NMR spectroscopy. To understand the conformational and dynamic differences in CAP-Gly induced by binding to microtubules, knowledge of its 3D structure in the free form solved by solid-state NMR methods is essential.
In this work, we present the 3D structure of CAP-Gly solved by MAS NMR spectroscopy, as our starting point for comprehensive structural and dynamics analysis of CAP-Gly/MT assembly by magic angle spinning methods. Analysis of MAS NMR spectra of CAP-Gly acquired at 19.9 and 14.1 T reveals that residues T43, V44, and A45 exhibit two distinct conformations. This stretch of residues is in close spatial proximity to G59, whose mutation to serine results in dSBMA disorder (see above).15; 19 Our results indicate the 3D MAS NMR CAP-Gly structures exhibit large positional and torsion angle deviations in the loops compared to the regions of the rigid secondary structure elements (α-helices and β-sheets), both within the ensemble of computed structures and with respect to the existing X-ray structures of CAP-Gly complexes with EB1 and ZnCLIP.6; 21; 22; 23 Considerable deviations between the torsion angles derived from the MAS NMR and X-ray structures are observed in the highly conserved GKNDG motif involved in CAP-Gly domain binding to microtubules5; 6; 9; 21; 24; 25; 26 and in residues spatially proximal to this motif, which indicates unique structural variability of this region.
Formation of the CAP-Gly complex with EB1 gives rise to multiple chemical shift perturbations, from which we infer the CAP-Gly residues constituting the binding interface. Our results are in excellent agreement with the X-ray diffraction studies.6; 27 Strikingly, the complexed CAP-Gly is locked into a single conformer around the T43-A45 stretch of residues. Furthermore, spectral resolution is dramatically enhanced in the complex compared with the free CAP-Gly, suggesting a more rigid structure.
The findings discussed in this work in conjunction with our recent report establishing conformational flexibility of loop regions of CAP-Gly28 suggest that inherent conformational plasticity of CAP-Gly and in particular of the GKNDG motif giving rise to these different conformations may be essential for the dynactin’s interactions with microtubules and its binding partners and for walking along the microtubules.
Results and Discussion
Distance Restraints
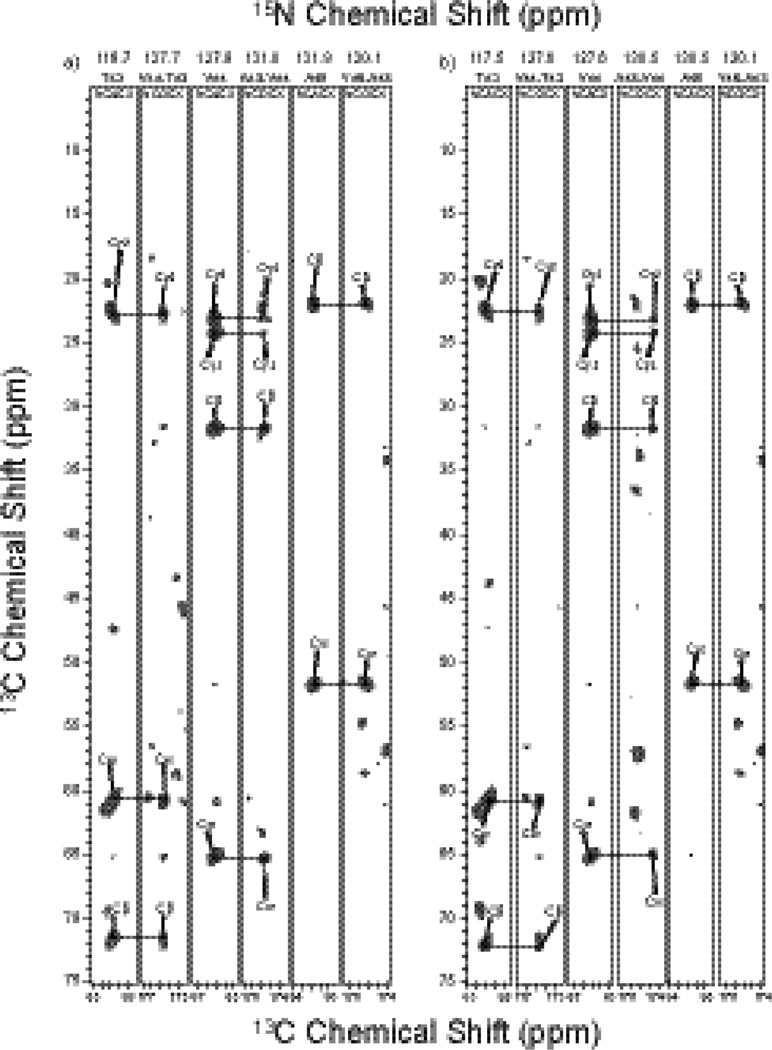
Protein structure determination by MAS NMR spectroscopy requires the measurement of a large number of distance restraints, most commonly, by detecting 13C-13C correlations. In order to extract 13C-13C distance restraints, R211 (PARIS) experiments29; 30 were performed with the mixing times of 50 ms, 200 ms, and 500 ms, on [2-13C]-glycerol/U-15N and [1,3-13C]-glycerol/U-15N CAP-Gly at T = −2 °C (the spectra are shown in Fig. 2, and Fig. S1 and S2 of the Supplementary). The backbone assignments of [U-13C, 15N]-CAP-Gly were reported by our group previously.14 We have used these assigned chemical shifts for the analysis of the various R211 spectra presented in this work. The details of the assignment strategy are described in and illustrated in Fig. S1 and S2 of the Supplementary.
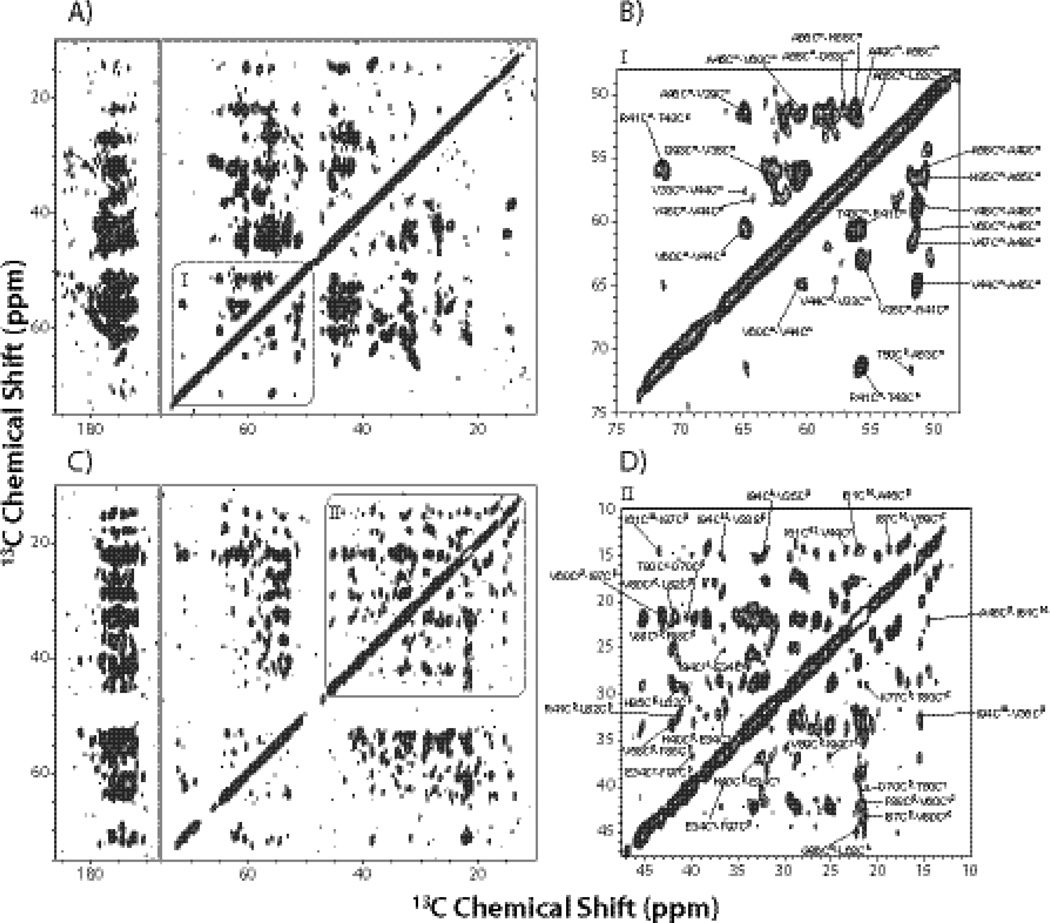
Fig. 2.
14.4 T R211 spectra of the [2-13C]-glycerol/U-15N labeled CAP-Gly (A and B), and the [1,3-13C]-glycerol/U-15N labeled CAP-Gly (C and D), acquired with the mixing time of 500 ms. In B) and D), the insets showing region I and II illustrate examples of assignments corresponding to medium- and long- range correlations.
In the 200 ms spectra, the inter-residue correlations represent mostly sequential contacts with several exceptions of some weak peaks corresponding to long-range correlations (see Fig. S1 and S2 of the Supplementary). As shown in the expanded regions of Fig. 2 depicting the 500 ms R211 spectra of the two CAP-Gly samples under investigation, a large number of inter-residue Cα-Cαbackbone correlations were assigned in [2-13C]-glycerol/U-15N CAP-Gly, and multiple inter-residue side-chain correlations were assigned in [1,3-13C]-glycerol/U-15N CAP-Gly as well. By combining all correlations from the six spectra, we obtained 917 distance restraints in total, including 197 long-range (|i−j| ≥ 5) distance restraints for the final structure refinement, as summarized in Table 1.
Table 1.
Summary of the CAP-Gly Structure Ensemble from the Final Refinementa.
| Restraintsb | Total number |
| Total distance restraints | 917 |
| Restraints with multiple assignments | 175 |
| Intra-residue | 436 |
| Sequential (|i−j|=1) | 187 |
| Medium range (1<|i−j|<4) | 97 |
| Long range (|i−j|>4) | 197 |
| H-bond distance restraints | 54 |
| Total (Φ,Ψ) torsional angle restraints | 138 |
| Energy terms | Average values (kcal/mol) |
| Total | −490.20 ± 34.60 |
| NOE | 28.00 ± 4.17 |
| ANGL | 43.95 ± 4.52 |
| BOND | 5.89 ± 1.82 |
| CDIH | 3.60 ± 1.26 |
| HBDB | −67.52 ± 4.21 |
| IMPR | 5.01 ± 1.62 |
| RAMA | −569.69 ± 26.94 |
| VDW | 60.55 ± 22.18 |
| Violations | Restraint name |
| NOEPot term (1 violation) | (62 LEU CA) (61 ILE CG1) |
| CDIH term (1 violation) | (37 GLY N) (37 GLY CA) (37 GLY C) (38 LYS N) |
| VDW term (1 violation) | (35 VAL HB) (39 GLY HA1) |
The final structure ensemble includes 10 lowest-energy structures.
Each distance restraint represents a unique correlation between a pair of atoms, and is counted once for each atomic pair.
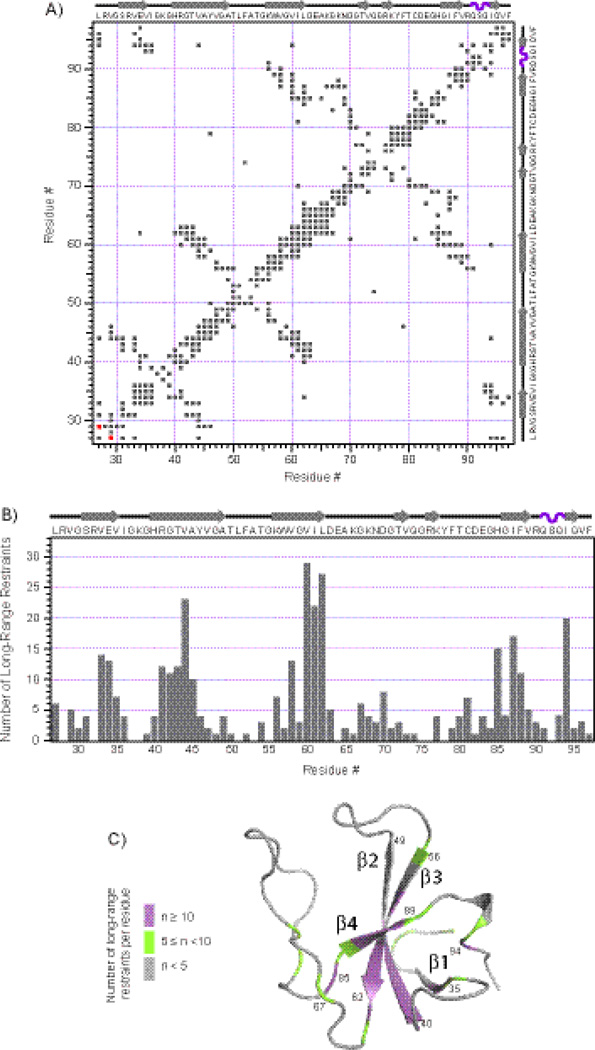
In Fig. 3 (A), a 2D contact map is shown in which a shaded square indicates that there is one or more distance restraint (corresponding to a unique final assignment) between the corresponding residues. In Fig. 3 (B), the number of long-distance restraints is plotted for each residue, illustrating that on average, 5.6 long-range restraints per residue were derived from MAS NMR spectra. Not surprisingly, the results indicate that significantly more long-range distance restraints (9.8 per residue on average) are present in the regions corresponding to rigid secondary structure elements (β-sheets and α-helices) compared to the loop regions (2.8 long-range restraints per residue on average). In Fig. 3 (C), the number of long-range restraints is mapped onto the 3D structure of CAP-Gly determined from the MAS NMR restraints. It is also instructive to identify the violations in the 917 MAS NMR-derived distance restraints with respect to the corresponding distances in the X-ray structure 2HQH.23 As shown in Fig. 3(A), there is only one medium-range restraint (L27Cδ-V29Cγ1 depicted in red) that is in violation with 2HQH. This finding is highly unlikely to bear any significance for either of the structures (our SSNMR or X-ray) in light of the fact that 2HQH represents a complex of CAP-Gly with ZnCLIP in contrast to the free CAP-Gly studied here.
Fig. 3.
Distance restraints contact map and statistics. A) A matrix representation of all the intra- and inter-residue contacts generated from the distance restraints corresponding to final assignments of the 2D MAS NMR spectra (in gray). The contact depicted in red represents the only one medium-range distance restraint that is in violation with the X-ray structure 2HQH. B) The number of long-range restraints per residue is plotted in the context of the secondary structure of CAP-Gly. C) The number of long-range restraints per residue is mapped onto the 3D structure of CAP-Gly calculated from the MAS NMR restraints (average from the ensemble of ten lowest-energy structures). Note that the N-terminus residues 19–26 and C-terminus residues 98–107 did not appear in any of the 2D MAS NMR spectra and were omitted from the structure calculation.
Backbone Dihedral Angles and Secondary Structure Prediction
In addition to the distance restraints, backbone dihedral angle restraints were also essential for CAP-Gly structure determination, and these not only helped define secondary structures but also improved the accuracy of calculated 3D structures. The backbone torsion angles were derived from the solid-state NMR chemical shifts using the TALOS+ program.31 The chemical-shift-constrained backbone torsion angles (Ф, Ψ) were mostly consistent with the secondary structures determined by X-ray crystallography, as illustrated in Fig. S3 of the Supplementary. As expected, for most of the residues comprising β strands the X-ray and MAS NMR torsion angles agree very well. The largest deviations between X-ray and MAS NMR torsion angles were found for residues belonging to flexible loop regions (G37, K38, A65, K66, K68, D70, G71, and H85) or termini of β strands (G48 and G86). Interestingly, the residues K68-G71 belong to the conserved GKNDG motif, and residues A65, K66, H85 and G86 are in close spatial proximity to the GKNDG motif. This result is not surprising in light of the following: i) as discussed above, the X-ray structures represent complexes of CAP-Gly with EB1 or ZnCLIP, and these complexes are heterotetrameric or heterooctameric species formed upon binding of CAP-Gly to its protein partners; ii) the X-ray data were collected at cryogenic temperatures, in contrast to the MAS NMR spectra acquired at temperatures between −2 °C and 4 °C (arguably, the NMR temperatures are closer to physiological conditions), and iii) our recent work indicates that loop regions of CAP-Gly are flexible on ns-us timescales, as revealed by lower H-C dipolar order parameters compared to the values observed in the β-sheet and α-helical regions of the protein.28 We also refer the reader to the previous reports on microcrystalline proteins from our group,14; 31; 32; 33; 34; 35; 36 and from others37; 38 where similar observations were made for torsion angles in loops vs. rigid secondary structure elements.
3D Structure
The ensemble of 3D structures of CAP-Gly was calculated ab initio from the combination of unambiguous and ambiguous 13C-13C distance as well as backbone torsion angle restraints. The correlations derived from MAS NMR experiments were assigned to different distance ranges based on the cross peak volume and the mixing times at which the correlations were recorded. For the correlations arising from a spectrum acquired with a given mixing time, several cross peaks from one-bond correlations were selected, and the average integrated peak intensity was calculated and used as calibrated peak intensity.39 For other peaks arising from intra- or inter-residue correlation, we assigned the correlations as strong, medium and weak, on the basis of the ratio of their integrated intensity to the calibrated peak intensity, following the strategy described by Franks et al.39 The distance boundaries were defined for each intensity class at each mixing time, from a minimum distance of 1 Å to a maximum distance, as shown in Table 2. All intra- or inter-residue correlations that we assigned in the R211 spectra and the resulting distance restraints were classified according to Table 2. The 917 distance restraints defined above and 138 (Ф, Ψ) torsion angle restraints were utilized in structure calculation using version 2.29 of the program Xplor-NIH.40; 41 Table 1 shows the statistics of distance and backbone torsion angle restraints used for the final structure refinement.
Table 2.
The Distance Boundaries Used in the CAP-Gly Structure Calculation, Classified by Weak, Medium and Strong Peaks in the R211 MAS NMR Spectra Acquired with Different Mixing Times.
| τmix, ms | Strong, Å | Medium, Å | Weak, Å |
|---|---|---|---|
| 50 | 1.0–3.3 | 1.0–4.5 | 1.0–5.6 |
| 200 | 1.0–4.8 | 1.0–5.7 | 1.0–7.0 |
| 500 | 1.0–5.8 | 1.0–6.7 | 1.0–8.2 |
In Fig. 4, the 3D structure calculation results are summarized. The ten lowest-energy structures out of five hundred structures calculated by the above refinement protocols are depicted in Fig. 4 (A). There is only one NOE violation, one dihedral angle (CDIH) violation and one VDW violation in the final refinement (shown in Table 1). As summarized in Table 3, the RMSD values within the ensemble of the above ten structures are 0.54 ± 0.12 Å for all backbone heavy atoms (N, Cα, and C), and 0.25 ± 0.05 Å for backbone heavy atoms located in rigid secondary structures (α-helices and β-strands). The summary of our structure quality validation is presented in the Supplementary, Table S4. The equivalent resolution according to ProCheck-NMR27 is 1.48 Å (with respect to average value), 1.7 Å (with respect to percentage of residues in helices, sheets, and loops), 1.9 Å (with respect to hydrogen bond energy standard deviation), 1.3 Å (with respect to χ-1 pooled standard deviation), and 1.0 Å (with respect to standard deviation of χ-2 trans angle).
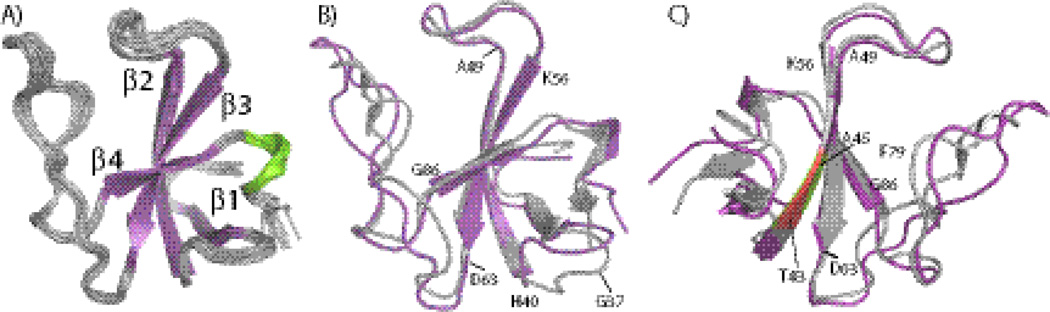
Fig. 4.
SSNMR structures of CAP-Gly compared with X-ray structure 2HQH. A) The ensemble of the ten lowest-energy structures of CAP-Gly determined from MAS NMR distance restraints and backbone dihedral angle restraints using the Xplor-NIH structure calculation protocol. B) The average structure from the ensemble shown in A) (purple) overlaid with the X-ray structure 2HQH (grey). C) The back view of B), where the stretch T43-V44-A45 is highlighted to illustrate its position in the two structures.
Table 3.
The RMSD between the Average MAS NMR Structure and Various X-ray and Solution NMR Structures of CAP-Gly.a
| Backbone, Åb |
Heavy atoms, Åb |
Backbone vs. 1TXQ, Å |
Backbone vs. 2COYc, Å |
Backbone vs. 2HKQ, Å |
Backbone vs. 2HQH, Å |
Backbone vs. 3E2U, Å |
|
|---|---|---|---|---|---|---|---|
| All residues | 0.54 ± 0.12 | 1.14 ± 0.10 | 1.66 | 1.79 | 1.68 | 1.70 | 1.71 |
| Residues in secondary structure | 0.25 ± 0.05 | 0.96 ± 0.12 | 0.58 | 0.70 | 0.60 | 0.56 | 0.60 |
Note that X-ray and solution NMR structures represent systems different from CAP-Gly under investigation: X-ray structures are those of complexes of CAP-Gly and their binding partners EB1 and ZnCLIP while solution NMR structure is of different CAP-Gly contruct.
Fit of the ensemble of the 10 lowest energy structures to the average structure.
This is a solution NMR structure of CAP-Gly deposited in the Protein Data Bank (but not yet published).
We have also examined our MAS NMR structure in the context of the existing p150Glued CAP-Gly structures. To date, several high-resolution structures have been reported of p150Glued CAP-Gly in complex with its interaction partners EB1 and Zn-knuckle domain of CLIP170 (ZnCLIP), determined by X-ray crystallography,6; 21; 22; 23 and another solution NMR structure of free p150Glued CAP-Gly has been deposited in the Protein Data Bank (PDB file 2COY). In these studies, slightly different CAP-Gly constructs were used compared to the CAP-Gly construct utilized in this work (see Fig. S6 of the Supplementary). The analysis indicates that the overall backbone atom RMSDs between the average NMR structure (see Materials and Methods) and the four X-ray structures of CAP-Gly in complex with EB1 or ZnCLIP, are in the range of 1.66–1.71 Å, as shown in Table 3. It is clear that the MAS NMR and the X-ray structures have the same overall topology. The average MAS NMR structure contains several rigid secondary structure elements: four well-defined β-strands and a short 310 helix. These rigid regions of the protein have an RMSD of 0.56–0.60 Å with respect to the X-ray structures, considerably lower than that for the entire backbone (see above). In Fig. 4 (B), the average CAP-Gly structure is overlaid onto the 2HQH X-ray structure of the CAP-Gly/ZnCLIP complex, and it is clear the deviations are generally small in the regions of the rigid secondary structure elements, whereas significant differences can be found for the loop regions (See Fig. S3 in Supplementary). Interestingly, there are two short β-stands (β4 and β5) located in the long loop regions connecting the β3 and β6 strands in the X-ray structures and a very short β-stand (β7) close to the C-terminus, but the average MAS NMR structures do not have these short β-strands.
There are two possible explanations for considerable positional and torsion angle deviations in the loop regions as well as for the missing short β-strands in the MAS structure compared to the X-ray structure 2HQH:
The loop regions (including residues G37-G39, T50-G55 and D63-G84) are far away from the β strands in the core of CAP-Gly, and the distances between the above loop residues and the closest β strand are usually greater than 7.0 Å according to the X-ray structures.6; 21; 22; 23 The upper detectable 13C-13C distance range is 7.0–8.0 Å for solid-state NMR spin diffusion experiments,37; 39; 42; 43 limiting the number of useful long-range restraints for the long loop region.
The loops in CAP-Gly are inherently conformationally flexible. This inherent flexibility of the loop regions in CAP-Gly on nano- to microsecond timescales has been evidenced by their reduced 1H-13C dipolar order parameters with respect to those of β-sheets and α-helices, reported in our recent work.28 Additional evidence of the loop flexibility comes from the X-ray crystallographic B factors. In Fig. S4 of the Supplementary, the plot of the B-factors of amide N for each residue against the residue number indicates that loop regions exhibit higher B-factors than β-sheets and α-helices and a significant variability in B factors across the different X-ray structures, indirectly suggesting different mobility among the various samples. Taken together, our 1H-13C dipolar order parameters and X-ray B factors consistently indicate that loops in CAP-Gly are mobile and can adopt different conformations depending on the conditions. As discussed below, we have conducted a MAS NMR study of the CAP-Gly/EB1 complex and detected significant conformational changes upon formation of the complex, consistent with the second possibility (vide infra).
Different Conformations of CAP-Gly Revealed by MAS NMR Spectroscopy
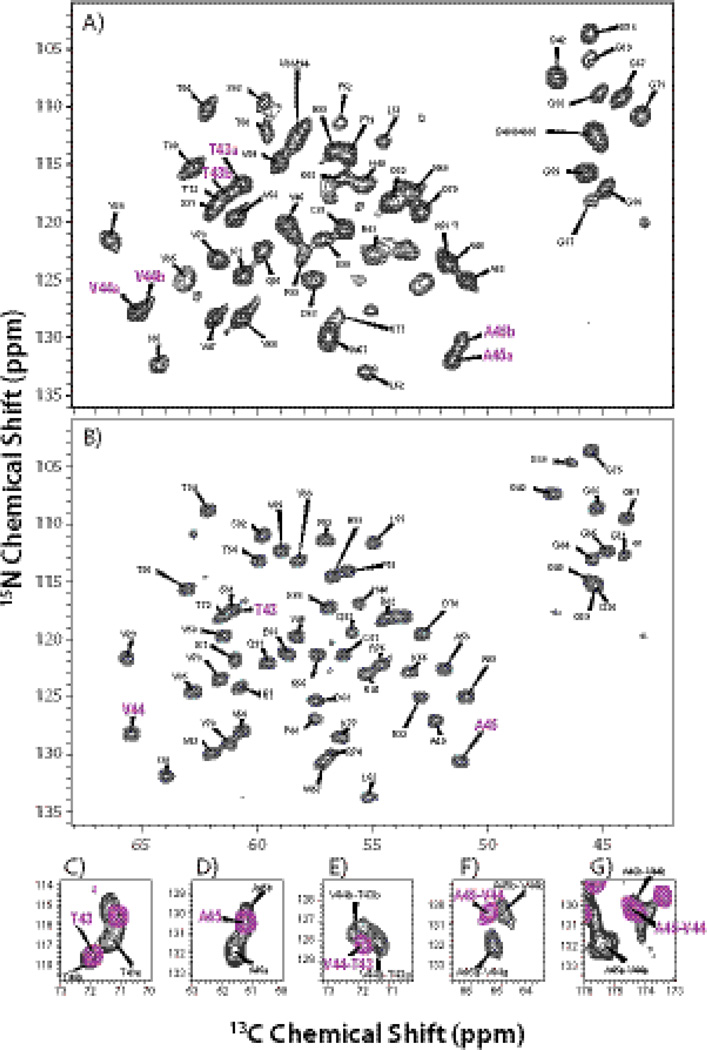
In several heteronuclear experiments conducted at 14.1 T, we found that one special residue, A45, exhibits double peaks. Fig. S5 displays the 2D NCA spectra of [2-13C]-glycerol/U-15N CAP-Gly sample at −2 °C and −20 °C, as well as the expanded regions of relevant planes of 3D NCACB and 3D NCACX data sets acquired at −20 °C. Interestingly, peak doubling in A45 is observed at both temperatures in the 2D and 3D heteronuclear spectra, while the chemical shifts of the corresponding peaks exhibit relatively small differences. The fact that there are two peaks for A45 indicates that CAP-Gly exists as two conformers, which we designate as conformer a and conformer b (A45a and A45b), as shown in Fig. S5.
At 19.9 T, the spectra of U-13C, 15N CAP-Gly show dramatically improved resolution and sensitivity compared to the 14.1 T datasets. As shown in Fig. 5 and 6 (A), in addition to A45, T43 and V44 also give rise to a pair of peaks with different chemical shifts, which could not be seen at 14.1 T due to their low intensity and overlap with other resonances. Backbone walks through this stretch of residues are shown in Fig. 5 for each of the two conformers; from these walks sequential resonance assignments were derived. Table 4 summarizes the chemical shifts of the two conformers of T43-V44-A45 detected at 19.9 T. We note that while the peaks associated with T43-V44-A45 are clearly doubled, the small chemical shift differences between peaks a and b do not result in significantly different TALOS+ torsion angle predictions. As a result, the calculated ensemble of structures does not retain a signature of this conformational heterogeneity. This may be not surprising also in light of the fact that chemical shifts are sensitive to environmental factors other than torsion angles. These include any electrostatic effects (e.g., hydrogen bonding with other residues or proximity to hydration water molecules or changes in protonation states of nearby residues), differences in hydrophobic environment in the vicinity of the residue (e.g., sidechain conformation of hydrophobic residues in the vicinity of the atom in question), as well as dynamic effects.
Fig. 5.
Backbone walk for residues T43-V44-A45 for conformer a (A) and conformer b (B) using 3D NCACX and NCOCX spectra acquired at 19.9 T.
Fig. 6.
The NCA spectra of free U-13C,15N CAP-Gly (A) and U-13C,15N CAP-Gly in complex with EB1 (B) acquired at 19.9 T. The labels of the cross peaks corresponding to T43, V44, and A45 are shown in purple. In (C) and (D), the overlaid 2D NCACX spectra of free CAP-Gly and CAP-Gly in complex with EB1 illustrate: (C) the two conformers of T43a and T43b in free CAP-Gly (black) and a single peak of T43 (purple) of CAP-Gly in complex; (D) the two conformers of A45a and A45b in free CAP-Gly (black) and a single peak of A45 (purple) of CAP-Gly in complex. In (E), (F) and (G), the overlaid inserts of NCOCX spectra of free CAP-Gly and CAPGly in complex are presented, illustrating the sequential cross peaks corresponding to two conformers in free CAP-Gly: V44a-T43a and V44b-T43b, A45a-V44a and A45b-V44b (all shown in black), and to a single conformer of CAP-Gly in complex with EB1 V44-T43 and A45-V44 (shown in purple).
Table 4.
13C and 15N Chemical Shifts of Conformer a and Conformer b of T43-V44-A45 for free CAP-Gly, and the single conformer of T43-V44-A45 for CAP-Gly in complex with EB1.
| Conformer a of free CAP-Gly | ||||
| N (ppm) | Cα (ppm) | Cβ (ppm) | C (ppm) | |
| T43 | 116.95 | 60.65 | 71.40 | 174.66 |
| V44 | 127.88 | 65.22 | 31.67 | 176.01 |
| A45 | 131.89 | 51.69 | 22.01 | 176.28 |
| Conformer b of free CAP-Gly | ||||
| N (ppm) | Cα (ppm) | Cβ (ppm) | C (ppm) | |
| T43 | 117.30 | 60.99 | 72.14 | 174.68 |
| V44 | 127.56 | 65.01 | 31.71 | 174.73 |
| A45 | 130.60 | 51.56 | 21.97 | 176.38 |
| CAP-Gly in complex with EB1 | ||||
| N (ppm) | Cα (ppm) | Cβ (ppm) | C (ppm) | |
| T43 | 117.51 | 61.05 | 71.94 | 174.33 |
| V44 | 128.16 | 65.44 | 31.54 | 174.38 |
| A45 | 130.59 | 51.37 | 22.38 | 176.63 |
The above three residues are located in the twisted β2 strand with A45 being in the center of this strand, as shown in Fig. 4 (C). These three residues are also close to two loops – the loop N-terminal to β1 and the dynamic long loop N-terminal to the β-strand consisting of G86-V89. This stretch of residues is also in spatial proximity to G59, which forms a bifurcated hydrogen bond with A45 and Y46, and whose mutation to serine is associated with a neurodegenerative disorder dSBMA.15; 19 As demonstrated below, when CAP-Gly forms a complex with EB1, this stretch of three residues is locked into a single conformer. Together with our prior findings,28 these observations lead us to speculate that conformational flexibility of CAP-Gly may be essential for its ability to bind to its various binding partners and walk along the microtubules.
CAP-Gly/EB1 Complex: Single Conformation and Binding Interface of CAP-Gly
Intrigued by the observation of two distinct conformers of CAP-Gly present in the free protein and by their absence in the reported X-ray structures of CAP-Gly in complexes with its binding partners EB1 and ZnCLIP, we pursued MAS NMR characterization of the CAP-Gly/EB1 complex. We note that this complex is a heterotetramer with two molecules of CAP-Gly bound to the EB1 dimer, and the total molecular weight is 36.5 kDa. In this work, we focused our attention on CAP-Gly by preparing the differentially labeled sample where U-13C,15N CAP-Gly is bound to natural abundance EB1 dimer.
In Fig. 6 (B), a 2D NCA spectrum of CAP-Gly/EB1 complex acquired at 19.9 T is shown. Strikingly, the spectral resolution is much higher than that of the free CAP-Gly shown in Fig. 6 (A). From 3D NCACX, NCOCX, CANCX and 2D DARR experiments, we have assigned the chemical shifts for most of the residues of CAP-Gly in complex with EB1. Interestingly, the results reveal substantial chemical shift perturbations in CAP-Gly upon binding to EB1, as summarized in Tables S2 and S3 of the Supplementary.
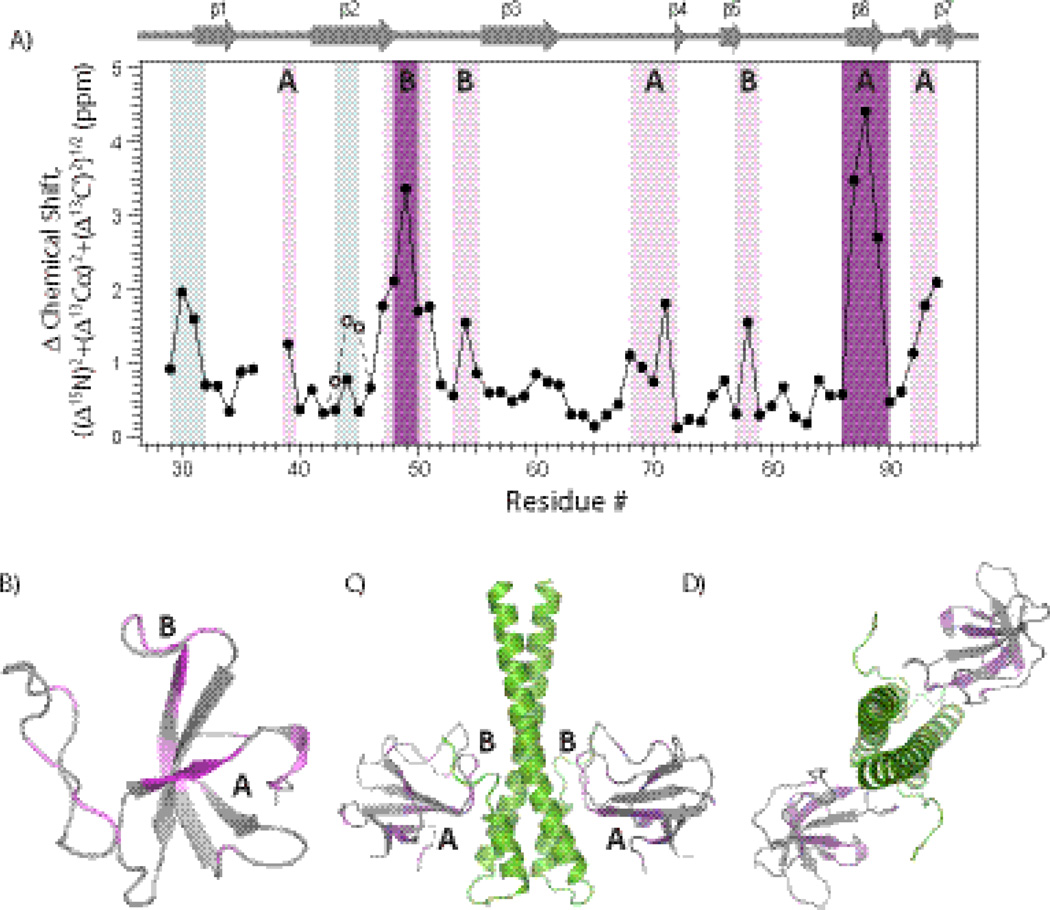
The chemical shift perturbations of backbone atoms (15N, 13Cα, and 13C’) are plotted against the residue number in Fig. 7 (A). For several stretches of residues in CAP-Gly, chemical shift differences greater than 1 ppm are observed upon binding to EB1, with seven residues showing perturbations between 2 and 4.5 ppm. These are 30GS31, 47VGATLFAT54, 68KNDG71, Y78, 87IFV89, and 92SQI94. Interestingly, resonances corresponding to residues G37 and K38 are not present in the 2D and 3D dipolar-based MAS spectra of the complex, possibly due to conformational exchange interfering with their detection.
Fig. 7.
Chemical shift perturbations of CAP-Gly upon binding to EB1. (A) Chemical shift differences (represented as ((Δ15N)2+(Δ13Cα)2+(Δ13C’)2)1/2) between CAP-Gly/EB1(193–268) complex and free CAP-Gly plotted versus the residue number. The residues exhibiting chemical shift perturbations >2 ppm and 1–2 ppm are labeled as dark purple and violet, respectively. The open and filled circles for the stretch of residues T43-A45 represent the chemical shift differences between CAP-Gly/EB1 complex and free CAP-Gly conformers a and b, respectively. The two regions with large chemical shift perturbations (>1 ppm) are labeled as regions A and B. (B) Chemical shift perturbations mapped onto the 3D MAS NMR structure of CAP-Gly. Residues K68-G71, I87-V89 and S92-I94 correspond to the CAP-Gly/EB1 interface A. Residues V47-L51 and T54, comprising the β2 strand and the β2–β3 loop, correspond to the CAP-Gly/EB1 interface B found in the X-ray structure 2HKQ. In (C) and (D), the chemical shift perturbations are mapped onto the X-ray structure 2HKQ of CAP-Gly/EB1 complex. In (C) and (D), EB1 is shown in light green color. The dynamic tail (residues P256-Y268) of EB1 could not be detected in the X-ray structure 2HKQ, but was resolved when synthesized peptide corresponding to this tail was co-crystallized with mutant CAP-Gly.6
Previously, Hayashi et al. reported a solution NMR study of the CAP-Gly domain in complex with the C-terminal tail of EB1 (residues 247–268).22 This investigation revealed significant chemical shift perturbations in CAP-Gly upon formation of the complex for the following residues: G37, A53, G71, 87IFV89, and 92SQI94. In our study, we observe chemical shift perturbations for the same residues as in Hayashi et al., as well as for additional regions of CAP-Gly encompassing residues 47VGATL51, T54, and Y78. We note that in our study, we utilized the full-length C-terminal domain of EB1 spanning residues 193–268, and therefore a more extensive CAP-Gly/EB1 intermolecular interface is formed.
The chemical shift perturbations observed in our MAS NMR work are displayed in the context of the 3D structure of CAP-Gly (Fig. 7 (B)). The results reveal two regions participating in the binding to EB1, designated as A and B in Fig. 7 (A) and (B). These regions correspond to the two intermolecular CAP-Gly/EB1 interfaces found in the previous X-ray studies of CAP-Gly/EB1 complex6 and the complex of EB1-EB3 heterodimer with two CAP-Gly molecules.27 Binding interface A is comprised of the GKNDG motif and several aromatic residues (F52, W57, and F88) of CAP-Gly. According to the X-ray results, this interface contains the EEY motif of the C-terminal tail of EB1, and is the same binding interface observed by Hayashi et al. in solution (see above). In our MAS NMR investigation, stretches 68KNDG71, 87IFV89, and 92SQI94 of CAP-Gly belonging to this interface exhibit large chemical shift perturbations upon binding to EB1. Residues G37 and K38, which are close to the hydrophobic contact, are not assigned in our spectra of the CAP-Gly/EB1 complex as the respective peaks are missing (see above).
Binding interface B consists of the β2–β3 loop (49ATLFAT54) of CAP-Gly. According to the X-ray structure, it is comprised of the EB-like motif of EB1, with the side chain of A49 of CAP-Gly being optimally oriented to insert itself into the hydrophobic cavity of EB1.6 In our MAS NMR investigation, residues 47VGATL51, T54, and Y78 exhibit large chemical shift perturbations in the CAP-Gly/EB1 complex, consistent with them either comprising or being in the vicinity of this binding interface B. For instance, Y78 is in close spatial proximity to A49 and T50, and its chemical shift is perturbed by 1.55 ppm upon formation of the complex. In Fig. 7 (C) and (D), chemical shift perturbations are mapped onto the X-ray structure of CAP-Gly/EB1 complex.6
Remarkably and in contrast to our findings in the free CAP-Gly, we observed that the T43-V44-A45 stretch of residues is locked into a single conformation upon binding with EB1, as shown in Fig. 6 (B). This is also illustrated in Fig. 6 (C–G) depicting the expansions of the heteronuclear correlation spectra around the regions containing cross peaks of these residues for the free CAP-Gly and the CAP-Gly/EB1 complex. The chemical shifts of T43, V44, and A45 for free CAP-Gly and CAP-Gly/EB1 are summarized in Table 4. The chemical shift values for T43, V44, A45 in the CAP-Gly/EB1 complex are intermediate between those for conformer a and conformer b in free CAP-Gly, and closer to those of conformer b.
That the T43-V44-A45 stretch of residues is locked into a single conformation in the CAP-Gly/EB1 complex, in conjunction with the fact that the NMR spectra for the complex exhibit higher resolution, suggests that binding to EB1 may attenuate the internal mobility of CAP-Gly domain. This finding inspires us to pursue comprehensive dynamics studies of CAP-Gly and CAP-Gly/EB1 complex.
Implications of Conformational Plasticity of CAP-Gly for Biological Function and Disease
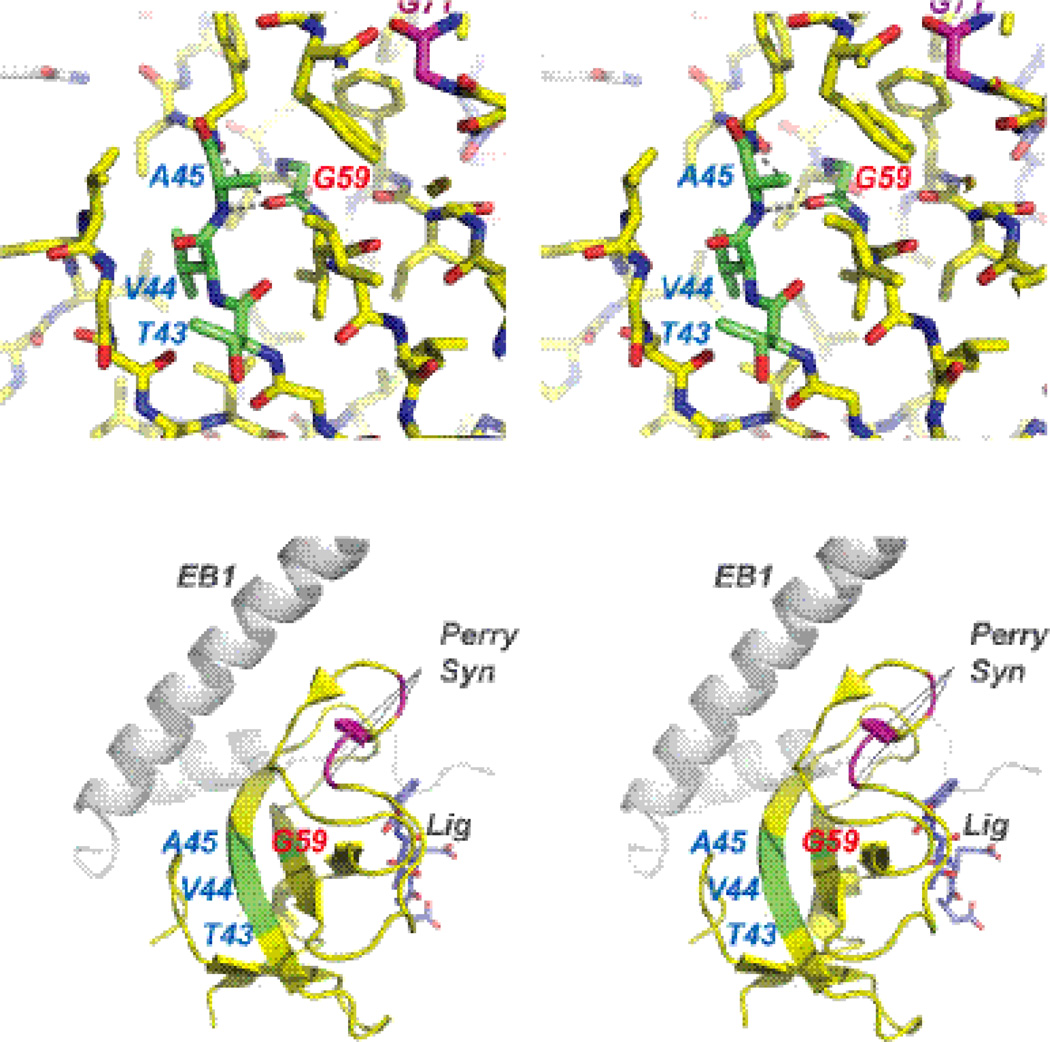
The discovery in this work of two distinct conformations around the T43-V44-A45 region of residues in the unligated CAP-Gly is very exciting and may have direct implications for understanding the molecular mechanisms of disease pathologies. In Fig. 8, we show the stereoview of the T43-V44-A45 region with respect to point mutations associated with neurodegenerative disorders. Residue G59 whose mutation to serine has been found in patients with dSBMA,15; 19 makes a bifurcated hydrogen bond to the backbone of A45 and Y46. In the G59S mutant, this essential interaction is likely disrupted (the structure of this mutant is not available as it is insoluble).
Fig. 8.
Stereoview of the T43-V44-A45 region with respect to point mutations associated with neurodegenerative disorders. A) Residue G59 whose mutation to serine is associated with dSBMA, makes a bifurcated hydrogen bond to the backbone of A45 and Y46. We observe that A45 possesses two distinct conformations in the unlighted form, which collapse to a single conformation upon binding to EB1. B) The dynamic region is comprised by residues T43-V44-A45, which do not directly interact with EB1, the EEY ligand, and are distant from the point mutations associated with Perry's Syndrome.
As discussed above, the two distinct conformations around A45 in the unlighted form of CAP-Gly collapse into a single conformation upon binding to EB1. Remarkably, the residues T43-V44-A45 in this region do not directly interact with EB1, the EEY ligand, and are distant from the point mutations associated with another neurodegenerative disorder, Perry's Syndrome (vide supra). Our previous work revealed that in Perry’s Syndrome mutant, the binding of CAP-Gly to EB1 is abrogated.20 Thus, taken together, the results of this work and of our previous studies point to distinct molecular mechanisms associated with dSBMA and Perry’s syndrome pathologies.
Our results also explain why the X-ray structures of CAP-Gly/EB1 and CAP-Gly/ZnCLIP complexes do not show any signs of conformational heterogeneity: binding of CAP-Gly to its partner proteins locks it into a single conformer. Furthermore, the data reported in this and our recent study28 indirectly suggest that in the free CAP-Gly, very slow motions (on timescales slower than milliseconds) might be present and responsible for the interconversion between the two conformers in the T43-V44-A45 region. It will be instructive to probe the possible presence of such motions in the forthcoming experiments.
Finally, we note that we have previously observed an unusually high degree of conformational changes in point mutants of CAP-Gly associated with a neurological disorder, Perry syndrome,20 as well as in CAP-Gly bound to microtubules.14 As discussed above, we have also observed systematically attenuated dipolar order parameters in the loop regions of CAP-Gly, at −5 °C.28 Furthermore, we have recently discovered (to be reported elsewhere) that CAP-Gly exhibits unusual temperature dependence of solid-state NMR spectra: at 10 °C and up to 25 °C, there is dramatic line broadening for many of the peaks and/or disappearance of the resonances corresponding to the loop regions. Above 25 degrees °C, narrow lines are recovered again. Thus the current study, in conjunction with our prior work and with the above recent unpublished temperature-dependent MAS NMR data, demonstrates that CAP-Gly has high mobility and unique dynamic signature, and suggests a hypothesis that this remarkable conformational plasticity may be associated with the intrinsically high loop content in CAP-Gly and may be necessary for this protein to be able to interact with its binding partners and to walk along microtubules. To test this intriguing hypothesis, full characterization of the structure and dynamics of different CAP-Gly complexes and point mutants is under way in our laboratory.
In summary, we have determined the 3D structure of the CAP-Gly domain of mammalian dynactin by MAS NMR spectroscopy, using distance and backbone torsion angle restraints acquired through a series of homo- and heteronuclear correlation experiments. Analysis of backbone torsion angles derived from SSNMR as well as from X-ray diffraction studies of CAP-Gly complexes with EB1 and ZnCLIP reveals that the conformation of the critical GKNDG motif in the long loop varies significantly among the different structures, and has unique conformation in the MAS NMR structure. Remarkably, two conformations of the T43-V44-A45 stretch of residues are observed in the free CAP-Gly, whereas the protein is locked into a single unique conformer in the CAP-Gly/EB1 complex. Extensive chemical shift perturbations observed in multiple stretches of residues of CAP-Gly upon formation of the complex with EB1, reveal the intermolecular binding interface. Taken together, the results presented in this report and in our prior studies suggest that unique conformational plasticity of CAP-Gly may be required for its biological function, inspiring further investigations of 3D structure and residue-specific backbone dynamics of CAP-Gly and its assemblies with binding partners and microtubules.
Materials and Methods
Materials
Common chemicals were purchased from Fisher Scientific or Sigma-Aldrich. 15NH4Cl, [2-13C]-glycerol, [1,3-13C]-glycerol and U-13C6 glucose were purchased from Cambridge Laboratories, Inc. The SMT3 fusion vector and Ulp1 protease expression system was a generous gift of Dr. Christopher Lima (Weill Medical College, Cornell University).
Sample Preparation
Sparsely 13C-enriched and uniformly 15N-enriched CAP-Gly (construct spanning residues 19–107) was expressed from E. coli grown on [2-13C]-glycerol and [1,3-13C]-glycerol (Cambridge Isotopes) as carbon source and 15NH4Cl as nitrogen source.42; 44; 45 In contrast to the original papers, the E. coli strain used here did not lack succinate dehydrogenase and malate dehydrogenase. Isolation and purification of the protein has been done as described previously.14 The U-13C,15N-enriched CAP-Gly was expressed from the same E. coli strain, using U-13C6 glucose as carbon and 15NH4Cl as the nitrogen sources.
The expression and purification for EB1(193–268) were performed according to the published protocol.20
The two CAP-Gly samples for distance restraints measurements were prepared by controlled precipitation from polyethylene glycol by slow addition of a solution of 30% PEG-3350 to the solution of 21.4 mg of [2-13C]-glycerol/U-15N CAP-Gly (42.8 mg/mL) and 24.3mg of [1,3-13C]-glycerol/U-15N CAP-Gly (38.5 mg/mL), both dissolved in 10 mM MES buffer (10 mM MgCl2, pH 6.0), following the protocol developed previously for thioredoxin.32; 33 This condition was chosen from a minimal hanging drop screen and yields CAP-Gly microcrystals in a hanging drop.14 The sample was centrifuged, the supernatant removed, and the pellet used for solid-state NMR experiments. More than 90% protein was precipitated as indicated by the Bradford assay. 14–15 mg of hydrated precipitate of each sample were packed into a 1.8 mm rotor, respectively. The U-13C, 15N CAP-Gly sample was prepared following the same procedure, and 24.4 mg of hydrated PEG precipitate was packed into a 3.2 mm Bruker rotor.
The CAP-Gly/EB1 complex was prepared by mixing 11.8 mg of U-13C, 15N CAP-Gly and 10.2 mg of natural abundance EB1 (molar ratio 1:1) in 20 mM phosphate buffer (pH=7.0) containing 50 mM NaCl and 1mM DTT. The complex stoichiometry and purity were verified by size-exclusion chromatography and SDS-PAGE, and the results are consistent with our previous study.20 The solid-state NMR sample of the complex was precipitated by slow addition of 50% PEG-3350 to the solution of the protein complex (137 mg/ml). 23.5 mg of hydrated precipitate of the sample were packed into a 3.2 mm Bruker rotor.
Solid-State NMR Spectroscopy
The NMR spectra were acquired at 14.1 and 19.9 T. The 14.1 T data sets for [2-13C]-glycerol,15N and [1,3-13C]-glycerol,15N CAP-Gly were collected using a narrow bore Varian InfinityPlus instrument equipped with a 1.8 mm MAS triple-resonance probe built in the laboratory of Ago Samoson (Tallinn University of Technology). All spectra were acquired at 10 kHz MAS frequency controlled to within ± 5 Hz by a Varian MAS controller. The temperature was calibrated for this probe using a PbNO3 temperature sensor,46 and the actual temperature at the sample was maintained at −2 °C ± 0.1 °C throughout the experiments using the Varian temperature controller. The RDSD R211 (PARIS) pulse sequence was used for 13C-13C distance restraints measurement.29; 30 Typical 90° pulse lengths were 3.5 µs for 1H and 4.2 ms for 13C. Two-pulse phase-modulation (TPPM) decoupling47 with the field strength of 85 kHz was applied during the acquisition and evolution periods in all experiments. The pulse lengths for the 90° pulses were 4 and 4.15 µs for 1H and 13C, respectively. During the mixing time (50–500 ms), the RF field on the proton channel was matched to the spinning speed of 10 kHz. The spectral width was set to 330 ppm in ω2 and to 179 ppm in ω1 with the carrier frequency positioned at 96 ppm. The maximum evolution time in the indirect t1 dimension was 5.0 ms, and 270 t1 points were acquired. The number of scans was 100, 128, and 800 for the experiments acquired with the mixing time of 50 ms, 200 ms and 500 ms, respectively.
The 2D NCA spectra were acquired for [2-13C]-glycerol/U-15N CAP-Gly sample at −2 °C and −20 °C, respectively. The typical 90° pulse lengths were 3.4–3.5 µs (1H). The 1H-15N cross-polarization (CP) employed a linear amplitude ramp of 80–100%, the 1H RF field was 45 kHz, and the center of the ramp on the 15N was Hartmann-Hahn matched the first spinning sideband. The band-selective magnetization transfer from 15N to 13Cα was realized using a 6 ms SPECIFIC-CP48 with a tangent amplitude ramp, and the RF field strengths were 25 kHz, 15 kHz, and 90 kHz for 15N, 13C, and 1H channels, respectively. The typical 1H decoupling power was 80–85 kHz during the acquisition and evolution periods in all experiments.
The 19.9 T data sets were acquired for U-13C,15N CAP-Gly and for U-13C,15N CAP-Gly/n.a. EB1 complex samples on a 19.9 T Bruker Avance III instrument equipped with 3.2 mm EFree HCN probe. The Larmor frequencies are 850.4 MHz (1H), 213.8 MHz (13C), and 86.2 MHz (15N). The MAS frequency was 14 kHz controlled to within ±5 Hz by a Bruker MAS controller. The temperature was calibrated for this probe using a PbNO3 temperature sensor,46 and the actual temperature at the sample was maintained within ± 0.1 °C throughout the experiments using the Bruker temperature controller.
For U-13C,15N-labeled CAP-Gly sample, 2D NCA, NCACX and NCOCX, and 3D NCACX, NCOCX, and CANCX experiments were acquired at 4°C ± 0.2°C. For U-13C,15N CAP-Gly/n.a. EB1 complex, 2D NCA, NCACX and NCOCX, and 3D NCACX, NCOCX, and CANCX spectra were collected at 4°C ± 0.2°C. The other experimental conditions were reported by us previously.14
All data were processed with NMRpipe.49 The chemical shifts were referenced to DSS, using the 13C methylene peak in solid adamantine as an external standard. Peak assignments were performed in Sparky.50 We note that the resonances corresponding to the N-terminal stretch of residues 19–26 and C-terminal stretch of residues 98–107 were not present in the 2D homo- and heteronuclear correlation spectra; the distance restraints for these residues could not be derived, and therefore, these regions were not included in the structure calculation.
Structure Calculation
The structure of CAP-Gly was calculated in Xplor-NIH version 2.29.40; 41 Input restraints included distance restraints and torsion angles (as shown in Table 1). The chemical shifts were first pre-checked for consistency and referenced by the TALOS+ built-in program using the empirical correlation between certain sets of chemical shift data.51 Torsion angles (Φ, Ψ) were then predicted by TALOS+,31 using 13Cα,13Cβ,13CO and 15N chemical shifts. In addition to the energy terms corresponding to the distance and torsion angle restraints standard Xplor-NIH energy terms were used. Knowledge-based energy terms included the torsion angle potential of mean force52 and the hydrogen bond potential of mean force.53 Standard bond, bond angle and improper torsion angle restraints were used to maintain proper covalent geometry, and a repulsive quartic van der Waals term was employed to prevent atomic overlap.
Assignment of ambiguous distance restraints and initial structure calculation was accomplished using a protocol consisting of three passes of torsion angle molecular dynamics simulated annealing. A final molecular dynamics simulated refinement step was then performed to generate an ensemble of high-quality structures for analysis. All calculations are summarized in Table S1. 500 structures were calculated during each assignment pass and final refinement. Each simulated annealing calculation consisted of torsion angle molecular dynamics at 3500 K, an annealing schedule during which the temperature was gradually reduced, and final gradient minimization. The duration of the high-temperature dynamics portion of the calculation was 4000 ps for the assignment passes, and 100 ps during structure refinement. The simulated annealing schedule for the first pass calculation had the temperature reduced in 25 K increments to 100 K, while the schedule used in all other calculations saw the temperature reduced to 25 K in 12.5 K increments. The input structures for pass 1 possessed randomized torsion angles, and each subsequent pass utilized the lowest energy structure of the previous pass, with randomized velocities. Assignment pass 1 utilized the •soft• form of the distance restraint potential energy such that incorrect assignments would not cause large distortions in the structures. Subsequent molecular dynamics simulated annealing calculations used the •hard• form of this potential with quadratic walls.
After each pass of structure calculation the ambiguous restraints were analyzed based on the 50 lowest energy structures. Restraints showing large violations were discarded. Restraint assignments present in 90% of the structures were labeled unique. All of the torsion angles predicted by TALOS+ were used as restraints in the structure calculation. The refinement pass also included 54 hydrogen bond distance restraints generated from the lowest energy pass 3 structure using Mark Gerstein's find-hbonds program (available online at Helix Systems Scientific Supercomputing at the NIH).54 The statistics on restraints, Xplor-NIH energy terms and violations of the final structure refinement are summarized in Table 1. Secondary structure predictions were carried out using the program Stride.55
The average MAS NMR structure was determined by numerically averaging the coordinates of each individual NMR structure in the ensemble; the resulting secondary structure was found to consistent with those of the individual structures from the ensemble,
Supplementary Material
The 3D structure of CAP-Gly domain of mammalian dynactin was determined by MAS NMR. Two conformations are found in T43-V44-A45, in proximity to critical residue G59. Binding interface in CAP-Gly/EB1 complex was determined. CAP-Gly is locked into a single conformer upon binding to EB1. Implications of conformational plasticity in biological function are discussed.
Acknowledgements
This work was supported by the National Institutes of Health (NIH Grants R01GM085396 and 8P30GM103519-03 from NIGMS, and 5P30RR031160-03 from NCRR). C. D. Schwieters was supported by the NIH Intramural Research Program of the Center for Information Technology. We acknowledge the support of the National Science Foundation (NSF Grant CHE0959496) for the acquisition of the 850 MHz NMR spectrometer at the University of Delaware.
Abbreviations
- SSNMR
solid-state NMR
- MAS
magic angle spinning
- MT
microtubule
- MAP
microtubule-associated proteins
- CAP-Gly
cytoskeleton-associated protein-glycine-rich domain
- EB1
end-binding protein 1
- dSBMA
distal spinal bulbar muscular atrophy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The chemical shifts and distance restraints for free CAP-Gly and CAP-Gly/EB1 complex have been deposited to the Biological Magnetic Resonance Bank (accession numbers for free CAPGly are 17937, 19025 and RCSB103038; for the CAP-Gly/EB1 complex- 19031). The p150Glued CAP-Gly MAS NMR structure coordinates have been deposited to the Worldwide Protein Data Bank (PDB ID: 2M02).
Contributor Information
Si Yan, Email: syan@udel.edu.
Guangjin Hou, Email: hou@udel.edu.
Charles D. Schwieters, Email: Charles.Schwieters@nih.gov.
Shubbir Ahmed, Email: SAhmed@coh.org.
John C. Williams, Email: JWilliams@coh.org.
Tatyana Polenova, Email: tpolenov@udel.edu.
REFERENCES
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Schroer TA. Dynactin. Annual Review of Cell and Developmental Biology. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 3.Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nature Reviews Molecular Cell Biology. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 6.Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic plus TIP networks. Molecular Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Molecular Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with alpha-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo M, Li SL, Finley J, Liu ZJ, Qiu SH, Chen HL, Luan CH, Carson M, Tsao J, Johnson D, Lin GD, Zhao J, Thomas W, Nagy LA, Sha BD, DeLucas LJ, Wang BC. Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. Journal of Biological Chemistry. 2002;277:48596-–48601. doi: 10.1074/jbc.M208512200. [DOI] [PubMed] [Google Scholar]
- 10.Scheel J, Pierre P, Rickard JE, Diamantopoulos GS, Valetti C, van der Goot FG, Haner M, Aebi U, Kreis TE. Purification and analysis of authentic CLIP-170 and recombinant fragments. J Biol Chem. 1999;274:25883–25891. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- 11.Perez F, Pernet-Gallay K, Nizak C, Goodson HV, Kreis TE, Goud B. CLIPR-59, a new trans-Golgi/TGN cytoplasmic linker protein belonging to the CLIP-170 family. J Cell Biol. 2002;156:631–642. doi: 10.1083/jcb.200111003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, Sakamoto A, Takagi T, Shirouzu M, Yabuki T, Nunokawa E, Seki E, Matsuda T, Aoki M, Miyata Y, Hirakawa N, Inoue M, Terada T, Nagase T, Kikuno R, Nakayama M, Ohara O, Tanaka A, Yokoyama S. The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure. 2004;12:1719–1728. doi: 10.1016/j.str.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Venkateswarlu K, Hanada T, Chishti AH. Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- 14.Sun S, Siglin A, Williams JC, Polenova T. Solid-state and solution NMR studies of the CAP-Gly domain of mammalian dynactin and its interaction with microtubules. J Am Chem Soc. 2009;131:10113–10126. doi: 10.1021/ja902003u. [DOI] [PubMed] [Google Scholar]
- 15.Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nature Genetics. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 16.Mojsilovic-Petrovic J, Jeong GB, Crocker A, Arneja A, David S, Russell D, Kalb RG. Protecting motor neurons from toxic insult by antagonism of adenosine A2a and Trk receptors. Journal of Neuroscience. 2006;26:9250–9263. doi: 10.1523/JNEUROSCI.1856-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, LaMonte BH, Harmison GG, Puls I, Fischbeck KH, Holzbaur EL. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. Journal of Cell Biology. 2006;172:733–745. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, Tsuboi Y, Yamada T, Gutmann L, Elibol B, Bhatia KP, Wider C, Vilarino-Guell C, Ross OA, Brown LA, Castanedes-Casey M, Dickson DW, Wszolek ZK. DCTN1 mutations in Perry syndrome. Nature Genetics. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, Finnegan K, Holzbaur EL, Fischbeck KH, Ludlow CL. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed S, Sun S, Siglin AE, Polenova T, Williams JC. Disease-Associated Mutations in the p150(Glued) Subunit Destabilize the CAP-gly Domain. Biochemistry. 2010;49:5083–5085. doi: 10.1021/bi100235z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nature Structural & Molecular Biology. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150(Glued) complex. Molecular Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi I, Plevin MJ, Ikura M. CLIP170 autoinhibition mimics intermolecular interactions with p150(Glued) or EB1. Nature Structural & Molecular Biology. 2007;14:980–981. doi: 10.1038/nsmb1299. [DOI] [PubMed] [Google Scholar]
- 24.Badin-Larcon AC, Boscheron C, Soleilhac JM, Piel M, Mann C, Denarier E, Fourest-Lieuvin A, Lafanechere L, Bornens M, Job D. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc Natl Acad Sci U S A. 2004;101:5577–5582. doi: 10.1073/pnas.0307917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, Wehland J, Andrieux A, Job D. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174:839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, Schweitzer A, Saoudi Y, Pointu H, Bosc C, Salin PA, Job D, Wehland J. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc Natl Acad Sci U S A. 2005;102:7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular Nmr. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. H-1-C-13/H-1-N-15 Heteronuclear Dipolar Recoupling by R-Symmetry Sequences Under Fast Magic Angle Spinning for Dynamics Analysis of Biological and Organic Solids. Journal of the American Chemical Society. 2011;133:18646–18655. doi: 10.1021/ja203771a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manna T, Honnappa S, Steinmetz MO, Wilson L. Suppression of microtubule dynamic instability by the plus TIP protein EB1 and its modulation by the CAP-Gly domain of p150(Glued) Biochemistry. 2008;47:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- 30.Hou GJ, Yan S, Sun SJ, Han Y, Byeon IJL, Ahn J, Concel J, Samoson A, Gronenborn AM, Polenova T. Spin Diffusion Driven by R-Symmetry Sequences: Applications to Homonuclear Correlation Spectroscopy in MAS NMR of Biological and Organic Solids. Journal of the American Chemical Society. 2011;133:3943–3953. doi: 10.1021/ja108650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bax A, Shen Y, Delaglio F, Cornilescu G. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular Nmr. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marulanda D, Tasayco ML, Cataldi M, Arriaran V, Polenova T. Resonance assignments and secondary structure analysis of E-coli thioredoxin by magic angle spinning solid-state NMR spectroscopy. Journal of Physical Chemistry B. 2005;109:18135–18145. doi: 10.1021/jp052774d. [DOI] [PubMed] [Google Scholar]
- 33.Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. Magic angle spinning solid-state NMR spectroscopy for structural studies of protein interfaces. Resonance assignments of differentially enriched Escherichia coli thioredoxin reassembled by fragment complementation. Journal of the American Chemical Society. 2004;126:16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Paramasivan S, Marulanda D, Cataidi M, Tasayco ML, Polenova T. Magic angle spinning NMR spectroscopy of thioredoxin reassemblies. Magnetic Resonance in Chemistry. 2007;45:S73–S83. doi: 10.1002/mrc.2092. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Tasayco ML, Polenova T. Dynamics of Reassembled Thioredoxin Studied by Magic Angle Spinning NMR: Snapshots from Different Time Scales. Journal of the American Chemical Society. 2009;131:13690–13702. doi: 10.1021/ja9037802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun SJ, Butterworth AH, Paramasivam S, Yan S, Lightcap CM, Williams JC, Polenova T. Resonance assignments and secondary structure analysis of dynein light chain 8 by magic-angle spinning NMR spectroscopy. Canadian Journal of Chemistry-Revue Canadienne De Chimie. 2011;89:909–918. doi: 10.1139/v11-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Doherty T, Li J, Lu WY, Barinka C, Lubkowski J, Hong M. Resonance Assignment and Three-Dimensional Structure Determination of a Human alpha-Defensin, HNP-1, by Solid-State NMR. Journal of Molecular Biology. 2010;397:408–422. doi: 10.1016/j.jmb.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker J, Ferguson N, Flinders J, van Rossum BJ, Fersht AR, Oschkinat H. A sequential assignment procedure for proteins that have intermediate line widths in MAS NMR spectra: amyloid fibrils of human CA150.WW2. Chembiochem. 2008;9:1946–1952. doi: 10.1002/cbic.200700706. [DOI] [PubMed] [Google Scholar]
- 39.Franks WT, Wylie BJ, Schmidt HLF, Nieuwkoop AJ, Mayrhofer RM, Shah GJ, Graesser DT, Rienstra CM. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 41.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 42.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 43.Zech SG, Wand AJ, McDermott AE. Protein structure determination by high-resolution solid-state NMR spectroscopy: Application to microcrystalline ubiquitin. Journal of the American Chemical Society. 2005;127:8618–8626. doi: 10.1021/ja0503128. [DOI] [PubMed] [Google Scholar]
- 44.Hong M. Determination of multiple ***φ***-torsion angles in proteins by selective and extensive (13)C labeling and two-dimensional solid-state NMR. J Magn Reson. 1999;139:389–401. doi: 10.1006/jmre.1999.1805. [DOI] [PubMed] [Google Scholar]
- 45.LeMaster DM, Kushlan DM. Dynamical Mapping of E. coli Thioredoxin via 13C NMR Relaxation Analysis. J Am Chem Soc. 1996;118:9255–9264. [Google Scholar]
- 46.Neue G, Dybowski C. Determining temperature in a magic-angle spinning probe using the temperature dependence of the isotropic chemical shift of lead nitrate. Solid State Nucl Magn Reson. 1997;7:333–336. doi: 10.1016/s0926-2040(96)01291-x. [DOI] [PubMed] [Google Scholar]
- 47.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear Decoupling in Rotating Solids. Journal of Chemical Physics. 1995;103:6951–6958. [Google Scholar]
- 48.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Molecular Physics. 1998;95:1197–1207. [Google Scholar]
- 49.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Goddard TDK, DG . Sparky. Vol. 3. San Francisco: University of California; Sparky 3. University of California, San Francisco. [Google Scholar]
- 51.Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbo-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- 52.Kuszewski J, Gronenborn AM, Clore GM. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Science. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grishaev A, Bax A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. Journal of the American Chemical Society. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 54.Gerstein M. A Resolution-Sensitive Procedure for Comparing Protein Surfaces and Its Application to the Comparison of Antigen-Combining Sites. Acta Crystallographica Section A. 1992;48:271–276. [Google Scholar]
- 55.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins-Structure Function and Genetics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.