Abstract
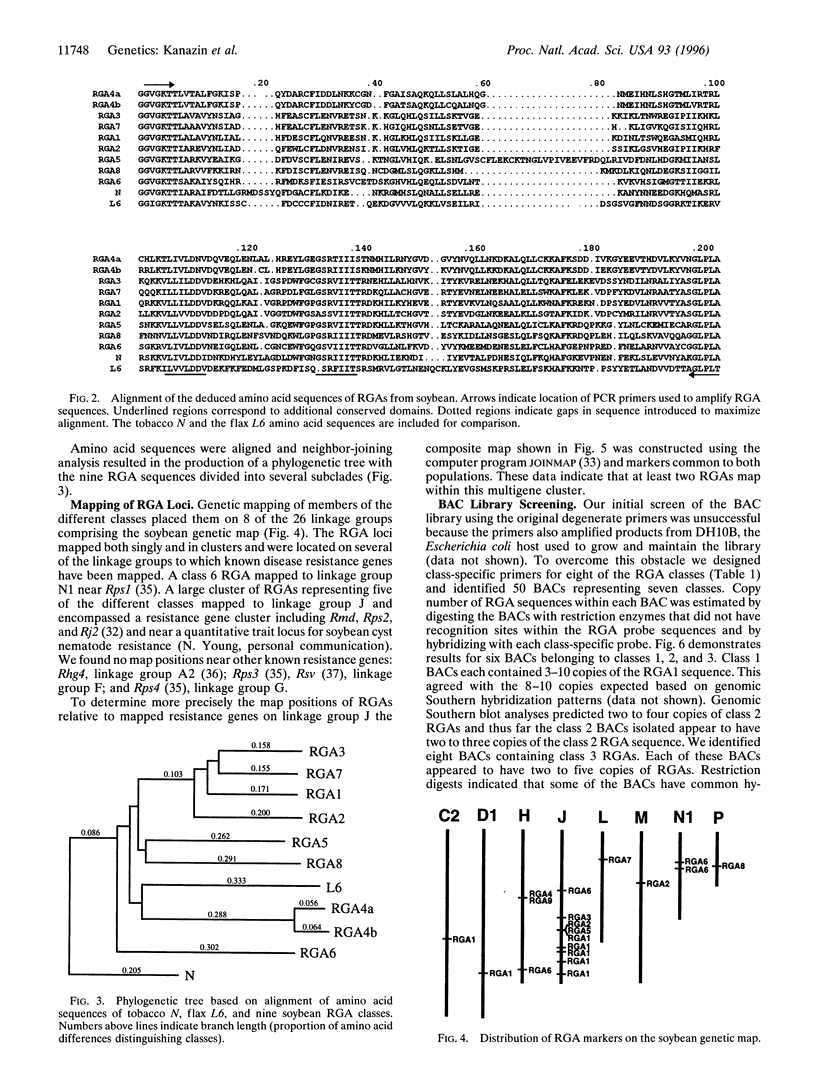
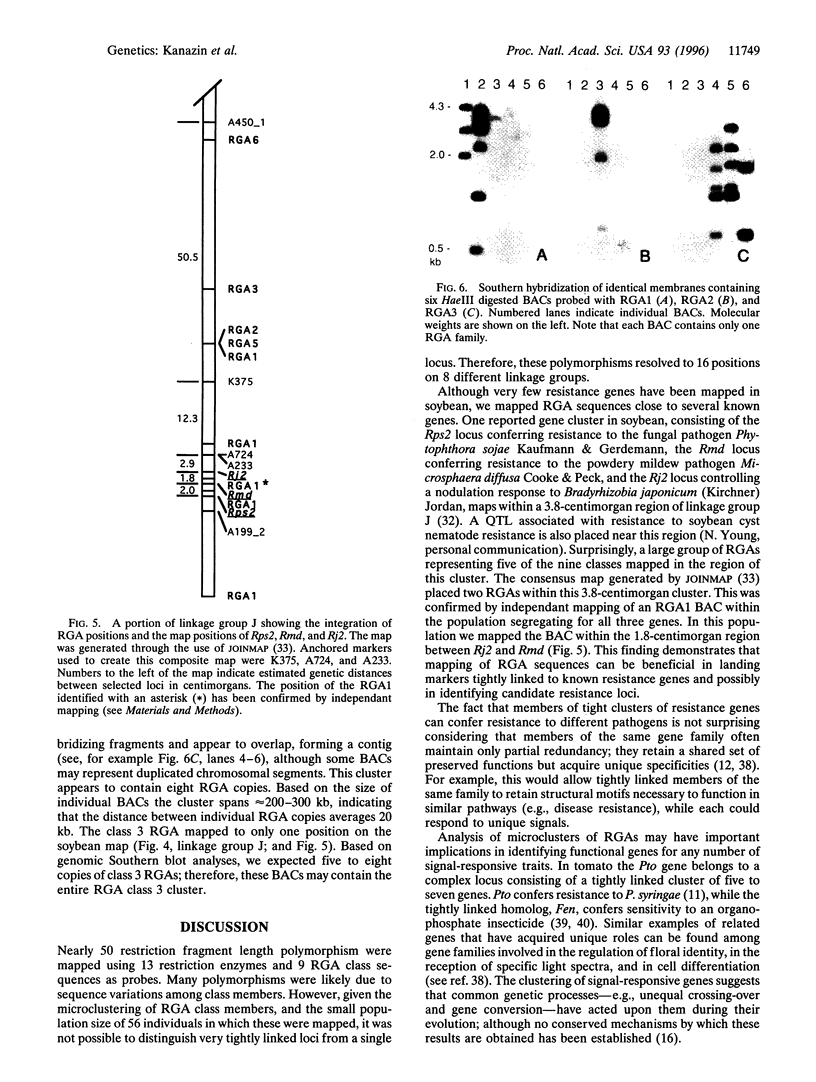
Sequences of cloned resistance genes from a wide range of plant taxa reveal significant similarities in sequence homology and structural motifs. This is observed among genes conferring resistance to viral, bacterial, and fungal pathogens. In this study, oligonucleotide primers designed for conserved sequences from coding regions of disease resistance genes N (tobacco), RPS2 (Arabidopsis) and L6 (flax) were used to amplify related sequences from soybean [Glycine max (L.) Merr.]. Sequencing of amplification products indicated that at least nine classes of resistance gene analogs (RGAs) were detected. Genetic mapping of members of these classes located them to eight different linkage groups. Several RGA loci mapped near known resistance genes. A bacterial artificial chromosome library of soybean DNA was screened using primers and probes specific for eight RGA classes and clones were identified containing sequences unique to seven classes. Individual bacterial artificial chromosomes contained 2-10 members of single RGA classes. Clustering and sequence similarity of members of RGA classes suggests a common process in their evolution. Our data indicate that it may be possible to use sequence homologies from conserved motifs of cloned resistance genes to identify candidate resistance loci from widely diverse plant taxa.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bent A. F., Kunkel B. N., Dahlbeck D., Brown K. L., Schmidt R., Giraudat J., Leung J., Staskawicz B. J. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994 Sep 23;265(5180):1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991 Apr 5;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Dangl J. L. Pièce de Résistance: novel classes of plant disease resistance genes. Cell. 1995 Feb 10;80(3):363–366. doi: 10.1016/0092-8674(95)90485-9. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S. P., Whitham S., Choi D., Hehl R., Corr C., Baker B. Transposon tagging of tobacco mosaic virus resistance gene N: its possible role in the TMV-N-mediated signal transduction pathway. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4175–4180. doi: 10.1073/pnas.92.10.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. G., Lawrence G. J., Finnegan E. J., Anderson P. A. Contrasting complexity of two rust resistance loci in flax. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4185–4188. doi: 10.1073/pnas.92.10.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M. R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R. W., Dangl J. L. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995 Aug 11;269(5225):843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Hulbert S. H., Bennetzen J. L. Recombination at the Rp1 locus of maize. Mol Gen Genet. 1991 May;226(3):377–382. doi: 10.1007/BF00260649. [DOI] [PubMed] [Google Scholar]
- Jones D. A., Thomas C. M., Hammond-Kosack K. E., Balint-Kurti P. J., Jones J. D. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994 Nov 4;266(5186):789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Keim P., Diers B. W., Olson T. C., Shoemaker R. C. RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics. 1990 Nov;126(3):735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. W., Moore M. J. Novel HOX, POU and FKH genes expressed during bFGF-induced mesodermal differentiation in Xenopus. Nucleic Acids Res. 1994 Sep 25;22(19):3990–3996. doi: 10.1093/nar/22.19.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B., Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994 Oct;19(10):415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., Lincoln S. E., Newberg L. A., Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987 Oct;1(2):174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lawrence G. J., Finnegan E. J., Ayliffe M. A., Ellis J. G. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell. 1995 Aug;7(8):1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y. T., Martin G. B. The disease-resistance gene Pto and the fenthion-sensitivity gene fen encode closely related functional protein kinases. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4181–4184. doi: 10.1073/pnas.92.10.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. B., Brommonschenkel S. H., Chunwongse J., Frary A., Ganal M. W., Spivey R., Wu T., Earle E. D., Tanksley S. D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993 Nov 26;262(5138):1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Martin G. B., Frary A., Wu T., Brommonschenkel S., Chunwongse J., Earle E. D., Tanksley S. D. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994 Nov;6(11):1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri F., Yu G. L., Ausubel F. M. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994 Sep 23;78(6):1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Pickett F. B., Meeks-Wagner D. R. Seeing double: appreciating genetic redundancy. Plant Cell. 1995 Sep;7(9):1347–1356. doi: 10.1105/tpc.7.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena K. M., Hooker A. L. On the structure of a gene for disease resistance in maize. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1300–1305. doi: 10.1073/pnas.61.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd K. W., Mayo G. M. Genes conferring specific plant disease resistance. Science. 1972 Jan 28;175(4020):375–380. doi: 10.1126/science.175.4020.375. [DOI] [PubMed] [Google Scholar]
- Song W. Y., Wang G. L., Chen L. L., Kim H. S., Pi L. Y., Holsten T., Gardner J., Wang B., Zhai W. X., Zhu L. H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995 Dec 15;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Staskawicz B. J., Ausubel F. M., Baker B. J., Ellis J. G., Jones J. D. Molecular genetics of plant disease resistance. Science. 1995 May 5;268(5211):661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Sudupak M. A., Bennetzen J. L., Hulbert S. H. Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics. 1993 Jan;133(1):119–125. doi: 10.1093/genetics/133.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S. P., Choi D., Hehl R., Corr C., Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994 Sep 23;78(6):1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]




