Live imaging reveals that muscle microtubules are highly dynamic and build a durable network nucleated by static Golgi elements.
Abstract
Skeletal muscle microtubules (MTs) form a nonclassic grid-like network, which has so far been documented in static images only. We have now observed and analyzed dynamics of GFP constructs of MT and Golgi markers in single live fibers and in the whole mouse muscle in vivo. Using confocal, intravital, and superresolution microscopy, we find that muscle MTs are dynamic, growing at the typical speed of ∼9 µm/min, and forming small bundles that build a durable network. We also show that static Golgi elements, associated with the MT-organizing center proteins γ-tubulin and pericentrin, are major sites of muscle MT nucleation, in addition to the previously identified sites (i.e., nuclear membranes). These data give us a framework for understanding how muscle MTs organize and how they contribute to the pathology of muscle diseases such as Duchenne muscular dystrophy.
Introduction
Microtubules (MTs) have recently been implicated in the pathology of Duchenne muscular dystrophy (DMD; Khairallah et al., 2012), the most common genetic disease of skeletal muscle. This puts a spotlight on muscle MTs, whose organization is poorly understood. During myogenesis, coordinated waves of subcellular remodeling affect MTs as well as MT-organizing centers (MTOCs), ER exit sites (ERES), and the Golgi complex (Tassin et al., 1985a,b; Lu et al., 2001; Musa et al., 2003; Bugnard et al., 2005; Srsen et al., 2009; Zaal et al., 2011). The resulting organization has little resemblance to that of proliferating cells. Muscle cultures can be used to study the first phase of MT reorganization that takes place during differentiation of myoblasts into multinucleated myotubes. But cultured myotubes do not mature into the fibers of which muscle is made. Therefore most of our knowledge of MTs in adult muscle comes from immunofluorescence images of single fibers, hand-teased from rodent muscles (Ralston, 1993; Ralston et al., 1999, 2001). Muscle fibers are shaped like flattened cylinders; their cytoplasm is mostly filled with actomyosin filaments. Between filaments and the plasmalemma there is a thin cytoplasmic layer that contains nuclei, other organelles, and what we refer to as surface MTs. Tubulin immunofluorescence shows that these MTs form a grid-like network with very few clear starting or ending points (Fig. S1 A). There are no clues as to their organization. Also lacking from these MTs are the asters that typically represent MT nucleation sites in images of proliferating cells (we call aster a flower-shaped figure formed by several MTs, which each have one end anchored to a central point and the other end free [Fig. S1 A, insets]). In addition, Golgi elements (the small but numerous Golgi complexes of muscle fibers) are positioned at the vertices of the MT lattice in a unique and unexplained organization. It became clear that we could only understand this organization by looking at live cells.
To do so, we have introduced GFP- and mCherry-tagged MT and Golgi markers into the mouse flexor digitorum brevis (FDB) muscle (Schertzer et al., 2006; Schertzer and Lynch, 2008; DiFranco et al., 2009; Fig. S1 G). We have characterized their dynamics ex vivo, in single fibers obtained by enzymatic digestion of the muscle (Bekoff and Betz, 1977; Rosenblatt et al., 1995), and in vivo, in muscles of live animals. We show that muscle MTs are highly dynamic and grow from static Golgi elements. Thus, like some other cells (Efimov et al., 2007; Rivero et al., 2009; Ori-McKenney et al., 2012), muscle fibers use the Golgi complex as a MTOC, while forming a MT network unlike any other.
Results and discussion
EB3-GFP reveals a dynamic network of MTs in live muscle, both ex vivo and in vivo
The MT plus tip end-binding protein EB3 is arguably the best marker of growing MTs (Stepanova et al., 2003) but its distribution in muscle fibers is unusual because it is found not only at the tips but anywhere along MTs (Fig. S1 B). Our first goal was therefore to clarify EB3 distribution by observing EB3-GFP in live fibers (see Materials and methods and Fig. S1 D2).
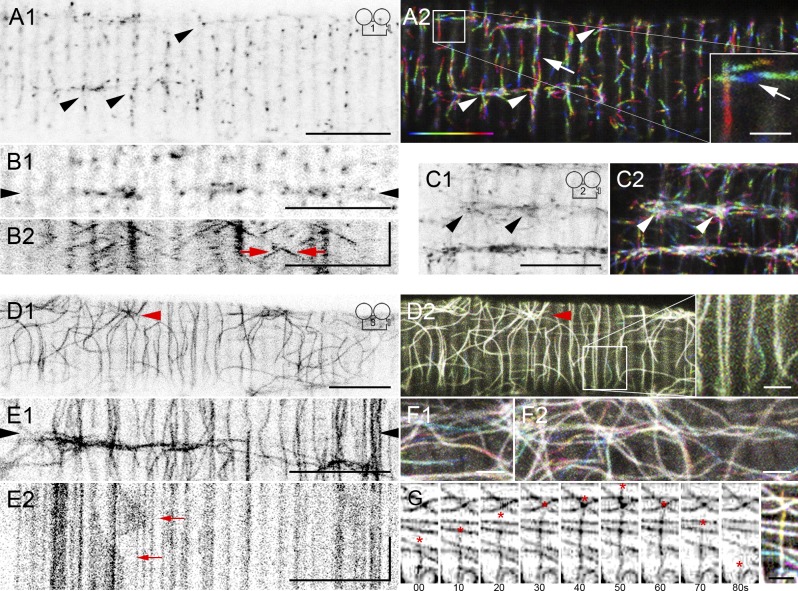
In plated fibers (Fig. 1, A1 and B1), each of the EB3-GFP puncta moves (Video 1) mostly longitudinally (parallel to the fiber axis) or transversely (perpendicular). To analyze EB3-GFP dynamics we generated color-coded time-lapse projections (Fig. 1 A2) and kymograph plots (Fig. 1 B). Puncta move indifferently left or right (56 vs. 44%; Fig. 1 A2) and up or down, and at similar speeds in all directions (Fig. 1 B, arrows). These results are consistent with EB3-GFP labeling the plus tips of MTs growing on parallel and antiparallel tracks. Puncta can be followed on average for 4.3 ± 0.1 s, i.e., ∼0.6 µm. Occasional spots (Fig. 1, A, C, and D, arrowheads) release EB3-GFP again and again, suggesting cytoplasmic MT nucleation sites. Projections show these spots to be at MT intersections. The mean growth rate of EB3-GFP in muscle fibers is 8.6 ± 0.1 µm/min (Table 1), similar to that in Drosophila melanogaster neuronal dendrites (Ori-McKenney et al., 2012).
Figure 1.
EB3-GFP shows steady MT dynamics ex vivo and in vivo, whereas GFP-tubulin highlights a durable MT frame. A single image, focused near the surface of a plated fiber expressing EB3-GFP (A1), shows typical puncta. The dynamics can be appreciated in the corresponding time-lapse series (Video 1) and in its projection (A2). Color coding of the projection helps to visualize movement: the first image of the series is colored blue and the last one magenta, as in the bar. EB3-GFP puncta mostly move longitudinally and transversely, as if along parallel and antiparallel tracks (A2, arrows). Some MT intersections (arrowheads) seem to behave as MT nucleation sites. An image from another fiber (B1) and the kymograph of the line between the arrowheads (B2; see Materials and methods) indicate that puncta move at the same speed in either direction on the longitudinal axis (oblique lines; red arrows). EB3-GFP dynamics in vivo (C1 and C2; Videos 2 and 5 and Fig. S2) validate plated fibers in all respects (arrowheads point to nucleation sites). In contrast, GFP-tubulin in plated fibers (D–G) appears static: the color-coded projections (D2, F1, and F2), with the same number of images and frame rate as A2, are practically white. An aster (arrowhead) indicates an MT nucleation site. The kymograph (E2) of the line between the arrowheads (E1) shows stationary MTs, with occasional local movement (arrows). Muscle MTs show dynamic instability (G); the asterisk shows the plus end of a MT growing and shrinking over 80 s. See Tables 1 and S1 for data quantitation and technical parameters. Bars: (A–E) 10 µm; (insets) 2 µm; (kymograph vertical time axes) 60 s.
Table 1.
Measurements of MT growth rate
| MT marker | Growth rate ± SEM | |
| Plated fibers | Intravital | |
| µm/min | µm/min | |
| EB3-GFP | 8.6 ± 0.1 (n = 463) | 5.0 ± 0.1 (n = 220) |
| GFP-tubulin | 5.6 ± 0.2 (n = 49) | 3.6 ± 0.2 (n = 31) |
To validate plated fibers as a model for muscle in vivo, we built a setup for intravital microscopy of the FDB and surrounding muscles (Fig. 1 C; Videos 2 and 5; and Fig. S2, A–C). EB3-GFP dynamics, the presence of antiparallel tracks, and the existence of cytoplasmic nucleation centers validate all ex vivo observations. The mean MT growth rate indicated by EB3-GFP in vivo is 5.0 ± 0.1 µm/min.
GFP-tubulin highlights a stationary, durable frame
To visualize the entire MTs we then examined GFP-tubulin. Compared with EB3-GFP, GFP-tubulin in plated fibers first appears immobile, highlighting a frame that remains unchanged for minutes (Fig. 1 D1 and Video 3); the color-coded projections are mostly white (Fig. 1 D2). However, there are pockets of growing and shrinking MTs (Fig. 1, D2 [inset] and E–G), reflecting dynamic instability (Mitchison and Kirschner, 1984). The mean growth rate is 5.6 ± 0.2 µm/min. GFP-tubulin, like EB3-GFP, grows indifferently left or right (45 ± 4 vs. 55 ± 4%), up or down. When two MTs are on the same stretch of the frame they grow as frequently in parallel as in antiparallel directions (48 vs. 52%).
In vivo recordings of GFP-tubulin also validated the ex vivo data (Fig. S2, D and E; and Video 6). GFP-tubulin–labeled MTs in vivo grow at 3.6 ± 0.2 µm/min. We ascribe the apparently slower growth rate of GFP-tubulin compared with that of EB3-GFP to the different methods of analysis (manual vs. PlusTipTracker; see Materials and methods) and to intrinsic differences between the two markers. Tubulin is the building block of MTs, whereas EB3 is in dynamic association with MTs from which it dissociates when MTs pause or shrink (explaining why EB3 puncta can only be followed on average for 0.6 µm).
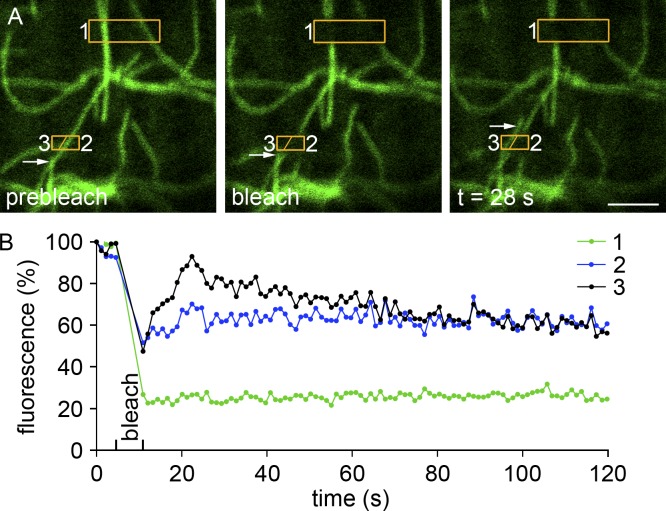
To learn more about the MT frame, we followed FRAP of regions of interest containing one to three MT tracks (Fig. 2). Because tubulin subunits are not in dynamic exchange with MTs, we did not expect true recovery, i.e., a uniform progressive return of the bleached track. Recovery was possible if new GFP-tubulin–labeled MTs grew along the bleached track, as occurred, but in only 7 out of 45 MT tracks. However, in half the regions of interest a new MT entered the box near the bleached track (Fig. 2 A, box 3 and arrow). Displacement of the bleached MT portion, which would indicate motor-induced MT transport, was not observed. The FRAP results are consistent with a frame of stable, immobile MTs and/or of anchored, growing MTs. Many of the observed MTs show dynamic instability, which results in repeated loss and gain of the EB3–tip complex. This can explain the apparent discrepancy between the low number of GFP-tubulin recoveries and the larger number of EB3-GFP puncta that would move in comparable fiber areas.
Figure 2.
FRAP showing only growth (or transport) of MTs restores fluorescence to the bleached area. (A) GFP-tubulin in a plated FDB fiber before (prebleach), just after (bleach), and 28 s after photobleaching of two regions of interest surrounded by orange boxes. For quantitation of recovery (B), the lower box was divided into parts 2 and 3. In seven independent photobleachings, more than half of the bleached boxes recover some fluorescence, as is the case in box 3, by growth or transport of a MT distinct from the original one (A, arrows). Bar, 2 µm.
The durable MT frame serves as a track for growing MTs
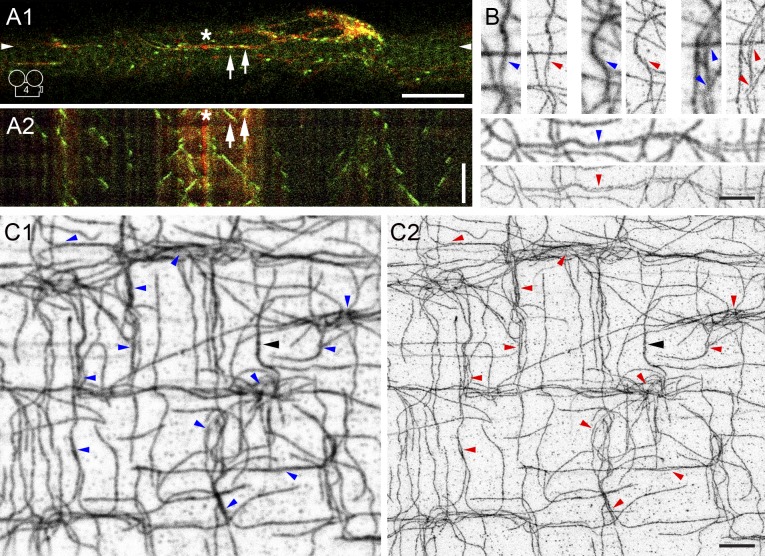
To clarify the apparent differences between EB3 and tubulin dynamics, we coexpressed EB3-GFP and mCherry-tubulin (Fig. 3 A). Each of the markers behaves as if alone, indicating that one does not interfere with the other and that neither affects the network as a whole (also see immunoblotting controls in Fig. S1, E and F). EB3-GFP moves along mCherry-tubulin–labeled tracks (Fig. 3 A1 [arrows] and Video 4). In addition, multiple EB3 dots move in the same or in opposite directions on a single track. The kymograph (Fig. 3 A2) emphasizes both the contrast in motility and the association of EB3-GFP with mCherry-tubulin.
Figure 3.
Dynamic MTs grow along MTs and are bundled. EB3-GFP and mCherry-tubulin were coexpressed and simultaneously imaged in plated fibers (A1; Video 4). EB3-GFP (green) moves along static mCherry-tubulin tracks (red; A2 kymograph). The asterisk indicates a nucleation spot and the arrows point to EB3-GFP–labeled MTs growing toward each other. G-STED superresolution microscopy of FDB fibers stained for α-tubulin resolves MTs into two or more components (B and C; panels with blue arrowheads show confocal images; panels with red arrowheads show the corresponding G-STED image at a resolution of 65 nm). A black arrowhead points to a probably unresolved MT. Bars: (A) 10 µm; (B and C) 2.5 µm; (A2, vertical time axis) 60 s.
These observations imply that muscle MTs are bundled. However, diffraction-limited microscopy, with a resolution of 250 nm at best, cannot resolve closely spaced MTs, which have outer diameters of 25 nm. Using G-STED superresolution microscopy (Vicidomini et al., 2013), we reached an improved resolution of 65 nm and resolved MT tracks into two to four strands, often torsading around each other (Fig. 3, B and C, red arrowheads). With a few exceptions (Fig. 3 C, black arrowhead) the strands are of even intensity, suggesting that they represent single MTs. Muscle MTs thus form small bundles with space between strands for MT-associated proteins (MAPs; Chen et al., 1992).
MT tracks may contain dynamic MTs growing alongside stable MTs, as suggested by the stationary character of the GFP-tubulin frame (Fig. 1 D). However, few muscle MTs have the curly shape and posttranslational tubulin modifications typical of stable MTs (Schulze et al., 1987). Muscle MTs could be stabilized by the muscle-specific isoform of MAP4 (Nguyen et al., 1997), which lines each muscle MT (Fig. S1 C) and whose function is not clear (Mangan and Olmsted, 1996; Casey et al., 2003). However, it is most likely that the stationary MT frame is entirely composed of dynamic MTs fasciculating along each other. Although MTs can bear compressive forces and support a cell structurally (Brangwynne et al., 2006), a dynamic frame of short MTs may be better suited to contracting fibers; indeed, excessive MT stabilization has been implicated in cardiac hypertrophy (Sato et al., 1997; Takahashi et al., 2003) and in DMD (Khairallah et al., 2012).
Muscle MTs nucleate from static Golgi elements
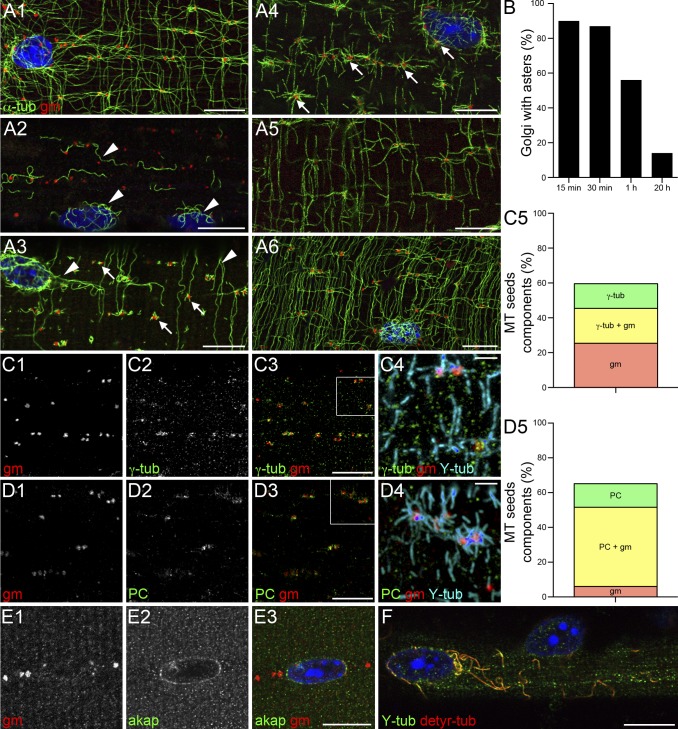
During muscle differentiation the MTOC redistributes from centrosomes to nuclear membranes (Tassin et al., 1985a; Lu et al., 2001; Bugnard et al., 2005; Zaal et al., 2011). EB3-GFP and GFP-tubulin, however, grow from additional nucleation centers (Fig. 1 and Videos 1–3), which are at MT intersections, where we also find Golgi elements (Fig. 4 A1). To assess whether Golgi elements serve as nucleation centers, we treated muscle fibers with nocodazole (NZ) to depolymerize MTs, washed NZ away, and analyzed MT recovery in relation to Golgi elements. After NZ treatment (Fig. 4 A2), only curly, NZ-resistant MTs are left (arrowheads), many of which surround nuclei. These MTs are detyrosylated, i.e., stable (Fig. 4 F). During the first minutes of recovery they elongate (Fig. 4 A3, arrowheads), whereas MT seeds appear around Golgi elements (arrows) and nuclei. These seeds rapidly grow into well-formed asters, strikingly centered on Golgi elements (Fig. 4 A4, arrows). MTs from different nucleation centers become interconnected (Fig. 4 A5) and an integrated network reforms (Fig. 4 A6). The percentage of Golgi elements associated with asters progressively decreases (Fig. 4 B), indicating that the asters become part of the reformed MT network. We conclude that Golgi elements (in addition to the nuclear membranes already described in myotubes) nucleate MTs in muscle fibers.
Figure 4.
MTs are nucleated on Golgi elements that concentrate γ-tubulin and pericentrin. To investigate MT nucleation, plated FDB fibers were treated with NZ to depolymerize MTs and fixed after different periods of recovery. They were then stained with anti–α-tubulin (α-tub) and anti-GM130 (gm) to label MTs and Golgi elements. In a control fiber (A1), Golgi elements are along MTs, especially at crossings, and around nuclei. After NZ, before recovery (A2), only a few curly MTs remain (arrowheads). These contain both detyrosylated and tyrosylated tubulins (detyr-tub and Y-tub; F), indicating the presence of both stable and dynamic MTs. In the first minute of recovery, NZ-resistant MTs elongate (A3, arrowheads) and MT seeds appear around Golgi elements (A3, arrows). A few minutes later, full asters centered on Golgi elements become prominent (A4, arrows). MTs then progressively reform a network (A5 and A6). During early recovery, ∼90% of Golgi elements are at the center of MT asters; at steady-state, only ∼10% are (B; data are from a single representative experiment out of three; n = 100, from five fibers, for each time point). Fibers at early stages of recovery (as shown in A3) were stained with anti-GM130 and with antibodies against γ-tubulin (γ-tub), pericentrin, and AKAP450. γ-Tubulin and pericentrin are concentrated on Golgi elements of MT seeds (C1–C4 and D1–D4); AKAP450 is only detected around nuclei (E1–E3). Most MT seeds are associated with Golgi elements ± γ-tubulin and some with γ-tubulin alone (C5; Table 2). Similar results are obtained for quantitation with pericentrin (D5; Table 3; two independent experiments, nine fibers, 400 MT seeds for C5, and 322 MT seeds for D5). Bars: (A–F) 10 µm; (C4 and D4) 2 µm.
In proliferating cells, the involvement of the Golgi complex in MT nucleation has been confirmed by treatment with brefeldin A (BfA), which redistributes Golgi components including Golgi-nucleated MTs to ERES (Rivero et al., 2009). In muscle fibers we found no convincing effect of BfA, likely because the Golgi elements are dispersed and colocalized with ERES even in the absence of BfA (Ralston et al., 1999; 2001; Lu et al., 2001).
Muscle fibers thus have at least two categories of MT nucleation sites, nuclei and Golgi elements. We do not know whether there are any biochemical or functional differences between MTs nucleated from these distinct sites, but one can hypothesize that perinuclear MTs are involved in the positioning of nuclei (Elhanany-Tamir et al., 2012; Metzger et al., 2012; Wilson and Holzbaur, 2012), whereas Golgi-nucleated MTs could play a role in GLUT4 translocation (Semiz et al., 2003) and lysosomal positioning (Fukuda et al., 2006; Korolchuk et al., 2011).
γ-Tubulin and pericentrin are associated with Golgi elements
The MTOC protein γ-tubulin is required for both centrosomal and noncentrosomal MT nucleation (Erhardt et al., 2002; Bugnard et al., 2005; Efimov et al., 2007; Rivero et al., 2009; Ori-McKenney et al., 2012; Zhu and Kaverina, 2013), but it is anchored by different proteins. In nonmuscle cells, Golgi γ-tubulin is linked either to the trans-Golgi by GCC185 and CLASP2 (Efimov et al., 2007) or to the cis-Golgi by GM130 and AKAP450 (Rivero et al., 2009).
By immunofluorescence we detected γ-tubulin on GM130-labeled Golgi elements of muscle fibers (Fig. 4 C and Table 2). CLASP2 could not be detected at all, and AKAP450 was only seen in the perinuclear area (Fig. 4 E). However, AKAP450 resembles another MTOC protein, pericentrin. AKAP450 and pericentrin are large coiled-coil proteins that share a centrosome-binding sequence (Gillingham and Munro, 2000). We found in silico that they also share sequences in the GM130-binding region of AKAP450 (Hurtado et al., 2011). Moreover, AKAP450 and kendrin, a pericentrin isoform, collaborate to bind γ-tubulin to centrosomes (Takahashi et al., 2002). Pericentrin could therefore replace or supplement AKAP450 in muscle fibers. Pericentrin was detected in fibers and found colocalized with Golgi elements (Fig. 4 D and Table 3). Although these results are consistent with cis-Golgi MT nucleation, we have not been able to confirm the involvement of GM130 because knocking it down by shRNA was inconclusive. It is possible that both cis- and trans-Golgi nucleations coexist or that other cis-Golgi proteins are involved.
Table 2.
Association of MT seeds with GM130 and γ-tubulin
| Associated with | Fraction of MT seeds ± SEM |
| % | |
| GM130 + γ-tubulin | 20.1 ± 2.8 |
| GM130 only | 25.4 ± 2.9 |
| γ-Tubulin only | 14.1 ± 1.7 |
| None | 40.4 ± 4.0 |
Based on two experiments and 322 MT seeds.
Table 3.
Association of MT seeds with GM130 and pericentrin
| Associated with | Fraction of MT seeds ± SEM |
| % | |
| GM130 + pericentrin | 45.5 ± 4.9 |
| GM130 only | 6.1 ± 0.8 |
| Pericentrin only | 13.6 ± 1.5 |
| None | 34.8 ± 4.5 |
Based on two experiments and 400 MT seeds.
Finally, we assessed the motility of Golgi elements. We coexpressed galactosyl-transferase-mCherry (GalT-mCherry), a marker of the Golgi complex, with GFP-tubulin or EB3-GFP (Fig. S3, Video 7, and Video 8). In both plated fibers (Fig. S3 A) and in vivo (Fig. S3 B) Golgi elements are static. Thus, the classic model that MTs position the Golgi complex is turned upside down in muscle, where, instead, static Golgi elements position MT nucleation. The lack of motility of muscle Golgi elements is consistent with their steady positioning along Z bands (Kaisto and Metsikkö, 2003).
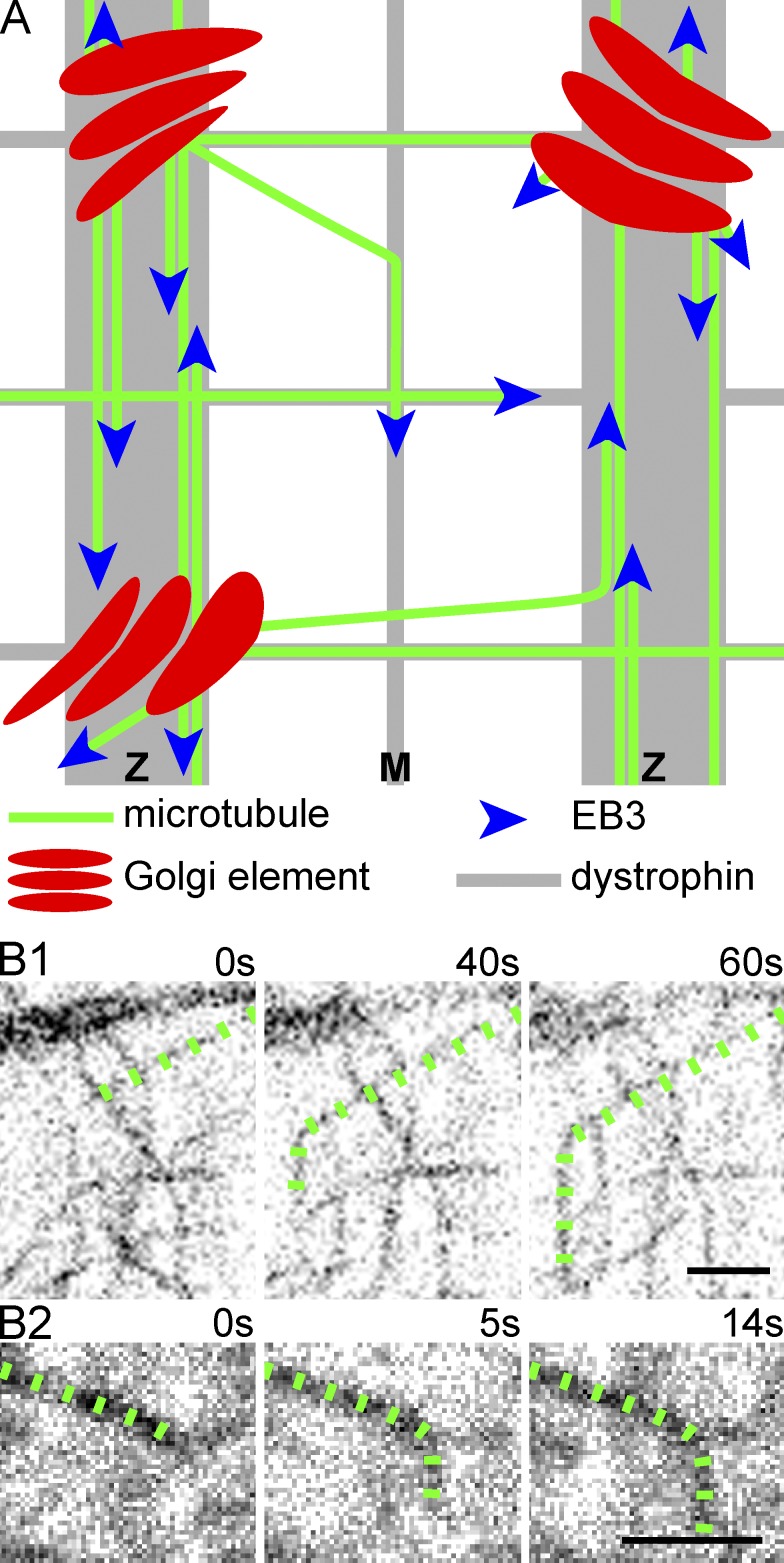
Building a model of muscle MT organization
At this point, we can start building a model (Fig. 5 A). At steady-state, MTs nucleated from Golgi elements grow along other dynamic and/or stable MTs to form bundles. To explain the orthogonal grid of MTs, we must involve dystrophin, the protein missing in DMD. Dystrophin is a MAP (Prins et al., 2009) and its absence in the mdx mouse prevents MTs from forming an orthogonal grid (Percival et al., 2007; Prins et al., 2009). However, dystrophin does not line MTs as MAP4 does; instead, dystrophin appears to define domains along which MTs grow preferentially (Prins et al., 2009). Dystrophin may capture MTs; those that start at an oblique angle (Fig. 1 D2, colored MTs) often abruptly change their orientation (Fig. 5 B) when they encounter the transverse bands that contain dystrophin. Studies of MT dynamics and directionality in mdx muscles will help us to further develop this model.
Figure 5.
Model of MT organization in skeletal muscle fibers at steady-state. (A) MTs nucleating from Golgi elements grow parallel or antiparallel to existing MTs, thereby forming small bundles, which are guided or restricted by the dystrophin bands positioned along Z lines, M bands, and longitudinal stripes. MTs starting at an oblique angle reorient upon contact with other MTs or dystrophin bands, as observed (B) in GFP-tubulin–expressing fibers. The MT lattice that results is both durable and dynamic. Bars, 2 µm.
Skeletal muscle MTs, like the proverbial canary in the mine, are affected by practically all physiological and pathological changes in muscle, most likely because they are sensitive to patterned contractions (Ralston et al., 2001). They reach and dynamically connect all domains of muscle fibers. MTs are moored by Golgi elements through ERES to the ER/sarcoplasmic reticulum, which extends longitudinally in the myofibrillar core (Kaisto and Metsikkö, 2003). They interact with the triad junctions of T-tubules and sarcoplasmic reticulum (Fourest-Lieuvin et al., 2012), with dystrophin, and also with muscle-specific protein networks involved in the maintenance of sarcomeric organization (Ayalon et al., 2008, 2011; Randazzo et al., 2013). For a long time, the relevance of muscle MTs and the consequences of their perturbations were not clear. Now muscle MTs are finally starting to receive long overdue attention.
Materials and methods
Antibodies and other reagents
Several antibodies were gifts: rabbit anti–mouse/human detyrosylated tubulin from G. Cooper IV and T. Galien (Veterans Affairs Medical Center, Charleston, SC; Sato et al., 1997); rabbit anti–mouse/human EB3 from A. Akhmanova (Utrecht University, Utrecht, Netherlands; Stepanova et al., 2003); rabbit anti–mouse mMAP4 from J. Olmsted (University of Rochester, Rochester, NY; Casey et al., 2003); and rabbit anti–human AKAP450/AKAP350 from J. Goldenring (Medical College of Georgia, Augusta, GA; Shanks et al., 2002). Other antibodies were purchased commercially: rat anti-tyrosylated tubulin from Accurate; rabbit anti–α-tubulin and mouse anti-GAPDH from Abcam; mouse anti–α-tubulin, rabbit anti-acetylated tubulin, and rabbit anti–γ-tubulin from Sigma-Aldrich; rabbit anti-detyrosylated tubulin from EMD Millipore; mouse anti-GM130 and mouse anti-pericentrin from BD; rabbit anti-GFP from Cell Signaling Technology; and rabbit anti-pericentrin from Covance. Goat anti–mouse and anti–rat conjugated with DyLight 488, 549, and 647 were obtained from Jackson ImmunoResearch Laboratories, Inc. and goat anti–rabbit conjugated with Alexa 488, 546, 568, and 647 were obtained from Molecular Probes. For Western blot analyses we used horseradish peroxidase–conjugated goat anti–rabbit and goat anti–mouse purchased from Bio-Rad Laboratories and Alexa 680–conjugated goat anti–rat and anti–mouse antibodies purchased from Invitrogen.
Plasmids
All plasmids expressed in this study are based on pEGFP-N1 or pEGFP-C1 vectors (Takara Bio Inc.). p-EB3-GFP-N1 cDNA was a gift from A. Akhmanova (Stepanova et al., 2003), p-EGFP-tubulin-C1 was obtained from Takara Bio Inc., and p-mCherry-tubulin-C1 was constructed from p-eGFP-tubulin-C1 and p-mCherry-C1. p-GalT-mCherry-N1 was constructed from p-GalT-eGFP-N1 (Zaal et al., 1999) and p-mCherry-N1 (a gift from G. Patterson [National Institute of Biomedical Imaging and Bioengineering, Bethesda, MD]). p-EGFP-EMTB-N1 was subcloned from p-EMTB-N1 (a gift from C. Bulinski [Columbia University, New York, NY]).
cDNA injection and electroporation into mouse muscles
All animal protocols were reviewed and approved by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Animal Care and Use Committee. Mice were C57BL/6 (The Jackson Laboratory), 6 to 8 wk old unless otherwise mentioned. To obtain cDNA expression we followed the protocol of DiFranco et al. (2009) with a few modifications. Mice were anesthetized with 4% isoflurane throughout the procedure and received a subcutaneous injection of 0.05 mg/kg buprenorphine-HCl to avoid pain. To loosen the extracellular matrix and allow the plasmid to reach the FDB muscle, which extends along the sole of the foot, 10 µl of 0.5 U/µl (0.36 mg/ml) hyaluronidase was injected through the skin at the heel. After 1 h, 20–50 µg of endotoxin-free plasmid was injected (Genewiz) at 5 mg/ml in sterile DPBS. After 15 min, acupuncture needles (0.20 × 25 mm, Tai Chi; Lhasa OMS) were placed under the skin at the heel and at the base of the toes and connected to an ECM 830 BTX electroporator (BTX Harvard Apparatus). Six pulses of 20 ms each at 1 Hz were applied to yield an electric field of ∼75 V/cm. 5–7 days later the animal was killed to collect muscles or prepared for intravital imaging. Expression of the injected cDNA is easily verified under fluorescence illumination on the dissection microscope (Fig. S1 G). The efficiency of expression ranges from 20 to 80% of the fibers depending on the plasmid.
Selection of EB3-GFP and GFP-tubulin constructs to track MTs in muscle fibers
In search of suitable MT markers we expressed GFP-tubulin (Fig. S1 D1), EB3-GFP (Fig. S1 D2), and the MT-binding domain of the MAP ensconsin (EMTB-GFP; Fig. S1 D3). Our criteria for accepting a construct were as follows: pattern and location indistinguishable from native MTs (Fig. S1 A), even at moderate expression levels, normal appearance of the whole MT network, and useful signal-to-noise ratio. EB3-GFP and GFP-tubulin satisfied all criteria. Regrettably, EMTB (Faire et al., 1999) caused abnormalities of MTs in all but the lowest expressing fibers. We verified by immunoblotting (Fig. S1 F) that GFP-tubulin undergoes detyrosylation and acetylation, the normal posttranslational modifications of muscle tubulin (Gundersen et al., 1989), and that EB3-GFP overexpression does not affect the posttranslational modifications of tubulin (Fig. S1 E). We also checked by immunofluorescence that the expression of the GFP constructs does not alter the respective patterns of detyrosylated and tyrosylated tubulin. Finally we verified that mCherry constructs gave results similar to those with the corresponding GFP construct.
Intravital imaging
Mice injected with cDNA and electroporated as described in a previous paragraph were anesthetized by intraperitoneal injection of 75 mg/kg of sodium pentobarbital. A flap of skin was removed from the plant of the foot to expose the FDB. The mouse was placed in a tub-shaped stage insert (custom designed at the National Institute of Arthritis and Musculoskeletal and Skin Diseases for the TCS SP5 confocal microscope [Leica]), the bottom of which was made of a no. 1.5 coverglass. The exposed FDB, facing the objective lens, was held on the coverglass by a small lever arm (Fig. S3, A and B). The mouse temperature was maintained at 37°C with a heating lamp, and breathing of the animal was monitored visually throughout the experiment.
FDB fiber preparation
Mice were killed by CO2 followed by cervical dislocation. FDB muscles were dissected in sterile DPBS under an MZ FLIII dissecting microscope (Leica), rinsed in sterile DMEM, and incubated with rotation for 3 h at 37°C in DMEM containing 1.5 mg/ml type I collagenase from Clostridium histolyticum (Sigma-Aldrich) and 1 mg/ml BSA. Fibers were then freed from the muscle by trituration and plated on Mattek dishes or Lab-Tek chambered coverglass that had been coated for 1 h with a 1:10 dilution of Matrigel (BD). Fibers were plated in 0.1 ml of growth media (GM), consisting of DMEM supplemented with 20% FBS and 0.2% chicken embryo extract. After 2 h they were fed with GM supplemented with penicillin-streptomycin. The fibers were used within 24 h, ahead of the reorganization of MT and other cytoskeletal components, which occurs after denervation or when patterned activity is modified (Ralston et al., 2001, 2006). The only damage directly linked to the collagenase treatment is a degradation of the fine structure of the neuromuscular junction. We have not searched for neuromuscular junction–specific MTs (Schmidt et al., 2012). FDB fibers are predominantly type IIA (intermediate type). Their MTs are organized similarly to those in fast-twitch fibers (Ralston et al., 1999).
Drug treatments
To prevent MT polymerization and induce loss of MTs, plated fibers were incubated for 4 h at 37°C in 4 µg/ml NZ (Sigma-Aldrich) in GM. After washing out the drug with GM at 37°C, MTs were left to recover at 37°C in GM for 2 min to 24 h. Fibers were then fixed for staining, either with methanol at −20°C or with 4% PFA (Electron Microscopy Sciences). The Golgi complex was disrupted with 5 µg/ml BfA (Sigma-Aldrich) at 37°C for 1 h. For unrecovered NZ or BfA controls, the drug was added to PFA; alternately, methanol was used for fixation. When combined with NZ treatment, BfA was added for the last hour of the 4-h incubation with NZ and throughout the washout and fixation process.
Immunofluorescence
Fixed fibers were blocked for 2 h at RT in PBS containing either 5% BSA, 1% normal goat serum, and 0.04% saponin or the blocking reagent from the Mouse On Mouse basic kit (Vector Laboratories). Fibers were then incubated with primary antibodies for 2 h at RT (or overnight at 4°C) and with secondary antibodies for 2 h at RT, counterstained with Hoechst 33342, and mounted in Vectashield (Vector Laboratories). Rinses between and after antibody incubations were three times 5 min with PBS containing 0.04% saponin.
Microscopy
Confocal images were collected using a 63× 1.4 NA or a 40× 1.25 NA oil immersion objective lenses on a TCS SP5 (Leica) driven by the LAS AF 2.6.1 software or on an LSM 780 confocal microscope (Carl Zeiss) driven by Zen 2011. In both cases, images were collected in sequential scanning to avoid cross talk. Live fibers were imaged in phenol red–free GM with 25 mM Hepes at 37°C, using a Tokai Hit heated stage insert on the SP5 and a Pecon Lab-Tek S1 stage insert on the LSM 780. We used a confocal pinhole between 1 and 3 Airy units (AU) to increase depth of field and signal intensity while limiting potential laser damage to live fibers.
For FRAP sequences on the LSM 780, we wanted to achieve close to complete photobleaching of target regions without damaging them. We tested different protocols and settled on 10 bleaching iterations with a 40-mW Argon laser at 80%. We were satisfied that no damage was done because MTs that had been photobleached continued to grow outside of the bleached area.
Unless otherwise mentioned, the images shown are single frames. Camera icons refer to the corresponding video, available online. Projections of images were done as “maximum” projections. Images were exported in 8-bit TIF format and linearly adjusted using Photoshop CS5, and then cropped and resized if needed for composing montages. Some movies were processed in ImageJ (National Institutes of Health) with an image stabilizer and Kalman filter. For black and white images, the grayscale was inverted to facilitate viewing. Settings used for still images and videos can be found in Table S1.
Superresolution microscopy
Fibers stained with anti-tubulin followed by Alexa 488 or 647 goat anti–mouse IgG were mounted in Prolong Gold. They were imaged using time-gated detection on the gated HyD detectors of a TCS SP8 X G-STED system (Leica) with pulsed white light laser excitation. The conditions for confocal imaging were identical to those for time-lapse series, i.e., pinhole opened to 1.5 AU to increase brightness and depth of field, whereas G-STED parameters were optimized for best resolution, i.e., pinhole between 0.5 and 1.0 AU.
Image rendering and analysis
Two techniques were used to make dynamics perceptible in single static images: color coding and kymograph plotting. Color coding of confocal time series was done with Photoshop CS5. Each image of the 38-s time series had its red, green, and blue levels set to obtain a progressive change of colors, the first frame being blue (red = 0, green = 0, and blue = 255) and the last frame magenta (red = 255, green = 0, and blue = 255). A moving object appears rainbow colored in projection whereas a stationary object is white. Kymographs were done on image stacks in ImageJ with the Reslice tool. The position of the line that is extracted from each image and repeated in the kymograph is indicated by two arrowheads on one of the images. An object that moves along the selected line during the recording of the series appears as an oblique dash in the kymograph, a stationary object on the line appears as a vertical line, and an object that crosses the line appears as a single dot.
EB3-GFP speed was analyzed with PlusTipTracker, a MatLab-based open source software (Applegate et al., 2011). The tracks were detected with PlusTip GetTracks, with the following parameters, adapted depending on the quality of the movie: gap length = 3, angle = 10–15, radius range = 1–8, fluctuation radius = 2, and maximum shrinkage factor = 1.5. Each track was checked by hand with PlusTip SeeTracks to avoid false positives. GFP-tubulin MT growth rate was analyzed with ImageJ. The path covered was tracked, measured manually, and converted to a growth rate in micrometers per minute.
To quantitate the association of nascent MTs with Golgi elements, γ-tubulin, and/or pericentrin (Fig. 4, C5 and D5; and Tables 2 and 3), we examined the tubulin staining of triple-stained immunofluorescence images while hiding the other channels, marked the position of presumed MT seeds (at least three MT fragments), and then examined whether Golgi and γ-tubulin or pericentrin staining were present. Because MT seeds at early recovery times are less organized than the later asters, it is possible to count as seeds the simple crossing of two MTs. It is likely that this explains the relatively high percentage of MT seeds not linked to any of the components.
Immunoblots
Fibers were prepared as described in a previous paragraph but instead of being plated they were rinsed three times in DPBS, lysed in 40 µl of loading buffer (National Diagnostics), and boiled. Protein concentration was assayed with the 2D Quant Kit (GE Healthcare). Proteins were separated on 10% acrylamide gels and immunoblotted according to standard procedures. Protein bands were detected either with the Odyssey infrared imaging system (Li-Cor) or with photographic film.
Online supplemental material
Fig. S1 presents background and quality control data: immunofluorescence of muscle fibers stained for tubulin, EB3, and MAP4, and characterization of muscle fibers expressing GFP constructs by fluorescence and immunoblotting. Fig. S2 illustrates the setup for and images from intravital imaging of EB3-GFP and GFP-tubulin. Fig. S3 illustrates simultaneous recordings of the Golgi marker GalT-mCherry and of a MT marker, ex vivo and in vivo. Videos provide examples of time-lapse recordings and are essential to perceive MT dynamics. Videos 1 and 2 present EB3-GFP dynamics ex vivo and in vivo, respectively. Video 3 shows GFP-tubulin ex vivo. Video 4 shows simultaneous EB3-GFP and mCherry-tubulin recordings ex vivo. Video 5 shows a more extensive muscle area expressing EB3-GFP in vivo. Video 6 shows GFP-tubulin in vivo. Video 7 shows simultaneous GFP-tubulin and GalT-mCherry recordings ex vivo, and Video 8 shows EB3-GFP and GalT-mCherry in vivo. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201304063/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201304063.dv.
Supplementary Material
Acknowledgments
We wish to acknowledge the late George Cooper IV for generous advice over the years. We thank all colleagues who shared reagents; Jonathan Boyd and Geoff Daniels for essential help with G-STED imaging; Gary Melvin for design and fabrication of hardware for intravital imaging; and Nina Raben, Rachel Myerowitz, Dan Sackett, and Andrew Milgroom for critical reading of the manuscript.
This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
Abbreviations used in this paper:
- AU
- Airy units
- BfA
- brefeldin A
- DMD
- Duchenne muscular dystrophy
- ERES
- ER exit sites
- GalT
- galactosyl-transferase
- GM
- growth media
- FDB
- flexor digitorum brevis
- MAP
- MT-associated protein
- MT
- microtubule
- MTOC
- MT-organizing center
- NZ
- nocodazole
- SR
- sarcoplasmic reticulum
References
- Applegate K.T., Besson S., Matov A., Bagonis M.H., Jaqaman K., Danuser G. 2011. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176:168–184 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon G., Davis J.Q., Scotland P.B., Bennett V. 2008. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell. 135:1189–1200 10.1016/j.cell.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Ayalon G., Hostettler J.D., Hoffman J., Kizhatil K., Davis J.Q., Bennett V. 2011. Ankyrin-B interactions with spectrin and dynactin-4 are required for dystrophin-based protection of skeletal muscle from exercise injury. J. Biol. Chem. 286:7370–7378 10.1074/jbc.M110.187831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekoff A., Betz W.J. 1977. Physiological properties of dissociated muscle fibres obtained from innervated and denervated adult rat muscle. J. Physiol. 271:25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., MacKintosh F.C., Kumar S., Geisse N.A., Talbot J., Mahadevan L., Parker K.K., Ingber D.E., Weitz D.A. 2006. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J. Cell Biol. 173:733–741 10.1083/jcb.200601060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E., Zaal K.J., Ralston E. 2005. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskeleton. 60:1–13 10.1002/cm.20042 [DOI] [PubMed] [Google Scholar]
- Casey L.M., Lyon H.D., Olmsted J.B. 2003. Muscle-specific microtubule-associated protein 4 is expressed early in myogenesis and is not sufficient to induce microtubule reorganization. Cell Motil. Cytoskeleton. 54:317–336 10.1002/cm.10105 [DOI] [PubMed] [Google Scholar]
- Chen J., Kanai Y., Cowan N.J., Hirokawa N. 1992. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 360:674–677 10.1038/360674a0 [DOI] [PubMed] [Google Scholar]
- DiFranco M., Quinonez M., Capote J., Vergara J. 2009. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J. Vis. Exp. 19:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 12:917–930 10.1016/j.devcel.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanany-Tamir H., Yu Y.V., Shnayder M., Jain A., Welte M., Volk T. 2012. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 198:833–846 10.1083/jcb.201204102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M., Stoppin-Mellet V., Campagne S., Canaday J., Mutterer J., Fabian T., Sauter M., Muller T., Peter C., Lambert A.M., Schmit A.C. 2002. The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115:2423–2431 [DOI] [PubMed] [Google Scholar]
- Faire K., Waterman-Storer C.M., Gruber D., Masson D., Salmon E.D., Bulinski J.C. 1999. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J. Cell Sci. 112:4243–4255 [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin A., Rendu J., Osseni A., Pernet-Gallay K., Rossi D., Oddoux S., Brocard J., Sorrentino V., Marty I., Fauré J. 2012. Role of triadin in the organization of reticulum membrane at the muscle triad. J. Cell Sci. 125:3443–3453 10.1242/jcs.100958 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Ewan L., Bauer M., Mattaliano R.J., Zaal K., Ralston E., Plotz P.H., Raben N. 2006. Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann. Neurol. 59:700–708 10.1002/ana.20807 [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G.G., Khawaja S., Bulinski J.C. 1989. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J. Cell Biol. 109:2275–2288 10.1083/jcb.109.5.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L., Caballero C., Gavilan M.P., Cardenas J., Bornens M., Rios R.M. 2011. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J. Cell Biol. 193:917–933 10.1083/jcb.201011014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisto T., Metsikkö K. 2003. Distribution of the endoplasmic reticulum and its relationship with the sarcoplasmic reticulum in skeletal myofibers. Exp. Cell Res. 289:47–57 10.1016/S0014-4827(03)00231-3 [DOI] [PubMed] [Google Scholar]
- Khairallah R.J., Shi G., Sbrana F., Prosser B.L., Borroto C., Mazaitis M.J., Hoffman E.P., Mahurkar A., Sachs F., Sun Y., et al. 2012. Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci. Signal. 5:ra56 10.1126/scisignal.2002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Joseph D., Bugnard E., Zaal K.J., Ralston E. 2001. Golgi complex reorganization during muscle differentiation: visualization in living cells and mechanism. Mol. Biol. Cell. 12:795–808 10.1091/mbc.12.4.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan M.E., Olmsted J.B. 1996. A muscle-specific variant of microtubule-associated protein 4 (MAP4) is required in myogenesis. Development. 122:771–781 [DOI] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., Baylies M.K. 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 484:120–124 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Musa H., Orton C., Morrison E.E., Peckham M. 2003. Microtubule assembly in cultured myoblasts and myotubes following nocodazole induced microtubule depolymerisation. J. Muscle Res. Cell Motil. 24:301–308 10.1023/A:1025477807393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.L., Chari S., Gruber D., Lue C.M., Chapin S.J., Bulinski J.C. 1997. Overexpression of full- or partial-length MAP4 stabilizes microtubules and alters cell growth. J. Cell Sci. 110:281–294 [DOI] [PubMed] [Google Scholar]
- Ori-McKenney K.M., Jan L.Y., Jan Y.N. 2012. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 76:921–930 10.1016/j.neuron.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival J.M., Gregorevic P., Odom G.L., Banks G.B., Chamberlain J.S., Froehner S.C. 2007. rAAV6-microdystrophin rescues aberrant Golgi complex organization in mdx skeletal muscles. Traffic. 8:1424–1439 10.1111/j.1600-0854.2007.00622.x [DOI] [PubMed] [Google Scholar]
- Prins K.W., Humston J.L., Mehta A., Tate V., Ralston E., Ervasti J.M. 2009. Dystrophin is a microtubule-associated protein. J. Cell Biol. 186:363–369 10.1083/jcb.200905048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E. 1993. Changes in architecture of the Golgi complex and other subcellular organelles during myogenesis. J. Cell Biol. 120:399–409 10.1083/jcb.120.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E., Lu Z., Ploug T. 1999. The organization of the Golgi complex and microtubules in skeletal muscle is fiber type-dependent. J. Neurosci. 19:10694–10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E., Ploug T., Kalhovde J., Lømo T. 2001. Golgi complex, endoplasmic reticulum exit sites, and microtubules in skeletal muscle fibers are organized by patterned activity. J. Neurosci. 21:875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E., Lu Z., Biscocho N., Soumaka E., Mavroidis M., Prats C., Lømo T., Capetanaki Y., Ploug T. 2006. Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J. Cell. Physiol. 209:874–882 10.1002/jcp.20780 [DOI] [PubMed] [Google Scholar]
- Randazzo D., Giacomello E., Lorenzini S., Rossi D., Pierantozzi E., Blaauw B., Reggiani C., Lange S., Peter A.K., Chen J., Sorrentino V. 2013. Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J. Cell Biol. 200:523–536 10.1083/jcb.201205118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S., Cardenas J., Bornens M., Rios R.M. 2009. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 28:1016–1028 10.1038/emboj.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J.D., Lunt A.I., Parry D.J., Partridge T.A. 1995. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim. 31:773–779 10.1007/BF02634119 [DOI] [PubMed] [Google Scholar]
- Sato H., Nagai T., Kuppuswamy D., Narishige T., Koide M., Menick D.R., Cooper G., IV 1997. Microtubule stabilization in pressure overload cardiac hypertrophy. J. Cell Biol. 139:963–973 10.1083/jcb.139.4.963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer J.D., Lynch G.S. 2008. Plasmid-based gene transfer in mouse skeletal muscle by electroporation. Methods Mol. Biol. 433:115–125 10.1007/978-1-59745-237-3_7 [DOI] [PubMed] [Google Scholar]
- Schertzer J.D., Plant D.R., Lynch G.S. 2006. Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 13:795–803 10.1016/j.ymthe.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Schmidt N., Basu S., Sladecek S., Gatti S., van Haren J., Treves S., Pielage J., Galjart N., Brenner H.R. 2012. Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J. Cell Biol. 198:421–437 10.1083/jcb.201111130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E., Asai D.J., Bulinski J.C., Kirschner M. 1987. Posttranslational modification and microtubule stability. J. Cell Biol. 105:2167–2177 10.1083/jcb.105.5.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiz S., Park J.G., Nicoloro S.M., Furcinitti P., Zhang C., Chawla A., Leszyk J., Czech M.P. 2003. Conventional kinesin KIF5B mediates insulin-stimulated GLUT4 movements on microtubules. EMBO J. 22:2387–2399 10.1093/emboj/cdg237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks R.A., Steadman B.T., Schmidt P.H., Goldenring J.R. 2002. AKAP350 at the Golgi apparatus. I. Identification of a distinct Golgi apparatus targeting motif in AKAP350. J. Biol. Chem. 277:40967–40972 10.1074/jbc.M203307200 [DOI] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C., Merdes A. 2009. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 10:28 10.1186/1471-2121-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C.C., Lansbergen G., Dortland B., De Zeeuw C.I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Yamagiwa A., Nishimura T., Mukai H., Ono Y. 2002. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell. 13:3235–3245 10.1091/mbc.E02-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Shiraishi H., Ishibashi Y., Blade K.L., McDermott P.J., Menick D.R., Kuppuswamy D., Cooper G., IV 2003. Phenotypic consequences of beta1-tubulin expression and MAP4 decoration of microtubules in adult cardiocytes. Am. J. Physiol. Heart Circ. Physiol. 285:H2072–H2083 [DOI] [PubMed] [Google Scholar]
- Tassin A.M., Maro B., Bornens M. 1985a. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100:35–46 10.1083/jcb.100.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A.M., Paintrand M., Berger E.G., Bornens M. 1985b. The Golgi apparatus remains associated with microtubule organizing centers during myogenesis. J. Cell Biol. 101:630–638 10.1083/jcb.101.2.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicidomini G., Schönle A., Ta H., Han K.Y., Moneron G., Eggeling C., Hell S.W. 2013. STED nanoscopy with time-gated detection: theoretical and experimental aspects. PLoS ONE. 8:e54421 10.1371/journal.pone.0054421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., Holzbaur E.L. 2012. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci. 125:4158–4169 10.1242/jcs.108688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal K.J., Smith C.L., Polishchuk R.S., Altan N., Cole N.B., Ellenberg J., Hirschberg K., Presley J.F., Roberts T.H., Siggia E., et al. 1999. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 99:589–601 10.1016/S0092-8674(00)81548-2 [DOI] [PubMed] [Google Scholar]
- Zaal K.J., Reid E., Mousavi K., Zhang T., Mehta A., Bugnard E., Sartorelli V., Ralston E. 2011. Who needs microtubules? Myogenic reorganization of MTOC, Golgi complex and ER exit sites persists despite lack of normal microtubule tracks. PLoS ONE. 6:e29057 10.1371/journal.pone.0029057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Kaverina I. 2013. Golgi as an MTOC: making microtubules for its own good. Histochem. Cell Biol. 140:361–367 10.1007/s00418-013-1119-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.