SUMMARY
Although essential for T cell function, the identity of the T cell receptor “inside-out” pathway for lymphocyte function-associated antigen 1 (LFA-1) adhesion has proved elusive. Here, we define the “inside-out” pathway mediated by N-terminal SKAP1 (SKAP-55) domain binding to the C-terminal SARAH domain of RapL. TcR induced Rap1-RapL complex formation and LFA-1 binding failed to occur in Skap1−/− primary T cells. SKAP1 generated a SKAP1-RapL-Rap1 complex that bound to LFA-1, whereas a RapL mutation (L224A) that abrogated SKAP1 binding without affecting MST1 disrupted component colocalization in vesicles as well as T cell-dendritic cell (DC) conjugation. RapL expression also “slowed” T cell motility in D011.10 transgenic T cells in lymph nodes (LNs), an effect reversed by the L224A mutation with reduced dwell times between T cells and DCs. Overall, our findings define a TCR “inside-out” pathway via N-SKAP1-C-RapL that regulates T cell adhesion, motility, and arrest times with DCs in LNs.
INTRODUCTION
Integrins regulate multiple aspects of immunity including migration to sites of inflammation, migration to in lymph nodes (LNs), and conjugate formation between T cells and antigen-presenting cells (APCs) (Dustin et al., 2004; Hogg et al., 2003; Germain et al., 2006). They comprise as many as 20 α-β heterodimers whose binding is affected by altered conformation and clustering (Hynes, 2002; Carman and Springer, 2003; Dustin et al., 2004; Hogg et al., 2003). T cell function is regulated by the β2 family members such as lymphocyte function-associated antigen [LFA-1; αL(CD11a)-β2 (CD18)] and α4 integrins such as α4β1 (VLA-4). LFA-1 binds to intercellular adhesion molecules (ICAM)-1 and -2 on major histocompatibility complex (MHC)-bearing APCs (Kupfer 2006; Dustin, 2005). Although LFA-1 is constitutively expressed in a low-affinity form in resting T cells, antigen-receptor complex (TcRζ-CD3) and chemokine receptor ligation generates “inside-out” signals that activate integrins (Kinashi, 2005). LFA-1 α and β subunits create a headpiece formed by a β-propeller domain of the α subunit that interacts with a dinucleotide fold domain in the β subunit termed the I-like domain (Takagi et al., 2002). Increased affinity involves exposure of the I domain ligand binding site, whereas clustering reduces the dissociation rate of binding (Sarantos et al., 2005).
Over the past decade, it has been of major interest to identify make-up of the “inside-out” signaling pathway. TcRζ-CD3 signaling mediators influence adhesion including CD4- and CD8-p56lck (Rudd et al., 1994), interleukin 2 (IL-2)-inducible T cell kinase (ITK) (Labno et al., 2003), the guanine nucleotide exchange factor Vav-1 (Collins et al., 1997; Krawczyk et al., 2002), phosphatidylinositol 3-kinase (PI 3K) (Shimizu et al., 1995), GTP binding proteins such as Rac-Rho (Collins et al., 1997; Mor et al., 2007) and Rap1 (Bivona et al., 2004; Kinashi, 2005) and its binding partner RapL (regulator of cell adhesion and polarization enriched in lymphoid tissues) (Katagiri et al., 2003; 2006), and RIAM (Rap1-interacting adaptor molecule) (Lafuente et al., 2004). In addition, adaptors SLP-76 (76-kD src homology 2 domain-containing leukocyte phosphoprotein) (Kuhné et al., 2003), ADAP (adhesion and degranulation–promoting adaptor protein) (official gene name designation: Fyb) (Griffiths et al., 2001; Peterson et al., 2001; Geng et al., 2001), and SKAP1 (src kinase-associated phosphoprotein 1: official gene name designation; also SKAP-55, src kinase-associated phosphoprotein-55) (Wang et al., 2007; Wang et al., 2003; Jo et al., 2005; Kliche et al., 2006) have been implicated. The challenge has been to distinguish specific effectors of LFA-1 adhesion from the generic events needed for cell integrity and function.
One model involves Rap1 and its binding partners RapL and RIAM (Kinashi, 2005). Rap1 was first implicated in adhesion by Rap1A overexpression (Bos et al., 2001; Kinashi, 2005) and in transgenic Rap1V12 T cells (Sebzda et al., 2002). RIAM contains a Ras association (RA) domain and a PH (pleckstrin homology) domain and interacts with talin, SKAP1, and Ena (VASP) proteins (Lafuente et al., 2004; Ménasché et al., 2007; Lee et al., 2009). RapL (also MAXP1, NORE1B, and RASSF3) is an immune cell isoform of the RASSF5 (Ras association domain family 5) family (Katagiri et al., 2003, 2006; Tommasi et al., 2002). It has an unique N-terminus followed by a RA and C-terminal coiled-coil (SARAH) domain (Katagiri et al., 2003). The RA domain binds GTP-bound active Rap1, whereas the SARAH domain binds the coiled-coil domain of the serine kinase MST-1 (Katagiri et al., 2006; Zhou et al., 2008).
A competing model involves immune cell-specific adaptors SLP-76, ADAP, and SKAP1 in which TCR induces SLP-76 binding to ADAP that in turns binds to SKAP1 (Rudd, 1999; Jordan et al., 2003; Samelson, 2002; Wang and Rudd, 2008). ADAP is composed of a unique NH2-terminus, an internal SH3 domain, two nuclear localization sequences (NLSs), and a C-terminal SH3-like domain (da Silva et al., 1997; Musci et al., 1997). p59fynT preferentially phosphorylates two YDDV motifs for SLP-76 SH2 domain binding (Veale et al., 1999; Raab et al., 1999). Loss of the YDDV sites reduces LFA-1-ICAM1 adhesion, T cell-APC conjugate formation, and assembly of the peripheral supramolecular activation complex (pSMAC) (Wang et al., 2004). ADAP binds to the adaptor SKAP1, which has a unique NH2-terminal region followed by a PH domain and a COOH-terminal SH3 domain (Marie-Cardine et al., 1997; Liu et al., 1998; Marie-Cardine et al., 1998; Kang et al., 2000). Binding is mediated primarily by SKAP1 SH3 domain binding to a proline-rich region in ADAP, and ADAP-SH3-like domain to a unique sequence in SKAP1 (Liu et al., 1998; Marie-Cardine et al., 1998; Kang et al., 2000). ADAP also protects SKAP1 from proteolytic degradation (Huang et al., 2005). Retroviral transduction and shRNA knockdown first implicated SKAP1 in adhesion (Jo et al., 2005; Geng et al., 1999, 2001; Duke-Cohan et al., 2006), whereas deletion of the SKAP1 SH3 domain or expression of a peptide encoding the ADAP proline region ablates adhesion (Wang et al., 2003; Kliche et al., 2006). Both Fyb−/− and Skap1−/− primary T cells show defective LFA-1 adhesion (Griffiths et al., 2001; Peterson et al., 2001; Wang et al., 2007). Unlike Fyb/− T cells that show the concurrent loss of SKAP1 expression, Skap1−/− T cells retain ADAP expression and yet still show defective adhesion (Wang et al., 2007). These observations identified SKAP1 as an effecter of the ADAP-SKAP1 module, although the molecular basis for this function has not been established. The requirement for SKAP1 varies with the strength of the TCR signal (Wang et al., 2007; Mueller et al., 2007; Wang and Rudd, 2008).
Although the Rap1-RapL-RIAM and SLP-76-ADAP-SKAP1 modules are needed for LFA-1 adhesion, the identity of the TcRζ-CD3 induced “inside-out” signaling pathway has not been resolved. In this study, we define a pathway to account for TcR “inside-out” signaling that integrates the SKAP1 and Rap1-RapL pathways. ADAP-SKAP1 and Rap1-RapL modules comprise a single pathway coordinated by SKAP1 in the regulation of LFA-1 adhesion leading to reduced T cell migration in LNs.
RESULTS
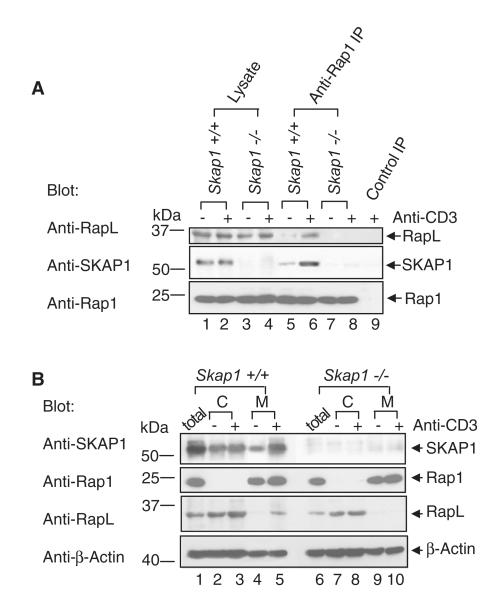
Rap-1-RapL Complex Does Not Form in Skap1−/− Primary T Cells
Rap1 binding to RapL is a necessary step in the upregulation of LFA1-mediated adhesion (Kinashi, 2005). At the same time, SKAP1-deficient T cells elicit defective adhesion (Wang et al., 2007). To test for a connection between these proteins, we initially assessed anti-CD3 induced Rap1-RapL complex formation in freshly isolated primary T cells from spleens of Skap1+/+ and Skap1−/− mice (Wang et al., 2007). T cells were stimulated with anti-CD3, lysed, and subjected to precipitation with anti-Rap1, followed by blotting with anti-RapL or SKAP1 (Figure 1A). Anti-Rap1 coprecipitated RapL from Skap1+/+ T cells (upper panel, lanes 5 and 6), and this increased with anti-CD3 ligation at 5 min (lane 6 versus 5) and 15 min postactivation (data not shown). This confirmed that anti-CD3 ligation induces binding of Rap1 to RapL in wild-type T cells (Katagiri et al., 2003). By contrast, anti-Rap1 failed to coprecipitate RapL from resting or activated Skap1−/− T cells (lanes 7 and 8). Immunoblotting confirmed the presence of RapL and Rap1 in lysates from Skap1+/+ and Skap1−/− T cells (upper and lower panels, lanes 1–4, respectively). These data showed that SKAP1 expression is needed for anti-CD3 induction of the Rap1-RapL complex in primary T cells.
Figure 1. RapL-Rap1 Complex Fails to Form in Skap1−/− Primary T Cells.
(A) T cells from wild-type Skap1+/+ and Skap1−/− mice were ligated with anti-CD3 and subjected to precipitation with anti-Rap1 followed by blotting with anti-RapL (upper panel), anti-SKAP1 (middle panel), or anti-Rap1 (lower panel).
(B) Cytosolic and membrane fractions of Skap1+/+ and Skap1−/− T cells were blotted with anti-SKAP1 (upper panel), anti-Rap1 (upper-middle panel), anti-RapL (lower middle panel), or anti-actin (lower panel).
Given this role, we then assessed whether SKAP1 bound to the Rap1-RapL complex. Indeed, anti-Rap1 coprecipitated SKAP1 from wild-type cells (middle panel, lanes 5 and 6), and this increased with anti-CD3 ligation (lane 6 versus 5). Similar results were obtained when anti-RapL was used for precipitating antigen (data not shown). Anti-SKAP1 blotting confirmed the presence of SKAP1 in lysates from Skap1+/+, but not Skap1−/− T cells (lanes 1 and 2 and lanes 3 and 4, respectively). These data indicated that anti-CD3 induces SKAP1 binding to the Rap1-RapL complex in primary T cells.
SKAP1 Is Needed for RapL Translocation to the Membrane to Interact with Rap1
To determine the mechanism by which SKAP1 regulates RapL-Rap1 binding, we isolated cytosolic and membrane fractions from untreated and anti-CD3 activated T cells and blotted them with anti-Rap1, anti-RapL, or anti-SKAP1 (Figure 1B). Although anti-CD3 ligation induced the translocation of a fraction of RapL from the cytosol to the membrane fraction of wild-type T cells (lower-middle panel, lane 5 versus 4), it failed to induce RapL translocation in Skap1−/− T cells (lanes 10 versus 9). SKAP1 also increased membrane binding in response to anti-CD3 ligation (upper panel, lane 5 versus 4). By contrast, Rap-1 was constitutively associated with the membranes of activated and nonactivated Skap1+/+ and Skap1−/− T cells (upper-middle panel). The presence of actin served as a loading control (lower panel). Identical results were observed in Fyb−/− (i.e., adap−/−) T cells (Figure S1A available online). These results showed that SKAP1 is needed for anti-CD3-induced RapL translocation to the membranes of cells where it could interact with Rap1.
SKAP1 Coordinates RapL-Rap1 Binding
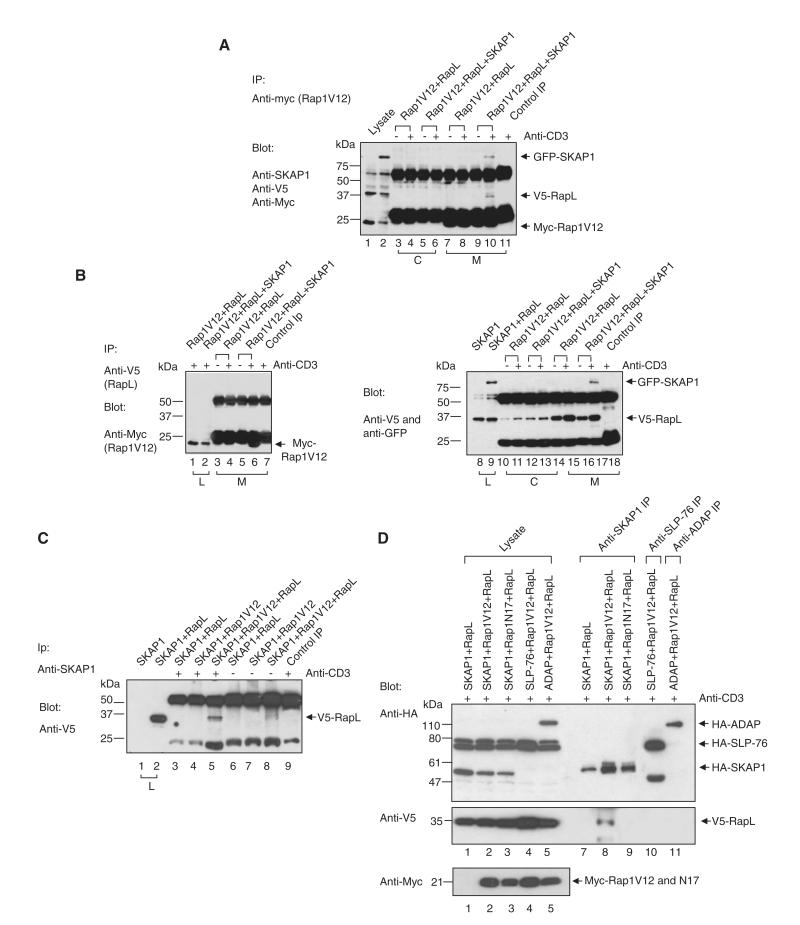
To further examine the mechanism of SKAP1 regulation of Rap1-RapL complex formation, we expressed V5-RapL, myc-Rap1V12 and GFP-SKAP1 in Jurkat T cells and precipitated them from cell lysates using anti-myc (Rap1V12) and subsequently blotted them with anti-SKAP1, anti-V5 (Rap1), or anti-myc (Rap1V12) (Figure 2A). Rap1V12 is the constitutively active form of Rap1. Contrary to a previous report (Katagiri et al., 2003; Katagiri et al., 2004), coexpression of RapL and Rap1 alone failed to allow for coprecipitation of V5-RapL with anti-myc Rap1. No coprecipitated RapL was seen from cytosolic (lanes 3 and 4) and membrane fractions (lanes 7 and 8) or from resting (lane 3 and 7) or anti-CD3-ligated cells (lanes 4 and 8). Coprecipitation was also not observed with different concentrations of CD3 antibody (1–10 μg/ml; data not shown). By contrast, the mere coexpression of SKAP1 with RapL and Rap1-V12 resulted in anti-myc (Rap1V12) coprecipitation of RapL as well as SKAP1 (lane 10). This occurred in the membrane (lane 10), but not in the cytosolic fraction (lane 6) and required anti-CD3 ligation (lane 10 versus 9). Rabbit anti-mouse served as a negative control (lane 11). The identity of each band was confirmed by blotting with individual antibodies, and the expression of endogenous SKAP1 was low in these cells (data not shown). These data confirmed that TCR-CD3-induced Rap1-RapL complex formation required SKAP1.
Figure 2. SKAP1 Coordinates RapL-Rap1V12 Complex Formation.
(A) Jurkat T cells transfected with RapL-V5, Rap1-myc, and/or SKAP1-GFP were ligated with anti-CD3, separated into cytosolic and membrane fractions, lysed, and precipitated with anti-myc (myc-Rap1V12). Samples were blotted with anti-V5 (RapL), anti-SKAP1, and anti-myc (Rap1V12).
(B) As in (A) except that anti-V5 (RapL) was used for precipitation and anti-myc (Rap1V12) was used for blotting.
(C) As in (A) except that lysates were precipitated with anti-SKAP1 and then blotted with anti-V5 (RapL).
(D) HA-SKAP1, HA-SLP-76, or HA-ADAP were coexpressed with V5-RapL and/or myc-Rap1V12 or myc-Rap1N17 in Jurkat cells, precipitated with anti-SKAP1, anti-SLP-76, or anti-ADAP, and blotted with anti-HA (upper panel), anti-V5 (middle panel), or anti-myc (lower panel).
The same result was obtained when anti-V5 (RapL) was used to precipitate protein (Figure 2B). Whereas coexpression of Rap1 and RapL failed to allow for anti-V5 (RapL) coprecipitation of Rap1 (lanes 3 and 4), the coexpression of SKAP1 led to the coprecipitation of Rap1V12 (lane 6) (lowest band under the light chain of IgG). Anti-CD3 ligation was needed (lane 6 versus 5). Anti-myc (Rap1V12) and anti-V5 (RapL) also coprecipitated SKAP1 as detected by anti-GFP or anti-SKAP1 (Figure 2A, lane 10 and Figure 2B, lane 17, respectively). This association also required anti-CD3 ligation (Figure 2A, lane 10 versus 9 and Figure 2B, lane 17 versus 16, respectively). Conversely, anti-SKAP1 was able to coprecipitate RapL (Figure 2C, lane 5) from V5-RapL-, myc-Rap1V12-, and GFP-SKAP1-expressing cells (lane 5 versus 8).
Importantly, active Rap1V12 but not inactive Rap1N17 supported anti-SKAP1 coprecipitation of RapL (Figure 2D, middle panel, lane 8 versus 9), and neither SLP-76 nor ADAP (the binding partner of SKAP1) could substitute for SKAP1 (lanes 10 and 11 versus 8). In this instance, SKAP1, ADAP and SLP-76 were tagged with HA. This observation indicated that SKAP1 plays a specific role in promoting Rap1-RapL complex formation that cannot be replaced by SLP-76 or ADAP. Moreover, this data indicate that complex formation depends on active Rap1. Expression of the various proteins was confirmed by blotting of cell lysates (all panels, lanes 1–5) and in anti-SKAP1 (upper panel, lanes 7–9), anti-SLP-76 (lane 10), or anti-ADAP precipitations (lane 11). Taken together, these observations confirmed with a different approach that SKAP1 was needed for Rap1-RapL complex formation in T cells and that SKAP1 becomes part of the complex in response to anti-CD3 ligation.
SKAP1 N-Terminal Domain Binds RapL C-Terminal Coiled-Coil Domain
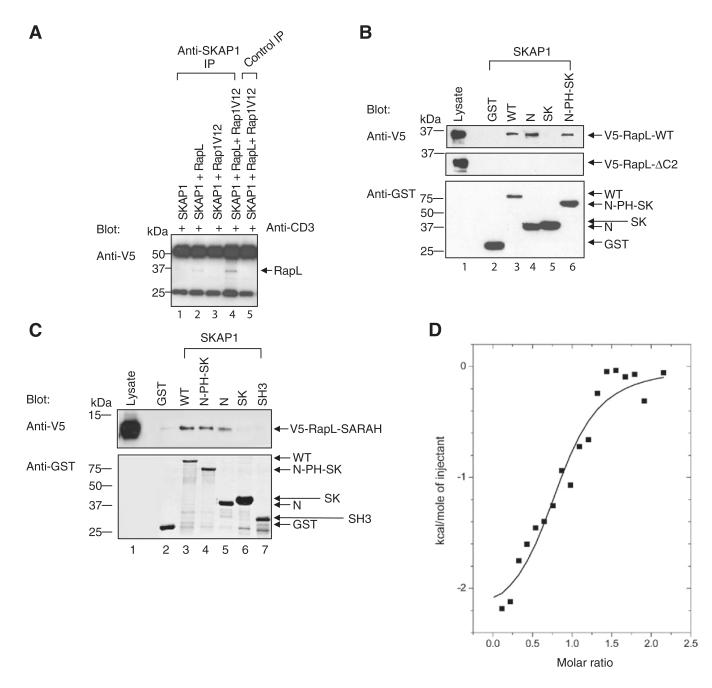
Given the requirement for SKAP1 expression for Rap1-RapL binding, it was important to define the binding site for SKAP1 in the Rap1-RapL complex. Unlike with Rap1V12-RapL coexpression, SKAP1 and RapL coexpression alone occasionally resulted in a low amount of coprecipitation (an example in Figure 3A, lane 2). Although this binding increased markedly with RapLV12 expression (lane 4), the observation suggested that SKAP1 might bind directly to RapL.
Figure 3. SKAP1 N Terminus, N-SKAP1, Binds to the C-RapL SARAH Domain.
(A) SKAP1-GFP, RapL-V5, and/or Rap1V12-myc expression in Jurkat cells, followed by precipitation with anti-SKAP1, and blotting with anti-V5 (RapL).
(B) GST-SKAP1 and domains were incubated with lysates of V5-RapL full-length or V5-RapLΔC2-transfected 293T cells for pull-down assays that were blotted with anti-V5. The upper and middle panel show an anti-V5-RapL blot; the lower panel shows an anti-GST blot.
(C). As in (B) except that cells were transfected with V5-RapL-SARAH and blotted with anti-V5 (upper panel) or anti-GST (lower panel).
(D) Isothermal titration calorimetry (ITC) of binding between N-SKAP1 and GST-C-RapL. Status of purified proteins detected by silver staining are shown in Figure S1.
To confirm this and define the sites of binding, we used GST fusion proteins encoding different regions of SKAP1 in a pull-down assay from lysates of cells transfected with wild-type V5-tagged RapL or a truncated version of RapL lacking the C-terminal coiled-coil (SARAH) domain (V5-RapL-Δc2) (Figure 3B). The pull-down assay with glutathione sepharose beads was followed by blotting with anti-V5 as previously described (Raab et al., 1999; Kang et al., 2000). The GST fusion proteins included full-length SKAP1, N-terminal (N-SKAP1; residues 1–104), SK region (SK; residues 209–285), or N plus PH and SK regions (N-PH-SK; residues 1–285). Although WT, N, and N-PH-SK readily precipitated V5-tagged RapL (upper panel, lanes 3,4, and 6), GST alone and GST-SK failed to precipitate the protein (lanes 2 and 5, respectively). Anti-GST blotting confirmed expression of the constructs (lower panel). These data indicated that the N-terminus of SKAP1 (N-SKAP1) interacts with RapL.
In this assay, deletion of the C-terminal SARAH domain of RapL (i.e., RapL-ΔC2) completely abrogated binding to WT, N, and N-PH-SK RapL (middle panel, lanes 3–6), suggesting that N-SKAP1 bound to the RapL SARAH domain (i.e., C-RapL). To test this directly, we incubated GST-SKAP1 domains with lysates of cells expressing V5-C-RapL and assessed them for binding (Figure 3C). Full-length SKAP1 WT, N, and N-PH-SK precipitated C-RapL (lanes 3, 4, and 5), whereas GST alone, GST-SK, or GST-SH3 failed to precipitate the protein (lanes 2, 6, and 7, respectively). Anti-GST blotting confirmed expression of the constructs (lower panel). These data indicated that N-SKAP1 interacts with coiled-coil C-RapL SARAH domain.
The structure of related SKAP2 shows an N-terminal four-helix bundle dimerization domain that packs against an inactive PH domain (Swanson et al., 2008). Many of the residues including those needed for dimerization are conserved in SKAP1 (Marie-Cardine et al., 1997; Liu et al., 1998). Dimerization will need to be released to facilitate PH domain activation. To therefore assess directly the interaction between N-SKAP1 and C-RapL, we purified individual N-SKAP1 and C-RapL domains as GST-fusion proteins, and purification was followed by Factor X cleavage (see Experimental Procedures). Silver staining of material showed the presence of individual subunits without contaminating material (Figure S1B). An interaction between these proteins was then examined with isothermal titration calorimetry (ITC), a method that measures stoichiometry, binding constants, and thermodynamic parameters in a single interaction (Figure 3D). The ITC data determined at 25°C clearly demonstrated that the SARAH RapL C-terminal coiled-coil domain interacts directly with N-SKAP1 with a stoichiometry that approximates to unity (n = 0.85). The equilibrium dissociation constant, Kd, of 0.6 uM corresponds to a change in free energy of binding (ΔG) of −8.5 kcal/mol. The change in enthalpy of binding is relatively small (ΔH = −2.3 kcal/mol) and hence, at the experimental temperature, the interaction is entropy driven (T ΔS = 6.2 kcal/mol). This is likely to indicate that the interaction is accompanied by a substantial contribution from the release of solvent molecules from the surface area that is buried on forming the complex. These observations indicated that the N-SKAP1 binds directly and on a one-to-one basis to the C-RapL SARAH domain.
RapL-L224A Mutation Selectively Disrupts SKAP1 without Affecting MST-1 Binding
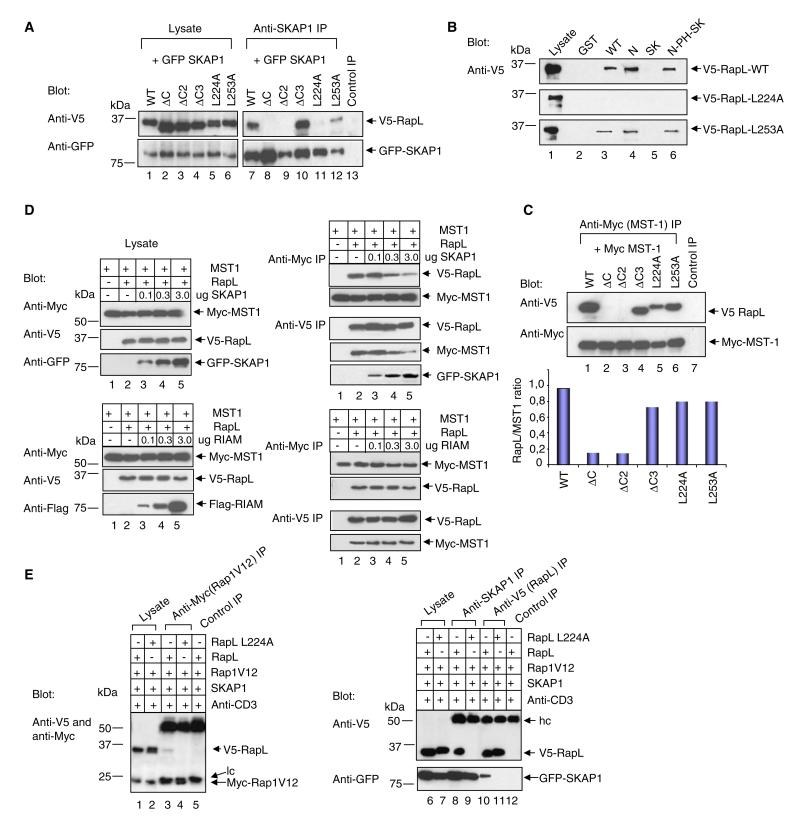
N-SKAP1 binding to C-RapL was next confirmed by deletion and mutation analysis (Figure 4A). RapL mutants tested included RapLΔC (residues 1–222; lacking the entire SARAH-coiled-coil domain), RapLΔC2 (residues 1–243; lacking part of the SARAH domain), RapLΔC3 (residues 1–254; C-terminal and outside the SARAH domain) or, alternatively, full-length RapL with point mutations at residues L224A (RapL-L224A) and L253A (RapL L253A) within the coiled-coil domain. Whereas anti-SKAP1 precipitated wild-type and the RapLΔC3 mutant (lanes 7 and 10, respectively), it failed to precipitate the RapLΔC2 and RapLΔC mutants (lanes 8 and 9, respectively). Each of the V5-tagged constructs and GFP-SKAP1 was expressed as seen in cell lysates (upper and lower panels, lanes 1–6).
Figure 4. The RapL-L224A Mutant Selectively Ablates SKAP1 Binding.
(A) V5-tagged RapL and mutant coexpression with GFP-SKAP1 in 293T cells, followed by precipitation with anti-SKAP1, and blotting with anti-V5 (upper panel) and anti-GFP (lower panel).
(B) GST-SKAP1 and domains were used to pull-down V5-RapL (upper panel), V5-RapL-L224A (middle panel), or V5-RapL-L253A (lower panel) and then they were blotted by anti-V5 (RapL).
(C) V5-tagged RapL and domains were coexpressed with Myc tagged-MST-1; this coexpression was followed by precipitation with anti-myc and blotting with anti-V5 (upper panel) or anti-myc (lower panel). The lower histogram shows the ratio of RapL/MST1.
(D) The upper-left panels show the expression of MST-1, RapL, and SKAP1 in cell lysates of transfected cells. The upper-right panels show precipitations from lysates and blotting as indicated. The lower-left panels show the expression of MST-1, RapL, and RIAM in cell lysates of transfected cells. The lower-right panels show precipitations from lysates and blotting as indicated.
(E) V5-RapL, V5-RapL-L224A, myc-Rap1, and SKAP1-GFP were coexpressed in Jurkat T cells; coexpression was followed by anti-CD3 ligation and precipitation with anti-myc (Rap1V12), anti-SKAP1, or anti-V5 as indicated. Samples were blotted with a combination of anti-V5-myc, anti-V5, and anti-GFP as indicated.
The same result was obtained in GST pull-down assays (Figure 4B). WT V5-RapL, V5-RapL-L224A, and V5-RapL-L253A were expressed and assessed for binding to the different regions of SKAP1. Although the N-SKAP1 precipitated WT RapL and the L253A mutant (upper and lower panels, lane 4), it failed to precipitate the L224A single-site mutant (middle panel, lane 4). These data showed by two independent approaches that the RapL-L224A mutation disrupts SKAP1 N-terminal binding.
Whereas N-SKAP1 bound to the SARAH domain, the serine-threonine kinase MST-1 has also been reported to bind to the same region in RapL (Katagiri et al., 2006). For defining the function of the SKAP1-RapL interaction, and distinguishing it from the MST-1-RapL interaction, it was therefore necessary to identify a RapL mutant that abrogated SKAP1 binding without affecting the binding of MST1. To assess MST-1 binding to RapL, we coexpressed myc-MST-1 and V5-tagged RapL, precipitated an anti-myc MST-1, and blotted with anti-V5 for RapL (Figure 4C). Coexpression of MST-1 and RapL showed clear binding (lane 1), consistent with previous reports (Katagiri et al., 2006). However, unlike SKAP1, MST-1 also bound to the L224A mutant (lane 5 versus 1, 4, and 6) (see normalized densiometric values). By contrast, as a control, deletion of the coiled-coil domain in RapLΔC and RapLΔC2 eliminated MST-1 binding, confirming binding to the SARAH domain (lanes 2 and 3 versus 1). Rabbit anti-mouse served as a negative control (lane 7).
Confirmation of the overlap in binding to the C-RapL SARAH domain was also seen in SKAP1 and MST1 coexpression studies (Figure 4D). V5-RapL and myc-MST1 were initially coexpressed in Jurkat cells with increasing levels of GFP-SKAP1 as titrated with 0.1, 0.3, and 3.0 ug/ml of plasmid DNA with additional GFP control plasmid for maintaining equal levels of DNA. As shown by blotting of cell lysates, GFP-SKAP1 expression increased with increasing amounts of the GFP-SKAP1 plasmid without affecting myc-MST1 or V5-RapL expression (upper left panel; lanes 2–5). From this lysate, anti-myc precipitates were then blotted with anti-V5 RapL and anti-myc-MST1 (upper-right panel). Although anti-myc readily precipitated MST1 and associated RapL (upper panels, lane 2), increasing SKAP1 expression reduced the amount of RapL coprecipitated MST1 (lanes 3–5). Conversely, although anti-V5 RapL coprecipitated MST1 (lower panels, lane 2), increasing SKAP1 expression reduced levels of coprecipitated MST1 (lanes 3–5). At the same time, SKAP1 showed an increased binding to RapL (lowest panel; lanes 3–5). As a control, FLAG-tagged RIAM (another protein involved in adhesion) failed to compete for MST1 binding to RapL with increasing levels of expression (lower-left and -right panels). These findings showed that SKAP1 can compete with MST1 for binding to RapL, a result consistent with binding to the same SARAH domain of RapL.
Given the identification of a L224A mutation that selectively affected SKAP1 binding, we next assessed whether the L224A mutation could prevent SKAP1 regulation of the Rap1-RapL complex. RapL or RapL-L224A was coexpressed with Rap1V12 in the presence or absence of SKAP1 and examined for Rap1 precipitation of RapL (Figure 4E). Whereas anti-myc Rap1 precipitated V5-RapL from SKAP1-, RapL-, and Rap1V12-transfected cells (lane 3), RapL-L224A failed to support this (lane 4). Blotting with the respective antibodies confirmed the presence of Rap1V12 and RapL in cell lysates (lanes 1 and 2). Anti-SKAP1 also coprecipitated RapL from cotransfected cells (lane 8), but not from cells with RapL-L224A (lane 9). Anti-RapL blotting confirmed the presence of RapL expression (lanes 6 and 7). Conversely, anti-V5 RapL precipitated GFP-SKAP1 from cotransfected cells (lower panel, lane 10), but not from cells expressing RapL-L224A with SKAP1 and Rap1 (lane 11). These data indicated that N-SKAP1 binding to the RapL-L224 site is the key event needed to regulate Rap1-RapL complex formation in T cells.
SKAP1-RapL Regulates RapL-Rap1 and SKAP1-RapL Colocalization
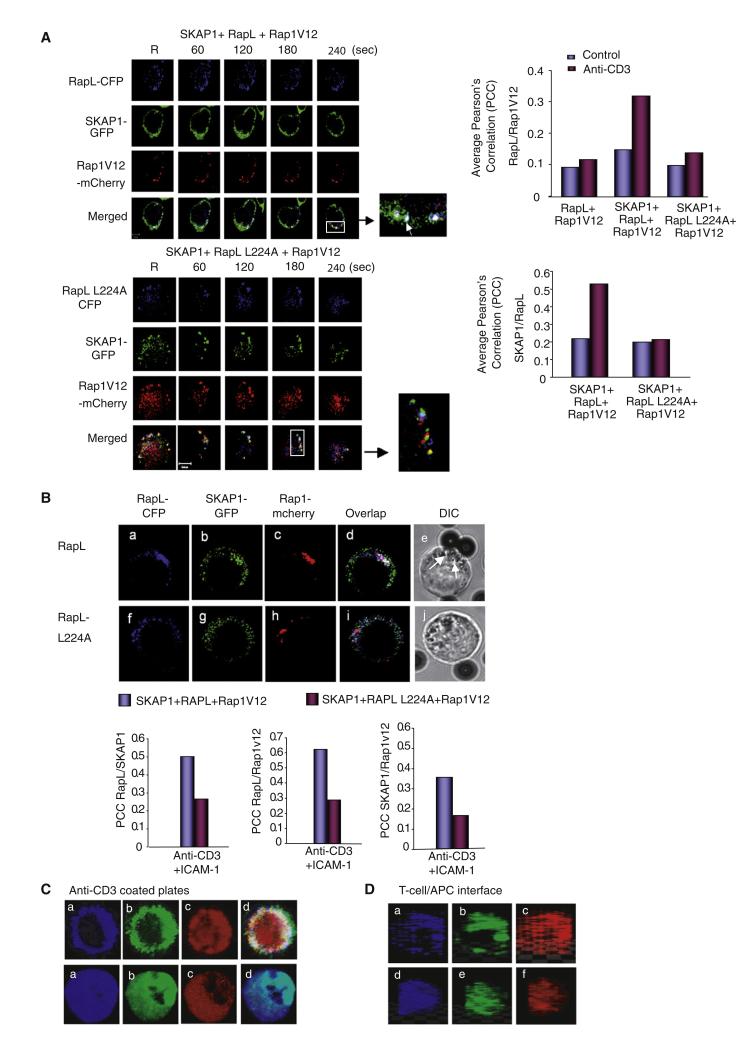
Identification of the RapL-L224 mutant allowed an examination of the role of N-SKAP1-C-RapL binding in intracellular localization (Figure 5). Cells transfected with RapL-CFP, SKAP1-GFP, and Rap1V12-mcherry were seeded on coverslips with immobilized anti-CD3 and ICAM-1 and imaged at the contact area by confocal microscopy (Bunnell et al., 2002; Schneider et al., 2008). Cells with only low-moderate intensities of expression were imaged so that problems with protein overexpression could be avoided. RapL-CFP, SKAP1-GFP, and Rap1V12-mcherry localized in clusters in resting and anti-CD3-ligated T cells over 0–240 s (Figure 5A, upper panels; Movie S1). Anti-CD3 did not alter the number or size of the clusters. Further, surprisingly, RapL-L224A, SKAP1-GFP, and Rap1V12-mcherry also were found in clusters in cells cotransfected with the RapL-L224A mutant (lower panels) (Movie S1). No obvious difference was noted in the average size or number of clusters in RapL or RapL-L224A transfected cells (data not shown). All clusters migrated with a similar pattern and speed (data not shown) along the peripheral contact region (Figure S2A).
Figure 5. RapL224A Disrupts Colocalization and pSMAC Formation.
(A) As shown in the left panels, RapL-CFP or RapL-L224A plus SKAP1-GFP and Rap1V12-mcherry were expressed in Jurkat cells and then incubated on anti-CD3 coated coverslips and imaged in real-time with a Zeiss confocal microscope. Insets showed examples of merged colors. The right panels show histograms of average PCC values for RapL-Rap1V12 and SKAP1-RapL colocalization.
(B) RapL-CFP, RapL-L224A-CFP, SKAP1-GFP, and/or Rap1V12-mcherry localization with beads coupled to anti-CD3/ICAM1 for 10 min. The lower panel shows PPC values for colocalization. (Be) and (Bj) show DIC images (arrows pointing to vesicles).
(C) RapL/RapL-L224A-CFP, SKAP1-GFP, and Rap1V12-mcherry were coexpressed in Jurkat cells responding to plate-bound anti-CD3 for 20–30 min. The upper subpanels (RapL, SKAP1; Rap1-coexpressing cells) show the following: RapL (a); SKAP1 (b); Rap1 (c); and merged (d). The lower subpanels (RapL-L224A, SKAP1; Rap1 coexpressing cells) show the following: RapL-L224A (a); SKAP1 (b); Rap1 (c); and merged (d).
(D) Localization of RapL-RapL-L224A-CFP, SKAP1-GFP, and Rap1V12-mcherry in T8.1 cells incubated with L625 cells and Ttox peptide. The subpanels show the following: RapL, SKAP1; Rap1 coexpressing cells (a–c): RapL (a); SKAP1 (b), Rap1 (c); RapL-L224A, SKAP1; Rap1 coexpressing cells (d–f): RapL (d); SKAP1 (e), Rap1 (f).
The one difference between RapL and RapL-L224A was in their colocalization with Rap1 and SKAP1 as determined by Pearson’s correlation coefficient (PCC) with Volocity software (right panels). RapL and Rap1V12 showed low PCC values in resting and anti-CD3 ligated cells (i.e., average PPC = 0.09 and 0.12, respectively) in the absence of SKAP1 (upper panel). SKAP1 coexpression had little effect on Rap1-RapL colocalization in resting cells (i.e., PCC = 0.14); however, it substantially increased RapL-Rap1V12 colocalization in anti-CD3-activated cells (i.e., average PCC value = 0.34). Similar values have been reported for other colocalized proteins (Huse et al., 2006). This indicated that SKAP1 is needed for RapL-Rap1V12 colocalization, an observation consistent with the coprecipitation results (Figures 1 and 2). In contrast, RapL-L224A failed to support RapL-Rap1V12 colocalization in resting and anti-CD3-ligated cells (i.e., average PCC = 0.1 and 0.13, respectively). This was also observed over the time course of imaging (Figure S2B, upper panel). Similar differences were also noted in the SKAP1-RapL colocalization (lower panel) in which the limited colocalization in resting cells increased with anti-CD3 ligation (i.e., average PCC = 0.23 and 0.55, respectively). However, only weak SKAP1 colocalization with RapL-L224A was observed in response to anti-CD3 (i.e., average PCC = 0.2), or over a time-course (Figure S2B).
Similar results were obtained when anti-CD3-CD18 were copresented on beads (Figure 5B). CD18 (or ITGB2) is the beta 2 subunit that pairs with CD11α of LFA-1. RapL, Rap1V12, and SKAP1 colocalized at the site of contact with the beads (upper panel, panels a-d). Much of the fluorescence signal was associated with vesicular-like structures at the contact region as seen by translucent microscopy (Figure 5Be; see arrows). By contrast, coexpressed RapLL224A, Rap1V12, and SKAP1, although still associated with vesicular structures, failed to colocalize (lower panel). Less colocalization of vesicles was also observed. Reduced cluster colocalization was confirmed by PCC values for Rap1-RapL-RapL224A, SKAP1-RapL-RapL224A, and SKAP1-Rap1V12 (lower panels). The nature of the vesicles was unclear. Staining of the vesicles with EEA1 and Rab7, markers of early endosomes, showed limited overlap with RapL-CFP clusters (Figure S3).
Incubation for longer periods of time (i.e., 5–15 min) resulted in the formation of a peripheral region that resembled a peripheral supra-molecular complex (pSMAC), as described by others (Kupfer, 2006) (Figure 5C, upper panels). The pSMAC is enriched with LFA-1, whereas the cSMAC is composed of the TcR, CD28, and other signaling proteins (Dustin et al., 2004; Kupfer, 2006). In this case, imaging was conducted at the contact area between the T cell and anti-CD3 covered coverslip surface. RapL and SKAP1 both formed a pericentric ring (Figures 5Ca and 5Cb, respectively) that closely overlapped (Figures 5Cd). Rap1V12 formed a similar pericentric overlapping region together with additional material in the inner region (Figures 5Cc). By contrast, coexpression of RapL-L224A with SKAP1 and Rap1V12 showed a loss of the SMAC-like region, instead showing a diffuse pattern for RapL-L224, SKAP1, and Rap1V12 at the interface (Figures 5Ca–5Cd, lower panels). N-SKAP1 binding to C-RapL therefore regulates the ability of the three components to colocalize in clusters and in a pSMAC-like formation.
Similar alterations in the formation of the IS as observed between T8.1 T cells and L625 APCs (Figure 5D). T8.1 cells express a TcR that reacts with Ttox peptide as presented by I-Ak on L625 cells (Blank et al., 1993). SKAP1 and RapL in the T cell colocalized in the circular pattern at the site of contact with the APC similar to a pSMAC (Figures 5Da and 5Db). Rap1 was found to partially colocalize with circular SKAP1 and RapL, but was also found in other regions of IS (Figures 5Dc). RapL-L224A expression disrupted the pSMAC-like structure for RapL L224A, SKAP1, and Rap1, leaving a more diffuse pattern at the IS (Figures 5Dd and 5De). These data showed that N-SKAP1-C-RapL determines the colocalization of RapL, SKAP1 and Rap1 in both newly formed clusters and in the more mature synapse of anti-CD3 activated T cells.
N-SKAP1-C-RapL Binding Regulates RapL Binding to LFA-1
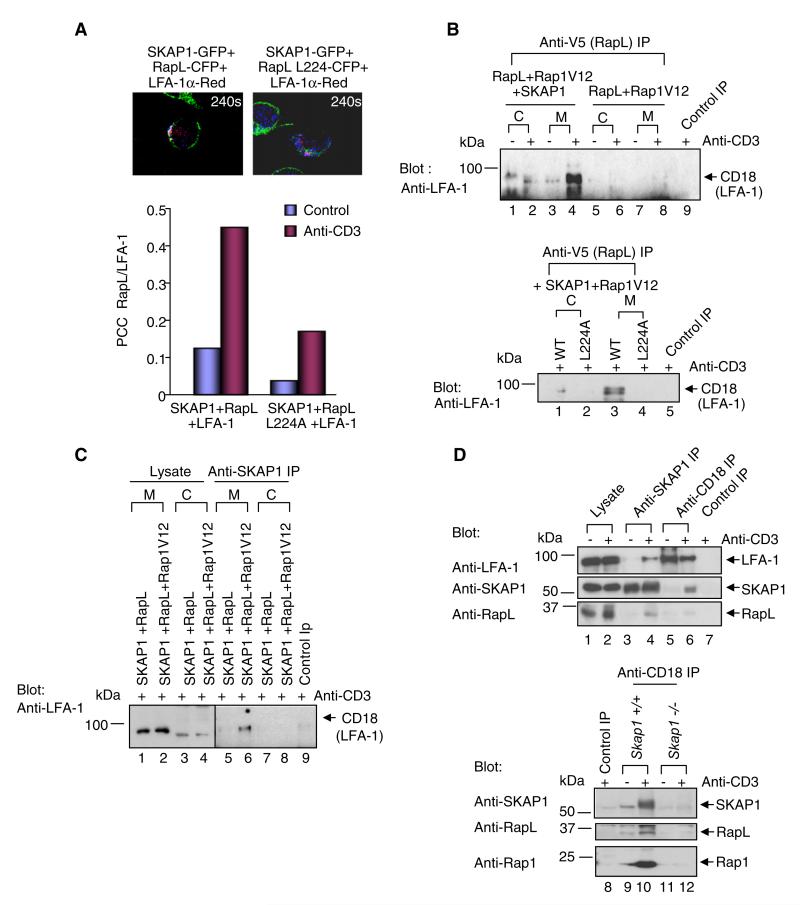
Given this, it was possible that N-SKAP1-C-RapL might control RapL binding to LFA-1. Jurkat cells were cotransfected with RapL-RapL-L224A-CFP, SKAP1-GFP, and LFA-1 (CD11a)-mcherry and imaged by immunofluoresent microscopy (Figure 6A). Transfected CD11a-mcherry increased ICAM-1 binding (data not shown). RapL colocalized with CD11a in response to anti-CD3 ligation as judged by PCC values (i.e., average from 0.14 to 0.45) (example in upper-left panel; lower histogram), whereas RapL-L224A showed reduced levels of colocalization (i.e., ~ 0.18) (upper-right panel; lower histogram).
Figure 6. SKAP1-Rap1-RapL Is Required for Binding to LFA-1.
(A) The upper panels show examples of images of cells at 240 s expressing SKAP1-GFP, RapL-CFP, and LFA-1a-red. The lower panel shows a histogram of average PCC values.
(B) SKAP1, RapL-Rap1V12, or RapL-Rap1V12 were expressed in Jurkat cells, precipiated with anti-V5 (RapL), and blotted with anti-CD18.
(C) SKAP1 and RapL or SKAP1, RapL, and Rap1V12 were expressed; this was followed by precipitation with anti-SKAP1 and blotting with anti-CD18.
(D) As shown in the upper panel, Jurkat cells untreated or ligated wtih anti-CD3 for 10 min were precipitated with anti-SKAP1 or anti-CD18 and then blotted as indicated. The lower panel shows T cell splenocytes from wild-type Skap1+/+ (WT) and Skap1−/− cells. Skap1+/+ or Skap1−/− T cells, unstimulated or ligated with anti-CD3 for 10 min, were precipitated with anti-CD18 and then blotted with anti-SKAP1 (upper panel), anti-RapL blotting (middle panel), and anti-Rap1 blotting (lower panel).
Binding between RapL and LFA-1 was then assessed biochemically by anti-V5-RapL precipitation from cells expressing SKAP1, RapL, or RapL-L224A and Rap1V12 followed by blotting with anti-CD18 (Figure 6B). CD18 is the beta 2 subunit that pairs with CD11a of LFA-1. Anti-V5-RapL failed to precipitate CD18 from cells transfected with RapL and Rap1V12 (lanes 5–8), in neither the cytosolic (lanes 5 and 6) nor membrane fractions (lanes 7 and 8). By contrast, SKAP1 coexpression with RapL and Rap1V12 allowed anti-RapL coprecipitation of CD18 from the membranes of cells in response to anti-CD3 (lane 4). Unlike with wild-type RapL (lower panel, lane 3), coexpression of RapL-L224A with SKAP1 and Rap1V12 failed to coprecipitate LFA-1 from the membrane fraction (lane 4). Similarly, anti-SKAP1 coprecipitated CD18 from membranes of anti-CD3 activated cells expressing RapL, Rap1, and SKAP1 (Figure 6C, lane 6).
Lastly, the complex could also be precipitated from primary Skap1+/+, but not from Skap1−/− T cells (Figure 6D). Anti-SKAP1 coprecipitated endogenous CD18 in a manner dependent on anti-CD3 ligation in Skap1+/+ T cells (lane 4 versus 3). Conversely, anti-CD18 coprecipitated SKAP1 (middle panel, lane 6 versus 5). Both anti-SKAP1 and anti-CD18 coprecipitated RapL with anti-CD3 ligation (lanes 4 versus 3 and 6 versus 5, respectively). Rabbit anti-mouse served as a negative control (lane 7).
Further, in a comparison of primary Skap1+/+ versus Skap1−/− primary T cells, anti-CD18 coprecipitated SKAP1 (upper panel, lanes 10 versus 9), RapL (middle panel, lanes 10 versus 9), and Rap1 (lower panel, lanes 10 versus 9). By contrast, anti-CD18 coprecipitated little if any protein from Skap1−/− cells (all panels, lanes 11 and 12). Collectively, these observations indicated that SKAP1 expression is needed for the binding of the SKAP1-RapLRap1 complex to LFA-1.
RapL224A Disrupts LFA-1-ICAM1 Binding and T Cell-Conjugate Formation
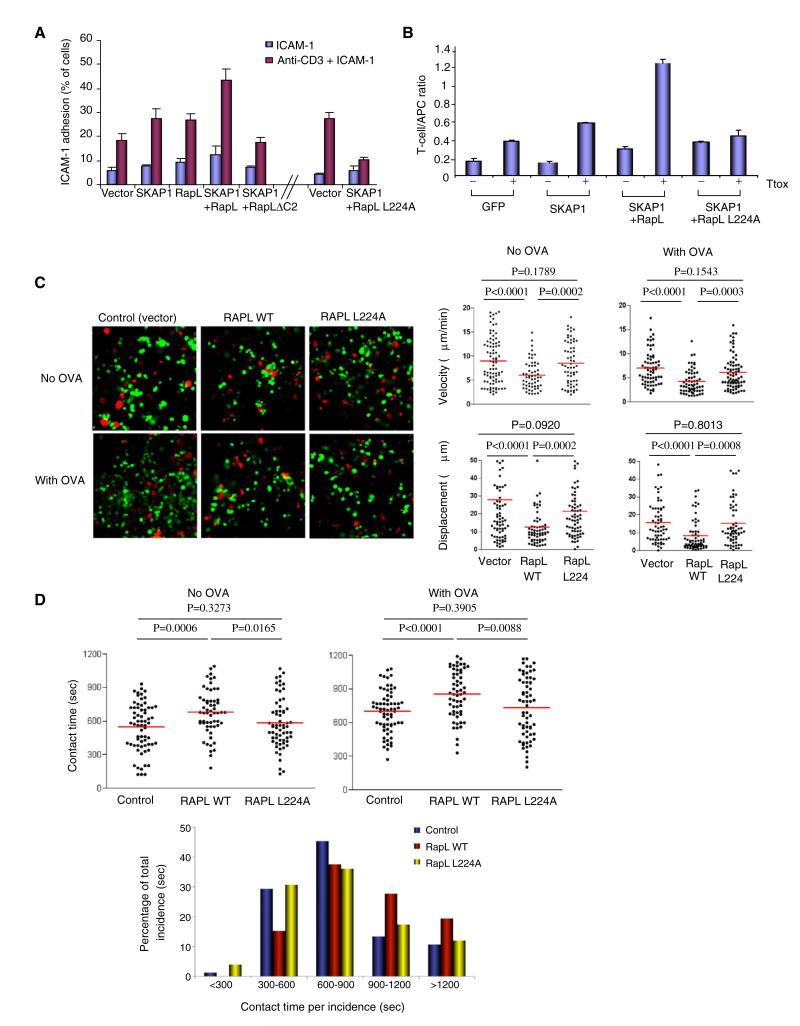
It was next important to examine whether SKAP1-RapL binding was needed for LFA-1-mediated adhesion. T cells were cultured on tissue culture plates coated with ICAM-1 in the presence or absence of soluble anti-CD3 and assessed for binding, as described (Wang et al., 2003) (Figure 7A). In these experiments, RapL and SKAP1 were used to transfect cells, although the additional coexpression of Rap1V12 gave similar results (data not shown). Anti-CD3 increased binding of vector-transfected cells to ICAM-1 cells by 3 to 4-fold. Expression of SKAP1 and RapL individually increased ICAM1 binding further, whereas SKAP1-RapL coexpression had a cooperative effect. Neither RapLΔC2 lacking RapL coiled-coil (SARAH) domain, nor the RapL-L224A mutant supported this increase in binding when coexpressed with RapL (see Figure S4A).
Figure 7. L224A Disrupts Anti-CD3-Induced ICAM-1 Binding and T cell-APC Conjugate Formation.
(A) Jurkat cells transfected with vector, SKAP1, RapL, RapLΔC2, or RapL-L224A were assessed for binding to ICAM-1 on micro-titer plates in response to anti-CD3 ligation. Values expressed as a percentage of the total number of overlaid cells. Standard deviations (SDs) are based on triplicate data points per condition in each experiment.
(B). T8.1 cells transfected with RapL-WT or RapL-L224A were cultured with L625 antigen-presenting cells in the presence or absence of tetanus toxoid (TTox) peptide and assessed for conjugate formation. Values were expressed as a function of the T cell/APC ratio. SDs are based on triplicate data points per condition in each experiment.
(C) DO.11 T cells were transfected with RapL or RapL-L224A were labeled with CFSE, whereas DCs were labeled with SNARP7. Cells were overlaid on congenic LN slices in the presence or absence of OVA peptide. The left panels show the migration of T cells (see Movie S2). The upper dot plots show velocity in the presence and absence of OVA peptide. The lower dot plots show displacement values.
(D) The upper panels show dot plots for contact times between transfected T cells and DCs in the presence and absence of OVA peptide. The lower histogram shows above data expressed as contact times per incidence.
Similarly, RapL-L224A failed to support increased conjugate formation with APCs (Figure 7B). The T cell/APC ratio corresponds to the number of adherent T cells in a given field (i.e., at least three fields with greater than 60 APCs) divided by the number of APCs in the same field. Although the addition of Ttox peptide increased conjugate formation by 2-fold (i.e., from 0.18 to 0.4 T cell/APC ratio) in the GFP-transfected control, SKAP1-RapL coexpression increased conjugate formation by 10-fold (i.e., from 0.18 to 1.2 T cell/APC ratio) (also see Figure S4B). By contrast, the RapL-L224A mutant failed to support an increase in conjugation. Overall, these findings showed that L224A mutant disrupted the cooperative effect of SKAP1 and RapL in increasing ICAM1 binding and conjugate formation.
RapL “Slows” T Cell Motility and Enhances T Cell-DC Conjugation in LNs
Next T cell motility was imaged in ex vivo isolated LN slices as previously described (Bajénoff et al., 2006; Asperti-Boursin et al., 2007). This technique allows the visualization of T cell movement and interaction with APCs in LNs, as has been observed by multiphoton microscopy (Bousso and Robey, 2003; Mempel et al., 2004; Miller et al., 2002; Hugues et al., 2004; Zinselmeyer et al., 2005; Schneider et al., 2006). Freshly purified DO11.10 T cells were labeled with CFSE and overlaid on congenic LN slices in the presence or absence of OVA peptide (Figure 7C; Movie S2). Movement was partially dependent on ICAM1 binding as shown by a 35%–40% reduction in number of bound cells and in the motility of those that adhered in the presence of anti-ICAM1 (Figure S5). Vector-transfected T cells were observed to move randomly over the surface where they would often enter and reappear along the nodal surface. In the presence of perfused OVA peptide, T cells reduced their speed from a mean average speed of 9 to 7 um/ml (Figure 7C, upper dot plots; p value = 0.005; also Figure S6) and displacement from the point of origin (i.e., from 28 to 15 μM) (Figure 7C, lower dot plots). Intriguingly, RapL transfection reduced T cell motility in the absence and presence of peptide from an average speed of 6 and 4 um/ml, respectively (Figure 7C, upper dot plots; Figure S6). The effect of RapL on antigen-reduced displacement was even more substantial (i.e., 28 to 8 μM) (lower dot plots). By contrast, the RapL-L224A mutant failed to reduce the velocity in the absence (p value = 0.1789) or in the presence of OVA peptide (p value = 0.1543). RapL and L224A mutant were expressed to a similar degree as detected by FACS analysis in 60%–70% of the T cells examined (data not shown). These observations show that RapL slowed T cell movement in a manner dependent on the L224A SKAP1 binding site.
Dwell times were also measured between T cells and mature DCs labeled with SNARF7 (Figure 7D; Movie S2). As expected, the addition of OVA peptide increased the contact times between T cells and APCs (i.e., from 550 to 700 s) (upper dot plots). Consistent with an effect on motility, RapL expression increased the contact times with APCs in the absence of peptide (i.e., from 550 to 700 s; p value = 0.0006), and prolonged even further the increased contact time observed in the presence of peptide (i.e., from 700 to 850 s; p value = 0.0001). By contrast, the L224A mutant did not increase the dwell times with APCs in the absence or presence of peptide (p values = 0.3273 and 0.3905, respectively). When scored on the basis of contact time per incidence (lower panel), control cells showed a majority of short-term contacts (30%: 300–600 s; 45%: 600–900 s), whereas RapL increased the percentage of cells to form longer-term interactions with DCs (39%: 600–900 s; 30%: 900–1200 s). The loss of SKAP1 binding skewed the majority of interactions back to shorter time periods (32%: 300–600 s; 38%: 600–900 s). These observations show that RapL can increase dwell times with APCs in LNs in a manner dependent on the L224A SKAP1 binding site.
DISCUSSION
Whereas TCR ligation generates “inside-out” signals for LFA-1 adhesion, the nature of the pathway has yet to be clarified. The challenge has been to distinguish specific effectors of LFA-1 adhesion from generic events. The pathway is central to immunity given that it regulates T cell migration in LNs and T cell-APC dwell times. In this study, we have identified an “inside-out” pathway that couples the TCR with the SKAP1 and Rap1-RapL pathways for adhesion. SKAP1 expression was found to be essential for TCR-driven Rap1-RapL complex formation in primary T cells, and the binding of the newly formed SKAP1-RapL-Rap1 complex to the cytoplasmic tail of LFA-1. N-SKAP1 domain binding to the C-RapL domain connected the pathways such that a RapL-L224A mutant that selectively disrupted SKAP1 binding impaired ICAM-1 binding and T cell-APC conjugate formation. RapL expression also reduced T cell motility (i.e., “slowing”) and increased the T cell-APC dwell times of D011.10 transgenic T cells with DCs in LN slices, an effect reversed in the L224A mutant. Overall, our findings unite the ADAP-SKAP1 and Rap1-RapL modules into a single TCR-driven “inside-out” pathway that activates LFA-1 adhesion, slows T cell motility, and enhances T cell-DC conjugation in LNs.
Previous studies identified two modules for “inside-out” signaling, Rap1-RapL-RIAM and another mediated by adaptors SLP-76, ADAP, and SKAP1 (Rudd, 1999; Kinashi, 2005; Wang and Rudd, 2008). We previously showed that Skap−/− T cells expressing ADAP still have reduced LFA-1 adhesion, thus identifying SKAP1 as an effecter of the SKAP1-ADAP module (Wang et al., 2007). Our present findings provide an underlying mechanism of this effecter function by showing that SKAP1 regulates RapL-Rap1 complex formation and its binding to LFA-1 for clustering. RapL failed completely to translocate to the membranes of Skap1−/− and Fyb−/− primary T cells. Further, RapL and Rap1 failed to form a complex in Skap1−/− T cells or in coexpression studies lacking SKAP1 expression. By contrast, most Rap1 was constitutively associated with the membranes of untreated and anti-CD3-treated cells in which anti-CD3 ligation can converts Rap1 into GTP-bound Rap1 (Bos et al., 2001; Bivona et al., 2004; Schneider et al., 2005). Specificity was shown by the requirement for anti-CD3 ligation and the fact that neither ADAP nor SLP-76 could substitute for SKAP1. Our findings therefore show that ADAP-SKAP1 and Rap1-RapL are not separate modules, but rather comprise a single “inside-out” pathway that couples the TCR complex with the upregulation of LFA-1 adhesion. By contrast, chemokine (i.e., SDF1 and CCL21) induced LFA-1 adhesion-migration can occur normally in Skap-1−/− cells (Wang et al., 2010).
The TCR “inside-out” pathway was mediated by SKAP1 N-terminal region (N-SKAP1) binding to the coiled-coil SARAH domain of RapL (C-RapL) as shown by mutational, GST pull-down studies and isothermal titration calorimetry. ITC measurements of the isolated domains showed a stoichiometry of 0.85 and equilibrium dissociation constant of 0.6 uM, a value in the range of other interactions in signal transduction (Pawson and Nash, 2000). RapL can accommodate Rap1-GTP binding to the RBD domain and SKAP1 binding to C-RapL. By contrast, SKAP1 competed for MST1 binding to C-RapL, indicating the existence of at least two types of Rap1-RapL complexes, one associated with SKAP1 and another with MST1. At the same time, optimal SKAP1 binding to RapL required active Rap1. In one scenario, GTP-Rap1 would change the conformation of RapL for more efficient SKAP1 binding. As a precedent, Rap1 binding to Nore1A is weakened by intramolecular C1 domain binding (Harjes et al., 2006). Although Rap1 constitutively bound to membranes, we cannot exclude that a small subset of cytosolic Rap1 interacts with SKAP1-RapL to help shuttle the SKAP1-RapL complex. SKAP1 may also facilitate Rap1 activation (Kliche et al., 2006). In either case, Rap1-RapL expression without SKAP1 failed to colocalize with or bind to LFA-1, and anti-CD18 failed to coprecipitate RapL-Rap1 from Skap1−/− primary T cells. Further, the RapL L224A mutation prevented LFA-1 binding and reduced ICAM-1 adhesion. Within the complex, RapL is likely to mediate a direct interaction with the αL tail of LFA-1(Katagiri et al., 2003; 2006). GST-fusion proteins of SKAP1 failed to precipitate LFA-1 (data not shown).
Confocal real-time imaging showed that these mediators formed dynamic clusters in response to anti-CD3 ligation. A substantial portion of these were localized in membrane vesicles that further colocalized at the point of contact with anti-CD3-ICAM1 on beads. Colocalization of RapL with SKAP1, RapL with Rap1, and SKAP1-RapL with LFA-1 depended on the L224A residue for SKAP1 binding, a result consistent with the inability of anti-RapL to coprecipitate CD18 from L224A-expressing cells, and of anti-CD18 to coprecipitate LFA-1 from Skap1−/− T cells. Although the nature of these vesicles are unclear, they partially labeled with endosomal markers EEA1 and Rab7 and could potentially recycle and release SKAP1-RapL-Rap1 to LFA-1. Inhibition of endosome recycling blocks integrin-mediated adhesion of T cells (Bivona et al., 2004).
The SKAP1-RapL “inside-out” pathway also modulated the motility of T cells in LNs and the dwell times with DCs. T cells moved randomly where they were often seen to enter and reappear along the nodal surface. Some 35%–40% of movement was dependent on ICAM1 binding as observed in anti-ICAM blocking experiments in which TCR engagement reduced movement (i.e., “stop signal”). RapL itself slowed T cell movement in nodes introducing the possibility it may also contribute to the “stop-signal” for conjugate formation (Dustin et al., 1997). It reduced the mean average velocity and displacement (i.e., movement from origin) of DO11.10 T cells, a result consistent with its ability to increase LFA-1 adhesion and T cell-APC dwell times. It further reduced motility induced by OVA peptide and had an antigen-independent or substitution effect in slowing cells in the absence of peptide. The level of reduced velocity is an underestimation given the normal limitation of transfection (i.e., 60%–70% expression). Both slowing and increased dwell times were SKAP1 dependent as shown by L224A reversal of the effects. The L224A mutant resembled the GFP control in both the absence and presence of OVA. Consistently, Skap−/− T cells also show impaired slowing in LNs in response to SEA (data not shown). Overall, our observations have identified an “inside-out” pathway that alters T cell behavior in lymph nodes and contributes to the slowing of T cells for more efficient conjugate formation and T cell responses to antigen. Chemokines and coreceptors such as CTLA-4 will then add to the mixture of signals that determine the ultimate behavior of cells (Rudd, 2008). Motility in the LN slices showed partial sensitivity to anti-CCR7 blockade (data not shown; Asperti-Boursin et al., 2007).
In the overall scheme, the “inside-out” pathway would be initiated by TCR-induced ADAP phosphorylation of YDDV motifs, sites that are preferentially phosphorylated by p59fyn and that bind the SLP-76 SH2 domain (Veale et al., 1999; Raab et al., 1999; Geng et al., 1999) (Figure S7). ADAP is in turn closely coupled to SKAP1 primarily via SKAP1 SH3 domain binding to ADAP (Liu et al., 1998; Marie-Cardine et al., 1998, Kang et al., 2000). In turn, SKAP1 regulates RapL translocation to membranes and formation of a SKAP1-RapL-Rap1 complex. N-SKAP1 mediates binding to C-RapL to form the SKAP1-RapL complex, whereas anti-CD3 activates Rap1-GTP to bind the RBD domain of RapL. A minimal trimeric complex would then bind to the LFA-1 via RapL binding to K1097-K1099 residues in αL tail (Tohyama et al., 2003; Katagiri et al., 2003). LFA-1 and SKAP1 are also recruited into lipid rafts (Krauss and Altevogt, 1999; Wang et al., 2003). At a minimum, ADAP prevents SKAP1 degradation to ensure sufficient levels of the effecter protein. However, it also has a G protein and VASP binding sites for actin remodeling and may modify adhesion events mediated by SKAP1. Further, whereas SKAP1-RapL-Rap1 binds the α chain of LFA-1, RIAM-talin and Kindlin 3 bind the β chain (Moser et al., 2008; Lee et al., 2009), leading to the possible formation of a large single complex or distinct regulatory complexes. Rap-1 is likely to exist in distinct pools, one pool engaged with RAPL and the second engaged with RIAM. It will be intriguing to know whether each pool directs a different modality of LFA-1 activation. RIAM-talin may increase affinity, whereas the SKAP1-RAPL could direct LFA-1 vesicle fusion and clustering. Similarly, SKAP-1 can compete for MST1 binding leading to potential antagonism between the two proteins and the formation of two distinct pools of RapL complexes. SKAP1 in turn intersects with RIAM via binding to its PH and RA domains (Kliche et al., 2006; Ménasché et al., 2007). Unlike in the case of SKAP1, coexpression of RIAM did not compete for MST binding to the SARAH domain of RapL. RIAM binding to SKAP1 could act to interlock α and β chains or help to terminate the response. Subsequent “outside-in” signaling for altered cell shape and motility is also dependent on SLP-76 binding to ADAP (Wang et al., 2009).
It is also important to note that an additional unidentified signal is needed from TCR ligation. This was shown by the fact that anti-CD3 ligation was needed for RapL-Rap1 complex formation even in the presence of constitutively active Rap1V12. One attractive possibility is that phosphatidylinositol 3-kinase (PI3K) provides this additional signal given that SKAP1 has a PH domain that binds PI-3,4,5-P3 (PIP3) for membrane localization (Marie-Cardine et al., 1997; Liu et al., 1998). In this model, TCR activation of PI3K would provide the shuttle signal for SKAP1 translocation of RapL to the plasma membrane and/or proximal vesicles. It is therefore interesting that SKAP2 dimerization via its N-terminal domain represses the function of its adjacent PH domain (Swanson et al., 2008). Similar residues are conserved in SKAP1 (Liu et al., 1998). RapL could promote the opening of the SKAP1 N-domain to allow for PH domain activation and membrane translocation. It is also tempting to propose that the BCR in B cells may promote the formation of an analogous SKAP2-RapL-Rap1 complex for LFA-1 adhesion.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
T cells were cultured in RPMI 1640 medium with 10% (vol/vol) fetal-calf serum and 1% (wt/vol) penicillin-streptomycin (Raab et al., 2001). Murine hybridoma T8.1-expressing TCR specific for Ttox (830–843) and L625.7 cells were gift of O. Acuto, Oxford University. (Wang et al., 2004; Blank et al., 1993). Anti-SKAP1 (Transduction Laboratories); anti-V5 (Invitrogen); anti-myc (Upstate Biotechnology); anti-GFP (Santa Cruz Biotechnology); anti-human CD3 (American Type Culture Collection); anti-mouse CD3 (2C11; hamster anti-mouse CD3) and anti-CD18 (anti-LFA-1) (BD Biosciences); and Alexa-488 and 568-conjugated secondary Abs (Molecular Probes), anti-EEA1 (Abcam), anti-rab7 (Cell Signaling), and anti-ICAM1 (R&D Systems; MN, USA) were purchased from the manufacturers indicated. Ttox (residues 830–843) was obtained from Research Genetics.
Generation of Plasmids and Mutagenesis
Full-length human SKAP1 cDNA were cloned into the pSRa expression vector and in-frame with the NH2 terminus of the GFP gene (Promega). N (residues 1–104), SK (residues 209–285), SH3 (residues 285–359) and N-PH-SK (residues 1–359) domains of SKAP1 were subcloned into the pGEX-2T vector (GE Healthcare). Human RapL was cloned into the pcDNA3.1-V5 expression vector (Invitrogen). RapL-L224A and L253A mutants were generated by site-direct mutagenesis (Quick Change protocol; Stratagene). Human Rap1V12 and Rap1N17 were cloned into myc-expression and pcDNA3.1-mCherry vectors. SKAP1 and RapL were also cloned into pSR alpha HAGFP and CFP (Stratagene). Plasmids for GST fusion proteins were transformed into the DH5α strain of Escherichia coli and induced with isopropyl-D-thiogalactopyranoside (IPTG) to produce GST fusion proteins as done previously (Kang et al., 2000). GST, GST-full-length SKAP1, and GST-fused subdomains of SKAP1 were adsorbed to Glutathione-Sepharose 4B (GE Healthcare).
Immunoprecipitation and Blotting
Immunoprecipitation and blotting were performed as previously described (Raab et al., 1999; 2001). For blotting, precipitates were separated by SDS-PAGE and transferred onto nitrocellulose filters (Schleicher and Schuell). Bound antibody was revealed with horseradish peroxidase-conjugated rabbit anti-mouse antibody with enhanced chemiluminescence (ECL, Amersham Biosciences). For purification of membrane fractions, Jurkat T cells were sheared in hypotonic buffer, nuclei were removed by low-speed centrifugation (1500 rpm, 10 min), and supernatant was recentrifugated at high speed (25,000 rpm) for 1 hr. The cytosolic fraction was found in the supernatant, whereas membranes remained in the pellet.
Confocal Microscopy
T cells were purified from Skap1+/+ and Skap1−/− mouse spleens as described (Wang et al., 2007; Wang et al., 2010). Jurkat T cells were cotransfected with expression plasmids (2–10 ug/106 cells) by microporation (Digital Bio Technology). T cells were transfected by microporation (Digital Bio Technology) or by Nucleofector Technology from Amaxa (Lonza,Germany). For live-cell imaging, poly-L-lysine (Sigma)-treated chambered cover slides (LabTek) were coated with 10 μg/ml mAb OKT3 while images of the contact area were acquired by resonance scanning confocal microscopy (TCS SP2 RS, Leica, Heidelberg, Germany) with excitation wavelengths of 514 nm for EYFP and 594 nm for mRFP and a 63× water-immersion objective (NA = 1.2). Images and Pearson’s correlation coefficients (PCCs) were processed with Volocity (Improvision) and ImageJ (National Institutes of Health). T cell conjugation assays were performed as previously described (Wang et al., 2003, 2007). T8.1 cells were cocultured with L625.7 adhesive cells that had been preincubated with Ttox peptide (830–943) (Blank et al., 1993). For this, transmitted light images were acquired every 10 s during 20 min with a 403 phase objective on a Zeiss LSM510 microscope. T cell-APC interactions were then monitored by ImageJ software.
Migration in LN Slices
Ex vivo imaging of T cells and APCs in LN slices was adapted as described (Asperti-Boursin et al., 2007; Bajénoff,et al., 2006). In brief, inguinal mouse LNs were embedded in low-gelling-temperature agarose (type VII-A; Sigma-Aldrich) and sliced with a vibratome (VT 1000S; Leica). Slices were transferred to 0.4 μm organotypic culture inserts (Millicell; Millipore). CD4+ T cells were purified by positive selection (Dynabeads mouse CD4 (L3T4). T cells incubated with CFSE (Sigma, Poole, UK) and matured bone marrow-derived dendritic cells (BMDCs) labeled with SNARF-1 (Invitrogen, Paisley, UK) were colayered on slices. BMDCs were generated from marrow by incubation with 20 ng/ml recombinant murine GM-CSF and 1000 IU/ml IL-4. On the day 7 of culture, BMDCs were induced to mature by addition of 1 μg/ml LPS to the cultures. Images were acquired every 10 s during 20 min with a 20× phase objective on a Zeiss LSM510 microscope. Cell motility was analyzed with Zeiss LSM confocal software and Volocity software (Improvision).
Isothermal Titration Calorimetry
For ITC analysis, recombinant GST-N-SKAP1 and GST-C-RapL was bacterially expressed as described previously (Prasad et al., 1993; Raab et al., 1995). ITC analysis was performed with the VP ITC (Microcal, Northampton, MA, USA) (Olsson et al., 2008; Ladbury, 2004). Protein was diluted from stock solution to the concentration required for the ITC experiment (4–10 μM) and dialyzed against the ITC buffer 7.5 (10 mM Na-K phosphate buffer [pH 7.5] and 150 mM NaCl). All binding data were analyzed by fitting the binding isotherm to a simple independent binding-site model with Origin software provided with the ITC (MicroCal).
ACKNOWLEDGMENTS
This work was supported by a programme grant from the Wellcome Trust. C.E.R. is the recipient of a Wellcome Trust Principal Research Fellowship, and J.E.L was the recipient of a Wellcome Trust Senior Research Fellowship. We thank H. Schneider (Cambridge University) for reviewing the manuscript and R. George (University College, London) for his help with the ITC experiments.
Footnotes
Supplemental Information includes seven figures and two movies and can be found with this article online at doi:10.1016/j.immuni.2010.03.007.
REFERENCES
- Asperti-Boursin F, Real E, Bismuth G, Trautmann A, Donnadieu E. CCR7 ligands control basal T cell motility within lymph node slices in a phosphoinositide 3-kinase-independent manner. J. Exp. Med. 2007;204:1167–1179. doi: 10.1084/jem.20062079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajénoff M, Egen JG, Koo LY, Laugier JP, Brau F, Glaichenhaus N, Germain RN. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Boitel B, Mège D, Ermonval M, Acuto O. Analysis of tetanus toxin peptide/DR recognition by human T cell receptors reconstituted into a murine T cell hybridoma. Eur. J. Immunol. 1993;23:3057–3065. doi: 10.1002/eji.1830231203. [DOI] [PubMed] [Google Scholar]
- Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: Adhering to new models. Nat. Rev. Mol. Cell Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- Bousso P, Robey E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat. Immunol. 2003;4:579–585. doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman CV, Springer TA. Integrin avidity regulation: Are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Collins TL, Deckert M, Altman A. Views on Vav. Immunol. Today. 1997;18:221–225. doi: 10.1016/s0167-5699(97)01037-2. [DOI] [PubMed] [Google Scholar]
- da Silva AJ, Li Z, de Vera C, Canto E, Findell P, Rudd CE. Cloning of a novel T-cell protein FYB that binds FYN and SH2-domain-containing leukocyte protein 76 and modulates interleukin 2 production. Proc. Natl. Acad. Sci. USA. 1997;94:7493–7498. doi: 10.1073/pnas.94.14.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke-Cohan JS, Kang H, Liu H, Rudd CE. Regulation and function of SKAP-55 non-canonical motif binding to the SH3c domain of adhesion and degranulation-promoting adaptor protein. J. Biol. Chem. 2006;281:13743–13750. doi: 10.1074/jbc.M508774200. [DOI] [PubMed] [Google Scholar]
- Dustin ML. A dynamic view of the immunological synapse. Semin. Immunol. 2005;17:400–410. doi: 10.1016/j.smim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Bromley SK, Kan Z, Peterson DA, Unanue ER. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc. Natl. Acad. Sci. USA. 1997;94:3909–3913. doi: 10.1073/pnas.94.8.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Bivona TG, Philips MR. Membranes as messengers in T cell adhesion signaling. Nat. Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- Geng L, Raab M, Rudd CE. Cutting edge: SLP-76 cooperativity with FYB/FYN-T in the Up-regulation of TCR-driven IL-2 transcription requires SLP-76 binding to FYB at Tyr595 and Tyr651. J. Immunol. 1999;163:5753–5757. [PubMed] [Google Scholar]
- Geng L, Pfister S, Kraeft SK, Rudd CE. Adaptor FYB (Fyn-binding protein) regulates integrin-mediated adhesion and mediator release: Differential involvement of the FYB SH3 domain. Proc. Natl. Acad. Sci. USA. 2001;98:11527–11532. doi: 10.1073/pnas.191378198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Dynamic imaging of the immune system: Progress, pitfalls and promise. Nat. Rev. Immunol. 2006;6:497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- Griffiths EK, Krawczyk C, Kong YY, Raab M, Hyduk SJ, Bouchard D, Chan VS, Kozieradzki I, Oliveira-Dos-Santos AJ, Wakeham A, et al. Positive regulation of T cell activation and integrin adhesion by the adapter Fyb/Slap. Science. 2001;293:2260–2263. doi: 10.1126/science.1063397. [DOI] [PubMed] [Google Scholar]
- Harjes E, Harjes S, Wohlgemuth S, Müller KH, Krieger E, Herrmann C, Bayer P. GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1. Structure. 2006;14:881–888. doi: 10.1016/j.str.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Hogg N, Laschinger M, Giles K, McDowall A. T-cell integrins: More than just sticking points. J. Cell Sci. 2003;116:4695–4705. doi: 10.1242/jcs.00876. [DOI] [PubMed] [Google Scholar]
- Huang Y, Norton DD, Precht P, Martindale JL, Burkhardt JK, Wange RL. Deficiency of ADAP/Fyb/SLAP-130 destabilizes SKAP55 in Jurkat T cells. J. Biol. Chem. 2005;280:23576–23583. doi: 10.1074/jbc.M413201200. [DOI] [PubMed] [Google Scholar]
- Hugues S, Fetler L, Bonifaz L, Helft J, Amblard F, Amigorena S. Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nat. Immunol. 2004;5:1235–1242. doi: 10.1038/ni1134. [DOI] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat. Immunol. 2006;7:247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Jo EK, Wang H, Rudd CE. An essential role for SKAP-55 in LFA-1 clustering on T cells that cannot be substituted by SKAP-55R. J. Exp. Med. 2005;201:1733–1739. doi: 10.1084/jem.20042577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MS, Singer AL, Koretzky GA. Adaptors as central mediators of signal transduction in immune cells. Nat. Immunol. 2003;4:110–116. doi: 10.1038/ni0203-110. [DOI] [PubMed] [Google Scholar]
- Kang H, Freund C, Duke-Cohan JS, Musacchio A, Wagner G, Rudd CE. SH3 domain recognition of a proline-independent tyrosine-based RKxxYxxY motif in immune cell adaptor SKAP55. EMBO J. 2000;19:2889–2899. doi: 10.1093/emboj/19.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K, Maeda A, Shimonaka M, Kinashi T. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- Katagiri K, Ohnishi N, Kabashima K, Iyoda T, Takeda N, Shinkai Y, Inaba K, Kinashi T. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat. Immunol. 2004;5:1045–1051. doi: 10.1038/ni1111. [DOI] [PubMed] [Google Scholar]
- Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 2006;7:919–928. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- Kinashi T. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 2005;5:546–559. doi: 10.1038/nri1646. [DOI] [PubMed] [Google Scholar]
- Kliche S, Breitling D, Togni M, Pusch R, Heuer K, Wang X, Freund C, Kasirer-Friede A, Menasche G, Koretzky GA, Schraven B. The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell. Biol. 2006;26:7130–7144. doi: 10.1128/MCB.00331-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss K, Altevogt P. Integrin leukocyte function-associated antigen-1-mediated cell binding can be activated by clustering of membrane rafts. J. Biol. Chem. 1999;274:36921–36927. doi: 10.1074/jbc.274.52.36921. [DOI] [PubMed] [Google Scholar]
- Krawczyk C, Oliveira-dos-Santos A, Sasaki T, Griffiths E, Ohashi PS, Snapper S, Alt F, Penninger JM. Vav1 controls integrin clustering and MHC/peptide-specific cell adhesion to antigen-presenting cells. Immunity. 2002;16:331–343. doi: 10.1016/s1074-7613(02)00291-1. [DOI] [PubMed] [Google Scholar]
- Kuhné MR, Lin J, Yablonski D, Mollenauer MN, Ehrlich LI, Huppa J, Davis MM, Weiss A. Linker for activation of T cells, zeta-associated protein-70, and Src homology 2 domain-containing leukocyte protein-76 are required for TCR-induced microtubule-organizing center polarization. J. Immunol. 2003;171:860–866. doi: 10.4049/jimmunol.171.2.860. [DOI] [PubMed] [Google Scholar]
- Kupfer A. Signaling in the immunological synapse: Defining the optimal size. Immunity. 2006;25:11–13. doi: 10.1016/j.immuni.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Labno CM, Lewis CM, You D, Leung DW, Takesono A, Kamberos N, Seth A, Finkelstein LD, Rosen MK, Schwartzberg PL, Burkhardt JK. Itk functions to control actin polymerization at the immune synapse through localized activation of Cdc42 and WASP. Curr. Biol. 2003;13:1619–1624. doi: 10.1016/j.cub.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury JE. Application of isothermal titration calorimetry in the biological sciences: Things are heating up! Biotechniques. 2004;37:885–887. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kang H, Raab M, da Silva AJ, Kraeft SK, Rudd CE. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA. 1998;95:8779–8784. doi: 10.1073/pnas.95.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Cardine A, Bruyns E, Eckerskorn C, Kirchgessner H, Meuer SC, Schraven B. Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J. Biol. Chem. 1997;272:16077–16080. doi: 10.1074/jbc.272.26.16077. [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A, Hendricks-Taylor LR, Boerth NJ, Zhao H, Schraven B, Koretzky GA. Molecular interaction between the Fyn-associated protein SKAP55 and the SLP-76-associated phosphoprotein SLAP-130. J. Biol. Chem. 1998;273:25789–25795. doi: 10.1074/jbc.273.40.25789. [DOI] [PubMed] [Google Scholar]
- Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- Ménasché G, Kliche S, Chen EJ, Stradal TE, Schraven B, Koretzky G. RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol. Cell. Biol. 2007;27:4070–4081. doi: 10.1128/MCB.02011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- Mor A, Dustin ML, Philips MR. Small GTPases and LFA-1 reciprocally modulate adhesion and signaling. Immunol. Rev. 2007;218:114–125. doi: 10.1111/j.1600-065X.2007.00538.x. [DOI] [PubMed] [Google Scholar]
- Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Thomas MS, Burbach BJ, Peterson EJ, Shimizu Y. Adhesion and degranulation-promoting adapter protein (ADAP) positively regulates T cell sensitivity to antigen and T cell survival. J. Immunol. 2007;179:3559–3569. doi: 10.4049/jimmunol.179.6.3559. [DOI] [PubMed] [Google Scholar]
- Musci MA, Hendricks-Taylor LR, Motto DG, Paskind M, Kamens J, Turck CW, Koretzky GA. Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J. Biol. Chem. 1997;272:11674–11677. doi: 10.1074/jbc.272.18.11674. [DOI] [PubMed] [Google Scholar]
- Olsson TS, Williams MA, Pitt WR, Ladbury JE. The thermodynamics of protein-ligand interaction and solvation: Insights for ligand design. J. Mol. Biol. 2008;384:1002–1017. doi: 10.1016/j.jmb.2008.09.073. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- Peterson EJ, Woods ML, Dmowski SA, Derimanov G, Jordan MS, Wu JN, Myung PS, Liu QH, Pribila JT, Freedman BD, et al. Coupling of the TCR to integrin activation by Slap-130/Fyb. Science. 2001;293:2263–2265. doi: 10.1126/science.1063486. [DOI] [PubMed] [Google Scholar]
- Prasad KV, Janssen O, Kapeller R, Raab M, Cantley LC, Rudd CE. Src-homology 3 domain of protein kinase p59fyn mediates binding to phosphatidylinositol 3-kinase in T cells. Proc. Natl. Acad. Sci. USA. 1993;90:7366–7370. doi: 10.1073/pnas.90.15.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Cai YC, Bunnell SC, Heyeck SD, Berg LJ, Rudd CE. p56Lck and p59Fyn regulate CD28 binding to phosphatidylinositol 3-kinase, growth factor receptor-bound protein GRB-2, and T cell-specific protein-tyrosine kinase ITK: Implications for T-cell costimulation. Proc. Natl. Acad. Sci. USA. 1995;92:8891–8895. doi: 10.1073/pnas.92.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Kang H, da Silva A, Zhu X, Rudd CE. FYN-T-FYBSLP-76 interactions define a T-cell receptor zeta/CD3-mediated tyrosine phosphorylation pathway that up-regulates interleukin 2 transcription in T-cells. J. Biol. Chem. 1999;274:21170–21179. doi: 10.1074/jbc.274.30.21170. [DOI] [PubMed] [Google Scholar]
- Raab M, Pfister S, Rudd CE. CD28 signaling via VAV/SLP-76 adaptors: Regulation of cytokine transcription independent of TCR ligation. Immunity. 2001;15:921–933. doi: 10.1016/s1074-7613(01)00248-5. [DOI] [PubMed] [Google Scholar]
- Rudd CE. Adaptors and molecular scaffolds in immune cell signaling. Cell. 1999;96:5–8. doi: 10.1016/s0092-8674(00)80953-8. [DOI] [PubMed] [Google Scholar]
- Rudd CE. The reverse stop-signal model for CTLA4 function. Nat. Rev. Immunol. 2008;8:153–160. doi: 10.1038/nri2253. [DOI] [PubMed] [Google Scholar]
- Rudd CE, Janssen O, Cai YC, da Silva AJ, Raab M, Prasad KV. Two-step TCR zeta/CD3-CD4 and CD28 signaling in T cells: SH2/SH3 domains, protein-tyrosine and lipid kinases. Immunol. Today. 1994;15:225–234. doi: 10.1016/0167-5699(94)90248-8. [DOI] [PubMed] [Google Scholar]
- Samelson LE. Signal transduction mediated by the T cell antigen receptor: The role of adapter proteins. Annu. Rev. Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- Sarantos MR, Raychaudhuri S, Lum AFH, Staunton DE, Simon SI. Leukocyte function-associated antigen 1-mediated adhesion stability is dynamically regulated through affinity and valency during bond formation with intercellular adhesion molecule-1. J. Biol. Chem. 2005;280:28290–28298. doi: 10.1074/jbc.M501662200. [DOI] [PubMed] [Google Scholar]
- Schneider H, Valk E, da Rocha Dias S, Wei B, Rudd CE. CTLA-4 up-regulation of lymphocyte function-associated antigen 1 adhesion and clustering as an alternate basis for coreceptor function. Proc. Natl. Acad. Sci. USA. 2005;102:12861–12866. doi: 10.1073/pnas.0505802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Downey J, Smith A, Zinselmeyer BH, Rush C, Brewer JM, Wei B, Hogg N, Garside P, Rudd CE. Reversal of the TCR stop signal by CTLA-4. Science. 2006;313:1972–1975. doi: 10.1126/science.1131078. [DOI] [PubMed] [Google Scholar]
- Schneider H, Smith X, Liu H, Bismuth G, Rudd CE. CTLA-4 disrupts ZAP70 microcluster formation with reduced T cell/APC dwell times and calcium mobilization. Eur. J. Immunol. 2008;38:40–47. doi: 10.1002/eji.200737423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebzda E, Bracke M, Tugal T, Hogg N, Cantrell DA. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 2002;3:251–258. doi: 10.1038/ni765. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Mobley JL, Finkelstein LD, Chan AS. A role for phosphatidylinositol 3-kinase in the regulation of beta 1 integrin activity by the CD2 antigen. J. Cell Biol. 1995;131:1867–1880. doi: 10.1083/jcb.131.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KD, Tang Y, Ceccarelli DF, Poy F, Sliwa JP, Neel BG, Eck MJ. The Skap-hom dimerization and PH domains comprise a 3′-phosphoinositide-gated molecular switch. Mol. Cell. 2008;32:564–575. doi: 10.1016/j.molcel.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–11. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Tohyama Y, Katagiri K, Pardi R, Lu C, Springer TA, Kinashi T. The critical cytoplasmic regions of the alphaL/beta2 integrin in Rap1-induced adhesion and migration. Mol. Biol. Cell. 2003;14:2570–2582. doi: 10.1091/mbc.E02-09-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Jin SG, Zhang XF, Avruch J, Pfeifer GP. RASSF3 and NORE1: Identification and cloning of two human homologues of the putative tumor suppressor gene RASSF1. Oncogene. 2002;21:2713–2720. doi: 10.1038/sj.onc.1205365. [DOI] [PubMed] [Google Scholar]
- Veale M, Raab M, Li Z, da Silva AJ, Kraeft SK, Weremowicz S, Morton CC, Rudd CE. Novel isoform of lymphoid adaptor FYN-T-binding protein (FYB-130) interacts with SLP-76 and up-regulates interleukin 2 production. J. Biol. Chem. 1999;274:28427–28435. doi: 10.1074/jbc.274.40.28427. [DOI] [PubMed] [Google Scholar]
- Wang H, Rudd CE. SKAP-55, SKAP-55-related and ADAP adaptors modulate integrin-mediated immune-cell adhesion. Trends Cell Biol. 2008;18:486–493. doi: 10.1016/j.tcb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moon EY, Azouz A, Wu X, Smith A, Schneider H, Hogg N, Rudd CE. SKAP-55 regulates integrin adhesion and formation of T cell-APC conjugates. Nat. Immunol. 2003;4:366–374. doi: 10.1038/ni913. [DOI] [PubMed] [Google Scholar]
- Wang H, McCann FE, Gordan JD, Wu X, Raab M, Malik TH, Davis DM, Rudd CE. ADAP-SLP-76 binding differentially regulates supramolecular activation cluster (SMAC) formation relative to T cell-APC conjugation. J. Exp. Med. 2004;200:1063–1074. doi: 10.1084/jem.20040780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu H, Lu Y, Lovatt M, Wei B, Rudd CE. Functional defects of SKAP-55-deficient T cells identify a regulatory role for the adaptor in LFA-1 adhesion. Mol. Cell. Biol. 2007;27:6863–6875. doi: 10.1128/MCB.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wei B, Bismuth G, Rudd CE. SLP-76-ADAP adaptor module regulates LFA-1 mediated costimulation and T cell motility. Proc. Natl. Acad. Sci. USA. 2009;106:12436–12441. doi: 10.1073/pnas.0900510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lu Y, Rudd CE. SKAP1 is dispensable for chemokine-induced migration of primary T-cells. Immunol. Lett. 2010;128:148–153. doi: 10.1016/j.imlet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Medoff BD, Chen L, Li L, Zhang XF, Praskova M, Liu M, Landry A, Blumberg RS, Boussiotis VA, et al. The Nore1B/Mst1 complex restrains antigen receptor-induced proliferation of naïve T cells. Proc. Natl. Acad. Sci. USA. 2008;105:20321–20326. doi: 10.1073/pnas.0810773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeyer BH, Dempster J, Gurney AM, Wokosin D, Miller M, Ho H, Millington OR, Smith KM, Rush CM, Parker I, et al. In situ characterization of CD4+ T cell behavior in mucosal and systemic lymphoid tissues during the induction of oral priming and tolerance. J. Exp. Med. 2005;201:1815–1823. doi: 10.1084/jem.20050203. [DOI] [PMC free article] [PubMed] [Google Scholar]